Abstract
High-throughput ‘omics’ data analysis via bioinformatics is one key component of the systems biology approach. The systems approach is particularly well-suited for the study of the interactions between nutrition and physiological state with tissue metabolism and functions during key life stages of organisms such as the transition from pregnancy to lactation in mammals, ie, the peripartal period. In modern dairy cows with an unprecedented genetic potential for milk synthesis, the nature of the physiologic and metabolic adaptations during the peripartal period is multifaceted and involves key tissues such as liver, adipose, and mammary. In order to understand such adaptation, we have reviewed several works performed in our and other labs. In addition, we have used a novel bioinformatics approach, Dynamic Impact Approach (DIA), in combination with partly previously published data to help interpret longitudinal biological adaptations of bovine liver, adipose, and mammary tissue to lactation using transcriptomics datasets. Use of DIA with transcriptomic data from those tissues during normal physiological adaptations and in animals fed different levels of energy prepartum allowed visualization and integration of most-impacted metabolic pathways around the time of parturition. The DIA is a suitable tool for applying the integrative systems biology approach. The ultimate goal is to visualize the complexity of the systems at study and uncover key molecular players involved in the tissue’s adaptations to physiological state or nutrition.
Keywords: bovine, dairy cow, bioinformatics, microarray, lactation, dynamic impact approach
Background
Systems biology: The comeback of an “Old” concept in the ‘Omics’ age
The application of methods allowing for integration of the individual components of an animal instead of the more reductionist approach is ideally suited for exploring the biological complexity of mammals. Modern systems biology is generally defined as an interdisciplinary field that concentrates on experimental and computational biology. From the experimental standpoint, systems biology concepts can and often are applied with a hypothesis or sets of hypotheses in mind, but also allow for a “discovery” type experimental approach (ie, without a specific hypothesis). Advancements in computational biology, genome sequencing, and high-throughput technologies in the last decade have provided the tools for approaching biological systems in an integrative fashion, ie, allow access to the functional capabilities of an individual organism en masse.
The origin of systems biology concepts can be traced to at least 1934 to Austrian biologist Ludwig von Bertalanffy who proposed the use of “general system theory” in biology as “a new approach to unity of science”.1 Cornish-Bowden and colleagues2,3 make the point that there has been interest in and efforts to apply systems research since the middle of the 20th century. Despite the wide variety of definitions of the term ‘systems biology’, eg, “a new name for old-fashioned reductionist biology practiced on an ever-larger scale, with ever-larger and more expensive machines”,3 there is recognition that “a genuine systemic view is not incompatible with gathering huge quantities of experimental data”; rather, the issue lies on whether proper application of high-throughput technologies and bioinformatics will allow continued advances towards understanding systems.3
Several examples from the non-ruminant literature underscore the need to analyze biological systems as systems and not as mere collections of parts.2 Ideally, integrating mRNA and protein expression responses with the global set of protein–protein and protein–DNA interactions will allow deducing underlying networks of genes/proteins. Several examples with model organisms underscore that genes/proteins act in concert with one another and with the environment.4,5 It is likely that such associations also play an important role in modern high-producing ruminants, eg, help coordinate efficient use of nutrients for milk or beef production. As an example, in a recent study Barendse et al6 reported that a large fraction of the genetic variation linked to feed efficiency occurred in promoter and microRNA motif regions of the bovine genome. Those findings suggested that at least some of the genetic variation in efficiency of nutrient use between animals could be due to differences in the regulation of gene expression.
In dairy production, management practices during the dry period and early lactation can essentially determine the productivity of the animal throughout lactation, ie, the peripartal period (ie, last 3 weeks through the first 3 weeks around parturition) is where the highest incidence of infectious and metabolic disease takes place. As such, we have argued previously7,8 that application of systems biology concepts during this physiological state would be valuable to uncover the underlying links (eg, pathways and networks) within and between tissues (eg, adipose and liver; liver and mammary), and also to discover new emergent properties that may arise from examining the interactions between all components of a system.8 This integrative approach will provide the means to arrive at a holistic view of how the organism function.9
Bioinformatics in systems biology
Bioinformatics entails the use of computational resources to analyze large-scale datasets from genome, transcriptome, and metabolome studies.10 One of the goals of bioinformatics is to accelerate the discovery of novel biological information from large amounts of ‘omics’ data. In terms of transcriptome data mining, the most widely-used approach is the so-called gene-enrichment approach (also known as overrepresented approach or ORA).11 Although this approach has been widely-used in the data mining process of transcriptome datasets, as previously highlighted7,11 and demonstrated recently,12 it has several limitations particularly associated with time-course experiments. In order to overcome the limitations associated with the ORA we have proposed a novel approach termed Dynamic Impact Approach (DIA).12 In the present review we will illustrate the capability of the DIA to allow for a simultaneously analysis of high-throughput data from multiple tissues in order to provide a more integrative analysis of the system.
Metabolic adaptations during late-pregnancy and early lactation in dairy cows uncovered by transcriptomics
The transition from pregnancy into lactation in mammals (a.k.a., “transition period”), and particularly in dairy cattle, is characterized by metabolic adaptations in major organs (eg, mammary, liver, adipose) that allow the animal to adjust to the need of synthesizing milk for the neonate.13–15 During the same time-frame, the animal experiences a general decrease in food intake.14,16,17 In modern high-producing dairy cows the dramatic increase in milk production and the concomitant decrease in food intake is the cause of marked negative energy balance (NEB), ie, an imbalance between dietary energy intake and output.17
Major metabolic changes reflected in blood during this period include an increase in non-esterified fatty acids (NEFA) and ketone bodies (β-hydroxybutyrate or BHBA is the major one), and a decrease in cholesterol and phospholipids.18,19 Several blood parameters not directly related to metabolism also are affected during this time, eg, increased of positive acute phase proteins (eg, haptoglobin and ceruloplasmin) and decreased negative acute phase proteins (eg, albumin, retinol binding protein) indicating that animals experience inflammatory-like conditions.18
It has been estimated that the liver of transition cows up-takes more than a quarter of circulating NEFA.20 The NEFA are partly oxidized to generate ATP or re-esterified to triacylglycerol (TAG) and stored as lipid droplets or assembled into very-low density lipoprotein (VLDL) to be release in the blood stream.14,20 The excess energy from NEFA oxidation is released as ketone bodies into blood and used by other tissues such as mammary gland, muscle, or nervous system. The accumulation of TAG in liver does not typically compromise liver function until it reaches more than 20% of cellular volume (ie, moderate fatty liver). Deleterious consequences occur when TAG reaches more than 30% of cellular volume.21 The incidence of fatty liver is determined by the blood NEFA level, but also appears to be a consequence of the inflammatory-like condition post-partum.13,22
The ruminant animal absorbs negligible amounts of glucose from the fore-stomach due to extensive bacterial fermentation in the rumen, as such, gluconeogenesis in liver also is a crucial metabolic outcome.20 This is particularly important in modern high-producing dairy cows, considering that the mammary gland of a Holstein cow can produce up to 2 kg of lactose per day. The glucose used by the mammary gland and by other tissues in ruminants is produced almost exclusively by the liver.20
As underscored by the brief overview above, several tissues play a prominent role in allowing cows to adapt successfully to the onset of lactation. However, it is evident that adipose, liver, and mammary gland play a central role in regulating overall metabolism. Our group has been studying the dynamic adaptations of those tissues during the transition period in dairy cows using transcriptomics in combination with bioinformatics tools. In the following sections we describe the main findings from those studies using mainly the DIA and focusing on metabolic regulation.
Metabolic demands in key tissues unveiled from DIA analysis of the transcriptome
The conceptual development and validation of the DIA and an in-depth discussion of its use have been presented elsewhere.7,12,23 Therefore, in this manuscript we wish to present a brief overview of the findings using the DIA, particularly for the KEGG pathways, of the mammary gland from pregnancy to end of subsequent lactation, and novel outcomes using DIA from transcriptomics of bovine adipose and liver during the transition from pregnancy to lactation. To demonstrate the capability of the DIA for integrative system biology, we have provided an example of inter-tissue physiological coordination of mammary, liver, and adipose tissue during the transition period. In addition, we present also the results of the DIA analysis of liver transcriptomics data from pregnancy to lactation in cows fed different level of energy prepartum.
Mammary tissue: what transcriptomics reveals during increased metabolic demand
In order to investigate the adaptation of the mammary gland to lactation, we have performed a transcriptomics analysis of the mammary tissue from one month prior parturition to 300 days in milk (ie, end of lactation) using a bovine specific microarray able to measure ca. 10,000 unique genes.23 The microarray analysis uncovered a tremendous transcriptome adaptation of the mammary tissue to lactation. For instance, compared to one month prior parturition we observed >4,000 DEG during maximal milk secretion.23 Those data indicated that mammary gland heavily relies on transcriptome change to initiate and maintain copious milk synthesis and secretion.
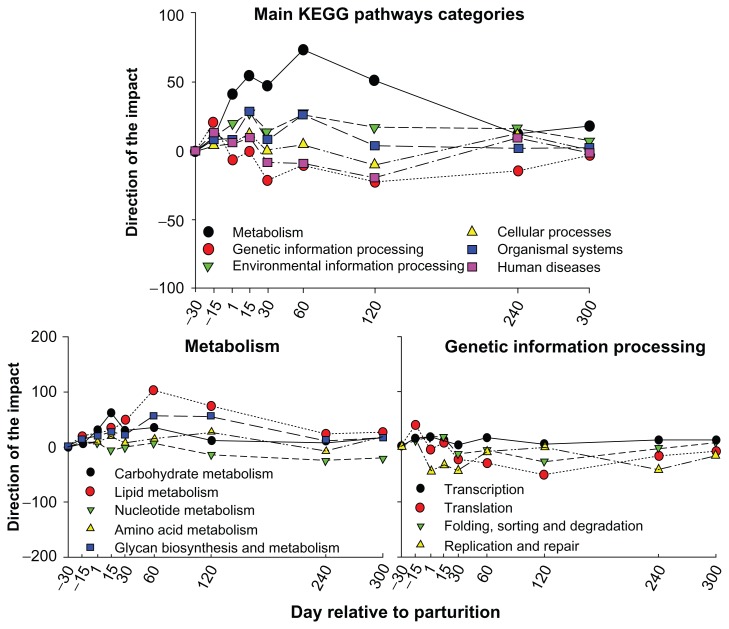
In Figure 1 are reported the direction of the impact of main KEGG pathway categories and subcategories related to ‘Metabolism’ and ‘Genetic information processing’ categories as calculated by the DIA12. The results clearly indicated that there was an overall induction of the metabolism in lactating bovine mammary but also an overall inhibition of the ‘Genetic information processing’. Except for ‘Nucleotide metabolism’ all the other sub-categories of KEGG pathways were induced during lactation, with ‘Lipid metabolism’, ‘Glycan biosynthesis and metabolism’, and ‘Carbohydrate metabolism’ being the most induced. The overall inhibition of ‘Genetic information processing’ was mostly due to the decrease of ‘Translation’ and ‘Replication and repair’ (Fig. 1).
Figure 1.
Metabolic adaptations of bovine mammary gland from end of pregnancy through end of subsequent lactation.
Notes: The data were analysed using the Dynamic impact Approach (DIA).12 The DIA results of the KEGG pathways analysis are reported. Shown are the direction of the impact12 for the main KEGG pathway categories and for several sub-categories related to ‘Metabolism’ and the sub-categories related to ‘Genetic information processing’. Thresholds for the analysis were false discovery rate ≤0.05 for the overall time effect, P-value ≤0.05 for comparisons, and a coverage of at least 30% of annotated genes in the pathways represented on the microarray platform.
Overall the data indicated a large increase in metabolism by the bovine mammary during lactation, with a large increase in demand of glucose, lipid (mostly long-chain fatty acid), and AA.23 The DIA analysis supported most of the previous finding about metabolism of the bovine mammary during lactation, indicating the reliability of the DIA for functional analysis of microarray data. However, as above discussed and previously known,24,25 the mammary gland is highly dependent and influenced by the biology of other organs, particularly liver and adipose.
Adipose tissue: transcriptomics adaptations during the peripartal period
The biological role of liver and mammary in the coordination of lactation in dairy cows has been known for a long-time.25 Until a decade ago adipose tissue was considered as a mere passive energy storage organ in the body, with some additional corollary functions such as providing cushion and thermoregulation. Currently, due to the rapid rise of obesity-related studies, adipose tissue appears to have an important degree of cross-talk with other tissues through the release of endocrine molecules,26,27 and it appears to be very sensitive to energy status of the whole organism.27
Temporal transcriptomics analyses of bovine adipose tissue during transition from pregnancy into lactation are scant. In a recent study, transcriptomics was applied to adipose tissue in first lactation Holstein cows during the transition period.28 The study highlighted the importance of several specific genes but did not provide functional analysis using a systems approach. We recently performed such analysis from the beginning of pregnancy through early lactation in multiparous cows fed diets designed to meet (~100% of net energy requirements) or exceed (~150%, ie, energy-overfed) energy requirements during the entire dry period.29 Overall, more than 3,000 genes were significantly affected (False discovery rate [FDR] ≤ 0.05) by time × energy status. In order to uncover the biological adaptations of adipose tissue in cows fed an energy-sufficient diet we performed KEGG pathway analysis using the DIA of the 956 differentially expressed genes (DEG; FDR ≤ 0.05 for the interaction and P-value between comparisons ≤ 0.01) affected by stage of lactation in the control group. The results (not shown) clearly indicated a general and large decrease of metabolism after parturition in the adipose tissue, with all the metabolic-related pathway sub-categories being inhibited at 14 vs. −14 days relative to parturition (d) with the exception of ‘Energy metabolism’ which was induced, mostly due to an induction of ‘Oxidative phosphorylation’. The most-inhibited pathway subcategories were ‘Lipid metabolism’, “Metabolism of other amino acids’ and ‘Xenobiotic biodegradation and metabolism’ (data not shown).
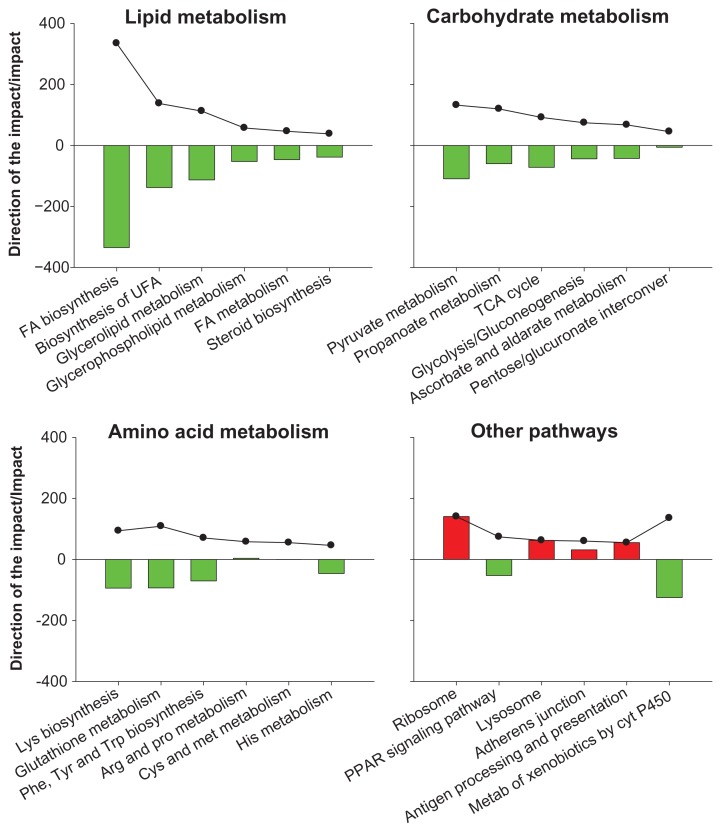
In Figure 2 are reported the most-impacted KEGG pathways among the ones within the ‘Metabolism’ category plus a few additional ones from other categories at two weeks post-partum relative to two weeks pre-partum. Within the ‘Lipid metabolism’ KEGG pathway sub-category all the most-inhibited pathways were related to the synthesis of lipid (Fig. 2); however, ‘Fatty acid metabolism’, a category which encompasses the catabolism of fatty acids, also was inhibited although with an overall lower impact compared to the pathways related to the synthesis of lipid. All the major pathways related to ‘Carbohydrate metabolism’ and ‘Amino acid metabolism’ were inhibited.
Figure 2.
Metabolic adaptations of adipose tissue from end of pregnancy through early lactation.
Notes: Sown are outputs (i.e., impact and direction of the impact) of selected KEGG pathways from the Dynamic Impact Approach analysis of genes differentially expressed in bovine subcutaneous adipose tissue at 14 days post-partum compared to 2 weeks pre-partum. The panels depict the impact (black line and dots) and the direction of the impact (bars; positive red bars denote activation while negative green bars inhibition) for the most impacted pathways in the KEGG subcategory ‘Lipid metabolism’, ‘Carbohydrate metabolism’, ‘Amino acid metabolism’, and other selected pathways. Threshold for the analysis were false discovery rate ≤0.05 for the overall time effect, P-value ≤0.05 for comparisons, and at least 30% of genes in the pathways represented on the microarray.
Overall, the data clearly showed a marked and general reduction of the metabolic activity of the adipose tissue after parturition, particularly for the synthesis of lipid. This makes sense considering that around parturition there is a large physiological change in order to initiate milk production. As indicated above, those changes include the reduction of food intake,16 the increase in energy demands for milk synthesis, the presence of energy-consuming inflammatory-like conditions,18,22,30 and the large decrease in plasma insulin associated with sever NEB.30,31 The decrease of plasma insulin also is accompanied by greater insulin insensitivity in all tissues with exception of the mammary gland.32 The reduction of plasma insulin on the one hand and the decrease of insulin sensitivity on the other hand are the major causes of adipose tissue lipolysis, leading to release of NEFA into the circulation with a concomitant inhibition of TAG synthesis.33 Overall, data from DIA analysis support the decrease of lipid synthesis, particularly TAG synthesis,33 as well the reduction of glucose32 and AA utilization in adipose tissue in early lactation. This adaptation of the adipose tissue might allow for more nutrients to be available for mammary gland.
Liver tissue: transcriptomics adaptations during the peripartal period
The metabolic rate of liver in dairy cows nearly doubles after parturition,34 a response closely associated with adjustments in lipid and glucose metabolism.34 Catabolism of fatty acids, through mitochondrial and peroxisomal oxidation, and esterification of fatty acids into TAG are enhanced after parturition. The TAG accumulate in the cytosol as lipid droplets or are packaged into lipoproteins to be exported into the blood.20 The rate of gluconeogenesis also increases dramatically after parturition,34 primarily to be exported to the mammary gland rather than stored as glycogen.20
We have used bovine liver transcriptomics data from two months prior parturition to forty-nine days into lactation to assess functional adaptations. The original statistical analysis19 uncovered a modest degree of transcriptome adaptations during the transition from pregnancy into lactation. The data, however, allowed proposing a qualitative model of physiological adaptation of liver also considering the adipose and mammary tissue.19
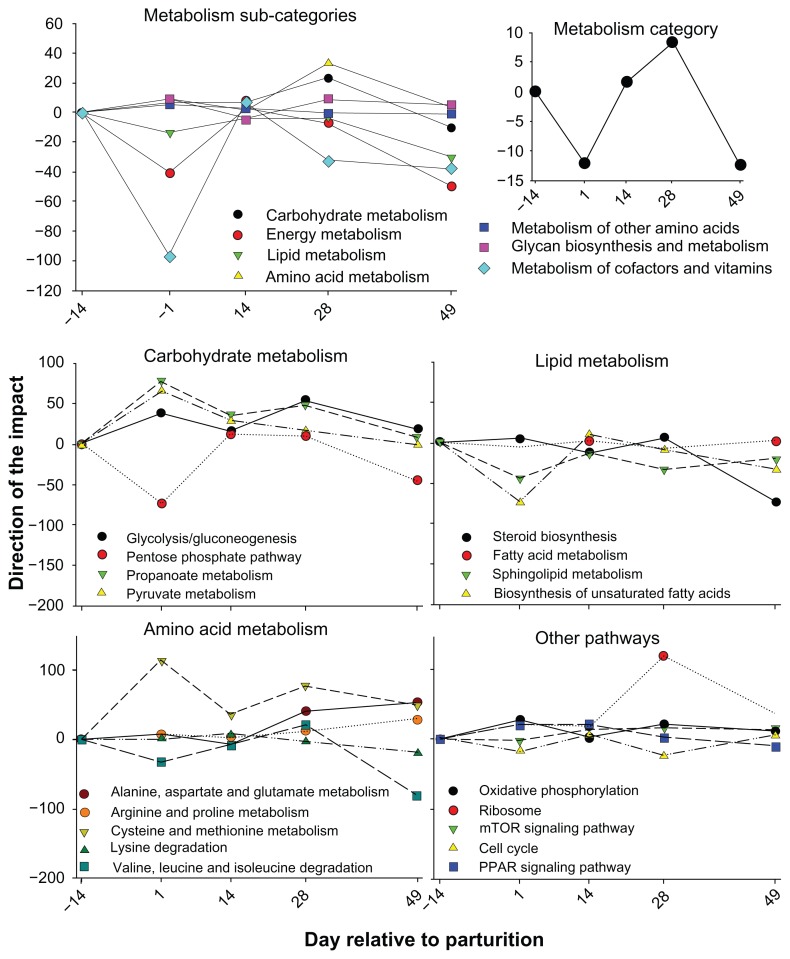
The same data (ie, control group) plus data from another set of animals fed different levels of energy prepartum35 were combined and re-analyzed statistically using a more powerful statistical approach.36 The new analysis uncovered 4,970 DEG with FDR ≤ 0.05 for time × treatment interaction. A DIA analysis was performed using data from the control group considering the postpartum time points relative to two weeks prior to parturition. The overall view of the KEGG pathway ‘Metabolism’ category and main sub- categories are reported in Figure 3. The DIA analysis revealed an overall, although modest, increase in metabolism during the first four weeks post-partum, with a drop just after parturition. The increase in metabolism was mostly due to ‘Amino acid metabolism’ and ‘Carbohydrate metabolism’ with an overall inhibition of ‘Metabolism of cofactors and vitamins’ (Fig. 3).
Figure 3.
Metabolic adaptations in bovine liver from end of pregnancy through peak lactation.
Notes: DIA functional analysis of bovine liver tissue microarray data from −14 to 49 days relative to parturition. The top panel depicts the direction of the impact as calculated by the DIA of main KEGG sub-categories pathways within the main ‘Metabolism’ category (depicted in the right side of the subcategories graph). The middle and bottom panels depict the direction of the impact of selected pathways (ie, among the most impacted as calculated by the DIA) in the sub-categories ‘Carbohydrate metabolism’, ‘Lipid metabolism’, and ‘Amino acid metabolism’. The bottom-right panel depicts the pattern of direction of the impact of other pathways not related to the above KEGG pathway sub-categories or not included in the main ‘Metabolism’ category but with indirect connection with metabolism. Threshold for the analysis were false discovery rate ≤0.05 for the overall time effect, P-value ≤0.05 for comparisons and at least 20% of genes in the pathways represented on the microarray.
The DIA analysis of the pathways in the ‘Carbohydrate metabolism’, ‘Lipid metabolism’, and ‘Amino acid metabolism’ sub-categories uncovered an overall increase of gluconeogenesis and propionate utilization (Fig. 3). Interestingly, the microarray data suggested that the utilization of propionate increased suddenly after parturition and remained high for the first month postpartum. Those data are supported by previous findings.20,34 None of the pathways related to lipid metabolism were increased after parturition. Previous data showed an increase in hepatic lipid metabolism, particularly an increase in fatty acid oxidation.20 Thus, our findings from DIA were apparently unexpected.
Interestingly, it has previously been proposed based on several lines of evidence that the main factors driving the increase in fatty acid oxidation in liver during the transition period are:20 (1) the increase in NEFA availability (ie, NEFA concentration in the blood); and (2) activity of carnitine palmitoyl transferase 1 (CPT-1), the rate-limiting enzyme for the entry of fatty acids into the mitochondria for oxidation after parturition. The latter was evidenced by an increase in CPT-1 activity37 and expression19 early postpartum. Our data support the previous proposal because they suggest that liver does not increase its ability to metabolize the fatty acids through greater expression of metabolic pathways; rather, the greater utilization of fatty acids is likely due to an increase in substrate and/or change in expression of few “key” molecules, such as CPT-1. Interestingly, our data also indicated a lack of induction of ketogenesis at the gene expression level (ie, the pathways was not impacted in the first month after parturition). The increase in ketogenesis after parturition is very important;20 therefore, the lack of induction of this pathway at the transcriptional level might indicate that ketone body production, as fatty acid catabolism, is mostly driven by concentration of substrates.
The metabolism of AA by liver postpartum has been discussed in detail previously.38,39 The liver of lactating bovine actively absorbs large amounts of AA and essentially determines their availability for other organs such as mammary gland.39 Results from the DIA analysis (Fig. 3) suggested that the AA metabolism-related pathways were the most-impacted in liver during the transition from pregnancy into lactation, particularly at the end of the first month of lactation. The increase in AA metabolism was mostly due to increase in ‘Cysteine and methionine metabolism’ and ‘Alanine, aspartate and glutamate metabolism’ (Fig. 3). Methionine is an essential AA, particularly limiting for milk synthesis.40 The surge in ‘Alanine, aspartate and glutamate metabolism’ observed with the DIA was mostly due to the increase expression of genes involved in utilization of aspartate for provision of TCA intermediates (ie, fumarate and oxaloacetate) and the formation of glutamine from glutamate. The biological significance of greater glutamine metabolism in liver is not readily apparent. Overall, the data indicated that most AA in the liver are used for production of TCA intermediates, ie, as energy sources likely to spare glucose for milk synthesis.
The DIA indicated an overall slight increase of ‘Oxidative phosphorylation’ despite an overall inhibition of energy production (the inhibition was mostly due to the ‘Sulfur metabolism’ pathway) coupled with a large increase in the protein synthesis machinery (ie, ‘Ribosome’) (Fig. 3). The increase in ATP production is probably also due to the greater need of protein synthesis. A substantial increase in bovine hepatic protein synthesis post-partum was reported previously.41 The increase in ‘Ribosome’ was not accompanied by an increase of “mTOR signaling pathway”, which is known to be the master regulatory of protein synthesis in non-ruminants.42 The data indicated that the liver had an increased capacity for protein synthesis overall, which might be important (among others) for the immune activity of the organ after parturition.13,22
Peroxisome proliferator-activated receptor alpha (PPARα) is a nuclear receptor highly-expressed in liver, with pivotal roles in controlling fatty acid metabolism at least in non-ruminant.43 Some recent data suggested that this nuclear receptor regulates lipid metabolism in ruminants as well.44 The importance of PPARα in dairy cow liver was inferred by up-regulation of its expression during early lactation19 and by the increase in expression of several target genes in neonatal calves treated with a specific PPARα agonist.45 Analysis of the liver microarray data around parturition using DIA uncovered an overall activation, although modest, of the ‘PPAR signaling’ pathway during the first two weeks postpartum (Fig. 3), which supports a role of PPAR in early lactation.
Overall, the data suggested that, from a metabolic point of view, the control of glucose metabolism, protein synthesis, production of energy, and amino acid metabolism are strongly regulated at the transcriptomics level; however, the liver transcriptome seems to respond modestly to the surge of NEFA, with only few key factors for fatty acid catabolism (not overall pathways) exhibiting changes in expression.
Integrative coordination of the transcriptome during the transition period
As alluded above in the introduction, the era of the reductionist approach in science experienced a setback with the advent of the “modern” systems biology approach.7 The understanding of the physiological or pathological adaptations of the animal to a change in physiological state requires an integrative view of metabolism. In addition, the use of a single time point is reductive and insufficient to capture the dynamism of the biological adaptations; thus, implementation of time-course experiments should be undertaken in a more routine basis. The DIA approach12 is well-suited for data mining in time-course studies involving multiple treatments. Comparisons can be performed between any dataset and the validity of such comparisons is more powerful if the same technology and the same statistical approach are used.
To illustrate the dynamics of tissue coordination during the transition period, we have compared the results from microarray experiments of mammary tissue, adipose tissue, and liver during the adaptation from pregnancy through early lactation. The microarray data were the same as used above for mammary gland, adipose, and liver.19,23,29 All datasets were analysed statistically using the same approach as described previously.23 Statistical results used for the analysis were from −14 days relative to parturition in liver and adipose or −15 days in mammary tissue. For simplicity we defined this prepartal time point as “−15 d” for all tissues.
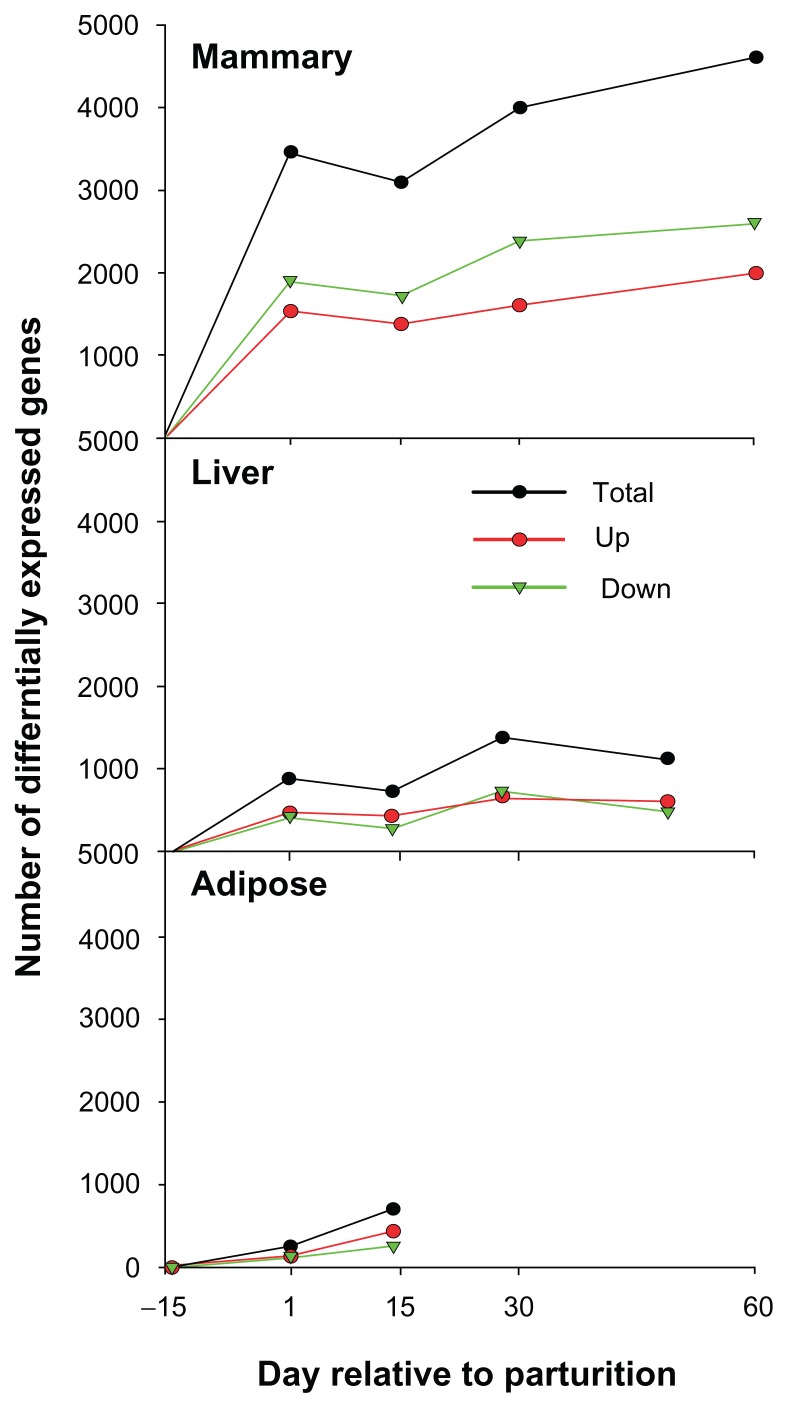
In Figure 4 is reported the number of DEG in mammary tissue, liver, and adipose tissues from 2 weeks prior parturition up to 60 days in milk. From the figure it is evident that the mammary gland experienced the larger transcriptomics change from pregnancy to lactation (between ca. 3,000 and ca. 5,000 DEG out of ca. 10,000 unique genes in the microarray platform; ie, 30%–50% of the transcriptome) compared with liver (between ca. 800 and ca. 1,400; the platform used for liver had only ca. 7,000 unique genes, ie, 11%–20% of the transcriptome) and adipose (between ca. 100 and ca. 900 out of 10,000 unique genes; ie, 1%–9% of the transcriptome) with also most of the DEG being down-regulated in mammary. As discussed above, those data emphasized the larger dependence of the lactating mammary gland from the transcriptome compared to other tissues.
Figure 4.
Number of differentially expressed genes in each time point relative to −15 in mammary tissue, liver, and adipose tissue.
Notes: The genes were deemed to be differentially expressed if the overall time effect was with a FDR < 0.05 and the comparison between each time point was with P-value < 0.05. Data from mammary and liver are from dataset prior deposited in GEO (GSE19055 for mammary tissue and GSE2692 for liver) while for adipose are from unpublished data. For all tissues the statistical analysis was as described previously.23
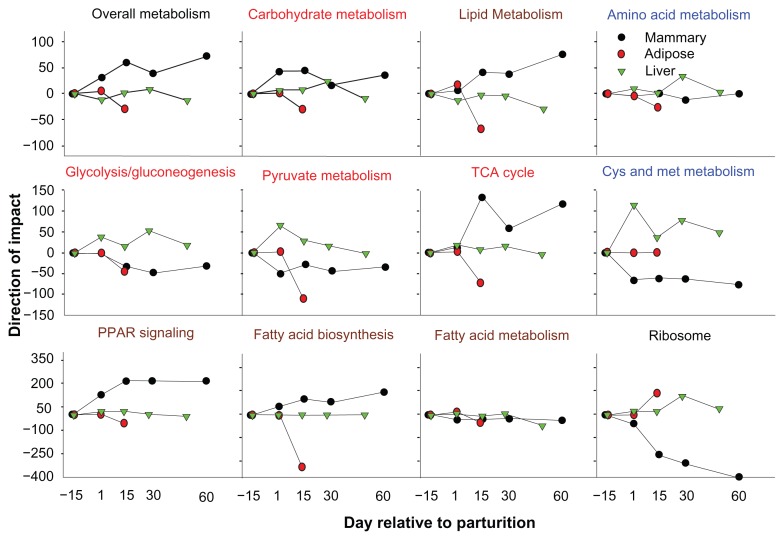
In Figure 5 are reported the direction of the impact as calculated by the DIA of overall ‘Metabolism’ KEGG category of pathways, the three main sub-categories of pathways related to metabolism and most interesting associated pathways. From a metabolic point of view, these data suggest that during early lactation the mammary gland has dominance over other tissues (Fig. 5) and, as such, these findings support well-established knowledge.46 The DIA also estimated that liver experiences a decrease in overall metabolism after the first month of lactation when peak milk yield is attained.23 In terms of coordinating adaptations to lactation, the data suggest that the metabolic importance of liver is more pronounced during the first month of lactation, which often coincides with the nadir of NEB.16,19 This in turn highlights that liver is critical during the period of extensive lipid mobilization due to NEB. However, the large prevalence of diseases in dairy cows during the first month of lactation13 also requires an “active” liver in order to participate in the immune response.18
Figure 5.
Direct comparison of metabolic adaptations in mammary tissue, adipose tissue, and liver during the two last weeks of pregnancy until peak lactation.
Notes: The panel with bold black title depicts the direction of the impact of the overall ‘Metabolism’ category of KEGG pathways in mammary gland, adipose tissue, and liver of dairy cows from 2 weeks pre-partum to 2 months postpartum. The panels with bold red, brown, and blue titles depict the direction of the impact for the ‘Metabolism’ sub-categories ‘Carbohydrate metabolism’, ‘Lipid metabolism’, and ‘Amino acid metabolism’, respectively. The panels with regular font title depict the direction of the impact for selected pathways in the above KEGG pathways sub-categories plus the ‘PPAR signaling’ and ‘Ribosome’ pathways.
The overview of ‘Metabolism’ KEGG pathway sub-categories (Fig. 5) clearly suggested that mammary gland experiences a large increase in lipid metabolism with a concomitant decrease of the same pathways in the adipose tissue, while in liver lipid metabolism remains quite stable with a slight decrease as lactation progresses. Specifically, synthesis of fatty acids and lipid was prevalent in mammary while strongly decreased in adipose (Fig. 5). Carbohydrate metabolism also was more prevalent in mammary (due mostly to ‘Galactose metabolism’) with an important role in liver and a considerable decrease in adipose (Fig. 5). The visualization of selected pathways related to carbohydrate metabolism (Fig. 5) revealed an increase in hepatic production of glucose via gluconeogenesis and use of lactate as energy source (eg, pyruvate metabolism, particularly in early lactation, Figure 5; KEGG pathway details are not shown). Those data are consistent with previous findings.14,20,34
Interestingly, the TCA cycle was induced during early lactation particularly in mammary gland but also in liver (Fig. 5). The increase of TCA cycle activity, together with an increase in ‘Oxidative phosphorylation’ (see above and previous data23), was indicative of an overall increase in energy production in both organs, with an apparent greater effect in mammary tissue. In liver and adipose tissue, glucose utilization is spared for the synthesis of lactose in mammary, eg, fatty acids in liver can provide a large amount of energy through oxidation (see above) but this metabolic pathway is not relevant for the lactating mammary gland.47,48 One common source of energy in both organs is the AA. From our data, AA metabolism appeared to be more important in liver compared to other tissues (Fig. 5). The liver has a greater degree of utilization of methionine and cysteine (as discussed above); whereas, the mammary gland appears to spare methionine, probably for the synthesis of milk proteins. In mammary gland, methionine is considered a limiting AA.40 The overall increase in AA metabolism by liver indicates a more predominant role of this tissue in utilization of AA. However, the present data do not allow speculating about differences in the degree of utilization of AA as energy sources in liver relative to mammary.
The large increase in the ‘Ribosome’ pathway was suggestive of an overall increase in protein synthesis in liver and adipose relative to mammary (Fig. 5). Previous studies have reported increases in protein synthesis in liver after parturition,41 while protein synthesis in adipose tissue during early lactation has been shown not to differ compared to pre-partum in rats49 and also ruminants (cited in33). The decrease of the protein synthesis machinery in mammary tissue during lactation is a novel finding, which has been discussed previously.50 We propose that this “unexpected” pattern is important to enhance translation of milk protein coding genes or genes coding for enzymes involved in milk synthesis over non-secreted or non-milk synthesis-related proteins;50 thus, the decrease of protein synthesis machinery indicated a remarkable change in functional specialization of the tissue. In this sense, the increase of the protein synthesis machinery in liver and adipose might be due to the acquisition and/or intensification of additional functions (namely for liver) with fairly specific tasks or a decrease in specialization of the tissue (namely for adipose). It also is possible that the change in physiological state in adipose tissue leads to enhanced translation of fewer mRNA, particularly in light of the observed decreases in expression of many genes29 (see above). If this is the case, we can expect instead an increase in specialization of the tissue.
Overall, the data suggest that at the onset of lactation the mammary gland becomes the prevalent metabolic tissue with a marked increase in anabolic activity. The liver appears to experience a slight increase in metabolism, particularly associated with carbohydrate (eg, gluconeogenesis) and AA. As reported previously14,20,34 this increase is likely devoted to respond to the mammary gland’s demands. Interestingly, our data showed that there is a concomitant and substantial decrease of adipose tissue metabolism, suggesting that the adipose tissue reduces its metabolism as a way to decrease energetic burden and spare resources for the mammary gland. The interaction between these three tissues is not only indirect (ie, through coordinated changes in metabolism) but as shown for liver and adipose tissue51 also direct.
Transcriptomics during transition: effect of nutrition analyzed by DIA
We have discussed previously the transcriptome adaptations in adipose tissue and liver in dairy cows consuming different levels of energy pre-partum.7 Briefly, the microarray data from adipose clearly showed that the effect of overfeeding energy prepartum is a transient one, ie, the greatest number of DEG were observed at two-weeks before parturition when cows were still consuming control or excess energy. Those data were analyzed via ORA and DIA and the most relevant pathways were discussed.7 All those pathways were related to lipid metabolism with DIA revealing ‘PPAR signaling’ as one of the most-impacted and indicating that overfeeding energy relative to energy-sufficient diets resulted in greater lipid synthesis, and this was likely regulated through PPARγ.
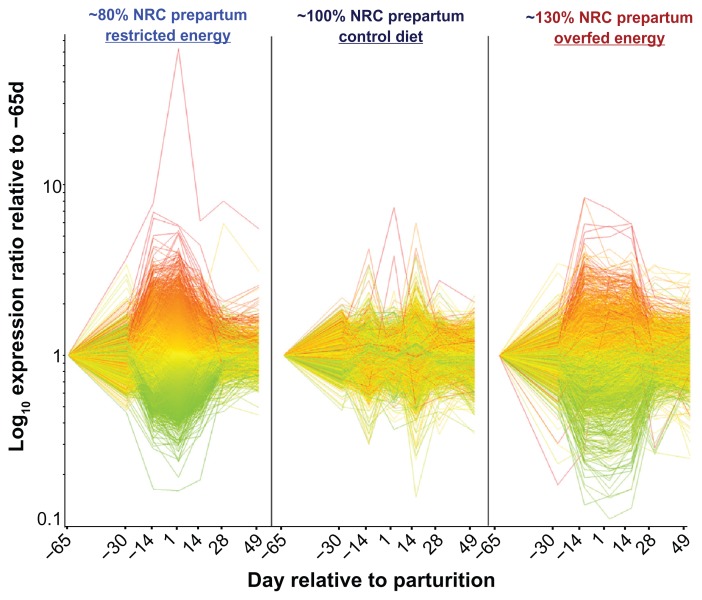
The analysis of liver was performed assembling and re-analyzing microarray data from two previous studies.19,35 The assemblage and the use of a more powerful statistical analysis clearly reveal a larger transcriptomics adaptation of liver experience either restricted or high energy prepartum compared to cows fed control diet (Fig. 6). It appears that the liver transcriptome is quite insensitive to the change of physiological phase but highly sensitive to the change in dietary energy; this might be interpreted as a “stress-associated” response.
Figure 6.
Overall transcriptomics adaptation to different level of prepartum energy in bovine liver.
Notes: Shown is the image generated by Genespring GX7 (Agilent) of the genes deemed to be differentially expressed with time × treatment with false discovery rate <0.05. Dataset was a combination of two previously published microarray data19,35 generated by experiments were cows were fed diets to meet 150% (ad-libitum), 80% (restricted), or 100% (control) of energy requirements during the dry period. All cows were fed the same diet after parturition. Statistical analysis was run using Mixed model as described previously23 with time × treatment as main effect. Green and red lines denote genes with an expression ratio lower or higher at 1 relative to −65 d in the cows fed restricted energy prepartum. This allows to quickly visualizing the pattern of same group of genes in the other treatments. From the image is evident that the control diet had a mild effect while either the restricted or the ad-libitum energy prepartum had a strong effect on the transcriptomics adaptation to lactation.
A k-means clustering analysis in association with ORA was previously performed in order to capture co-regulated functions.7 We observed very few enriched functions in the resulting clusters, with protein synthesis being one of the most coordinated.7 In this manuscript we present results of DIA analysis of the same dataset in liver to provide an integrative visualization of the metabolic adaptations to prepartum level of dietary energy.
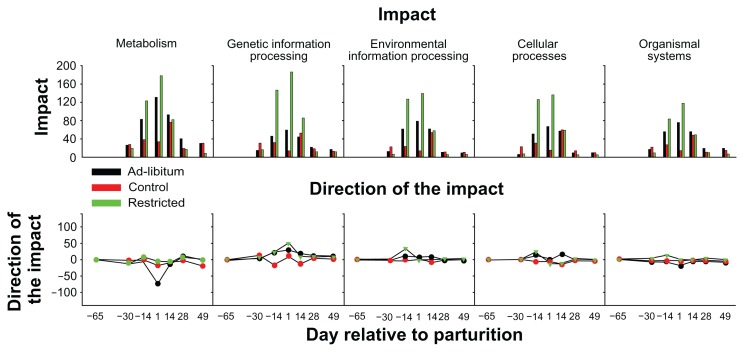
The DIA results of main categories of KEGG pathways are shown in Figure 7. The data clearly indicated that energy restriction prepartum had the strongest impact on all main categories of pathways, followed by high energy prepartum. The most impacted pathways were related to ‘Metabolism’ and ‘Genetic Information Processing’. When the direction of the impact results were considered it appeared clear that dynamics of metabolism were not affected overall by prepartum diet. The only exception being the evident reduction of metabolism the day after parturition in cows overfed energy prepartum.
Figure 7.
Effect of different levels of dietary energy prepartum on KEGG pathways: overall view.
Notes: Reported are 5 out of 6 main categories of KEGG pathways as provided by the Dynamic Impact Approach (DIA) analysis of the dataset in Figure 6. The upper panel report the impact and the lower panel the direction of the impact as previously described.12 The DIA analysis was run using the following cut-off: false discovery rate ≤0.05 for the time × treatment effect, P ≤0.05 for each comparison, and at least 20% of genes in the pathways represented on the microarray.
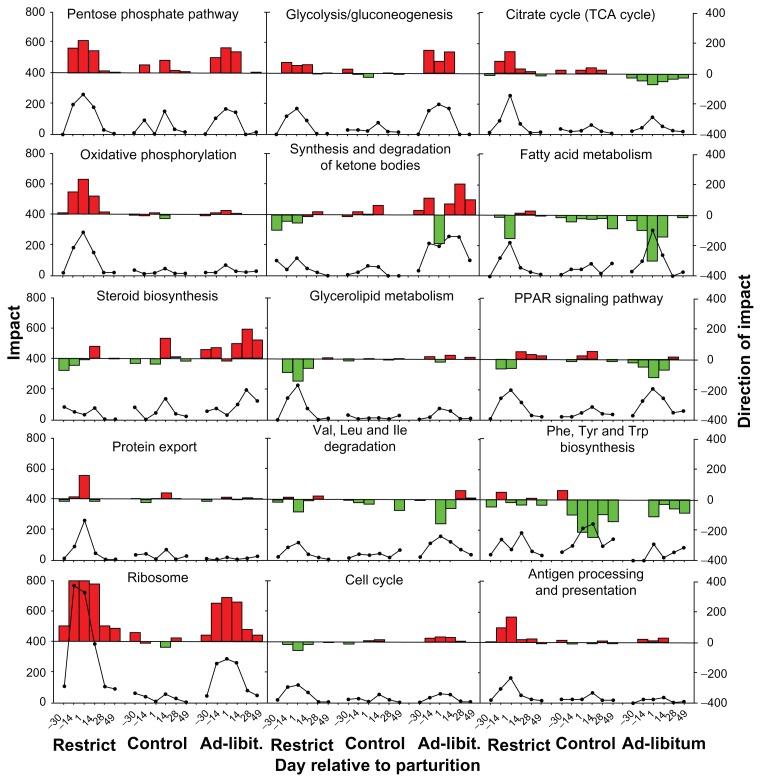
From this point of view it appears that KEGG sub-categories ‘Carbohydrate metabolism’, ‘Lipid metabolism’, ‘Amino acid metabolism’, and ‘Energy metabolism’ were inhibited overall early post-partum (Supplemental file 1). When pathways within those sub-categories were investigated (Fig. 8), ‘Fatty acid metabolism’ and ‘Synthesis and degradation of ketone bodies’ were among the most inhibited and potentially regulated by PPARα, as suggested by the ‘PPAR signaling’ pathway.
Figure 8.
Metabolic adaptations of bovine liver to different levels of dietary energy prepartum.
Notes: Reported are among the most impacted pathways related to metabolism in liver of cows fed different level of energy prepartum as described in caption of Figures 6 and 7. The lines in graphs depict the impact and the bars the direction of the impact as previously described.12 The red positive bars denote activation while the negative green bars inhibition of the pathway in each time point relative to −65 day relative to parturition as calculated by the DIA. Thresholds for analysis were false discovery rate ≤0.05 for the time × treatment effect, P ≤0.05 for each comparison, and at least 20% of genes in the pathways represented on the microarray.
Interestingly, ‘Fatty acid metabolism’ as well as ‘PPAR signaling’ were reduced already two weeks prior to parturition in the overfed cows prepartum and the pathway was slowly induced afterwards compared to the other groups. In addition, the prepartum energy-restricted cows experienced an apparent inhibition of fatty acid metabolism, but this inhibition was only transient with a quick recovery after parturition. The overall result was a slightly higher lipid metabolism among the three groups during the first month of lactation (Fig. 8). The pattern of ‘PPAR signaling’ was similar to ‘Fatty acid metabolism’ (Fig. 8). The cows overfed energy prepartum had a more pronounced increase in ketone body metabolism after parturition, suggesting a greater degree of ketogenesis relative to the other groups (Fig. 8). Those data are supported by the greater blood BHBA in the overfed vs. underfed cows.35
Another interesting revelation by the DIA analysis was the apparent greater increase in production of cholesterol in overfed cows, both before and after parturition (see ‘Steroid biosynthesis’ pathway in Figure 8), a response that we have confirmed in recent studies from our laboratory (Graugnard et al, Khan et al, unpublished results). The overfed cows had a greater accumulation of lipid in liver compared to the underfed cows.35 Cholesterol is an important substrate for the formation of lipoproteins in liver, with a potential, but unresolved, role in the regulation of VLDL synthesis and secretion.52 From this point of view a greater need for cholesterol synthesis in overfed cows might be associated with the greater blood NEFA, particularly post-partum,35 some of which is re-esterified into TAG and stored or repacked with lipoproteins. Because the amount of NEFA prepartum was similar regardless of diet,35 this mechanism does not explain the greater steroid synthesis prepartum. In addition, it has been observed that fatty liver in dairy cows is associated with a decrease in liver cholesterol.53 Therefore, those additional observations leave unexplained the apparently greater steroidogenesis in overfed cows.
Remarkably, the ‘Energy metabolism’ sub-category suggested that the energy production in both underfed and overfed animals prepartum was lower compared to control during the transition from pregnancy to lactation (Fig. 8). However, this pattern was mostly due to the ‘Sulfur metabolism’ pathway (data not shown); but the evaluation of ‘Oxidative phosphorylation’, the most important pathway in this subcategory, revealed an increase in energy production in liver of underfed cows. This also is supported by the increase in the ‘TCA cycle’ pathway (Fig. 8). The ‘Glycolysis/Gluconeogenesis’ pathway appeared to be more activated in both underfed and overfed animals compared to the control group from prepartum through two weeks postpartum (Fig. 8). The detailed visualization of the pathways (data not shown) indicated a greater utilization of lactate in the underfed group and likely for gluconeogenesis, while the overfed cows appeared to have used more glucose and less lactate as energy sources. This is in agreement with previous observations of a large decrease in % liver glycogen in cows experiencing substantial lipid accumulation.53
What appeared quite evident from Figure 8 was the greater increase during the transition period in ‘Ribosome’ KEGG pathway in both underfed and overfed cows relative to control, with a greater increase in the former. Those results were unexpected and we have no explanation of their likely biological meaning. Few studies regarding the relationship between energy in the diet and protein synthesis in the liver are available in the scientific literature. For instance, in lactating rats the restriction of energy or protein decreased the absolute protein synthesis rate in both liver and, in greater amount, mammary gland.54 In obese humans the reduction of dietary energy caused an overall reduction of protein synthesis.55 Chronic high dietary energy also failed to increase protein synthesis.56 Based on those previous findings we conclude that the large increase in ribosome is a novel finding that requires further investigation.
Overall, use of the DIA indicated that lipid metabolism in liver was highly sensitive to level of energy prepartum and PPAR signaling playing an important role in regulating it. The large increase in ribosome and, thus, the protein synthesis machinery due to overfeeding and underfeeding remains a puzzling discovery that should be studied in more detailed in the future. We are tempted to speculate that if such large increase in ribosome was translated into an overall greater protein synthesis it would have a dramatic effect on energy utilization for protein synthesis and, as such, could have placed an additional toll on the organ during this important physiological stage. What is clear from the analysis is that the postpartum responses represent a carryover effect due to chronic under or overfeeding during the dry period.
Perspectives
The breadth of knowledge that has been acquired on ruminant metabolism over the last half a century has allowed us to form a clearer picture of the key biochemical pathways, their “main” control points, and their response to nutrition in different tissues of the animal. The advent of genome-enabled technologies was a breakthrough, and groups across the world have already invested substantial amounts of resources in genome sequencing, annotation, and functional genomics (eg, transcriptomics), particularly in bovine. Availability of bioinformatics tools also has accelerated the interpretation of data from large-scale datasets. The development of the DIA was an attempt to provide a tool for analysis of time-course experiments in an integrative fashion. This tool has proven valuable for generating biologically- meaningful data from time-course transcriptomics experiments and has shown the capability for integrative analysis; thus, it is a suitable platform for systems biology. The development of web-accessible resources of integrated data related to dairy cattle nutrition and physiology will be of value to comparative biologists.
Supplementary Data
Excel file with results for the KEGG pathway DIA analysis of liver from −65 to +49 day relative to parturition in dairy cows fed restricted (80% NRC recommendation), control (100% NRC recommendation), or ad-libitum (130% NRC recommendation) pre-partum.
Footnotes
Portions of this work were presented (J.J. Loor and M. Bionaz) at an invited lecture during the Oskar Kellner Symposium, September 9–11, 2011, Warnemunde, Germany.
Funding sources
None.
Competing Interests
JJL and MB received patent-related funding from the University of Illinois.
Author Contributions
Conceived and designed the experiments: JJL. Conceived and performed the analyses: MB. Wrote the manuscript: MB, JJL. Agree with manuscript results and conclusions: JJL, MB. Jointly developed the structure and arguments for the paper: JJL, MB. Made critical revisions and approved final version: JJL, MB. All authors reviewed and approved of the final manuscript.
Disclosures and Ethics
Author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
References
- 1.von Bertalanffy L. General system theory: a new approach to unity of science. 1. Problems of general system theory. Human Biol. 1951;23(4):302–12. [PubMed] [Google Scholar]
- 2.Cornish-Bowden A. Making systems biology work in the 21st century. Genome Biol. 2005;6(4):317. doi: 10.1186/gb-2005-6-4-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornish-Bowden A, Cardenas ML. Systems biology may work when we learn to understand the parts in terms of the whole. Biochem Soc Trans. 2005 Jun;33(Pt 3):516–9. doi: 10.1042/BST0330516. [DOI] [PubMed] [Google Scholar]
- 4.Mo ML, Palsson BO. Understanding human metabolic physiology: a genome-to-systems approach. Trends Biotechnol. 2009 Jan;27(1):37–44. doi: 10.1016/j.tibtech.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Dobrin R, Zhu J, Molony C, et al. Multi-tissue coexpression networks reveal unexpected subnetworks associated with disease. Genome Biol. 2009;10(5):R55. doi: 10.1186/gb-2009-10-5-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barendse W, Reverter A, Bunch RJ, Harrison BE, Barris W, Thomas MB. A validated whole-genome association study of efficient food conversion in cattle. Genetics. 2007 Jul;176(3):1893–905. doi: 10.1534/genetics.107.072637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loor JJ, Bionaz M, Invernizzi G. Systems biology and animal nutrition: insights from the dairy cow during growth and the lactation cycle. In: te Pas MFW, Woelders H, Bannink A, editors. Systems Biology and Livestock Science. Wiley-Blackwell; 2011. pp. 215–46. [Google Scholar]
- 8.Loor JJ. Genomics of metabolic adaptations in the peripartal cow. Animal. 2010;4:1110–39. doi: 10.1017/S1751731110000960. [DOI] [PubMed] [Google Scholar]
- 9.Bruggeman FJ, Westerhoff HV. The nature of systems biology. Trends Microbiol. 2007 Jan;15(1):45–50. doi: 10.1016/j.tim.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Luscombe NM, Greenbaum D, Gerstein M. What is bioinformatics? A proposed definition and overview of the field. Methods Inf Med. 2001;40(4):346–58. [PubMed] [Google Scholar]
- 11.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009 Jan;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bionaz M, Periasamy K, Rodriguez-Zas SL, Hurley WL, Loor JJ. A novel dynamic impact approach (DIA) for functional analysis of time-course omics studies: validation using the bovine mammary transcriptome. PloS One. 2012;7(3):e32455. doi: 10.1371/journal.pone.0032455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drackley JK, Dann HM, Douglas GN, et al. Physiological and pathological adaptations in dairy cows that may increase susceptibility to periparturient diseases and disorders. Ital J Anim Sci. 2005 Oct-Dec;4(4):323–44. [Google Scholar]
- 14.Drackley JK. ADSA Foundation Scholar Award. Biology of dairy cows during the transition period: the final frontier? J Dairy Sci. 1999 Nov;82(11):2259–73. doi: 10.3168/jds.s0022-0302(99)75474-3. [DOI] [PubMed] [Google Scholar]
- 15.Bauman DE, Currie WB. Partitioning of nutrients during pregnancy and lactation: a review of mechanisms involving homeostasis and homeorhesis. Journal of Dairy Science. 1980 Sep;63(9):1514–29. doi: 10.3168/jds.s0022-0302(80)83111-0. [DOI] [PubMed] [Google Scholar]
- 16.Ingvartsen KL, Andersen JB. Integration of metabolism and intake regulation: a review focusing on periparturient animals. J Dairy Sci. 2000 Jul;83(7):1573–97. doi: 10.3168/jds.S0022-0302(00)75029-6. [DOI] [PubMed] [Google Scholar]
- 17.Grummer RR, Mashek DG, Hayirli A. Dry matter intake and energy balance in the transition period. Vet Clin North Am Food Anim Pract. 2004 Nov;20(3):447–70. doi: 10.1016/j.cvfa.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Bionaz M, Trevisi E, Calamari L, Librandi F, Ferrari A, Bertoni G. Plasma paraoxonase, health, inflammatory conditions, and liver function in transition dairy cows. Journal of Dairy Science. 2007 Apr;90(4):1740–50. doi: 10.3168/jds.2006-445. [DOI] [PubMed] [Google Scholar]
- 19.Loor JJ, Dann HM, Everts RE, et al. Temporal gene expression profiling of liver from periparturient dairy cows reveals complex adaptive mechanisms in hepatic function. Physiol Genomics. 2005 Oct 17;23(2):217–26. doi: 10.1152/physiolgenomics.00132.2005. [DOI] [PubMed] [Google Scholar]
- 20.Drackley JK, Overton TR, Douglas GN. Adaptations of glucose and long-chain fatty acid metabolism in liver of dairy cows during the periparturient period. J Dairy Sci. 2001;84(E Suppl):E100–12. [Google Scholar]
- 21.Reid IM, Collins RA. The pathology of post-parturient fatty liver in high-yielding dairy cows. Invest Cell Pathol. 1980 Jul-Sep;3(3):237–49. [PubMed] [Google Scholar]
- 22.Bertoni G, Trevisi E, Lombardelli R. Some new aspects of nutrition, health conditions and fertility of intensively reared dairy cows. Ital J Anim Sci. 2009;8(4):491–518. [Google Scholar]
- 23.Bionaz M, Periasamy K, Rodriguez-Zas SL, et al. Old and new stories: revelations from functional analysis of the bovine mammary transcriptome during the lactation cycle. PloS one. 2012;7(3):e33268. doi: 10.1371/journal.pone.0033268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mepham TB. Biochemistry of Lactation. Amsterdam: Elsevier Science Publishers B.V; 1983. Physiological aspects of lactation; pp. 3–28. [Google Scholar]
- 25.Bauman DE, Elliot JM. Control of nutrient partitioning in lactating ruminants. In: Mepham TB, editor. Biochemistry of Lactation. Amsterdam: Elsevier Science Publishers B.V; 1983. pp. 437–68. [Google Scholar]
- 26.Hausman GJ, Dodson MV, Ajuwon K, et al. Board-invited review: the biology and regulation of preadipocytes and adipocytes in meat animals. J Anim Sci. 2009 Apr;87(4):1218–46. doi: 10.2527/jas.2008-1427. [DOI] [PubMed] [Google Scholar]
- 27.Lee DE, Kehlenbrink S, Lee H, Hawkins M, Yudkin JS. Getting the message across: mechanisms of physiological cross talk by adipose tissue. Am J Physiol Endocrinol Metab. 2009 Jun;296(6):E1210–29. doi: 10.1152/ajpendo.00015.2009. [DOI] [PubMed] [Google Scholar]
- 28.Sumner-Thomson JM, Vierck JL, McNamara JP. Differential expression of genes in adipose tissue of first-lactation dairy cattle. J Dairy Sci. 2011 Jan;94(1):361–9. doi: 10.3168/jds.2010-3447. [DOI] [PubMed] [Google Scholar]
- 29.Janovick NA, Loor JJ, Ji P, et al. Overfeeding energy prepartum dramatically affects peripartal expression of mRNA transcripts in subcutaneous adipose tissue compared with controlling energy intake prepartum. J Dairy Sci. 2009;92(Suppl 1):709. [Google Scholar]
- 30.LeBlanc S. Monitoring metabolic health of dairy cattle in the transition period. J Reprod Dev. 2010 Jan;56(Suppl):S29–35. doi: 10.1262/jrd.1056s29. [DOI] [PubMed] [Google Scholar]
- 31.Grummer RR, Wiltbank MC, Fricke PM, Watters RD, Silva-Del-Rio N. Management of dry and transition cows to improve energy balance and reproduction. J Reprod Dev. 2010 Jan;56(Suppl):S22–8. doi: 10.1262/jrd.1056s22. [DOI] [PubMed] [Google Scholar]
- 32.Bell AW, Bauman DE. Adaptations of glucose metabolism during pregnancy and lactation. J Mammary Gland Biol Neoplasia. 1997 Jul;2(3):265–78. doi: 10.1023/a:1026336505343. [DOI] [PubMed] [Google Scholar]
- 33.Vernon RG, Pond CM. Adaptations of maternal adipose tissue to lactation. J Mammary Gland Biol Neoplasia. 1997 Jul;2(3):231–41. doi: 10.1023/a:1026380220364. [DOI] [PubMed] [Google Scholar]
- 34.Reynolds CK, Aikman PC, Lupoli B, Humphries DJ, Beever DE. Splanchnic metabolism of dairy cows during the transition from late gestation through early lactation. Journal of Dairy Science. 2003 Apr;86(4):1201–17. doi: 10.3168/jds.S0022-0302(03)73704-7. [DOI] [PubMed] [Google Scholar]
- 35.Loor JJ, Dann HM, Guretzky NA, et al. Plane of nutrition prepartum alters hepatic gene expression and function in dairy cows as assessed by longitudinal transcript and metabolic profiling. Physiol Genomics. 2006 Oct 3;27(1):29–41. doi: 10.1152/physiolgenomics.00036.2006. [DOI] [PubMed] [Google Scholar]
- 36.Bionaz M, Drackley JK, Rodriguez-Zas SL, et al. Uncovering adaptive hepatic gene networks due to prepartum plane of dietary energy and physiological state in periparturient Holstein cows. Journal of Dairy Science. 2007;90:678–8. [Google Scholar]
- 37.Dann HM, Drackley JK. Carnitine palmitoyltransferase I in liver of periparturient dairy cows: effects of prepartum intake, postpartum induction of ketosis, and periparturient disorders. J Dairy Sci. 2005 Nov;88(11):3851–9. doi: 10.3168/jds.S0022-0302(05)73070-8. [DOI] [PubMed] [Google Scholar]
- 38.Hanigan MD, Crompton LA, Reynolds CK, Wray-Cahen D, Lomax MA, France J. An integrative model of amino acid metabolism in the liver of the lactating dairy cow. J Theor Biol. 2004 May 21;228(2):271–89. doi: 10.1016/j.jtbi.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 39.Reynolds CK, Harmon DL, Cecava MJ. Absorption and delivery of nutrients for milk protein synthesis by portal-drained viscera. J Dairy Sci. 1994 Sep;77(9):2787–808. doi: 10.3168/jds.S0022-0302(94)77220-9. [DOI] [PubMed] [Google Scholar]
- 40.Bequette BJ, Backwell FR, Crompton LA. Current concepts of amino acid and protein metabolism in the mammary gland of the lactating ruminant. J Dairy Sci. 1998 Sep;81(9):2540–59. doi: 10.3168/jds.S0022-0302(98)70147-X. [DOI] [PubMed] [Google Scholar]
- 41.Bell AW. Regulation of organic nutrient metabolism during transition from late pregnancy to early lactation. J Anim Sci. 1995 Sep;73(9):2804–19. doi: 10.2527/1995.7392804x. [DOI] [PubMed] [Google Scholar]
- 42.Wang X, Proud CG. The mTOR pathway in the control of protein synthesis. Physiology (Bethesda) 2006 Oct;21:362–9. doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- 43.Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006 Apr;86(2):465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- 44.Bionaz M, Thering BJ, Loor JJ. Fine metabolic regulation in ruminants via nutrient-gene interactions: saturated long-chain fatty acids increase expression of genes involved in lipid metabolism and immune response partly through PPAR-alpha activation. Br J Nutr. 2011 Jul 6;:1–13. doi: 10.1017/S0007114511002777. [DOI] [PubMed] [Google Scholar]
- 45.Litherland NB, Bionaz M, Wallace RL, Loor JJ, Drackley JK. Effects of the peroxisome proliferator-activated receptor-alpha agonists clofibrate and fish oil on hepatic fatty acid metabolism in weaned dairy calves. J Dairy Sci. 2010 Jun;93(6):2404–18. doi: 10.3168/jds.2009-2716. [DOI] [PubMed] [Google Scholar]
- 46.Bauman DE, Mather IH, Wall RJ, Lock AL. Major advances associated with the biosynthesis of milk. J Dairy Sci. 2006 Apr;89(4):1235–43. doi: 10.3168/jds.S0022-0302(06)72192-0. [DOI] [PubMed] [Google Scholar]
- 47.Annison EF. Metabolite utilization by the ruminant mammary gland. In: Mepham TB, editor. Biochemistry of Lactation. Amsterdam: Elsevier Science Publishers B.V; 1983. pp. 399–436. [Google Scholar]
- 48.Davis CL, Bauman DE. Mammary gland metabolism. In: Mepham TB, editor. Biochemistry of Lactation. Amsterdam: Elsevier Science Publishers B.V; 1983. pp. 3–30. [Google Scholar]
- 49.Sinnett-Smith PA, Vernon RG, Mayer RJ. Enzyme and protein turnover in adipose tissue in pregnancy and lactation. Biochim Biophys Acta. 1982 Jan 12;714(1):58–64. doi: 10.1016/0304-4165(82)90126-x. [DOI] [PubMed] [Google Scholar]
- 50.Bionaz M, Loor JJ. Gene networks driving bovine mammary protein synthesis during the lactation cycle. Bioinform Biol Insights. 2011;5:83–98. doi: 10.4137/BBI.S7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vernon RG. Lipid metabolism during lactation: a review of adipose tissue-liver interactions and the development of fatty liver. J Dairy Res. 2005 Nov;72(4):460–9. doi: 10.1017/S0022029905001299. [DOI] [PubMed] [Google Scholar]
- 52.Shelness GS, Ingram MF, Huang XF, DeLozier JA. Apolipoprotein B in the rough endoplasmic reticulum: translation, translocation and the initiation of lipoprotein assembly. J Nutr. 1999 Feb;129(2S Suppl):456S–62. doi: 10.1093/jn/129.2.456S. [DOI] [PubMed] [Google Scholar]
- 53.Bobe G, Young JW, Beitz DC. Invited review: pathology, etiology, prevention, and treatment of fatty liver in dairy cows. J Dairy Sci. 2004 Oct;87(10):3105–24. doi: 10.3168/jds.S0022-0302(04)73446-3. [DOI] [PubMed] [Google Scholar]
- 54.Jansen GR, Hunsaker H. Effect of dietary protein and energy on protein synthesis during lactation in rats. J Nutr. 1986 Jun;116(6):957–68. doi: 10.1093/jn/116.6.957. [DOI] [PubMed] [Google Scholar]
- 55.Garlick PJ, Clugston GA, Waterlow JC. Influence of low-energy diets on whole-body protein turnover in obese subjects. Am J Physiol. 1980 Mar;238(3):E235–44. doi: 10.1152/ajpendo.1980.238.3.E235. [DOI] [PubMed] [Google Scholar]
- 56.Adechian S, Giardina S, Remond D, et al. Excessive energy intake does not modify fed-state tissue protein synthesis rates in adult rats. Obesity (Silver Spring) 2009 Jul;17(7):1348–55. doi: 10.1038/oby.2009.35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Excel file with results for the KEGG pathway DIA analysis of liver from −65 to +49 day relative to parturition in dairy cows fed restricted (80% NRC recommendation), control (100% NRC recommendation), or ad-libitum (130% NRC recommendation) pre-partum.