Abstract
Background and Aims
Plants modulate defence signalling networks in response to different biotic stresses. The present study evaluated the effect of a phloem-sucking aphid on plant defence mechanisms in pepper (Capsicum annuum) during subsequent pathogen attacks on leaves and rhizosphere bacteria on roots.
Methods
Plants were pretreated with aphids and/or the chemical trigger benzothiadiazol (BTH) 7 d before being challenged with two pathogenic bacteria, Xanthomonas axonopodis pv. vesicatoria (Xav) as a compatible pathogen and X. axonopodis pv. glycines (Xag) as an incompatible (non-host) pathogen.
Key Results
Disease severity was noticeably lower in aphid- and BTH + aphid-treated plants than in controls. Although treatment with BTH or aphids alone did not affect the hypersensitive response (HR) against Xag strain 8ra, the combination treatment had a synergistic effect on the HR. The aphid population was reduced by BTH pretreatment and by combination treatment with BTH and bacterial pathogens in a synergistic manner. Analysis of the expression of the defence-related genes Capsicum annum pathogenesis-related gene 9 (CaPR9), chitinase 2 (CaCHI2), SAR8·2 and Lipoxygenase1 (CaLOX1) revealed that aphid infestation resulted in the priming of the systemic defence responses against compatible and incompatible pathogens. Conversely, pre-challenge with the compatible pathogen Xav on pepper leaves significantly reduced aphid numbers. Aphid infestation increased the population of the beneficial Bacillus subtilis GB03 but reduced that of the pathogenic Ralstonia solanacearum SL1931. The expression of defence-related genes in the root and leaf after aphid feeding indicated that the above-ground aphid infestation elicited salicylic acid and jasmonic acid signalling throughout the whole plant.
Conclusions
The findings of this study show that aphid feeding elicits plant resistance responses and attracts beneficial bacterial populations to help the plant cope with subsequent pathogen attacks.
Keywords: Aphid, foliar feeding, Capsicum annuum, pepper, rhizosphere bacteria, plant defence, PGPR, Xanthomonas axonopodis
INTRODUCTION
Plants have survived diverse biotic and abiotic stresses by mounting defence mechanisms (Pieterse et al., 2009) in nature. To overcome biotic stresses caused by the attack of pathogens and herbivorous insects, the development of a more specific, targeted resistance machinery was required (Smith et al., 2009). Among the mechanisms of plant resistance, induced resistance was studied intensively due to its similarity to animal innate immunity, which is different from the so-called constitutive resistance (Pieterse et al., 2009). Recent reports have defined innate immunity in plants as an old and generalized defence response that includes the perception of pathogen-derived molecules, referred to as pathogen-associated molecular patterns (PAMPs) (Nurnberger et al., 2004; Zeidler et al., 2004; Zipfel et al., 2004). PAMPs include bacterial flagellin, elongation factor, lipopolysaccharide and others. In addition to innate immunity, plants have more sophisticated defence mechanisms such as the ‘gene-for-gene’ model. This hypersensitive response (HR) is a highly specific interaction between a plant resistance protein and a pathogen-mediated avirulent protein that results in programmed cell death to arrest pathogen growth in the infected plant tissue (Dangl et al., 1996; Mysore and Ryu, 2004; Zeidler et al., 2004). The majority of plant pathogens display strict host specificity and cannot infect non-host species. The resistance of plants to most potential pathogens is referred to as non-host resistance (Heath, 2000; Kamoun, 2001; Thordal-Christensen, 2003; Mysore and Ryu, 2004; Nurnberger et al., 2004). Plant immunity can be induced in response to feeding by herbivores or infection by pathogens and is mainly regulated by three signalling molecules, salicylic acid (SA), jasmonic acid (JA) and ethylene (ET), which are interconnected by complex signalling networks and crosstalk phenomena (Pieterse et al., 2009). Generally, JA-mediated responses are directed against herbivores and necrotrophic pathogens, whereas SA-mediated systemic acquired resistance (SAR) responses are active against biotrophic pathogens (Heil and Bostock, 2002; Bostock, 2005).
Among several insects known to elicit immune responses against plant pathogens, aphids (order Homoptera) are an important insect pest for most major crops, causing serious economic losses (Dedryver et al., 2010). Unlike other chewing herbivores, aphids are distinguished by feeding on phloem sap from the host plants via narrow piercing-sucking mouthparts referred to as stylets (Powell et al., 2006). Use of the stylets minimizes tissue damage to plant surface structures such as epidermal, mesophyll and parenchyma cells (Powell et al., 2006). For direct protection against the aphid's sucking, plants produce compounds that are toxic to some species of aphids. In the family Brassicaceae, which includes Arabidopsis thaliana, plants accumulate glucosinolates, a family of secondary metabolites that are a source of thiocyanates (Rask et al., 2000; Halkier and Gershenzon, 2006). Cleavage of indole glucosinolates by plant myrosinase is triggered by Myzus persicae feeding on Arabidopsis leaves. A recent study showed that aphid-mediated diindolylmethylcysteines that are generated by the reaction of indole-3-carbinol, a cleavage product of indole glucosinolates, with ascorbate, glutathione and cysteine have strong anti-feedant activity against M. persicae (Mi et al., 2008). Gene expression responses elicited by M. persicae attack most closely resemble SA-mediated gene induction with regard to defence signalling, despite the early up-regulation of JA- and ET-related genes.
However, results obtained with A. thaliana are not always comparable or relevant to crop plants. Aphid feeding experiments with JA- and SA-insensitive Arabidopsis mutant or transgenic lines have indicated that JA inhibits aphid population growth, whereas the involvement of SA signalling on aphid resistance was neutral or even positive (Thompson and Goggin, 2006). However, in tomato, SA plays a critical role in basal resistance and R-gene-mediated gene-for-gene resistance to aphids. More interestingly, a study with tomato jai1-1 (jasmonic acid insensitive 1) mutant plants, which are impaired in JA perception, showed no difference in potato aphid survival or fecundity (Bhattarai et al., 2007). Unfortunately, mechanistic studies of insect–crop interactions, with the exception of rice, are not available due to limited genetic and molecular information about crop species. Pepper is an emerging model system for studying insect–plant interactions because, in addition to expressed sequence tag (EST) data, studies on the defence signalling mechanisms and knockdown of target genes by virus-induced gene silencing have been reported in this plant (Ryu et al., 2004; Chung et al., 2006; Mi et al., 2008). However, studies aimed at understanding plant defence against aphids in pepper have not yet been reported, despite the economic importance of aphids as pests in many countries, including Korea.
In recent studies, resistance was induced above-ground (AG) by whitefly infestation to study the biological effects on both leaf- and root- (below-ground, BG) infecting bacterial pathogens. The induction of systemic resistance was confirmed by the significant up-regulation of the SA and JA defence signalling pathway marker genes, Capsicum annuum pathogenesis-related protein (CaPR)1, CaPR4, CaPR10 and Ca protease inhibitor (CaPIN) in both leaves (AG) and roots (BG) after whitefly feeding. Interestingly, AG whitefly feeding significantly increased the population density of beneficial BG micro-organisms, including Gram-positive bacteria, actinomycetes and saprophytic fungi that may induce systemic resistance (Yang et al., 2011). Among BG microbial groups, several Gram-positive Bacillus sp. strains significantly elicited plant systemic defences against the whitefly population in the tomato field (Murphy et al., 2000).
Here we provide new evidence that aphids, which are similar sucking insects to the whitefly, increase plant systemic immunity against the biotrophic bacterial pathogen Xanthomonas axonopodis. Furthermore, because both biotrophic pathogens and aphids are known to induce SA signalling, we evaluated whether pre-challenge with compatible and incompatible pathovars of X. axonopodis increased plant resistance to aphid feeding in pepper. The activation of signal transduction pathways by aphid infestation was investigated by assessing the transcriptional expression of pepper marker genes for SA and JA after aphid feeding. Conversely, pre-challenge with the compatible pathogen X. axonopodis pv. vesicatoria (Xav) on pepper leaves significantly reduced aphid numbers. The bacterial populations of the beneficial plant growth-promoting rhizobacteria Bacillus subtilis GB03, the saprophyte Pseudomonas fluorescens Pf-5 and the pathogenic Ralstonia solanacearum SL1931 were evaluated in the roots after aphid infestation in the leaf (Kloepper et al., 2004; Haas and Defago, 2005). Our studies provide a new understanding of tritrophic (insect–plant–beneficial root bacteria) interactions and their role in the induction of defence mechanisms.
MATERIALS AND METHODS
Plant preparation and disease assay
Pepper (Capsicum annuum L. ‘Bukang’) was used as the study plant because it interacts with multiple enemies and mutualists representing different guilds, and because the availability of genetic tools allows the analysis of gene expression patterns under different conditions. Seeds of C. annuum were surface-sterilized with 6 % sodium hypochlorite, washed four times with sterile distilled water, and then maintained at 25 °C for 3 d until germination on Murashige and Skoog medium (Duchefa, Haarlem, the Netherlands). The germinated seeds were then planted on soilless media (Punong Horiculture Nursery Media LOW, Punong Co. Ltd, Gyeongju, Korea). Plants were grown at 25 ± 2 °C under fluorescent light (12 h/12 h day/night cycle, approx. 7000 lx light intensity) in a controlled-environment growth room for seeding growth and transferred to the KRIBB greenhouse facility in Daejeon, South Korea, for aphid treatment. Two-week-old pepper plants were drenched with either 10 mL of a solution of 0·5 mm benzo (1,2,3) thiadiazole-7-carbothioic acid S-methyl ester (benzothiadiazole = BTH) (Syngenta, Research Triangle Park, NC, USA) or sterile water. At the same time, the aphid Myzus persicae Sulzer, a naturally occurring insect in the greenhouse in Daejeon in 2010–2011 (referred to as ‘green peach aphid’) was treated as the biological inducer. The aphid was maintained on pepper plants. Cross-phyla-induced plant immunity against bacteria or aphids was investigated by using Xav (Yang et al., 2009), a causal pathogen of bacterial leaf spot disease, as a compatible pathogen and X. axonopodis pv. glycines (Xag), a causal pathogen in the leaves of soybean (Lee et al., 2004), as an incompatible pathogen. One week after aphid, BTH and BTH + aphid treatments, all plants were inoculated with Xav and Xag on Luria-Bertani (LB; Duchefa) agar. For experimental use, bacteria were scraped from plates and resuspended in sterile water. The bacterial suspensions of both strains were adjusted to 106 c.f.u. mL−1 based on optical density and injected into pepper leaves using a 1-mL needle-less syringe (Doo Won Meditec Co., Kim Je, Korea). Disease severity was measured 7 d after pathogen challenge as described previously (Yang et al., 2009). Briefly, the severity of symptoms was scored from 0 to 5 as follows: 0, no symptoms; 1, yellowish colour; 2, chlorosis only; 3, necrosis and chlorosis; 4, partial necrosis of the inoculated area; and 5, complete necrosis of the inoculated area. The experiment had a completely randomized design with ten replications and was independently repeated four times.
Effect of leaf pathogens on the number of aphids
To investigate whether compatible and incompatible bacteria elicit plant immunity to aphid feeding with or without BTH treatment, Xav and Xag were infiltrated into fully developed leaves of 2-week-old pepper seedlings (Lee et al., 2004; Yang et al., 2009). A total of 0·5 mm BTH was drench-applied to the roots of pepper plants 7 d after pathogen challenge. One week after BTH treatment, the plants were transferred to the KRIBB greenhouse facility in Daejeon. The total number of adult aphids was counted 10 d after aphid exposure.
Quantification of root bacteria
The three bacteria, B. subtilis GB03, P. fluorescens Pf-5 and R. solanacearum SL1931, were generated as spontaneous mutants resistant to 100 µg mL−1 rifampicin in the media before the root colonization experiment. The number of introduced bacteria on the roots was counted at 0 and 7 d after drench application, as described previously (Ryu et al., 2003). In brief, pepper roots were incubated in sterile water for 30 min in a shaking incubator at 30 °C and the wash off was diluted and spread on trypticase soy broth agar containing 100 µg mL−1 rifampicin. The bacterial population was calculated from antibiotic-resistant colonies appearing 2–3 d after spreading.
Quantitative RT-PCR (qRT-PCR)
Molecular evidence for aphid-elicited expression of bacterial resistance-related genes in pepper was obtained using qRT-PCR. The relative mRNA expression of Capsicum annum Chitinase 2 (CaCHI2), Capsicum annum Pathogenesis-related gene 9 (CaPR9) and Capsicum annum Lipoxygenase1 (CaLOX1), which are expressed during incompatible pathogen/chemical-elicited SAR and plant growth-promoting rhizobacteria (PGPR)-elicited induced systemic resistance (ISR), was measured in leaves and roots (Hong et al., 2000; Park et al., 2001, 2002, 2004; Yang et al., 2009; Yi et al., 2009). Total RNA was isolated from leaf and root tissues treated with aphids, water, BTH + aphids and BTH 1 week after each treatment using the RNeasy plus mini kit (Qiagen) according to the manufacturer's instructions. To evaluate the expression of defence-related genes induced by aphid infestation, expression of CaSAR8·2 and CaLOX1 was investigated by collecting pepper leaf and root tissues at 0, 1 and 3 d after aphid treatment (see Fig. 4). For assessing the priming of defence-related genes, pepper leaf or root tissues were collected at 0 and 6 h post-inoculation of the bacterial pathogens Xav and Xag and at 0 and 2 days post-inoculation of the rhizosphere bacteria B. subtilis GB03, P. fluorescens Pf-5 and R. solanacearum SL1931. First-strand cDNA synthesis was carried out with 1 µg DNase-treated total RNA, oligo-dT primers and Moloney murine leukaemia virus reverse transcriptase (MMLV-RT; Enzynomics, Daejeon, Korea). The expression of candidate priming genes was analysed using the following primers: 5′-ATTGGACGATGGAAGCCATCACCAG-3′ and 5′-ATATTCCGAATGTCTAAAGTGGTAC-3′ for CaCHI2, 5′-GACTAGTTTCAAGAGCATCA-3′ and 5′-AATTGTATAGCCTGTAGCTG-3′ for CaPR9, 5′-TGCAGGTTACCTCCCAAATCGCCCA-3′ and 5′-CTATATCGACACACTGTTGGGTATTCCTT-3′ for CaLOX1, and 5′-TAGTGAGACTAAGAAAGTTGGACG-3′ and 5′-AAGAGTGCATGCAGTATCACAAAG-3′ for CaSAR8·2. CaActin was used as a control and analysed using the primers 5′-TTGGACTCTGGTGATGGTGTG-3′ and 5′-AACATGGTTGAGCCACCACTG-3′. A Chromo4 real-time PCR system (Bio-Rad) was used for qRT-PCR. Reaction mixtures consisted of cDNA, iQTM SYBR® Green Supermix (Bio-Rad) and 1 pm of each primer. The thermocycle parameters were as follows: initial polymerase activation, 10 min at 95 °C, and then 40 cycles of 30 s at 95 °C, 60 s at 55 °C and 30 s at 72 °C. Conditions were determined by comparing threshold values in a series of dilutions of the RT product, followed by a non-RT template control and a non-template control for each primer pair. Relative RNA levels were calibrated and normalized to the level of CaActin mRNA (GenBank accession no. AY572427).
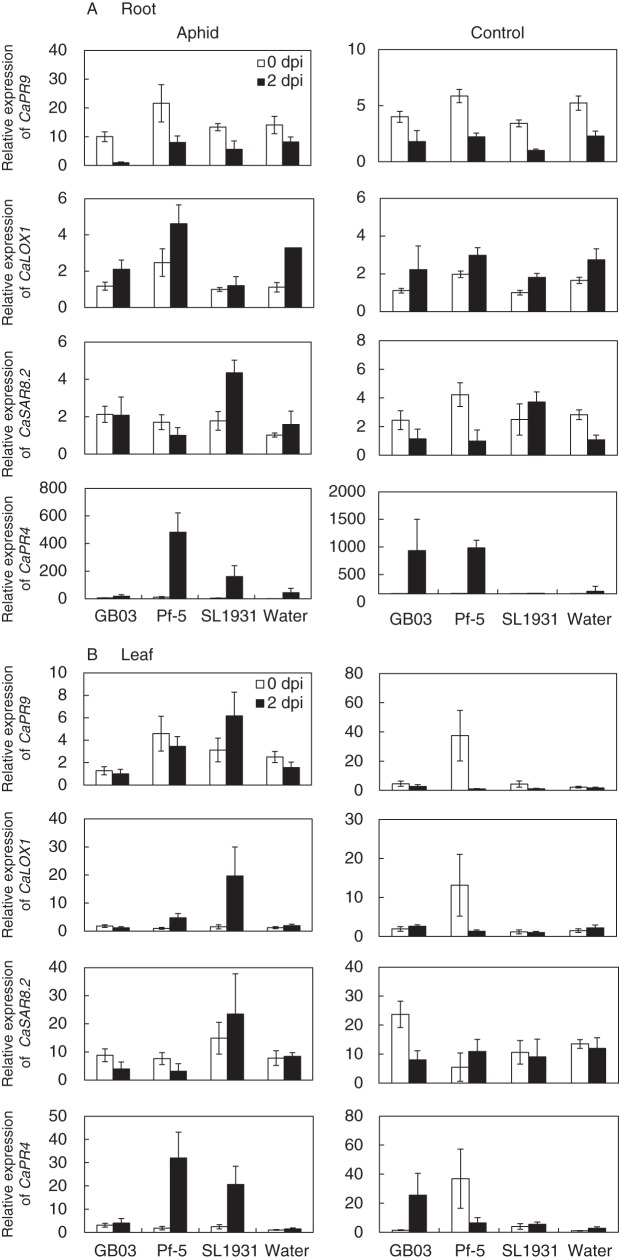
Fig. 4.
Induction of the CaPR9, CaCHI2 and CaLOX1 genes in aphid-, BTH-, water control- and aphid + BTH combination-treated plants. Expression levels of pepper resistance genes CaPR9, CaCHI2, CaSAR 8·2 and CaLOX1 were quantified by qRT-PCR at 0 and 2 d after drenching of 108 c.f.u. mL−1 suspension of B. subtilis GB03, P. fluorescens Pf-5, R. solanacearum SL1931 and water on plant root. Values are means ± s.e.m. with four replications per treatment. Different letters indicate significant differences between treatments (P < 0·05 according to the LSD test). The experiment was repeated three times with similar results.
Statistical analysis
Data were subjected to analysis of variance using JMP software ver. 4·0 (SAS Institute Inc., Cary, NC, USA; www.sas.com). The significance of direct and indirect biological or chemical treatment effects was determined by the magnitude of the F value at P = 0·05. When a significant F value was obtained for treatments, separation of means was accomplished using Fisher's protected least significant difference (LSD) test at P = 0·05. The results of repeated trials of each experiment outlined above were similar. Hence, one representative trial of each experiment is reported.
RESULTS
Induction of plant immunity against bacterial pathogens by aphid infestation
All experiments were conducted in soilless media-grown pepper plants that were freely exposed to the aphid population in the greenhouse. Typically, aphids colonized newly developed leaves and stems (data not shown). Assessment of disease resistance against the compatible pathogen Xav, which is a bacterial spot pathogen of pepper, showed reduced disease symptoms after two bacterial challenges in aphid- and BTH-treated plants compared with water controls (Fig. 1). Water control plants developed severe necrosis 7 d after pathogen challenge on leaves, while plants treated with aphids, BTH or BTH + aphids did not show any visible symptoms (Fig. 1A). Statistical analysis of disease severity revealed a significant level of plant immunity induced by aphid feeding. Post-hoc analysis revealed that the greatest resistance against Xav was elicited by aphid alone and BTH + aphid treatment, indicating an additive effect of these treatments (Fig. 1A). The quantification of bacteria showed similar patterns (data not shown).
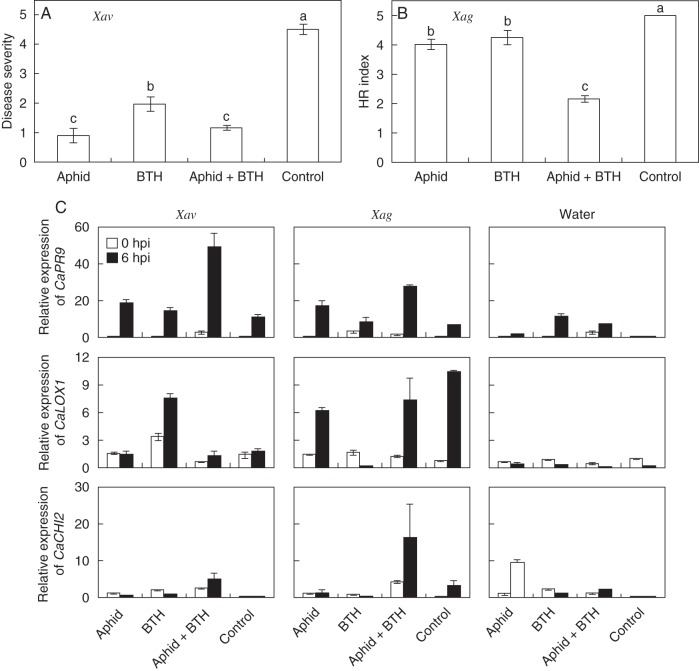
Fig. 1.
Induction of plant defence responses against compatible and incompatible pathovars of Xanthomonas axonopodis and defence priming of resistance genes by aphid infestation. (A) Induction of plant resistance against a compatible X. axonopodis pv. vesicatoria (Xav). Disease severity was measured 7 d after Xav challenge, on a scale of 1–5. (B) Induction of plant resistance against an incompatible X. axonopodis pv. glycines (Xag). The hypersensitive response (HR) index was measured 48 h after Xag inoculation, on a scale of 1–5. Values are means ± s.e.m., sample size n = 10 plants per treatment. Different letters indicate significant differences between treatments (P = 0·05), according to the least-significant difference (LSD) test. (C) Real-time qRT-PCR analysis of the expression levels of marker genes for SA (CaPR9), JA (CaLOX1) and ET (CaCHI2) in aphid, 0·5 mm BTH, aphid + BTH and water control at 0 and 6 h post-inoculation (hpi). The housekeeping gene CaActin was used as a control. Values are means ± s.e.m. The experiment was repeated four times with similar results.
Assessment of the effect of aphid feeding on plant defence responses against the soybean pathogen X. axonopodis pv. glycines strain 8ra (Xag), an incompatible (non-host) pathogen, showed that aphid pre-inoculation significantly delayed the HR index in response to the infiltration of Xag into pepper leaves within 36 h. Control plant leaves challenged with the pathogen developed significant necrosis within 36 h (Fig. 1B). By contrast, all three resistance-induction treatments (aphid, BTH and aphid + BTH) caused a significant reduction in the HR index compared with controls. Although no significant differences were observed among the individual treatments, aphid + BTH treatment reduced the HR index to 50 % that of the control-treated plants (Fig. 1B).
Induction of SA- and JA-related genes during aphid-elicited plant immunity
qRT-PCR was used to investigate the possible activation of a whitefly-infestation-mediated plant defence signalling pathway conferring resistance against bacterial pathogens. Increased expression of CaPR9, CaCHI2 and CaLOX1 under incompatible pathogen-induced SAR conditions and in response to treatment with defence signalling molecules such as SA, JA, ET and abscisic acid was reported previously (Kim and Hwang, 2000; Min et al., 2005; Park et al., 2001, 2004; Shin et al., 2001). The transcriptional expression of both CaPR9 and CaLOX1 was significantly up-regulated in Xav- and Xag-treated aphid infested pepper leaves (Fig. 1C). These results suggest that AG feeding by aphids elicited SA and JA/ethylene (ET)-dependent defence signalling pathways. BTH only induced the transcription of CaLOX1 in leaves challenged by Xav. However, the BTH + aphid combination treatment had a synergistic effect on the activation of CaPR9 in response to Xav and Xag infiltration and of CaCHI2 in response to Xag infiltration in the AG parts of the plants (Fig. 1C). In contrast, under the same conditions, the ET response gene CaCHI2 and JA response gene CaLOX1 were significantly repressed compared with BTH or aphid treatment alone (Fig. 1C). Assessment of the early responses to aphid treatment showed that the transcriptional expression of CaSAR8·2 and CaLOX1 in aphid-infested pepper leaves increased 2·98- and 3·5-fold, respectively, compared with control treatment on day 1, while no changes in CaSAR8·2 and decreased CaLOX1 expression were detected on day 3 (Fig. 2). In root, the CaLOX1 gene was induced 2·3-fold by aphid treatment on day 1, while no changes were observed on day 3. In contrast, the mRNA level of CaSAR8·2 in the root did not change on day 1, while a four-fold increase was detected on day 3 compared with the control treatment (Fig. 2A).
Fig. 2.
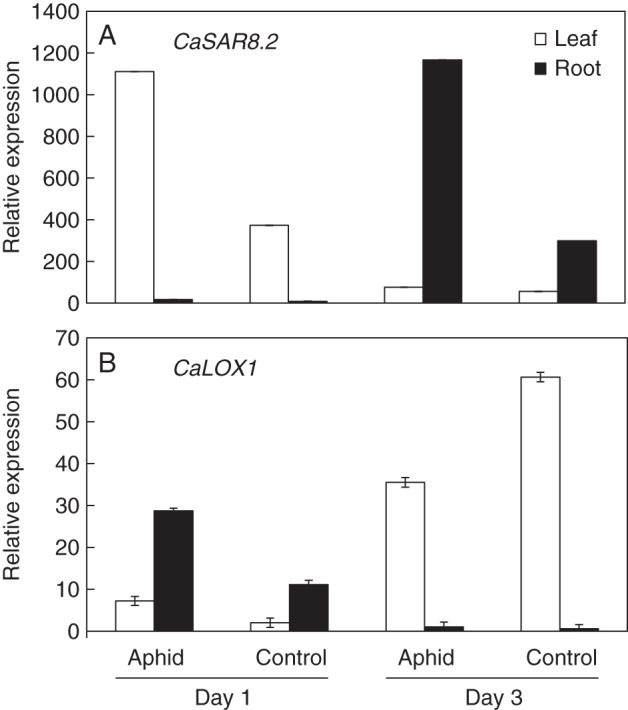
Plant defence gene expression after aphid infestation. The expression of defence-related genes was quantified using qRT-PCR 1 and 3 d after exposure to aphids. (A) CaSAR8·2 expression. (B) CaLOX1 expression. Values are means ± s.e.m.
Population dynamics of rhizosphere bacteria by aphid infestation
The effect of aphid infestation on the population densities of three rhizosphere bacteria (rhizobacteria), B. subtilis GB03, P. fluorescens Pf-5 and R. solanacearum SL1931, was evaluated. Seven days after root inoculation, the number of cells of strain GB03 in root was significantly (P = 0·05) higher in AG aphid-infested than control plants (Fig. 3A). In contrast, aphid infestation significantly (P = 0·05) reduced the number of cells of the pathogenic strain SL1931 (Fig. 3C), while the population of strain Pf-5 was not affected by aphid feeding (Fig. 3B).
Fig. 3.
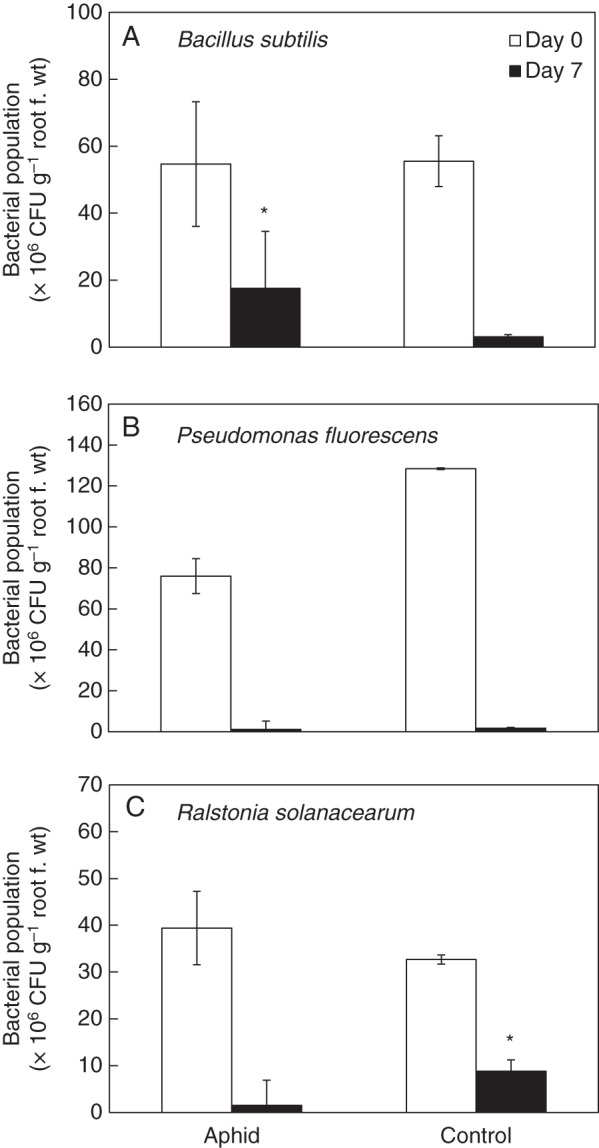
Effects of aphid infestation on bacterial populations in the pepper rhizosphere. Bacterial populations were quantified at the day of drenching application on pepper roots and 7 d after application. Population dynamics of (A) B. subtilis GB03, (B) P. fluorescens and (C) R. solanacearum 1931. Values are means ± s.e.m. *P < 0·05 according to the LSD test.
Plant defence priming by beneficial and pathogenic rhizosphere bacteria
Defence priming is elicited by beneficial root-associated bacteria (PGPR). In root, PGPR strains GB03 and Pf-5 caused a four-fold up-regulation in the transcription of CaPR4 in pepper seedlings without aphid treatment 2 d after rhizobacteria inoculations (Fig. 4A). CaPR4 and CaLOX1 showed increased transcription in Pf-5-treated aphid-infested plants while the transcriptional expression of CaPR9 and CaPR4 was slightly reduced in response to inoculation of strain GB03 in aphid-infested plants (Fig. 5A). The pathogenic bacterium R. solanacearum strain SL1931 induced only CaPR4 in the aphid-infested plant (Fig. 5A). In the AG (leaf) tissue, within 2 d after application of root bacteria, strain SL1931 significantly induced transcription of CaPR9, CaLOX1 and CaPR4 in aphid-infested plants (Fig. 4B). The beneficial bacterial strain Pf-5 increased only CaPR4 expression under aphid infestation conditions, while GB03 did not induce significant differences, except in expression of CaPR4 without aphid treatment.
Fig. 5.
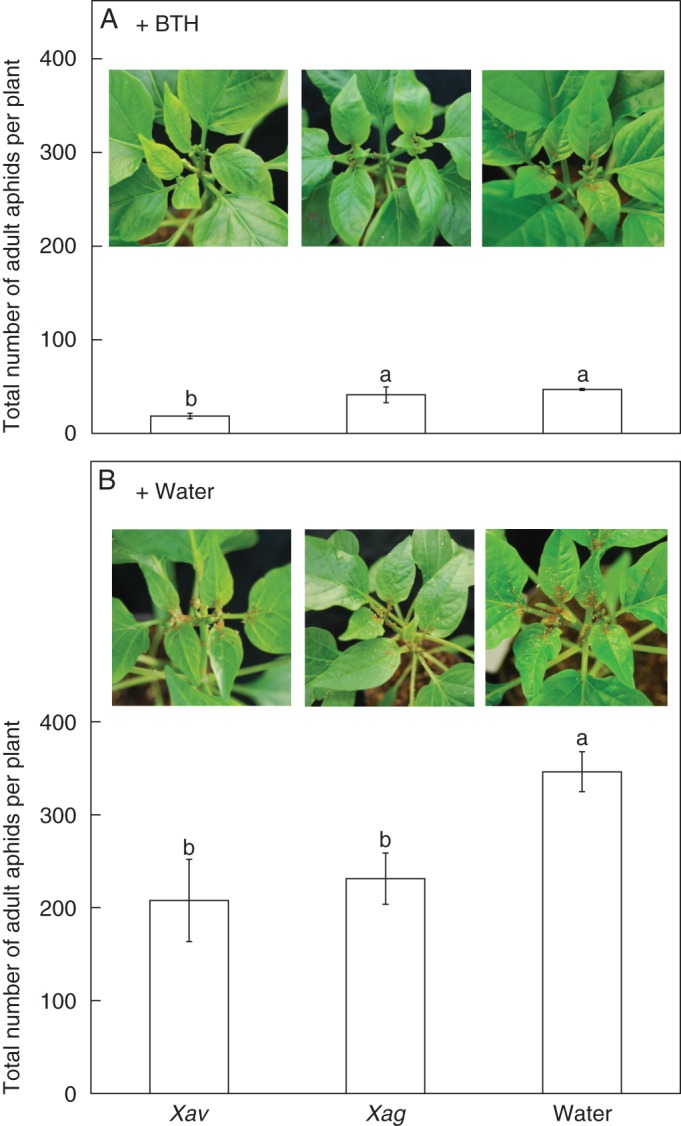
Effects of pathogen infection on aphid infestation. The total number of aphids per plant was counted on the leaves with pre-challenge of X. axonopodis pv. vesicatoria (Xav) and X. axonopodis pv. glycines (Xag) with and without BTH treatment. Representative photographs were taken 7 d after 0·5 mm BTH (A) and water (B) treatments of plants. Numbers of aphids per plant were counted 7 d after 0·5 mm BTH (A) and water (B) treatments of plants pre-inoculated with Xav and Xag 7 d previously. Values Re means ± s.e.m. with five replications per treatment. Different letters indicate significant differences between treatments (P = 0·05 according to the LSD test). The experiment was repeated four times with similar results after pathogen challenge.
Induced resistance to aphid infestation by pre-inoculation of bacterial pathogens
Quantification of the numbers of aphids in plants challenged with compatible and incompatible leaf bacterial pathogens 1 week after treatment with aphids, BTH or both revealed a decrease in the total number of aphids in pepper seedlings infiltrated with Xav and Xag of 45 % compared with water-treated control plants (Fig. 5B). Moreover, the combination Xav + BTH treatment showed the greatest reduction of aphid numbers among the three treatments, with a 46·3 % reduction compared with Xag + BTH or BTH treatment alone (Fig. 5A). Assessment of the effect of BTH showed that the number of aphids in BTH-treated plants was 46 and in water-treated control plants was 348, indicating a significant induction of plant immunity against aphid infestation by BTH treatment (Fig. 5). The mean (± s.e.m) number of aphids per plant was 342 ± 23·5 (Fig. 5).
DISCUSSION
The induction of resistance to insect attacks after infestation with the same or a different insect species has been reported previously. Studies on rice (Kanno and Fujita, 2003; Kanno et al., 2005) and tomato (Mayer et al., 2002) demonstrated that insect feeding can also change plant resistance responses against microbial pathogens. The present results demonstrate the induction of cross-resistance among phyla by showing that insects feeding on leaves can induce resistance to compatible and incompatible pathogens on the leaves. Conversely, pathogen pre-inoculation increased plant defence responses to subsequent insect feeding. Our results thus show the crosstalk between defence signalling pathways against insect pests and microbial pathogens, and introduce a new plant model system for the analysis of crop–aphid interactions, such as plant immunity to insects and pathogens. In addition, foliar aphid infestation recruited PGPR but reduced root colonization of pathogenic bacteria. To our knowledge, this is the first study demonstrating an aphid-specific increase in cross-plant resistance against an insect and a pathogen and attract beneficial bacteria in pepper.
The present study demonstrates that phloem-feeding insects can induce local resistance against both compatible and incompatible bacterial pathogens. A similar study using whitefly as a leaf feeding insect to test the induction of plant defences reported specific effects only against leaf pathogens (Mayer et al., 2002; Yang et al., 2011). In our study, aphid infestation elicited significant defence responses against bacterial spot disease caused by Xav and the non-host pathogen Xag (Fig. 1). Although previous studies did not report the effect of aphid infestation on host plant pathogen resistance, infestation with whitefly has been shown to increase the resistance of plants against Erysiphe cichoracearum, a casual pathogen of the biotrophic fungus powdery mildew and Xav (Mayer et al., 2002; Yang et al., 2011). Aphid infestation has been shown to enhance the transcription of SA- and JA-dependent signalling genes by transcriptome and RT-PCR analysis. SA, one of the key chemical signals produced in response to pathogen attack on resistant plants, is required for the induction of SAR (Dempsey et al., 1999), and SA signalling plays an important role in plant responses to aphid attacks (Pegadaraju et al., 2005; Louis et al., 2010). The production of SA and induction of SAR occur following activation of the HR, which is governed by resistance genes encoding receptors that recognize specific pathogens (Staskawicz et al., 1995). The induction of SAR results from a complex signal transduction process (Pickett and Poppy, 2001) and leads to the accumulation of pathogenesis-related proteins. The role of SA on aphid resistance is being studied.
Infestation of Arabidopsis plants with Myzus persicae (the green peach aphid) up-regulated mostly the SA-dependent signalling markers pathogenesis-related (PR) 1 and PR2 (also called BGL2) (Moran et al., 2002). Intriguingly, these two genes are induced by plant pathogenic bacteria and fungi, which are classified as biotrophs. A similar study showed that aphid reproduction is inhibited in npr1 and NahG plants deficient in SA signalling and production (Pegadaraju et al., 2005). Later, further study revealed that aphid resistance was not affected in NahG transgenic Arabidopsis plants and in plants with a deficiency in npr1, a global regulator of SA-dependent signalling pathways (Louis et al., 2010). However, marker genes for SA and JA signalling in pepper have not been studied in detail. In our study, aphid feeding on leaves had a priming effect by promoting the expression of three pepper defence genes, CaPR9, CaCHI2 and CaLOX1, as early as 6 h after compatible pathogen challenge (Fig. 2B). qRT-PCR analysis of these molecular marker genes for SA (CaPR9), ET (CaCHI2) and JA (CaLOX1) in pepper revealed that the CaPR9 gene was significantly up-regulated by the infiltration of compatible and incompatible pathogens. Interestingly, the other SA marker gene, CaCHI2, was not primed by aphid infestation alone but responded to aphid + BTH combination treatment, suggesting that the CaCHI2 gene can be synergistically primed by amplifying the SA signal with other SA-related molecules such as BTH (Fig. 1C). The JA-signalling pathway-associated gene CaLOX1 was only induced by aphids in response to incompatible pathogen inoculation, but its expression level did not change between combination-treated and control plants, suggesting that JA may not play an important role in the aphid-mediated priming of defence genes in pepper (Fig. 2B). A recent transcriptome analysis identified 200 up-regulated and 95 down-regulated genes within 48 h of aphid infestation (Delp et al., 2009). A validation study assessed the transcriptional expression of selected genes and identified several defence signalling-related genes, including those associated with the shikimate pathway, which provides chorismic acid that can be converted into salicylic acid (Delp et al., 2009). In another study, the mutant pad4 was selected due to its lack of resistance to M. persicae, which was mostly caused by delayed aphid-mediated leaf senescence. In contrast, overexpression of PAD4 (PHYTOALEXIN DEFICIENT4), which encodes a nucleo-cytoplasmic protein with similarity to lipases, significantly augmented aphid resistance. EDS1 directly interacts with PAD4 and is required for PAD4-dependent plant defence against pathogens but is not essential for resistance against M. persicae (Pegadaraju et al., 2007). Collectively, aphid feeding may enhance defence signalling mRNA levels similar to biotrophic pathogens, but the effect of aphid infestation may be mediated by an as-yet unknown branch of the Arabidopsis defence signalling network. Therefore, aphid-induced plant defences share similarities to plant reactions against biotrophic microbial pathogens mediated by SA-dependent pathways (Walling, 2000; Kaloshian and Walling, 2005; Pieterse et al., 2009). It is also remarkable that the combined treatment of aphid + BTH significantly activated the bSA-responsive CaPR9 and CaCHI2 genes but did not affect the JA-responsive CaLOX1 gene, suggesting that aphid infestation additively induces SA signalling independently of JA signalling (Pieterse et al., 2009).
Confirming our previous results, aphid infestation recruited the beneficial bacterium PGPR strain GB03 but inhibited pathogenic bacterial populations (Fig. 3A, C; Yang et al., 2011). Previous studies showed the induction of ISR against insect infestation by several PGPR strains (Mayer et al., 2002; Kloepper et al., 2004; Shoresh et al., 2010). We hypothesize that AG aphid infestation may enhance the secretion of root exudates, which may recruit beneficial PGPR strains and inhibit pathogenic bacteria. Whether the same or different root exudates act on recruitment and inhibition is not clear. It is also noteworthy that the Gram-negative PGPR strain Pf-5 increased the expression of defence-related genes such as CaPR9 and CaPR4 in roots and leaves (Fig. 5A, B), but the Gram-positive PGPR strain GB03 suppressed CaPR9 expression under aphid infestation conditions, indicating that PGPR modulate aphid infestation-induced plant defence responses locally and systemically.
In summary, the present study demonstrated that foliar attack by a sap-sucking insect elicited pathogen resistance not only against compatible Xav, but also against the non-host pathogen Xag. Systemic defence signalling elicited by aphid feeding mainly involved SA pathways, based on qRT-PCR analyses of the expression of different hormone-dependent genes. Aphid feeding on leaves primed plants for the up-regulation of the CaPR9, CaLOX1 and CaCHI2 genes after pathogen challenge. Our results provide new insight into the molecular basis of aphid-mediated plant immunity against pathogen infection, particularly the priming of defence responses against different phylum pathogens, which may help prepare the plant for subsequent pathogen attacks.
ACKNOWLEDGEMENTS
We thank Drs Joseph W. Kloepper, Joyce Loper, Doil Choi and Seon Woo Lee for providing the bacterial strains B. subtilis GB03, P. fluorescens Pf-5, X. axonopodis and R. solanacearum, respectively. Financial support was obtained from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-0011655), the Industrial Source Technology Development Program of the Ministry of Knowledge Economy (TGC0281011) of Korea, the Next-Generation BioGreen 21 Program (SSAC grant no. PJ008170), Rural Development Administration, South Korea, and the KRIBB initiative programme, South Korea.
LITERATURE CITED
- Bhattarai KK, Xie QG, Pourshalimi D, Younglove T, Kaloshian I. Coil-dependent signaling pathway is not required for Mi-1-mediated potato aphid resistance. Molecular Plant-Microbe Interactions. 2007;20:276–282. doi: 10.1094/MPMI-20-3-0276. [DOI] [PubMed] [Google Scholar]
- Bostock RM. Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annual Review of Phytopathology. 2005;43:545–580. doi: 10.1146/annurev.phyto.41.052002.095505. [DOI] [PubMed] [Google Scholar]
- Chung E, Ryu CM, Oh SK, et al. Suppression of pepper SGT1 and SKP1 causes severe retardation of plant growth and compromises basal resistance. Physiologia Plantarum. 2006;126:605–617. [Google Scholar]
- Dangl JL, Dietrich RA, Richberg MH. Death don't have no mercy: cell death programs in plant-microbe interactions. The Plant Cell. 1996;8:1793–1807. doi: 10.1105/tpc.8.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedryver CA, Le Ralec A, Fabre F. The conflicting relationships between aphids and men: a review of aphid damage and control strategies. Comptes Rendus Biologies. 2010;333:539–553. doi: 10.1016/j.crvi.2010.03.009. [DOI] [PubMed] [Google Scholar]
- Delp G, Gradin T, Ahman I, Jonsson LM. Microarray analysis of the interaction between the aphid Rhopalosiphum padi and host plants reveals both differences and similarities between susceptible and partially resistant barley lines. Molecular Genetics and Genomics. 2009;281:233–248. doi: 10.1007/s00438-008-0409-3. [DOI] [PubMed] [Google Scholar]
- Dempsey DA, Shah J, Klessig DF. Salicylic acid and disease resistance in plants. Critical Reviews in Plant Sciences. 1999;18:547–575. [Google Scholar]
- Haas D, Defago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nature Reviews. Microbiology. 2005;3:307–19. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J. Biology and biochemistry of glucosinolates. Annual Review of Plant Biology. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- Heath MC. Nonhost resistance and nonspecific plant defenses. Current Opinion in Plant Biology. 2000;3:315–319. doi: 10.1016/s1369-5266(00)00087-x. [DOI] [PubMed] [Google Scholar]
- Heil M, Bostock RM. Induced systemic resistance (ISR) against pathogens in the context of induced plant defences. Annals of Botany. 2002;89:503–512. doi: 10.1093/aob/mcf076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JK, Jung HW, Kim YJ, Hwang BK. Pepper gene encoding a basic class II chitinase is inducible by pathogen and ethephon. Plant Science. 2000;159:39–49. doi: 10.1016/s0168-9452(00)00312-5. [DOI] [PubMed] [Google Scholar]
- Kaloshian I, Walling LL. Hemipterans as plant pathogens. Annual Review of Phytopathology. 2005;43:491–521. doi: 10.1146/annurev.phyto.43.040204.135944. [DOI] [PubMed] [Google Scholar]
- Kamoun S. Nonhost resistance to Phytophthora: novel prospects for a classical problem. Current Opinion in Plant Biology. 2001;4:295–300. doi: 10.1016/s1369-5266(00)00176-x. [DOI] [PubMed] [Google Scholar]
- Kanno H, Fujita Y. Induced systemic resistance to rice blast fungus in rice plants infested by white-backed planthopper. Entomologia Experimentalis et Applicata. 2003;107:155–158. [Google Scholar]
- Kanno H, Satoh M, Kimura T, Fujita Y. Some aspects of induced resistance to rice blast fungus, Magnaporthe grisea, in rice plant infested by white-backed planthopper, Sogatella furcifera. Applied Entomology and Zoology. 2005;40:91–97. [Google Scholar]
- Kim YJ, Hwang BK. Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible. Physiologia Plantarum. 2000;108:51–60. [Google Scholar]
- Kloepper JW, Ryu CM, Zhang S. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology. 2004;94:1259–66. doi: 10.1094/PHYTO.2004.94.11.1259. [DOI] [PubMed] [Google Scholar]
- Lee S, Kim SY, Chung E, et al. EST and microarray analyses of pathogen-responsive genes in hot pepper (Capsicum annuum L.) non-host resistance against soybean pustule pathogen (Xanthomonas axonopodis pv. glycines) Functional & Integrative Genomics. 2004;4:196–205. doi: 10.1007/s10142-003-0099-1. [DOI] [PubMed] [Google Scholar]
- Louis J, Leung Q, Pegadaraju V, Reese J, Shah J. PAD4-dependent antibiosis contributes to the ssi2-conferred hyper-resistance to the green peach aphid. Molecular Plant-Microbe Interactions. 2010;23:618–627. doi: 10.1094/MPMI-23-5-0618. [DOI] [PubMed] [Google Scholar]
- Mayer RT, Inbar M, McKenzie CL, et al. Multitrophic interactions of the silverleaf whitefly, host plants, competing herbivores, and phytopathogens. Archives of Insect Biochemistry and Physiology. 2002;51:151–169. doi: 10.1002/arch.10065. [DOI] [PubMed] [Google Scholar]
- Mi SK, Song MC, Eun YK, et al. Galactinol is a signaling component of the induced systemic resistance caused by Pseudomonas chlororaphis O6 root colonization. Molecular Plant-Microbe Interactions. 2008;21:1643–1653. doi: 10.1094/MPMI-21-12-1643. [DOI] [PubMed] [Google Scholar]
- Min SS, Dong GK, Lee SH. Isolation and characterization of a jasmonic acid carboxyl methyltransferase gene from hot pepper (Capsicum annuum L.) Journal of Plant Biology. 2005;48:292–297. [Google Scholar]
- Moran PJ, Cheng Y, Cassell JL, Thompson GA. Gene expression profiling of Arabidopsis thaliana in compatible plant-aphid interactions. Archives of Insect Biochemistry and Physiology. 2002;51:182–203. doi: 10.1002/arch.10064. [DOI] [PubMed] [Google Scholar]
- Murphy JF, Zehnder GW, Schuster DJ, Sikora EJ, Polston JE, Kloepper JW. Plant growth-promoting rhizobacterial mediated protection in tomato against Tomato mottle virus. Plant Disease. 2000;84:779–784. doi: 10.1094/PDIS.2000.84.7.779. [DOI] [PubMed] [Google Scholar]
- Mysore KS, Ryu CM. Nonhost resistance: how much do we know? Trends in Plant Science. 2004;9:97–104. doi: 10.1016/j.tplants.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Nurnberger T, Brunner F, Kemmerling B, Piater L. Innate immunity in plants and animals: striking similarities and obvious differences. Immunological Reviews. 2004;198:249–266. doi: 10.1111/j.0105-2896.2004.0119.x. [DOI] [PubMed] [Google Scholar]
- Park CJ, Shin R, Park JM, Lee GJ, Yoo TH, Paek KH. A hot pepper cDNA encoding a pathogenesis-related protein 4 is induced during the resistance response to tobacco mosaic virus. Molecules and Cells. 2001;11:122–127. [PubMed] [Google Scholar]
- Park CJ, Shin R, Park JM, Lee GJ, You JS, Paek KH. Induction of pepper cDNA encoding a lipid transfer protein during the resistance response to tobacco mosaic virus. Plant Molecular Biology. 2002;48:243–254. doi: 10.1023/a:1013383329361. [DOI] [PubMed] [Google Scholar]
- Park CJ, Kim KJ, Shin R, Park JM, Shin YC, Paek KH. Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. The Plant Journal. 2004;37:186–198. doi: 10.1046/j.1365-313x.2003.01951.x. [DOI] [PubMed] [Google Scholar]
- Pegadaraju V, Knepper C, Reese J, Shah J. Premature leaf senescence modulated by the Arabidopsis PHYTOALEXIN DEFICIENT4 gene is associated with defense against the phloem-feeding green peach aphid. Plant Physiology. 2005;139:1927–1934. doi: 10.1104/pp.105.070433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegadaraju V, Louis J, Singh V, et al. Phloem-based resistance to green peach aphid is controlled by Arabidopsis PHYTOALEXIN DEFICIENT4 without its signaling partner ENHANCED DISEASE SUSCEPTIBILITY1. The Plant Journal. 2007;52:332–341. doi: 10.1111/j.1365-313X.2007.03241.x. [DOI] [PubMed] [Google Scholar]
- Pickett JA, Poppy GM. Switching on plant genes by external chemical signals. Trends in Plant Science. 2001;6:137–139. doi: 10.1016/s1360-1385(01)01899-4. [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. Networking by small-molecule hormones in plant immunity. Nature Chemical Biology. 2009;5:308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- Powell G, Tosh CR, Hardie J. Host plant selection by aphids: behavioral, evolutionary, and applied perspectives. Annual Review of Entomology. 2006;51:309–330. doi: 10.1146/annurev.ento.51.110104.151107. [DOI] [PubMed] [Google Scholar]
- Rask L, Andreasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J. Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Molecular Biology. 2000;42:93–113. [PubMed] [Google Scholar]
- Ryu CM, Hu CH, Reddy MS, Kloepper JW. Different signaling pathways of induced resistance by rhizobacteria in Arabidopsis thaliana against two pathovars of Pseudomonas syringae. New Phytologist. 2003;160:413–420. doi: 10.1046/j.1469-8137.2003.00883.x. [DOI] [PubMed] [Google Scholar]
- Ryu CM, Anand A, Kang L, Mysore KS. Agrodrench: a novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse Solanaceous species. The Plant Journal. 2004;40:322–331. doi: 10.1111/j.1365-313X.2004.02211.x. [DOI] [PubMed] [Google Scholar]
- Shin R, Lee GJ, Park CJ, et al. Isolation of pepper mRNAs differentially expressed during the hypersensitive response to tobacco mosaic virus and characterization of a proteinase inhibitor gene. Plant Science. 2001;161:727–737. [Google Scholar]
- Shoresh M, Harman GE, Mastouri F. Induced systemic resistance and plant responses to fungal biocontrol agents. Annual Review of Phytopathology. 2010;48:21–43. doi: 10.1146/annurev-phyto-073009-114450. [DOI] [PubMed] [Google Scholar]
- Smith JL, De Moraes CM, Mescher MC. Jasmonate- and salicylate-mediated plant defense responses to insect herbivores, pathogens and parasitic plants. Pest Management Science. 2009;65:497–503. doi: 10.1002/ps.1714. [DOI] [PubMed] [Google Scholar]
- Staskawicz BJ, Ausubel FM, Baker BJ, Ellis JG, Jones JD. Molecular genetics of plant disease resistance. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- Thompson GA, Goggin FL. Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. Journal of Experimental Botany. 2006;57:755–766. doi: 10.1093/jxb/erj135. [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H. Fresh insights into processes of nonhost resistance. Current Opinion in Plant Biology. 2003;6:351–357. doi: 10.1016/s1369-5266(03)00063-3. [DOI] [PubMed] [Google Scholar]
- Walling LL. The myriad plant responses to herbivores. Journal of Plant Growth Regulation. 2000;19:195–216. doi: 10.1007/s003440000026. [DOI] [PubMed] [Google Scholar]
- Yang JW, Yu SH, Ryu CM. Priming of defense-related genes confers root-colonizing bacilli-elicited induced systemic resistance in pepper. The Plant Pathology Journal. 2009;25:303–440. [Google Scholar]
- Yang JW, Yi HS, Kim H, et al. Whitefly infestation of pepper plants elicits defence responses against bacterial pathogens in leaves and roots and changes the below-ground microflora. Journal of Ecology. 2011;99:46–56. [Google Scholar]
- Yi HS, Heil M, Adame-Alvarez RM, Ballhorn DJ, Ryu CM. Airborne induction and priming of plant defenses against a bacterial pathogen. Plant Physiology. 2009;151:2152–2161. doi: 10.1104/pp.109.144782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler D, Zahringer U, Gerber I, et al. Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15811–15816. doi: 10.1073/pnas.0404536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]