Abstract
Background
Arabinogalactan proteins (AGPs) are complex proteoglycans of the cell wall found in the entire plant kingdom and in almost all plant organs. AGPs encompass a large group of heavily glycosylated cell-wall proteins which share common features, including the presence of glycan chains especially enriched in arabinose and galactose and a protein backbone particularly rich in hydroxyproline residues. However, AGPs also exhibit strong heterogeneities among their members in various plant species. AGP ubiquity in plants suggests these proteoglycans are fundamental players for plant survival and development.
Scope
In this review, we first present an overview of current knowledge and specific features of AGPs. A section devoted to major tools used to study AGPs is also presented. We then discuss the distribution of AGPs as well as various aspects of their functional properties in root tissues and pollen tubes. This review also suggests novel directions of research on the role of AGPs in the biology of roots and pollen tubes.
Keywords: Arabinogalactan proteins, cell wall, root, pollen tube, growth, development, plant–microbe interactions, sexual reproduction, plant proteoglycans, Arabidopsis, Nicotiana
GENERAL INTRODUCTION
Arabinogalactan proteins (AGPs) are a family of non-enzymatic cell surface hydroxyproline-rich glycoproteins (HRGPs) (see Table 1 for a list of abbreviations used in the text). These proteoglycans, analogous to animal proteoglycans, are found throughout the entire plant kingdom from bryophytes (e.g. Physcomitrella patens; Lee et al., 2005) to angiosperms (e.g. Arabidopsis thaliana; Schultz et al., 2000). AGPs have been implicated in a large number of biological functions throughout the plant life cycle and many are specifically associated with development and reproduction. AGPs encompass a large group of cell-wall proteins which share common features, including their ability to bind to β-d-glucosyl Yariv reagent (Yariv et al., 1967; see Fig. 1A), their typical arabino-galactosylated glycomodules, and many other features associated with their protein and nucleic sequences such as the presence of numerous hydroxyproline (HyP)-based sites of O-glycosylation, the possibility of being anchored to the plasma membrane, or the existence of many functional domains (often putative) that confer several possible biological functions (for reviews, see Seifert and Roberts, 2007; Ellis et al., 2010).
Table 1.
List of abbreviations
| AG | Arabinogalactan |
| AGPs | Arabinogalactan proteins |
| AGPE | Arabinogalactan protein-extensins |
| AM | Arbuscular mycorrhizas |
| ENOD | Early nodulin |
| FLA | Fasciclin-like AGP |
| GPI | Glycosylphosphatidylinositol |
| GTs | Glycosyltransferases |
| HRGPs | Hydroxyproline-rich glycoproteins |
| HyP | Hydroxyproline |
| mAb | Monoclonal antibodies |
| nsLTP | Non-specific lipid transfer protein |
| P4-Hs | Proline-4-hydroxylases |
| reb1-1 | root epidermal bulger |
| PELPIII | Pistil-specific extensin-like protein III |
| TTS | Transmitting-tract-specific |
| XTH | Xyloglucan endotransglucosylase hydrolase |
Fig. 1.

The Yariv reagent. (A) chemical structures of different phenylglycosides including active Yariv (β-d-glucosyl and β-d-galactosyl) and inactive Yariv (α-d-galactosyl and α-d-mannosyl), commercially available at www.biosupplies.com.au. (B) Histochemical staining of arabidopsis root with inactive Yariv. (C) Histochemical staining of arabidopsis root with active Yariv (note the reddish colour of the root). (D) Rocket electrophoresis of AGPs extracted from the roots of wild-type (WT) Arabidopsis thaliana, from the reb1-1 mutant or from the reb1-1 mutant grown in the presence of 10 mm galactose. The numbers at the top of rockets correspond to AGP quantities (in mg). CTR = 0 mg. Note that AGP content is recovered after addition of galactose (Gal) to reb1-1 mutant (0·45 mg). Standard AGPs were extracted and purified from red wine. Scale bars in (B) and (C) = 200 µm.
The protein backbone of AGPs is synthesized by members of a large multigene family consisting of 52–82 members in Arabidopsis thaliana (Schultz et al., 2002; Johnson et al., 2003; Showalter et al., 2010) and 69 members in rice (Oryza sativa) (Ma and Zhao, 2010). AGPs are characterized by the extensive O-glycosylation of the protein backbone which takes place post-translationally in the Golgi apparatus (see also Fig. 2). Typically the carbohydrate moiety accounts for >90 % w/w of the mass of the glycoprotein and consists predominantly of arabinose and galactose residues, although other ‘minor’ sugars including rhamnose, fucose, glucuronic acid and xylose are also present (Nothnagel, 1997; Showalter, 2001). The glycan composition of AGPs can vary greatly between species, between organs within the same species and may even be developmentally regulated within the same organ in different cell-types (Tsumuraya et al., 1988; Pennell et al., 1991). The two glycans most commonly found in AGPs are (1) a short oligo-arabinoside chain of three to four residues, and (2) a larger β-1,3-linked galactan backbone with 1,6-linked side chains containing galactose, arabinose and, often, rhamnose and glucuronic acid (Kieliszewski and Shpak, 2001; Tan et al., 2010; Tryfona et al., 2010). AGP glycomodules have been proposed to adopt different spatial configurations that modulate AGP functionality (Fincher et al., 1983; Qi et al., 1991; Showalter, 2001). Although AGPs are known as O-glycosylated proteins (Showalter, 2001; Seifert and Roberts, 2007) discrete N-glycosylation sites have also been reported to be present in the protein backbone (Du et al., 1996). The O-glycosylation of AGPs occurs predominantly on HyP residues, and less often on serine and threonine residues (Showalter, 2001). Proline hydroxylation is performed by a multigene family of enzymes, proline-4-hydroxylases (P4-Hs) (Vlad et al., 2007) that are believed to act within the endoplasmic reticulum. Using a synthetic gene strategy and engineering, a variety of consensus motifs to study glycosylation patterns in living plant cells (Kieliszewski and Shpak, 2001; Shpak et al., 2001; Tan et al., 2003, 2004; Estevez et al., 2006; Xu et al., 2008) it was shown that O-glycosylation leading to arabinosylation occurs preferentially on clustered Ser-(Hyp)4 contiguous sequences (contigs), whereas O-glycosylation leading to arabinogalactosylation occurs on Hyp-alanine, Hyp-serine, Hyp-threonine and Hyp-valine contigs. Based on the amino acid sequence and composition, AGPs were initially categorized into classical AGPs (consisting of a P/Hyp-rich domain heavily O-glycosylated, a hydrophobic C-terminal (C-ter) domain required for anchorage to the plasma membrane, and a signal peptide sequence) and non-classical AGPs (sometimes N-glycosylated and lacking the C-ter domain). However, this classification has subsequently been altered based on a better knowledge of plant genomes [Arabidopsis, The Arabidopsis Genome Initiative (2000); rice, Yu et al. (2005)], along with the development of several algorithms using a biased amino acid composition and the presence of signal peptides (Schultz et al., 2002; Showalter et al., 2010) or BLAST-based alignment (Borner et al., 2002; Schultz et al., 2002; Johnson et al., 2003). Thus AGPs are now classified into classical AGPs (characterized by a signal peptide, a P/Hyp rich domain and C-ter domain), AG peptides (short classical AGPs), fasciclin-like AGPs (FLA), Lys-rich AGPs, non-specific lipid transfer protein (nsLTP)-like AGPs and early nodulin (ENOD)-like AGPs (Schultz et al., 2000, 2002; Johnson et al., 2003; Sun et al., 2005; Yang et al., 2005; Mashiguchi et al., 2009; Ma and Zhao, 2010). The term chimeric AGPs was used to describe classical AGPs that harbour an additional protein domain such as FLAs, Lys-rich, nsLTP-like AGPs and plastocyanin-like AGPs. Some chimeric AGPs were found to lack the C-ter domain responsible for the glycosylphosphatidylinositol (GPI) anchorage as shown for FLAs (Johnson et al., 2003). All the chimeric AGPs and several other GPI-anchored proteins were predicted (though mostly not experimentally proven) to contain arabinogalactan type II (AG-II) glycomodules (Borner et al., 2002, 2003).
Fig. 2.
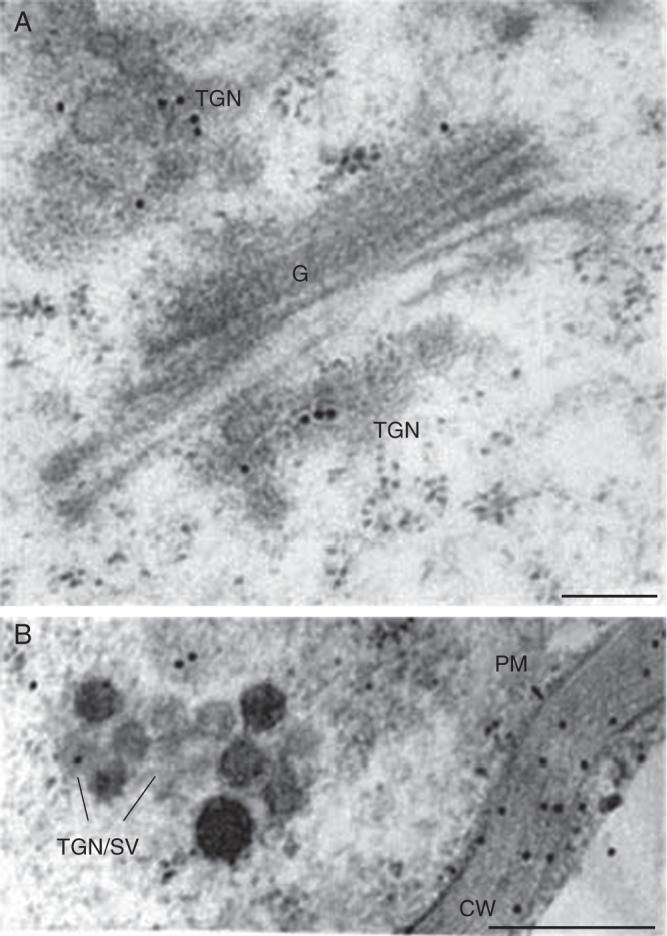
Electron micrographs of arabidopsis root cells labelled with the anti-AGP antibody JIM14. Note the immuno-gold labelling of Golgi cisternae (G) and the trans-Golgi network (TGN) in (A), and secretory vesicles (SV), the plasma membrane (PM) and the cell wall (CW) in (B). AGPs are known to be assembled within the endomembrane system. Roots were prepared by the high-pressure freezing technique and the immunogold labelling was performed as described in Driouich et al. (1993). Scale bars = 0·2 µm.
At the subcellular level, AGPs can be found in the cell wall, in the apoplast or anchored to the plasma membrane via a GPI anchor attached to the C-ter domain of the AGP backbone (Youl et al., 1998; Svetek et al., 1999). Cleavage of the GPI-anchor by phospholipases results on the release of the glycoprotein into the cell wall (Schultz et al., 1998; Borner et al., 2002, 2003). Interestingly, some GPI-anchored AGPs were shown to be associated with specific microdomains on the plasma membrane, the so-called lipid rafts (Borner et al., 2005; Grennan, 2007; Simon-Plas et al., 2011). Finally, AGPs can also be found in secretions, including root exudates and the extracellular media of cultured cells.
AGPs occur ubiquitously in land plants and have been isolated from a wide range of plant organs and cell types, from the flowers to root caps and border cells, and from pollen tubes to root hairs. They have been implicated to be involved in a variety of functions (Seifert and Roberts, 2007) including cell proliferation (Serpe and Nothnagel, 1994), cell expansion (Willats and Knox, 1996), programmed cell death (Gao and Showalter, 1999), pollen-tube growth (Wu et al., 1995; Coimbra et al., 2010), xylem differentiation (Motose et al., 2004), somatic embryogenesis (van Hengel et al., 2002), zygotic division and embryo development (Hu et al., 2006; Qin and Zhao, 2006). Recent reviews have described the structure and biology of AGPs (Seifert and Roberts, 2007; Ellis et al., 2010) highlighting the importance of these glycoproteins in plant survival and development. Here, we focus on the distribution and functions of AGPs in root tissues and pollen tubes.
PROBES AND TOOLS TO STUDY AGPS IN PLANTA
Several tools and probes have been developed and used to study AGP distribution and function in various plant species and organs. The most well-known and commonly used reagent is β-d-glucosyl Yariv that was generated 50 years ago (Yariv et al., 1962, 1967). Yariv and co-workers discovered that a phenylglycoside containing β-d-glucosyl residues (active Yariv) was able to bind and precipitate AGPs whereas related phenylglycosides containing α-d-mannosyl or α-d-galactosyl units (non-active Yariv) were not (Fig. 1A). Since then, Yariv reagents (both active and non-active) are commonly used in most, if not all, plant laboratories working on AGPs worldwide. Active Yariv has been largely used to isolate and quantify AGPs from a number of species [pear, Chen et al. (1994); arabidopsis, Schultz et al. (2000); tobacco, Du et al. (1994)]. It has also been widely used to investigate the function of AGPs in vivo based on its ability to bind AGPs and to interfere with their dynamics within the cell wall and plasma membrane. For instance, it has been well documented that active Yariv inhibits expansion and morphogenesis of root and pollen-tube cells (Willats and Knox, 1996; Mollet et al., 2002, Nguema-Ona et al., 2007).
AGP-directed monoclonal antibodies (mAbs) (Table 2) are another class of valuable probes that have been pivotal for deciphering AGP localization and function (Puhlmann et al., 1994; Knox, 1997). Most of the available mAbs reactive to AGPs are specific for epitopes associated with the carbohydrate moieties; although a few have specificity toward the protein backbone (Table 2). Similarly to active Yariv, some of the anti-AGP mAbs (e.g. JIM13) have been used to unravel the involvement of specific epitopes in controlling cell growth and morphogenesis in arabidopsis (van Hengel and Roberts, 2002; Nguema-Ona et al., 2007). Furthermore a large body of information regarding the spatial distribution of AGPs in plant tissues has been obtained employing mAbs in combination with a variety of imaging technologies (see below).
Table 2.
List of reagent and antibodies directed against AGP carbohydrate epitopes and proteic backbone
| Probe | Antigen | Epitope recognized | References |
|---|---|---|---|
| mAb | |||
| JIM4 | AGP from carrot | β-d-GlcA-(1,3)-α-d-GalA-(1,2)-α-l-Rha | Yates et al. (1996) |
| JIM8 | AG from sugar beet | Unknown | Pennell et al. (1991) |
| JIM13 | AGP from carrot | β-d-GlcA-(1,3)-α-d-GalA-(1,2)-α-l-Rha | Yates and Knox (1994); Yates et al. (1996) |
| JIM14 | AGP from carrot | Unknown | Yates and Knox (1994); Yates et al. (1996) |
| JIM15 | AGP from carrot | Unknown | Yates and Knox (1994); Yates et al. (1996) |
| JIM16 | AGP from carrot | Unknown | Yates and Knox (1994); Yates et al. (1996) |
| LM2 | AGP from rice | β-Linked GlcA | Smallwood et al. (1994); Yates et al. (1996) |
| MAC204 | AGP from pea | Unknown | Bradley et al. (1988); Pennell et al. (1989) |
| MAC207 | AGP from pea | β-GlcA-(1,3)-α-GalA-(1,2)-Rha | Bradley et al. (1988); van den Bosch et al. (1989) |
| MAC265 | AGP fom pea | Unknown | van den Bosch et al. (1989) |
| PCBC3 | AGP from Nicotiana alata style | Unknown | Fergusson et al. (1999) |
| CCRC-M7 | Rhamnogalacturonan I | β-(1,6)-Gal chain carrying one or more Ara residues | Steffan et al. (1995) |
| PN 16·4B4 | AG from Nicotiana glutinosa | Unknown | Norman et al. (1986) |
| MH4·3E5 | AG from Nicotiana tabacum | Unknown | Hahn et al. (1987) |
| LM6 | Sugar beet arabinan | α-(1,5)-l-linked arabinosyl heptasaccharide | Lee et al. (2005) |
| LM14 | AGP from carrot | Arabinose- and galactose-enriched carbohydrate chains | Moller et al. (2008) |
| Antiserum | |||
| Le-AGP1 | Proteic backbone from tomato | Lysine-rich subdomain of Le-AGP1 proteic backbone | Gao et al. (1999) |
| Reagent | |||
| Yariv reagent | AGP | n.a. | Yariv et al. (1967) |
AG, Arabinogalactan; AGP, arabinogalactan protein; GalA, galacturonic acid; GlcA, glucuronic acid; Rha, rhamnose; mAb, monoclonal antibody; n.a., not applicable.
For more information see the Plant Cell Wall Monoclonal Antibody Database at http://glycomics.ccrc.uga.edu/wall2/jsp/abIndex.jsp
Other chemical agents including inhibitors of AGP biosynthesis are 3,4-dehydroproline (Cooper and Varner, 1983; Vicré et al., 2005), ethyl 3,4 dihydroxy benzoate and α,α-dipyridyl (Velasquez et al., 2011). These have been used to investigate AGP functions. It is noteworthy that these agents are not specific for AGPs but also interfere with the biosynthesis of other HRGPs including extensin. 3,4-Dehydroproline is known to interfere with the hydroxylation of proline residues and hence the production of Hyp residues leading to a reduction in O-glycosylation of AGPs among other HRGP (Cooper and Varner, 1983). Ethyl 3,4 dihydroxy benzoate binds to the active site of P4-Hs while α,α-dipyridyl chelates a co-factor required for P4-Hs activity (Velasquez et al., 2011).
While chemical reagents and mAbs often recognize the carbohydrate moiety of AGPs, the cloning of a single AGP gene followed by its functional characterization, as well as the generation of mutants, has helped to assign functions to single AGP genes (Shi et al., 2003; Seifert and Roberts, 2007; Coimbra et al., 2009, 2010). In this context, genomic resources have proven highly valuable in the identification and cloning of AGP genes (Schultz et al., 2002; Ma and Zhao, 2010; Showalter et al., 2010). In addition, the development of publicly available microarray analyses, as well as bioinformatic tools to ‘mine’ such datasets, has enabled prediction of the expression and possible physiological roles of a given AGP in specific organs and tissues under specific experimental conditions. Using this approach (Schultz et al., 2002; Johnson et al., 2003; Showalter et al., 2010) it has also been shown that AGPs are most frequently expressed in floral tissues and pollen tubes, as well as in root tissues. More interestingly, Showalter et al. (2010) have shown that different AGP genes are co-expressed (AGPs with other AGPs including FLAs, AG peptides and chimeric AGPs) and are also often concurrently expressed with other HRGPs such as extensin. AGPs may also be co-expressed with genes involved in their post-translational modification including P4-Hs, and wall peroxidases, as well as with certain glycosyltransferases (GTs) genes involved in the synthesis of other cell-wall components (e.g. homogalacturonans that are suggested to be co-secreted with some AGPs; Showalter et al., 2010). Finally, fusion proteins such as Lycopercison esculentum AGP1:GFP have also been generated and used to investigate localization and function of AGPs (Zhao et al., 2002; Sun et al., 2004; Sardar et al., 2006; Nguema-Ona et al., 2007).
AGPS IN ROOTS: DISTRIBUTION AND FUNCTIONAL ASPECTS
Distribution of AGPs in roots: localization of AGPs in root tissues of arabidopsis and others species
Staining and localization of AGPs in plant tissues was initially made possible by the use of active Yariv as a histochemical probe (Fig. 1B, C). This led to the discovery of AGPs in taro mucilage (Colocasia esculenta) (Harris et al., 1992), tomato roots (Solanum lycopercisum) (Pogson and Davies, 1995), styles of tobacco (Nicotiana alata) (Gane et al., 1994) and Brassica napus microspores (Tang et al., 2006). However, major progress in mapping AGP distribution in root tissues was made using the various AGP-directed mAbs and polyclonal antibodies generated over the years by several laboratories (Knox, 1997; Pattathil et al., 2010; Hervé et al., 2011). The vast majority of these antibodies are directed against carbohydrate epitopes (Table 2). An antiserum which recognizes the protein core of LeAGP1 has also been generated (Gao et al., 1999).
Several anti-AGP mAbs raised against AGP fractions isolated from carrot cell cultures at different stages of development have been generated [MAC207, Pennell et al. (1989); JIM14-16, Knox et al. (1991); LM2, Smallwood et al. (1996)]. Some of these AGP-associated epitopes were shown to accompany differentiation of cambium cells during the secondary thickening in Arabidopsis thaliana root (Dolan and Roberts, 1995). Epitopes recognized by the JIM14 mAb were associated with sieve tubes of the phloem in the secondary thickened roots, while the JIM13 mAb was associated with young differentiated xylem cells (Dolan and Roberts, 1995; Dolan et al., 1995). JIM13 has also been shown to specifically localize to the root cap and border-like cells in seedlings of Arabidopsis thaliana (Fig. 2) (Vicré et al., 2005). JIM4 is another AGP-directed mAb recognizing a plasma membrane-associated epitope which has also been used for root tissue labelling (Knox et al., 1989, 1991). The epitope appears at a very early stage in the formation of the vascular pattern in carrot and has been proposed as a marker of cell identity in the developing pericycle cells of carrot (Knox et al., 1991). In maize roots, the AGP epitopes recognized by the mAb LM2 have been localized to the growing tips of root hairs, while a β-(1,6)-galactan epitope associated with AGPs (recognized by a polyclonal antiserum Gal4; Kikuchi et al., 1993) has been found over the entire root surface including root hairs (Samaj et al., 1999). Together, these observations indicate that AGP-associated epitopes are developmentally regulated in plant roots and that modulation of AGP expression occurs during cell development and positioning of cells within the apex. Along with AGPs, other HRGPs including extensins and chimeric AGPs are distributed over plant cell surfaces of growing and differentiating tissues and are believed to play a role in plant cell morphogenesis (Knox, 1995).
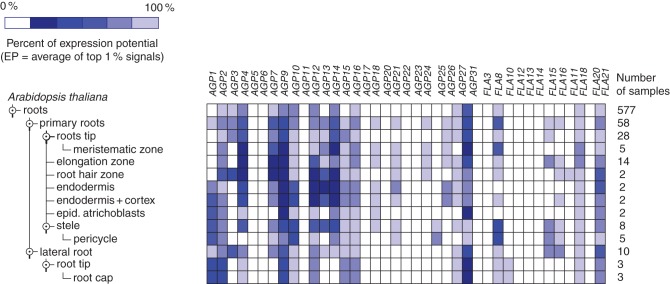
Sequencing of plant genomes including Arabidopsis thaliana (The Arabidopsis Genome Initiative, 2000) and Oryza sativa (Yu et al., 2005), along with the development of bioinformatics and molecular techniques, has allowed the mapping of AGP genes in the roots of arabidopsis based on their expression patterns. Using Genevestigator (Zimmermann et al., 2004) (Fig. 3), 15–20 AGP genes were found to be expressed in roots under different environmental conditions. However, few of them have been subjected to in-depth analyses. Screening of arabidopsis mutants has also allowed identification of patterns of expression for specific genes. A number of genes reflecting the diversity of AGP types has been found to be specifically expressed in root tissues including: the non-classical AGP AtAGP30 (At2g33790; van Hengel and Roberts, 2003) expressed in the arabidopsis root elongating zone and AtAGP31 (At1g28290; Liu and Mehdy, 2007); FLA genes, SOS5 (AtFLA4; At3g46550; Shi et al., 2003) and AtFLA1 (At5g55730; Johnson et al., 2011), also expressed in growing arabidopsis roots; nsLTP-like AGP family member AtXYP2 (At2g13820; Kobayashi et al., 2011) the closest homologue of Zinnia elegans ZeXYP previously shown to play a role in xylem differentiation (Motose et al., 2004) and a Lys-rich AGP gene, AtAGP17, expressed in arabidopsis roots (Gaspar et al., 2004). The functions of these AGPs are discussed below.
Fig. 3.
Relative level of expression of AGP and FLA genes in different root tissues of Arabidopsis thaliana. The figure was generated using Genevestigator (Zimmermann et al., 2004).
Functional properties of AGPs in roots
Role in root growth and morphogenesis
A number of studies have shown that AGPs are important for root development. Willats and Knox (1996) showed that treatment of Arabidopsis thaliana seedlings with active Yariv causes a disruption in root growth and abnormal morphology. Both cell elongation and expansion were affected, leading to short and swollen roots. Swelling (e.g. radial expansion) was mostly associated with epidermal cells. Indeed, the epidermal cells more exposed to the reagent exhibited radial expansion. Interestingly, root cap and meristem did not appear to be affected by the treatment (Vicré et al., 2005) suggesting that sensitivity to active Yariv varies between different tissues probably because of differences in reactivity of different AGPs to the reagent. Interestingly, the arabidopsis mutant reb1-1 (root epidermal bulger) displayed a root morphology similar to that obtained by active Yariv treatment. The root of the reb1-1 mutant was clearly shown to lack a subpopulation of AGPs normally present in wild-type root (Baskin et al., 1992; Ding and Zhu, 1997) confirming that AGPs are required for the control of oriented cell expansion in elongating roots (Willats and Knox, 1996). Extending on this, Andème-Onzighi et al. (2002) have shown that the swelling phenotype of the mutant is restricted to trichoblasts and that certain AGP epitopes (recognized by the mAbs LM2 and JIM14) are absent from the swollen cells.
As postulated by Ding and Zhu (1997), the REB1-1 gene was later shown to encode for a UDP d-glucose 4-epimerase that converts d-glucose into d-galactose (Seifert et al., 2002), providing galactose to AGPs and xyloglucans (Nguema-Ona et al., 2006), and supporting a link between the normal carbohydrate structure of AGPs and the control of cell expansion. Supplementation of the growth media of the reb1-1 mutant with 10 mm galactose restored the wild-type phenotype and AGP content in the root of the mutant (Fig. 1D). The importance of an unaltered carbohydrate moiety of AGPs in controlling root cell elongation was also confirmed for another arabidopsis mutant, mur1 (murus 1), deficient in a mannose 4,6 dehydratase, involved in the biosynthesis of fucose (van Hengel and Roberts, 2002). The mur1 mutant contains fewer terminal fucose residues on AGP glycans than the wild type and leads to changes in the cross-electrophoresis pattern of root AGPs and alterations of root morphology and elongation (van Hengel and Roberts, 2002). The addition of an eel (Anguila anguila) lectin that specifically binds to terminal fucose residues associated with AGPs to the media used to grow wild-type seedlings caused their roots to phenocopy those of mur1 mutants. This led to the conclusion that fucosylated AGPs are important for controlling root growth and development.
Further investigations of the reb1-1 mutant showed that swollen trichoblasts also displayed disorganized cortical microtubules (Andème-Onzighi et al., 2002; Nguema-Ona et al., 2006) suggesting a link between the cytoskeleton and plasma membrane/cell wall AGPs. To validate and extend on this finding, arabidopsis seedlings were treated with active Yariv or anti-AGP mAbs and cortical microtubules organization examined. Interestingly, within 30 min of treatment, cortical microtubules were shown to lose their transverse organization and detach from the inner side of the plasma membrane (Nguema-Ona et al., 2007). Concomitantly, disruption of AGP distribution at the outer side of the plasma membrane was observed (Nguema-Ona et al., 2007). Tobacco BY-2 suspension-cultured cells also exhibited microtubule and actin networks disorganization in response to Yariv treatment (Sardar et al., 2006).
Whether and how AGP and cytoskeleton elements are interconnected is yet to be elucidated. Nevertheless, it has been proposed that the flow of information could be directed from the cell wall to the cytoplasm or reversely, from the cytoplasm to the cell wall (Nguema-Ona et al., 2007). Here, AGP could interact with molecular factors such as receptor-like kinases for signal transduction into the cytoplasm (Ringli, 2010; Boisson-Dernier et al., 2011). The rapid effect of active Yariv treatment on cortical microtubules organization (within a few minutes) pleads for the closeness of AGPs and elements of the cytoskeleton. The ability of AGPs to be inserted in the external leaflet of the plasma membrane via a GPI-anchor (Schultz et al., 1998) places them in proximity to microtubules and suggests that this property may play a role in the interconnection. Sardar et al. (2006) showed that both microtubule and actin-disorganizing drugs had an effect on AGP localization, and active Yariv could disorganize, in a reversible manner, both actin and microtubules in BY-2 cells, supporting the possibility of bidirectional flow of information. They also suggested the implication of phospholipase D, wall-associated kinases and lectin receptor kinases as potential candidates contributing to the cell wall–plasma membrane–cytoskeleton continuum. These potential interactors could act as linkers between AGPs and microtubules (Sardar et al., 2006) possibly at specific plasma membrane domains such as lipid rafts (Mongrand et al., 2004; Grennan, 2007). However, Driouich and Baskin (2008) suggested the potential interactors could be part of a scaffold where cortical microtubules are essentially required for controlling the orientation of cellulose microfibrils via GPI-anchored partners such as COBRA protein (Roudier et al., 2005) or AGPs.
In addition to pharmacological studies that target several AGP populations (above), investigations focused on a particular AGP gene and its product have also linked biological functions to a given AGP. To date, about a dozen AGP-encoding genes have been shown to be expressed in root tissues of Arabidopsis thaliana and other species (e.g. Oryza sativa). The expression of some of these genes is regulated by factors including phytohormones. AGP30 is the first non-classical AGP shown to be specifically expressed in roots. It is a histidine-rich AGP that plays a role in root regeneration and seed germination (van Hengel and Roberts, 2003). Similarly to AtAGP30, Daucus carota AGP1 (DcAGP1), a close homologue of AtAGP30, is specifically expressed in roots and found abundantly in carrot cell cultures (Baldwin et al., 2001). Disruption of AGPs with active Yariv at the root surface of an arabidopsis agp30 mutant deficient in AtAGP30 displays a less severe phenotype than the wild type, supporting the lack of some AGPs in the roots of this mutant. More interestingly, the lack of AtAGP30 caused a general decrease in the perception of the hormone abscissic acid. This led to the proposal by Gens et al. (2000) and Wagner and Kohorn (2001) that AtAGP30 may be part of a signalling pathway involving cell wall-associated kinases (Gens et al., 2000; Wagner and Kohorn, 2001) or leucine-rich repeat receptor kinases (Tang et al., 2002) at the cell surface. AtAGP30 is expressed in non-hairy epidermal cells (atrichoblasts) of the meristematic and elongating zones where they might be involved in maintaining the shape of these cell lines (van Hengel et al., 2004). Its co-localization with the GLABRA2 supports a role in the early stage of root epidermal patterning in an abscissic acid-dependent manner. AtAGP30 is also expressed in cortical, endodermal and vascular tissues of the mature part of the roots (van Hengel et al., 2004). AtAGP31 is a close homologue of AtAGP30 and is a non-classical AGP with a cysteine-rich C-terminal PAC (proline-rich protein and AGP, containing cysteine) domain, a proline-rich domain and a histidine-rich domain. AtAGP31 gene is strongly expressed in vascular tissues including the phloem and primary xylem (Liu and Mehdy, 2007) and has been suggested to play a role in root development. Interestingly, AtAGP31 expression is repressed in the presence of a wounding stress or wounding-associated factors such as methyl-jasmonate, implicating a role in abiotic stress responses. Another subgroup of chimeric AGPs, the FLA, has also been shown to play a role in the control of morphogenesis. Under salinity stress, the lack of the SOS5/FLA4 gene in the sos5/fla4 mutant leads to a reduction in root growth and abnormal expansion of epidermal, cortical and endodermal cells (Shi et al., 2003). SOS5/FLA4 protein is strongly expressed in the root cortex and vascular tissues but weakly in epidermal cells and root hairs. It is a plasma membrane-associated AGP that contains two fasciclin-like domains and a C-terminal GPI anchor (Shi et al., 2003). Fasciclin domains are known to play a role in cell adhesion in animals, insects and algae (Kawamato et al., 1998). Interestingly, root cells of sos5 mutant have thinner and loosened cell walls compared with wild type. Shi et al. (2003) speculated that SOS5 proteins may aggregate or interact through fasciclin domains with other cell-wall components to form a network capable of maintaining cell-wall structure and proper cell expansion under salt stress. AtFLA1 is another FLA which has recently been shown to play a role in lateral root development and shoot regeneration from root tissues in Arabidopsis thaliana (Johnson et al., 2011). It is expressed in the mature vasculature of lateral roots and the elongation zone of the primary root. The function of AtFLA1 gene is unclear but one possibility is that it is involved in regulating cell differentiation and expansion in the newly formed lateral roots (Johnson et al., 2011) but may also have a role in defining cell fate and identity, a role that has been already proposed based on differential expression of specific AGP-associated epitopes as markers of cell fate (McCabe et al., 1997). Such a possibility remains to be demonstrated with the generation and use of anti-AtFLA1 mAbs.
Role in microbe interaction with roots
Apart from their important roles in root development, AGPs present in roots, specifically in root-released border cells/border-like cells and in exudates are important for plant–microorganism interactions within the rhizosphere. The role of root exudates in interactions between plant roots and microbes has been reviewed by Bais et al. (2006). The interactions are described as positive, when they lead to symbiotic associations with mycorrhizal fungi and soil bacteria, or biocontrol agents. In contrast, they are classified as negative when they lead to plant parasitism or pathogenesis (Bais et al., 2006). Exudates (containing mucilage and metabolites) are root secretions that contribute significantly to defining the microenvironment surrounding root surfaces wherein specific populations of microbes are attracted and can develop (Walker et al., 2003). Mucilage contains high molecular-weight cell-wall components including AGPs (Bacic et al., 1988; Moody et al., 1988) as observed in cowpea (Vigna unguiculata) (Knee et al., 2001), and maize root mucilage (Ma et al., 2010).
The unexpected finding that AGPs influence root interactions with microbes was found by studying an arabidopsis mutant resistant to transformation by Agrobacterium tumefaciens, the rat1 (resistant to agrobacterium transformation 1) (Nam et al., 1999). Gaspar et al. (2004) showed that absence of AtAGP17/RAT1 expression in the mutant suppresses Agrobacterium tumefaciens ability to colonize arabidopsis roots. Pre-treatment of wild-type roots with active Yariv prior to transformation with agrobacterium also prevents root colonization (Gaspar et al., 2004) and led Gaspar et al. (2004) to suggest that AtAGP17 deficiency may either affect a physical association between the root cell surface and bacteria at the initial stage of infection, or interfere with a signalling cascade in which AtAGP17 acts as an elicitor-like signalling molecule (Gaspar et al., 2004). The later assumption is supported by the finding that under-expression/lack of AtAGP17 prevents modulation of the content of salicylic and of certain pathogenesis-related proteins (e.g. PR1) leading to the resistant phenotype. It is well known that during infection by Agrobacterium tumefaciens and subsequent nodule formation, some plant defence mechanisms are repressed (Pitzschke and Hirt, 2010). Also, it is interesting to note that other Lys-rich AGPs such as LeAGP-1 and NaAGP4 (Gilson et al., 2001) have been found to respond to wounding and to pathogen attack. For instance, NaAGP4, a close homologue of AtAGP17, is repressed following leaf infection with the necrotrophic pathogen Botrytis cinerea, contrasting with extensin overexpression under the same conditions (Gilson et al., 2001). It is possible that AGP genes required for growth and development such as LeAGP1 or NaAGP4 are repressed under stress conditions to allow extensin-dependent cell wall cross-linking/strengthening to take over as a defence response.
Additionally, studies of interactions between symbiotic bacteria such as Rhizobium leguminosarum and pea roots during nodule formation have provided further evidence for the role of AGPs during root colonization by micro-organisms. Nodule formation starts with entrapping of rhizobia between legume root hairs and development of an infection thread which grows inward from the root hair toward epidermal and cortical cells of the root (reviewed by Gage and Margolin, 2000). A chimeric population of AGPs [called arabinogalactan protein-extensins (AGPE)] has been identified as the major component of the infection thread lumen (Rathbun et al., 2002). Immuno-localization with the MAC265 mAb (van den Boesch et al., 1989) revealed that AGPE are located both at the root surface and in the infection treads, and can be cross-linked by peroxidases (see also Kjellbom et al., 1997 for oxidative cross linking of plasma membrane AGPs). Rathbun et al. (2002) hypothesized that physical and biochemical properties of AGPE may have an important influence on the progess of tissue and cell colonization by Rhizobium, probably by surrounding the bacteria in the infection thread or by regulating the growth of the infection thread itself. Symbiotically defective mutants of pea (Pisum sativum) can fail to induce formation of infection threads. Tsyganova et al. (2009) showed that, in addition to recognizing new infection threads in the infection zone and mature infection threads in the nitrogen-fixing zone of root nodule in wild type and symbiotically deficient strains MAC265 labelled the intercellular spaces of infected nodule tissue and small cytoplasmic vesicles in symbiotically deficient strains. This suggests that the targeted secretion of the matrix glycoprotein recognized by the MAC265 mAb is closely correlated with growth of the infection thread (Tsyganova et al., 2009).
In addition to their role during development of infection threads, AGPs also play a role during early stages of nodule formation between Rhizobia and legumes. Downie (2010) reviewed the steps leading to a successful nodule formation prior to infection. While nod (nodulation) factors remain the major determinant for host specificity and nodule initiation, other factors such as bacterial exopolysaccharides and plant lectins can influence legume nodulation. Indeed, it was shown that attraction could also be mediated by bacterial cell wall glucomannan and plant lectins from pea (Pisum sativum) and favour a polar attachment in vitro or an attraction in vivo (Laus et al., 2006; Williams et al., 2008), in the absence of nod factors. It is known that rhizobia and agrobacteria attach in a polar manner to plant root surface (Matthysse and Kijne, 1998). Recently, Xie et al. (2012) identified an AGP in root exudates of pea that is able to induce a polar attachment of Rhizobium. Interestingly, the purified glycoprotein is recognized by the anti-AGP mAbs JIM13 and LM2, but not by MAC265, specific for AGPE. Treatments of the glycoprotein with proteases or glycosidases suppress the polar attachment, suggesting that both the carbohydrate moiety and the protein backbone are required for function (Xie et al., 2012). Xie et al. (2012) proposed that AGP could provide a source of nutrients for soil bacteria (Knee et al., 2001) and indirectly act in a complementary manner to the previously described legume-specific polar attachment mediated by specific plant lectins and bacterial glucomannans (Laus et al., 2006).
Much more ancient symbioses have been established between plants and fungi, forming arbuscular mycorrhizas (AM) and actinorhizes. After spore germination, establishment of the symbiosis includes hyphal branching, apressorium development after contacting the root, colonization of the root cortex, formation of intracellular arbuscules and, concomitantly, production of an extraradical mycelium from which spores are eventually formed (Balestrini and Lanfranco, 2006). The molecular similarity between the legumes/rhizobia and legumes/AM has suggested a common genetic programme for both symbioses (Gianinazzi-Pearson and Denarié, 1997). Van Buuren et al. (1999) also showed that, among several cell-wall genes required for wall metabolism activities necessary to favour AM colonization, AGPs and HRGP transcripts were identified in cells containing arbuscules (see also Balestrini and Lanfranco, 2006). The presence of AGPs at the symbiotic interface was clearly demonstrated by Berry et al. (2002). More recently, Schultz and Harrison (2008) proposed that plant AGPs could physically interact with the mycorrhizal fungus Glomus intraradices AGP-like proteins in the apoplastic compartment hosting the arbuscules. In this context, early nodulin ENOD11, a putative cell-wall proline-rich protein, having a low overall tyrosine content (Journet et al., 2001; Chabaud et al., 2002), is inducible upon simple adhesion of the AM fungus to the root surface. This protein was shown to play a role during the early stages of root nodulation and root colonization by AM in epidermal and cortical cells, including reorganization of host cytoplasm and construction of a novel apoplastic compartment, the pre-penetration apparatus where the fungus is destined to penetrate (Genre et al., 2005). Interestingly, ENOD11 is also expressed during root hair nodule formation, as well as in non-symbiotic root tissues such as root cap and root border cells (Journet et al., 2001), supporting the suggestion that ENOD11 may be responsible for the elastic properties of cell walls under these conditions.
In a pathogenesis context, few data on the role of AGPs at the root surface are available, while other HRGPs (e.g. extensins) (Esquerré-Tugaye and Mazau, 1974) are known to be involved in defence mechanisms. Xie et al. (2011) compared the distribution of HRGPs, including AGPs, in the roots of resistant and susceptible wax gourds prior to and after infection by Fusarium oxysporum or treatment with fusaric acid. In the absence of infection, the resistant cultivar constitutively expresses more extensin-associated epitopes than the susceptible cultivar. Upon infection, expression of extensin-associated epitopes decreased significantly in epidermal cells of the susceptible cultivar, while it remained unchanged in the resistant one. Interestingly, the anti-AGP-associated epitope recognized by the mAb CCRC-M7 (Steffan et al., 1995) was enhanced in the resistant cultivar, indicating that this epitope is likely to contribute to resistance. In contrast, other AGP-associated epitopes (recognized by LM2 and JIM16) were equivalently expressed in both cultivars before and after infection.
Further evidence for a role of AGPs in interactions with the rhizosphere was provided by arabidopsis root border-like cells. Border cells and border-like cells are released from the root tip either individually or as a group of attached cells (Hawes et al., 2000; Driouich et al., 2007). They play a major role in plant–microbe interactions within the rhizosphere and provide protection to the root (Hawes et al., 2000; Vicré et al., 2005; Driouich et al., 2012). In addition to root cap-secreted exudates which contain high molecular components including AGPs (Knee et al., 2001; Xie et al., 2012), border-like cells also synthesize and secrete significant amount of AGPs at their surface (Fig. 4) (Vicré et al., 2005; Wen et al., 2007; Cannesan et al., 2012). The proteome of border cell exudates plays a role in protecting the cap against infection (Wen et al., 2007). Hawes et al. (2000) listed functions associated with border cells and it is likely that many are AGP-dependent. Recently, it was shown that AGPs secreted by pea roots are capable of attracting zoospores of the pathogenic oomycete Aphanomyces euteiches and then inhibiting their germination (Cannesan et al., 2012).
Fig. 4.
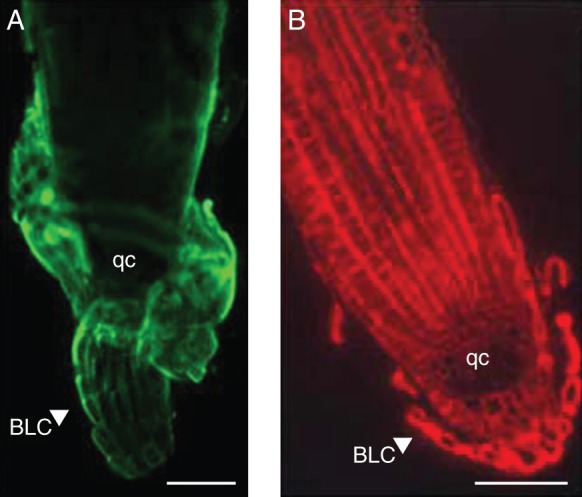
Arabidopsis root tips and associated border-like cells labelled with anti-AGP antibodies: JIM13 (A) and JIM14 (B). Note the strong labelling associated with border-like cells in (A) (secondary antibody conjugated to FITC) and the lack of labelling in the quiescent centre in (B) (secondary antibody conjugated to TRITC). The image in (A) is of an intact root while the image in (B) is of a sectioned root (as described in Nguema-Ona et al., 2006). Abbreviations: BLC, border-like cells; qc, quiescent centre. Scale bars = 100 µm.
In addition to AGPs, root cap-secreted exudates, root- and border cell-released enzymes involved in degradation and remodelling of cell wall glycoproteins and polysaccharides such as xyloglucan endotransglucosylase hydrolase (XTH) and beta-galactosidase (Wen et al., 2007) have been described. Their presence opens the possibility for novel modes of action. Thus, it is of interest to establish whether specific oligosaccharides released from AGPs in the rhizosphere could act as signals of infection. Such a function has been already proposed in the oligosaccharins theory (Albersheim et al, 1992; Creelman and Mullet, 1997) and was demonstrated for pectin-derived oligosaccharides (Cervone et al., 1989) and xyloglucan-derived oligosaccharides from plant cell walls (Fry et al., 1993). Root border-like cells of arabidopsis were shown to be highly enriched in AGPs that are likely to be released in the rhizosphere. In addition, the association of a rhizobacterium with root cells was shown to be AGP-dependent since the use of drugs acting on AGP abolished the interaction (Vicré et al., 2005). More interestingly, the homogalacturonan-deficient qua1-1 arabidopsis mutant has an altered release pattern of border-like cells that is accompanied by the secretion of abundant mucilage particularly enriched in AGPs and xylogalacturonan (Durand et al., 2009; Driouich et al., 2010). These molecules are suggested to enhance the protective capacity of border cells of the qua1-1 mutant (Driouich et al., 2010, 2012).
AGPS IN POLLEN TUBE AND OTHER FLOWER ORGANS: DISTRIBUTION AND FUNCTIONAL ASPECTS
Double fertilization in flowering plants requires the targeted delivery of sperm by the pollen tube. After landing on compatible stigmatic cells, pollen germinates and forms a tube that grows deep inside the specialized transmitting tissue of the pistil to arrive precisely at a specific target, the embryo sac. Typically, one pollen tube terminates its journey by entering a synergid cell and bursting to release the sperm nuclei, in a process called pollen tube reception. Unlike most plant cells, pollen tubes grow in a polarized way, restricted to the tip area. During pollen-tube growth, callose plugs are laid down at regular intervals behind the growing tip, and the area adjacent to the plug becomes vacuolated, which serves to maintain a region of concentrated cytoplasm containing organelles, the vegetative nucleus and the two sperm cells, near the tip (Wang et al., 2010). Tip growth is made possible through co-ordinated cellular activities, including a dynamic actin cytoskeleton system, targeted exocytosis, and regulated endocytosis (Hepler et al., 2001). One of the main features of growing pollen tubes is a tip-focused calcium gradient which is thought to be maintained by influx of extracellular calcium through calcium channels active at the extreme end of the growing tip (Feijó et al., 1995; Malhó et al., 1995). The grow trajectory is adjusted and apparently dependent on an intricate network of signalling events, largely unidentified, and probably involving molecules of different kinds (Preuss, 2002; Johnson and Lord, 2006; Mollet et al., 2007; Palanivelu and Johnson, 2010; Wang et al., 2010). Among them, AGPs have attracted much attention. Indeed, AGPs prevail in stigma exudates, style transmitting tissues, pollen grains and pollen tubes in many species (Cheung et al., 1995; Wu et al., 1995) and their implications in pollen formation, pollen-grain hydration, pollen-tube growth, cell-wall deposition, pollen-tube guidance and pollen-tube rejection during the gametophytic S-incompatibility are described.
Biochemical evidence for AGP occurrence in pollen tubes
Due to difficulties of extracting sufficient cell-wall material, only a few studies have focused on the biochemical characterization of the pollen-tube cell wall and more specifically on AGPs. In Nicotiana alata pollen-tube cell walls, the content of HyP is low (2·4 mol%) and linkage analyses of a total cell-wall extract did not reveal any type-II arabinogalactosyl residues generally found in AGPs (Rae et al., 1985), suggesting that AGPs are a minor component of pollen-tube cell wall. The total cell-wall extract was mainly composed of α-1,5-linked arabinan, a large amount of callose and low amount of cellulose (Rae et al., 1985). A more recent study on Arabidopsis thaliana pollen-tube cell walls revealed that typical type-II arabinogalactan residues of AGPs (i.e. 1,6-linked Gal; 1,3,6-linked Gal; 1,3-linked Gal; t-Ara and t-Gal) accounted for 47·5 % of the total galactosyl residues (Dardelle et al., 2010). The remaining galactosyl residues (1,4-linked Gal and 1,4,6-linked Gal) are typical of type-I arabinogalactan side chains of the pectic polysaccharide rhamnogalacturonan-I (Dardelle et al., 2010). Extraction and fractionation of Golgi vesicles from Japanese camellia (Camelia japonica) pollen tubes also revealed that the vesicles are enriched in protein with significant amount of galactosyl and arabinosyl residues. Moreover, the content of the vesicles reacted strongly with JIM13, indicating that they were enriched in AGPs (Hasegawa et al., 1998). Similarly, in Lilium longiflorum, SDS–PAGE and immuno-blot analyses of the total protein from pollen tubes probed with JIM13 or stained with the active Yariv displayed several bands greater than 97 kDa (Jauh and Lord, 1996). Together these studies indicate that AGPs occur in pollen-tube cell walls and that variation in AGP content, structure and size exists.
Microarray data of arabidopsis pollen development (Honys and Twell, 2004; Pina et al., 2005) clearly showed that pollen expresses a unique subclass of genes. The sperm cell transcriptome also indicated that male gametes have different gene expression from pollen grains (Borges et al., 2008). Recently, the pollen-tube gene-expression profile was evaluated (Wang et al., 2008; Qin et al., 2009) and a significantly different set of genes found to be expressed. Intriguingly, some genes were only expressed if the pollen tubes were growing through the pistil tissues, meaning that this interaction is central and that pistil-dependent gene expression exists. Pollen-tube gene-expression data showed high expression levels for some AGPs in pollen grains and pollen tubes (Fig. 5). Four of these genes, two classical AGPs (AGP6 and AGP11) and two AG-peptides (AGP23 and AGP40), are present only in pollen grains and tubes. These two AGP groups exhibit a high degree of similarity, although AG-peptides are very different molecules, typically consisting of fewer than 30 amino acid residues (Fig. 6).
Fig. 5.
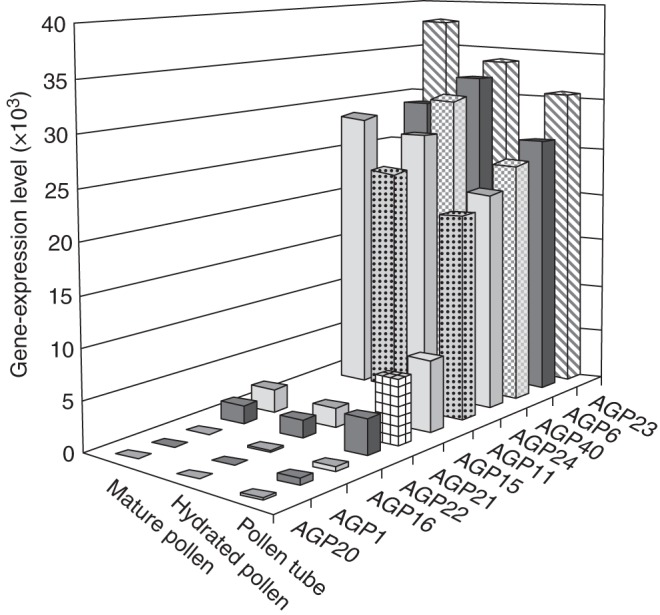
ATH1 genome array showing the gene-expression level for the different AGPs, at three stages of development: mature pollen, hydrated pollen and pollen tube (according to Wang et al., 2008).
Fig. 6.
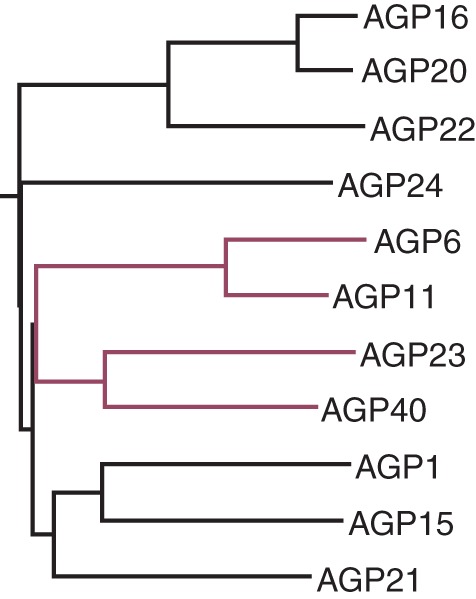
Clustal W sequence homology alignment for the 11 pollen AGPs. AGP6, AGP11, AGP23 and AGP40 are the only four pollen-specific AGPs in arabidopsis. AGP6 and AGP11 present the highest similarity scores and are closely related to AGP23 and AGP40 (highlighted in red)
Distribution of AGPs at different stages of pollen formation and pollen-tube growth
Use of AGP-specific mAbs to study AGP distribution in pollen and pollen tubes
The cell wall of pollen grains is composed of two layers, the intine, the inner wall, and the exine, the outer wall, (Wang et al., 2010). The pollen-tube cell wall in plants such as arabidopsis, tobacco and lily, with the exception of the tip is composed of two layers; the inner cell wall is enriched in callose and the outer cell wall is composed mostly of pectins, hemicellulose, cellulose and AGPs (Dardelle et al., 2010). At the tip, only the outer layer is present (Geitmann and Steer, 2006; Dardelle et al., 2010). In Arabidopsis thaliana, during pollen-grain formation in the anther at the beginning of meiosis, the labelling obtained with the mAbs JIM8 and JIM13 is selectively found in the cell walls of microsporocytes (Coimbra et al., 2007). After meiosis, the four haploid microspores called tetrads are surrounded by a thick wall composed of callose, pectin and hemicelluloses. Following degradation of the thick wall with β-(1,3)-glucan hydrolases (Hird et al., 1993), pectin methylesterases (Francis et al., 2006), polygalacturonases (Rhee et al., 2003) and other enzymes secreted by the tapetal cells, the microspores are released and AGP epitopes recognized by JIM8 and JIM13 are strongly detected in the cytoplasm and the outer surface of the microspores (Coimbra et al., 2007). After the first pollen mitosis, the bicellular pollen contains the generative and the vegetative cells. At this stage, the membrane of the generative cell is strongly labelled. After the second mitosis, the generative cell produces the two sperm cells with their membranes still strongly labelled (Coimbra et al., 2007). The specific labelling of the two male gametes is maintained in the mature pollen grains (van Aelst and van Went, 1992), and in pollen tubes grown in vitro (Coimbra et al., 2007) and in vivo (Lennon and Lord, 2000). In addition, a subset of AGP epitopes recognized by MAC207 is also present in the intine wall of the mature pollen grain close to the plasma membrane (van Aelst and van Went, 1992). Other mAbs such as LM2 do not show any cell-specific labelling during pollen formation (Coimbra et al., 2007). The cell wall of in vitro-grown pollen tubes displays different specificities towards the mAbs. The pollen-tube tip is more strongly labelled than the shank with LM2 and MAC207 antibodies (Fig. 7A, B, E) (Pereira et al., 2005; Dardelle et al., 2010). In contrast, no labelling is detected over the pollen-tube cell wall with JIM13 and JIM8 mAb (Dardelle et al., 2010).
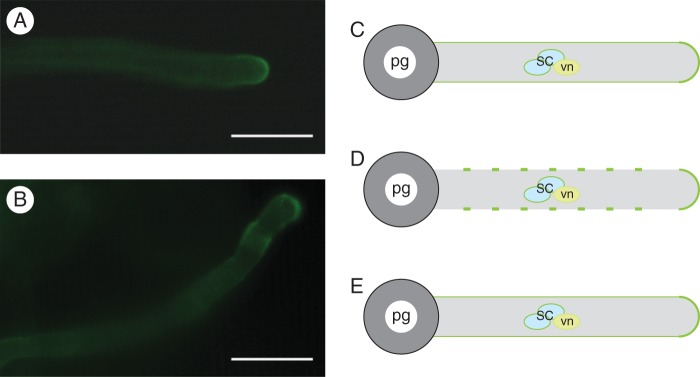
Fig. 7.
Immunolocalization of AGPs in Arabidopsis thaliana pollen tube and the most-common AGP-labelling patterns observed in pollen tubes. (A) Cell surface immuno-labelling of A. thaliana pollen tube with LM2 and (B) with MAC207. Immuno-labelling patterns of AGP epitopes (in green) found in (C) Lilium longiflorum, (D) Picea meyeri, Actinidia deliciosa and Lilium longiflorum after enzyme treatments, and (E) Podocarpus nagi, Picea wilsonii, Pinus densiflora, Annona cherimola, Arabidopsis thaliana and Nicotiana tabacum with JIM13. Abbreviations: pg, pollen grain; sc, sperm cells; vn, vegetative nucleus. Scale bars in (A) and (B) = 20 µm. The drawings in (C–E) are not to scale. The detection of AGP epitopes in the sperm cell membranes is generally deduced from electron microscopy observations (see also Dardelle et al., 2010).
In other species investigated including the gymnosperms (Cycas revoluta, Ginkgo biloba, Picea wilsonni and Pinus densiflora), the angiosperm monocots (Lilium longiflorum and Lolium perenne) and eudicots (Brassica napus, Beta vulgaris, Nicotiana tabacum, Nicotiana alata and Solanum peruvianum), AGP epitopes are detected in the intine wall of the pollen grains close to the plasma membrane (Table 3). In the pollen-tube cell wall, several AGP labelling patterns have been described (Table 3 and Fig. 7). In spruce (Picea meyeri) and kiwifruit (Actinidia deliciosa), a periodic ring-like deposition of AGP epitopes along the pollen tube length is observed using LM2 and JIM13, respectively (Fig. 6D) (Chen et al., 2007; Speranza et al., 2009). With MAC207, no visible labelling is detected in A. deliciosa pollen tubes (Speranza et al., 2009). In L. longiflorum, JIM13-16, MAC207 and LM2 labelling is restricted to the pollen-tube tip (Fig. 7C) (Jauh and Lord, 1996; Mollet et al., 2002). Whereas in N. tabacum, no visible labelling is observed in the pollen-tube cell wall using the MAC207 (Li et al., 1992) or PCBC3 (Fergusson et al., 1999) and uniform labelling of the entire pollen-tube cell wall is observed with JIM13 (Qin et al., 2007). Interestingly, treatments of the pollen tubes of N. tabacum and L. longiflorum with pectinase and/or cellulase, uncover AGP epitopes organized as a periodic deposition of ring-like structure along the entire pollen tube (Fig. 7D) (Li et al., 1992; Jauh and Lord, 1996). These results have been confirmed at sub-cellular level where AGP epitopes are found unevenly distributed over the plasma membrane along the pollen tube (Roy et al., 1997; Fergusson et al., 1999). Finally, within the pollen-tube cell, AGP epitopes are also detected, in many species, associated with secretory vesicles, the generative and the sperm cell membranes (Table 3). Differences in the labelling patterns observed with JIM13 and MAC207 antibodies are not easy to explain as the recognized epitopes are supposed to be identical (Table 3). By comparing the labelling patterns, the differences might be due to (a) the species, (b) the organization of the epitopes in the pollen-tube cell wall and/or the structure of the AGPs, (c) the accessibilities of the epitopes, (d) the type of pistil (wet versus dry stigma, hollow versus solid style), (e) the in vitro culture conditions and (f) probably the growth rate of the pollen tubes observed in vitro and in vivo. The pollen-tube growth in gymnosperms is generally slower (0·01–0·05 µm min−1 based on the length of the pollen tubes after 72 h) (Yatomi et al., 2002; Fernando et al., 2010) than that observed for angiosperms such as L. longiflorum (approx. 10·1 µm min−1 based on the length of the pollen tubes after 6 h) (Jauh and Lord, 1996), N. tabacum (average of 2·7 µm min−1 based on the length of the pollen tubes after 5 h) (Mollet et al., 2002) or arabidopsis (average of 1·3 µm min−1 based on the length of the pollen tubes after 16 h) (Dardelle et al., 2010).
Table 3.
Overview from several plant species of AGP localization during pollen-grain formation, and in pollen tubes grown in vitro and in vivo
| Species | Probe | Microscopy* | Culture | Cell type† | Labelling pattern† | References |
|---|---|---|---|---|---|---|
| Gymnosperms | ||||||
| Cycadales | ||||||
| Cycas revoluta | JIM13, LM2 | EM/CF | In vitro | PG and PT | JIM13: strong labelling of the intine and PT; LM2: strong labelling PT, no labelling of the intine wall | Yatomi et al. (2002) |
| Ginkgoales | ||||||
| Ginkgo biloba | JIM13, LM2 | EM/CF | In vitro | PG and PT | JIM13 and LM2: strong labelling of the intine and PT wall | Yatomi et al. (2002) |
| Pinales | ||||||
| Podocarpus macrophyllus | JIM13, LM2 | EM/CF | In vitro | PG and PT | JIM13: labelling of the intine and PT wall | Yatomi et al. (2002) |
| Podocarpus nagi | LM2 : weak labelling | |||||
| Abies veitchii | JIM13, LM2 | EM/CF | In vitro | PG and PT | JIM13: labelling of the intine and PT wall | Yatomi et al. (2002) |
| LM2 : no labelling of the PT, labelling of the intine wall | ||||||
| Cedrus deodara | JIM13, LM2 | EM/CF | In vitro | PG and PT | JIM13: strong labelling of the intine and PT wall | Yatomi et al. (2002) |
| Chamaecyparis obtusa | LM2 : no labelling | |||||
| Picea wilsonii | LM2 | FM | In vitro | PT | Whole PT wall | Chen et al. (2008) |
| LM2 | FM | In vitro | De-exined PG | Intine surface | Fang et al. (2008) | |
| Picea meyeri | LM2 | FM | In vitro | PT | Periodic ring-like pattern along the whole PT wall | Chen et al. (2007) |
| Pinus densiflora | JIM13, LM2 | EM/CF | In vitro | PG and PT | JIM13 and LM2: intine wall of PG. Whole PT wall (tip and back) and secretory vesicles | Mogami et al. (1999); Yatomi et al. (2002) |
| LM2: generative cell wall | ||||||
| Pinus bungeana | LM2 | FM | In vitro | De-exined PG | Intine surface | Fang et al. (2008) |
| Pinus banksiana | JIM13, LM2 | EM/CF | In vitro | PG and PT | JIM13and LM2: labelling of the intine and PT wall | Yatomi et al. (2002) |
| Pinus rigida | ||||||
| Cryptomeria japonica | ||||||
| Pinus elliottii | JIM13, LM2 | EM/CF | In vitro | PG and PT | JIM13: labelling of the intine and PT wall | Yatomi et al. (2002) |
| LM2: weak labelling of the PT, no labelling of the intine | ||||||
| Angiosperms | ||||||
| Monocot | ||||||
| Liliales | ||||||
| Lilium longiflorum | JIM13 | FM | In vitro | PG protoplast | Cell surface | Zhao et al. (2004) |
| JIM13 | EM/HPF | In vivo | PT | PM, inner cell wall and secretory vesicles | Roy et al. (1997) | |
| JIM13-16, LM2, MAC207 | FM | In vitro and in vivo | PT | PT tip wall | Jauh and Lord (1996) Mollet et al. (2002) | |
| JIM13, LM2, MAC207 | FM | In vitro and in vivo | Pectinase treated PT | Ring-like pattern along the whole PT wall | Jauh and Lord (1996) | |
| βGlcY (30 µm) | LM | In vitro | PT | PT tip wall, arrest of growth | Mollet et al. (2002) | |
| JIM13, LM2, MAC207 | EM/CF | In vivo | PT | PM, secretory vesicles and generative cell membrane | Jauh and Lord (1996) | |
| Poales | ||||||
| Lolium perenne | JIM13 | EM/CF | In vivo | PG formation | Microspore wall | Wisniewska and Majewska-Sawka (2006) |
| Eudicot | ||||||
| Magnoliales | ||||||
| Annona cherimola | JIM13 | FM | In vitro | PT | Whole PT wall | Mollet et al. (2002) |
| βGlcY (30 µm) | LM | In vitro | PT | Expanded region from the tip, arrest of growth | ||
| Brassicales | ||||||
| Arabidopsis thaliana | LM2, JIM13, | FM | In vitro | PT | LM2 and MAC207: stronger labelling at the tip than the back | Dardelle et al. (2010), Pereira et al. (2005) |
| MAC207 | JIM13: no labelling | |||||
| JIM8, JIM13, | FM | In vivo | PG formation | JIM8 and JIM13: strong labelling of microsporocyte wall, microspore wall, generative membrane and then sperm cells from microspore stage to mature pollen and in the pollen tube | Coimbra et al. (2007) | |
| MAC207, LM2 | MAC207 and LM2: no specific labelling pattern | |||||
| MAC207, JIM8 | EM/PF | In vitro | PG | MAC207: Intine wall close to the PM | van Aelst and van Went (1992) | |
| JIM8: sperm cell membrane | ||||||
| LM2, JIM13, MAC207 | EM/PF | In vivo | PG and PT | PG: intine | Lennon and Lord (2000) | |
| PT: sperm cell membrane | ||||||
| Brassica napus | JIM8 | FM | In vivo | PG formation | Tetrad wall, PM of mature PG, sperm cell membrane during anthesis | Pennel et al. (1991) |
| Caryophyllales | ||||||
| Beta vulgaris | MAC207 | FM | In vitro | PG | PM | Pennel et al. (1989) |
| JIM4, JIM8 | FM | PG formation | In the callose wall of the tetrad | Majewska-Sawka and Rodriguez-Garcia (2006) | ||
| JIM4: no labelling | ||||||
| JIM13: strong gold labelling in the callose wall of the tetrad | ||||||
| JIM13, LM2 | EM/CF | JIM8 and LM2: weak gold labelling in the callose wall of the tetrad | ||||
| Solanales | ||||||
| Nicotiana tabacum | PCBC3, MAC207, JIM8, JIM13 | EM/CF | In vitro | PG and PT | PCBC3: PG: PM | Fergusson et al. (1999) |
| PT: discontinuous labelling at the PM along the PT, between the inner and outer cell wall layers, in secretory vesicles and callose plugs | ||||||
| MAC207, JIM8, JIM13 : no labelling | ||||||
| MAC207 | FM | In vitro | PT | No labelling | Li et al. (1992) | |
| Cellulase treated PT | No tip labelling, strong periodic ring-like pattern back from the tip | |||||
| Pectinase treated PT | No tip labelling, weak periodic ring-like pattern back from the tip | |||||
| MAC207, JIM8 | EM/CF | In vitro | PG and PT | PG: uniform in the intine | Li et al. (1995) | |
| PT: discontinuous labelling along the outer wall of the PT. Labelling in the secretory vesicles and generative cell membrane. No tip labelling | ||||||
| βGlcY (30 µm) | LM | In vitro | PT | No labelling, no arrest of PT growth | Mollet et al. (2002) | |
| JIM4, JIM13, LM2, | FM | In vitro | PG formation and PT | JIM13: microspore wall and whole PT | Qin et al. (2007) | |
| JIM4 and LM2: weak labelling | ||||||
| βGlcY (50–100 µm) | LM | In vitro | PT growth | Slow down of PT growth | ||
| JIM13 | EM/CF | In vivo | PG and PT | PG: intine wall and vesicles | ||
| PT: cell wall, generative and sperm cell membranes | ||||||
| Lycopersicum peruvianum | PCBC3 | EM/CF | In vitro | PG and PT | PG: PM and generative cell membrane | Fergusson et al. (1999) |
| PT: between inner and outer cell wall layers. No labelling at the tip | ||||||
| Ericales | ||||||
| Actinidia deliciosa | JIM13 | FM | In vitro | PT | Whole PT with periodic brighter ring-like pattern along the PT and at the tip | Speranza et al. (2009) |
| JIM13, JIM8 | EM/CF | In vitro | PT | JIM8, 13 and 14: cytoplasm, PM and the outer cell wall | ||
| JIM14, MAC207 | JIM13: tube tip wall | |||||
| MAC207: no labelling | ||||||
* EM, Electron microscopy; FM, fluorescence microscopy; LM, light microscopy; CF, chemical fixation of the sample; PF, plunge freezing fixation; HPF, high-pressure freezing fixation.
† PT, Pollen tube, PG, pollen grain; PM, plasma membrane.
It is interesting to notice the recurrent occurrence of the LM2 and JIM13 epitopes associated with AGPs in pollen tubes of all the plant families analysed, and the occurrence of additional AGP-associated epitopes in higher plants, as shown in Table 2 in the different taxa. This observation prompts two interesting thoughts on AGPs diversity, occurrence and biology. First, AGP-associated epitopes in pollen tubes are more complex and diverse in higher plants when compared with other taxa. This reflects the complexity of the AGP-glycans' biosynthetic machinery or their evolutionary relationships (see recent review on evolution of plant cell walls; Sørensen et al, 2010). Second, in the case of AGPs, and contrasting with other polysaccharide-rich cell-wall components, the assembly of AGPs within the endomembrane system and their transport to the cell wall are still poorly understood.
AGP genes and proteins expressed in angiosperm pollen
Pollen development requires expression of early genes after meiosis, and late genes encoding products involved in cell-wall formation, pollen maturation, pollen recognition, pollen germination and pollen-tube growth. The late genes are expressed after pollen mitosis and represent the pollen-specific genes (Raghavan, 1997). In arabidopsis, several AGP genes including AtAGP6 (At5g14380) and AtAGP11 (At3g01700) were shown to be associated with pollen-tube development (Levitin et al., 2008; Coimbra et al., 2010). Their functions are described below. AGP genes expressed during pollen development were also studied in other plant species. Brassica campestris BcMF8 is a pollen-specific gene. Its deduced protein backbone presents features of a classical AGP (Huang et al., 2008). By differential screening, Gerster et al. (1996) characterized two highly homologous AGP genes from B. napus, Sta 39-4 and Sta 39-3 which were shown to be pollen-specific. BcMF8 shares high sequence identity with those two putative pollen-expressed AGP genes, and a lower similarity with the classical AGP genes AtAGP11 and AtAGP6. Anther pollen-specific AGP gene PO2 from alfalfa (Medicago sativa) was also characterized by Qiu et al. (1997) using differential screening technology. BAN102 from Chinese cabbage, another pollen-preferential gene which shared sequence similarity to AtAGP23, was isolated by Park et al. (2005). In rice (Oryza sativa), several AGP-encoding genes were dominantly expressed in anthers (Ma and Zhao, 2010) and the two classical AGP-encoding genes, OsAGP7 and OsAGP10, were highly expressed in pollen (Ma and Zhao, 2010). Although expression of OsAGP7 and OsAGP10 parallels that of AtAGP6 and AtAGP11 they do not have the closest phylogenetic relationship. In fact the phylogenetically closest rice genes exhibiting higher homology with AGP6 and AGP11 have a different expression pattern. Anand and Tyagi (2010) isolated an AGP (OsAGP) cDNA (586 bp) from rice preferentially expressed in the inflorescence and the deduced gene and protein sequence showed homology with the AGP-encoding gene AGP23 from arabidopsis. An effort was made to correlate the expression and promoter activity data with the possible function of OsAGP. The OsAGP promoter shows activity after the microspores have been separated from the callose wall and therefore is categorized as a late pollen-expressing gene with maximum level of expression in pollen grains from mature flowers. The OsAGP promoter is also active during germination and pollen-tube growth (Anand and Tyagi, 2010).
Functional aspects of AGPs in pollen tubes and styles
AGPs in pollen-grain and pollen-tube development
Pollen-tube growth is coupled with deposition of new wall material at the tip resulting in an increase in pollen tube length. It has been recently proposed that pollen-tube AGPs are sent to the apex, where they function in signalling processes related to pollen-tube guidance (Lamport et al., 2006; Zang et al., 2011). With pollen-tube continuous growth, AGPs are recycled by endocytosis, to be reused or to be sent for degradation in the multivesicular bodies (M. L. Costa et al., F.C. Universidade do Porto, unpubl. res.). Cell-wall components such as pectins or glycosylated proteins (e.g. AGPs) are known to carry complex and structurally diverse carbohydrate decorations. In addition, it is believed that different signalling molecules could bind to specific cell-wall carbohydrates (including those associated with AGPs) to modulate cell wall integrity and function. AGPs could act as co-receptors to sense extracellular signals and interact with transmembrane proteins, possibly receptor kinases or ion channels, to initiate signalling by triggering various intracellular events (Boisson-Dernier et al., 2011; Zang et al., 2011).
Treatments of N. tabacum pollen tubes with 30 µm active Yariv and the control, inactive Yariv, do not reveal red staining at the pollen-tube tip and do not affect growth (Mollet et al., 2002). In contrast, active Yariv reagent stains the pollen-tube tip of L. longiflorum and causes the growth of the pollen tube to be arrested (Table 3) (Mollet et al., 2002) without stopping vesicular secretion at the tip but disorganizing the cell wall architecture and composition due to abnormal callose deposition (Roy et al., 1998). Upon removal of the active Yariv from the culture medium, pollen-tube growth resumes by emergence of a new pollen-tube tip behind the arrested tip (Mollet et al., 2002). These data suggest that AGPs are important in the deposition of new cell-wall material during pollen-tube growth. Using higher concentrations of active Yariv (50 and 100 µm) and different culture conditions Qin et al. (2007) have also shown a significant reduction in pollen-tube growth in N. tabacum.
AGP6 and AGP11 are two arabidopsis genes which are strongly and specifically expressed in pollen grains and pollen tubes (Figs 5 and 6) (Levitin et al., 2008; Coimbra et al., 2009). To determine whether the agp6 agp11 mutant affected pollen-tube growth and seed set, female gametophytes at the pollination stage were analysed in emasculated flowers pollinated with wild-type and double-mutant pollen, using confocal laser-scanning microscopy with the conclusion that AGP6 and AGP11 are necessary for proper pollen-tube growth as well as for preventing untimely pollen-grain germination (Coimbra et al., 2010). The level of expression of all AGPs expressed in pollen tubes was monitored in agp6 agp11 and in single knockout mutants for agp6 and for agp11 by real-time PCR. Based on the expression levels of the pollen-specific AGPs, and depending on the pollen-tube growth medium conditions (M. L. Costa et al., F.C. Universidade do Porto, unpubl. res.), a consistent up-regulation of AGP40, a pollen-specific AG-peptide, was obtained in pollen grains and pollen tubes of the double null mutant agp6 agp11, and a more moderate over-expression of AGP23 (also a pollen-specific AG-peptide) (Figs 5 and 6). Early results from agp40 null-mutant characterization show no alteration in pollen grain development but illustrate a reduction in pollen-grain fitness, making it the ideal candidate for multiple knockouts. The triple mutant agp6 agp11 agp40 was obtained and characterized (Fig. 8A, B) (Costa et al., 2011). It showed an even more penetrant phenotype than the double mutant, with a significant reduction in seed production (Fig. 8B) and a higher number of early germinating pollen tubes inside the anthers (Fig. 8A). The strong distortion obtained in the germination percentage of agp6 agp11 pollen and pollen-tube length, together with an early germination inside the anther, for some of these double-mutant pollen tubes, encouraged another experiment to further analyse this phenotype. So, in order to understand the mode of action of these AGPs, an Affymetrix ATH1 genome array in the agp6 agp11 double null-mutant pollen tube was performed (Coimbra et al., unpubl. res.). An unexpectedly high number of genes showed altered expression levels in pollen tubes that lack the two cell-wall proteoglycans, AGP6 and AGP11, strengthening the idea that these molecules are involved in complex phenomena. Both calcium and signalling-related genes were altered, in agreement with the known roles of such phenomena in pollen-tube growth. The presumed involvement of AGPs in signalling cascades was also reinforced. Cysteine-rich proteins have been proposed to play a role in recognition and fertilization, and it was thus quite relevant that such genes were found to be differentially expressed. This microarray study was complemented with experiments using two hybrids of yeast which revealed that some AGP interactors are present during pollen-tube growth. These results are being presented and discussed elsewhere by S. Coimbra and colleagues. It is known that molecules involved in pollination and stress/defence responses are evolutionarily related (Dresselhaus and Márton, 2009; Kessler et al., 2010) and one of the most altered gene ontology groups on the agp6 agp11 array is stress, with several PR genes and heat-shock gene expression significantly distorted. Members of these families are involved in reproductive processes in other systems. It is thus of interest to investigate the role of these two AGPs and/or their partners, in the last stages of pollen–pistil interaction, sperm recognition and double fertilization.
Fig. 8.
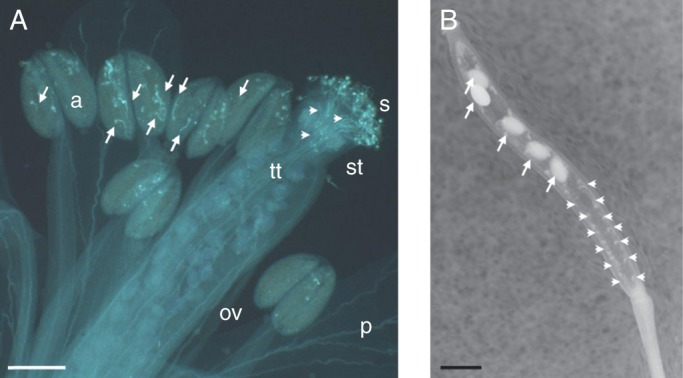
Triple-null arabidopsis mutant plant for AGP6, AGP11 and AGP40. (A) Decolourized aniline blue-stained flower. The anthers show a significant number of pollen tubes growing inside (arrows). Nevertheless some pollen grains maintain their normal germination behaviour and pollen tubes are visibly growing through the transmitting tissue (arrowheads). (B) The mature silique of the triple null mutant presents a reduce number of seeds (arrows) and many ovules remain attached to the placenta unfertilized and degenerated (arrowheads). Abbreviations: a, anther; ov, ovary; p, petal; s, stigma; st, style; tt, transmitting tract. Scale bars = 150 µm.
It is well known that certain AGPs are GPI-anchored to the plasma membrane (Youl et al., 1998; Svetek et al., 1999). The collective importance of the GPI-anchor of arabidopsis proteins has been addressed through the study of insertional mutations disrupting SETH1 and SETH2 genes, which encode homologues of known components of the GPI anchor synthesis complex (Lalanne et al., 2004). Interestingly, the mutant plants showed drastic impairment of pollen germination and pollen-tube growth probably because the mutations affect a large number of GPI-anchored proteins simultaneously. Classical AGPs, AG-peptides, FLAs, phytocyanins and lipid transfer proteins, as well as β-1,3-glucanases, are among the GPI-anchored proteins identified in arabidopsis pollen tubes by transcriptomic and proteomic analyses (Lalanne et al., 2004). The question remains, however, to what extent the GPI-anchor of a particular protein affects its function. In arabidopsis, previous studies demonstrated that proteins with FAS domains may function as adhesion molecules (Kim et al., 2002). FLA3 is involved in microspore development and may affect pollen intine formation, possibly by impacting on cellulose deposition. In FLA3-overexpressing transgenic plants, defective elongation of the stamen filament and reduced female fertility led to short siliques with low seed set, suggesting that ectopic expression of FLA3 may reduce or disrupt cell growth (Li et al., 2010).
Role of stylar AGPs during pollen-tube growth
During pollen-tube growth through the style, navigation is controlled at the surface of the specialized transmitting-tract epidermis in plants with a hollow style such as L. longiflorum or in the intercellular matrix of the transmitting tract tissue in plants with a solid style like tobacco, tomato, Arabidopsis, Actinidia and Amaranthus where AGPs are abundant (Jauh and Lord, 1996; Li and Showalter, 1996; Lennon et al., 1998; Cheung et al., 2000; Coimbra and Duarte, 2003; de Graff et al., 2003; Coimbra et al., 2007). In lily (L. longiflorum), analysis of the stylar exudates by SDS–PAGE and Western blot indicates that the electrophoretic mobility of AGPs ranged from 97 to 200 kDa as revealed with JIM13 and active Yariv staining (Jauh and Lord, 1996). Similarly, AGPs from solid styles of tobacco ranged from 45 to 140 kDa (Cheung et al., 2000; de Graff et al., 2003). Several tobacco AGPs [Transmitting-Tract Specific (NtTTS) and the Pistil-specific Extensin-Like Protein III (PELPIII)] from N. tabacum (AGPNa1, 120-kDa and NaTTS) and N. alata have been purified from the stylar transmitting-tract tissue and characterized (Wang et al., 1993; Du et al., 1994, 1996; Lind et al., 1994; Sommer-Knudsen et al., 1996; Bosch et al., 2001). The 120-kDa, PELPIII and NtTTS are not classical AGPs. The 120-kDa and PELPIII have chimeric structures with both AGP and extensin properties (Lind et al., 1994; Schultz et al., 1997; Bosch et al., 2001) whereas NtTTS has an unusual protein to sugar ratio, 65 : 35 by weight, and a basic pI (Cheung and Wu, 1999). The AGPNa1, 120-kDa and PELPIII are composed mainly of arabinose (45 %, 55 % and 48 %) and galactose (52 %, 45 % and 50 %), respectively, with the main linkages found in AGPs (t-Ara; 1,3,6-linked Gal in AGPNa1 and t-Ara; t-Gal; 1,3-linked Gal; 1,6-linked Gal and 1,3,6-linked Gal in 120-kDa and PELPIII) but additionally the 1,2-linked arabinosyl residues generally found in extensin are found in the 120-kDa and PELPIII (Lind et al., 1994; Gane et al., 1995; Du et al., 1996; Bosch et al., 2001).
Studies on N. tabacum and N. alata have shown that the glycosylated stylar TTS are able to attract and promote the growth of the pollen tubes in vitro (Wu et al., 2000) and transgenic tobacco plants with reduced level of NtTTS mRNA display a reduced fertility (Cheung et al., 1995). In contrast, deglycosylated NtTTS looses these properties, suggesting an important biological role of glycosylation (Cheung et al., 1995). During the growth of the pollen tubes in the extracellular matrix of the transmitting tissue, the loosely associated NtTTS is detected at the pollen-tube cell wall surface, in the inner callose wall and at the pollen-tube tip. The guidance of the pollen tubes is presumably promoted by a gradient of increasing glycosylation of the NtTTS from the top of the style to the ovary. Moreover, the pollen tubes are able to deglycosylate the NtTTS proteins and may incorporate the sugars (Wu et al., 1995). Together, these studies suggest that the carbohydrate moieties of AGPs may have several functions including signalling and/or as nutritional molecules (Cheung et al., 2000).
In N. alata unpollinated styles, the 120-kDa protein epitopes are evenly distributed in the extracellular matrix of the transmitting tissue (Lind et al., 1996). In pollinated styles with compatible pollen, the 120-kDa is concentrated in the inner wall adjacent to the pollen-tube cell wall and can be incorporated by the growing pollen tubes as shown by immuno-labelling in the cytoplasm and vesicle/vacuole-like structures (Lind et al., 1996; Goldraij et al., 2006). With incompatible pollen, similar labelling was observed (Lind et al., 1996). PELPIII is also localized in the intercellular matrix of the transmitting tissue in unpollinated styles. In pollinated styles, PELPIII epitopes are no longer detected in the transmitting tract but found associated with the pollen-tube callose wall and callose plugs (de Graff et al., 2003). Unlike NtTTS, PELPIII is apparently unmodified by the expanding pollen tubes (de Graff et al., 2003) and is not involved in promoting pollen-tube growth and attraction (Bosch et al., 2003). Interestingly, the stylar NaTTS, 120-kDa and PELPIII are able to bind in vitro to S-RNases (Cruz-Garcia et al., 2005), the stylar ribonucleases responsible for the degradation of the pollen-tube rRNA and growth arrest of the S-incompatible pollen in the Solanaceae gametophytic system (McCubbin and Kao, 2000). These data suggest that AGPs may be involved indirectly in the S-pollen rejection by forming a complex with the S-RNase that may promote the initial interaction with the growing pollen tubes (Cruz-Garcia et al., 2005). Among these three glycoproteins, the 120-kDa is the more polymorphic in size, ranging from 80 kDa to 120 kDa between different compatible and incompatible plants, and suppression of the 120-kDa by RNAi resulted in a deficient S-specific pollen rejection. Thus, the 120-kDa is probably one of the partners required for the gametophytic S-rejection (Hancock et al., 2005). More recently, Lee et al (2008) showed using yeast two-hydrid and pull-down assays, that three pollen-tube proteins can interact with the C-terminal domain of the stylar 120-kDa and NaTTS: (1) an S-RNase binding protein, a putative E3 ubiquitin ligase, (2) a C2 domain-containing protein (NaPCCP) that can bind lipids and regulates cellular processes and (3) a putative cysteine protease that may degrade vacuolar proteins (Lee et al., 2008). The NaPCCP is associated with the pollen tube plasma membrane and may function in the internal transport of the 120-kDa in the endomembrane system toward the vacuoles where the 120-kDa and S-RNase have been localized (Lee et al., 2009; reviewed by Kumar and McClure, 2010).
Mostly studied in tobacco, the results indicate that stylar AGPs play important roles in pollen-tube growth, guidance, nutrition and pollen-tube rejection. Similar functions may be found in other species but experimental data are required in order to draw a general conclusion.
CONCLUSIONS AND PERSPECTIVES
AGPs are fascinating cell wall molecules because of their diversity in terms of glycan structures and functional properties. In the last decade, major advances have been made in understanding some of the functions of these proteoglycans in roots, pollen tubes and other organs. However, future challenges are numerous and diverse with structural and functional characterization of individual AGPs as outstanding questions. The role of AGPs in plant interaction with microorganisms, particularly in roots, is relatively poorly understood, although AGPs were known for a while to be present in roots and secreted in the rhizosphere. Interesting studies have been recently reported on the implication of AGPs in the interaction between the root and microorganisms. They offer enthusiastic and exciting new research directions to study the biology of AGPs. More particularly, the recent finding that AGPs in root exudates of pea are able to induce a polar attachment of Rhizobium (Xie et al., 2012) support an active role of AGPs in plant–microbe interaction (see also Gaspar et al., 2004). Apart from this role in symbiotic interaction, it has been shown recently that AGPs isolated from pea root tip are also capable of attracting zoospores of the pathogenic oomycete Aphanomyces euteiches responsible for root rot disease then inhibiting their germination (Cannesan et al., 2012). Although investigation of AGP distribution at the cellular and tissue levels has been made possible through the use of several AGP-directed mAbs, most, if not all, of the mAbs are directed toward AG-glycan epitopes which are heterogeneous and structurally complex. Such complexity makes structural determination difficult, but efforts should be concentrated towards detailed characterization of the O-glycans of AGPs, including root and pollen-tube AGPs. This is a key issue as few complete glycan structures for native AGPs have been reported thus far (Kieliszewski and Shpak, 2001; Tan et al., 2010; Tryfona et al., 2010). Furthermore, if we were to relate structure to function, it would be of importance to elucidate AG-structures using appropriate analytical tools as has been performed for other cell-wall components, e.g. xyloglucan (Lerouxel et al., 2002; Nguema-Ona et al., 2006, 2012). AG-moieties are composed of up to 100 sugar residues, and oligosaccharides consisting of various sugar residues could be released by specific glycosidases (Takata et al., 2006; Tryfona et al., 2010). The released oligosaccharides could be then tested to assess their potential role in controlling development, morphogenesis, or protection of root cells against pathogens. Also, mutant analysis has proven fundamental to functional analysis of many cell-wall components and is also likely to be a useful avenue for investigating the functions of AGPs.
Another highly challenging issue is to investigate the biosynthesis of AG-glycans. First, the GTs involved will have to be identified and characterized. AGP-associated glycan epitopes have already been found in association with Golgi cisternae, the trans-Golgi network and secretory vesicles (Fig. 2). Efforts to identify GTs (Qu et al., 2008; Ellis et al., 2010; Wu et al., 2010) involved in the biosynthesis of these carbohydrate chains have been undertaken recently. Secondly, studies will have to be conducted on how these enzymes are organized within the endomembrane system and how such an organization leads to structural diversity. The third line of research would be to study whether and how the AG-moieties are remodelled by glycohydrolases in the apoplastic compartment.
Clearly advanced structural understanding, elucidation of mechanisms of AG biosynthesis, assessment of their potential role in cell interaction with microorganisms, their functions in pollen-tube growth and pollen–pistil interaction are all exciting questions for future research on AGPs.
ACKNOWLEDGEMENTS
The authors are thankful to Dr Zoë Popper (NUI Galway) for critical reading and comments on the manuscript. A.D and S.C wish to thank all the students who are working or have worked with them on the biology of arabinogalactan proteins for their contributory discussions. This work was supported by the research network VASI, the region of Haute Normandie and the European Funds (FEDER) to E.N.-O., M.V.-G, J.-C.M. and A.D., and by the FCT (Fundação para a Ciência e Tecnologia, Portugal) within the project PTDC/AGR-GPL/67971/2006 to S.C. This work was also financially supported by the bilateral PESSOA program No. 22823UF to A.D and S.C. This paper is dedicated to our colleague Alain Pierre Balangé, Professor of Plant Physiology at the Université de Rouen, who passed away on 15 January 2012.
LITERATURE CITED
- van Aelst AC, van Went JL. Ultrastructural immuno-localization of pectins and glycoproteins in Arabidopsis thaliana pollen grains. Protoplasma. 1992;168:14–19. [Google Scholar]
- Albersheim P, Darvill A, Augur C, et al. Oligosaccharins: oligosaccharides regulatory molecules. Accounts of Chemical Research. 1992;25:77–83. [Google Scholar]
- Anand S, Tyagi AK. Characterization of a pollen-preferential gene OSIAGP from rice (Oryza sativa L. subspecies indica) coding for an arabinogalactan protein homologue, and analysis of its promoter activity during pollen development and pollen tube growth. Transgenic Research. 2010;19:385–397. doi: 10.1007/s11248-009-9319-3. [DOI] [PubMed] [Google Scholar]
- Andème-Onzighi C, Sivaguru M, Judy-March J, Baskin TI, Driouich A. The reb1-1 mutation of Arabidopsis alters the morphology of trichoblasts, the expression of arabinogalactan proteins and the organisation of cortical microtubules. Planta. 2002;215:949–958. doi: 10.1007/s00425-002-0836-z. [DOI] [PubMed] [Google Scholar]
- Bacic A, Moody SF, Clarke AE. Structural analysis of secreted root slime from maize. Plant Physiology. 1988;80:771–777. doi: 10.1104/pp.80.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. The role of root exudates in rhizosphere interactions with plants and others organisms. Annual Review in Plant Biology. 2006;57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- Baldwin TC, Domingo C, Schindler T, Seetharaman G, Steacey N, Roberts K. DcAGP1, a secreted arabinogalactan protein, is related to a family of basic proline-rich proteins. Plant Molecular Biology. 2001;5:421–435. doi: 10.1023/a:1010637426934. [DOI] [PubMed] [Google Scholar]
- Balestrini R, Lanfranco L. Fungal and plant gene expression in arbuscular mycorrhizal symbiosis. Mycorrhiza. 2006;16:509–524. doi: 10.1007/s00572-006-0069-2. [DOI] [PubMed] [Google Scholar]
- Baskin TI, Bertzner AS, Hoggart R, Cork A, Williamson RE. Root morphology mutants in Arabidopsis thaliana. Australian Journal of Plant Physiology. 1992;19:427–437. [Google Scholar]
- Berry AM, Rasmussen U, Bateman K, Huss-Danell K, Lindwall S, Bergman B. Arabinogalactan proteins are expressed at the symbiotic interface in root nodule of Alnus spp. New Phytologist. 2002;155:469–479. doi: 10.1046/j.1469-8137.2002.00466.x. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Kessler SA, Grossniklaus U. The walls have ears: the role of plant CrRLK1Ls in sensing and transducing extracellular signals. Journal of Experimental Botany. 2011;62:1581–1591. doi: 10.1093/jxb/erq445. [DOI] [PubMed] [Google Scholar]
- Borges F, Gomes G, Gardner R, et al. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiology. 2008;148:1168–1181. doi: 10.1104/pp.108.125229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GHH, Sherrier DJ, Stevens TJ, Arkin IT, Dupree P. Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis.: a genomic analysis. Plant Physiology. 2002;129:486–499. doi: 10.1104/pp.010884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GHH, Lilley KS, Stevens TJ, Dupree P. Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: a proteomic and genomic analysis. Plant Physiology. 2003;132:568–577. doi: 10.1104/pp.103.021170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GHH, Sherrier DJ, Weimar T, et al. Analysis of detergent-resistant membranes in Arabidopsis: evidence for plasma membrane lipid rafts. Plant Physiology. 2005;137:104–116. doi: 10.1104/pp.104.053041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bosch KA, Bradley DJ, Knox JP, Perotto S, Butcher GW, Brewin NJ. Common components of the infection thread matrix and the intercellular space identified by immunocytochemical analysis of pea nodules and uninfected roots. EMBO Journal. 1989;8:335–341. doi: 10.1002/j.1460-2075.1989.tb03382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Knudsen JS, Derksen J, Mariani C. Class III pistil-specific extensin-like proteins from tobacco have characteristics of arabinogalactan proteins. Plant Physiology. 2001;125:2180–2188. doi: 10.1104/pp.125.4.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Derksen J, Mariani C. A functional study of stylar hydroxyproline-rich glycoproteins during pollen tube growth. Sexual Plant Reproduction. 2003;16:87–98. [Google Scholar]
- Bradley DJ, Wood EA, Larkins AP, Galfre G, Butcher GW, Brewin NJ. Isolation of monoclonal antibodies reacting with peribacteroid membranes and other components of pea root nodules containing Rhizobium leguminosarum. Planta. 1988;173:149–160. doi: 10.1007/BF00403006. [DOI] [PubMed] [Google Scholar]
- van Buuren ML, Malonado-Mendoza IE, Trieu AT, Blaylock LA, Harrison MJ. Novel genes induced during an arbuscular mycorrhizal (AM) symbiosis formed between Medicago truncatula and Glomus versiforme. Molecular Plant–Microbe Interactions. 1999;12:171–181. doi: 10.1094/MPMI.1999.12.3.171. [DOI] [PubMed] [Google Scholar]
- Cannesan MA, Durand C, Burel C, et al. Effect of arabinogalactan proteins from the root caps of Pisum sativum and Brassica napus on Aphanomyces euteiches zoospores chemotaxis and germination. Plant Physiology. 2012 doi: 10.1104/pp.112.198507. in press. http://dx.doi.org/10.1104/pp.112.198507 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervone F, Hahn GH, De Lorenzo G, Darvill A, Albersheim P. Host–pathogen interactions. XXXIII. A plant protein converts a fungal pathogenesis factor into an elicitor of plant defense responses. Plant Physiology. 1989;90:542–548. doi: 10.1104/pp.90.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud M, Venard C, Defaux-Petras A, Bécard G, Barker DG. Targeted inoculation of Medicago truncatula in vitro root cultures reveals MtENOD11 expression during early stages of infection by arbuscular mycorrhizal fungi. New Phytologist. 2002;156:265–273. doi: 10.1046/j.1469-8137.2002.00508.x. [DOI] [PubMed] [Google Scholar]
- Chen C-G, Pu Z-Y, Moritz RL, et al. Molecular cloning of a gene encoding an arabinogalactan-protein from pear (Pyrus communis) cell suspension culture. Proceedings of the National Academy of Sciences of the USA. 1994;91:10305–10309. doi: 10.1073/pnas.91.22.10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KM, Wu GL, Wang YH, et al. The block of intracellular calcium release affects the pollen tube development of Picea wilsonii by changing the deposition of cell wall components. Protoplasma. 2008;233:39–49. doi: 10.1007/s00709-008-0310-2. [DOI] [PubMed] [Google Scholar]
- Chen T, Teng N, Wu X, et al. Disruption of actin filaments by latrunculin B affects cell wall construction in Picea meyeri pollen tube by disturbing vesicle trafficking. Plant Cell Physiology. 2007;48:19–30. doi: 10.1093/pcp/pcl036. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wu HM. Arabinogalactan proteins in plant sexual reproduction. Protoplasma. 1999;208:87–98. [Google Scholar]
- Cheung AY, Wang H, Wu HM. A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell. 1995;82:383–393. doi: 10.1016/0092-8674(95)90427-1. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wu HM, Di Stilio V, et al. Pollen–pistil interactions in Nicotiana tabacum. Annals of Botany. 2000;85:29–37. (Suppl. A) [Google Scholar]
- Coimbra S, Duarte C. Arabinogalactan proteins may facilitate the movement of pollen tubes from the stigma to the ovules in Actinidia deliciosa and Amaranthus hypochondriacus. Euphytica. 2003;133:171–178. [Google Scholar]
- Coimbra S, Almeida J, Junqueira V, Costa M, Pereira LG. Arabinogalactan proteins as molecular markers in Arabidopsis thaliana sexual reproduction. Journal of Experimental Botany. 2007;58:4027–4035. doi: 10.1093/jxb/erm259. [DOI] [PubMed] [Google Scholar]
- Coimbra S, Costa ML, Jones BJ, Mendes MA, Pereira LG. Pollen grain development is compromised in Arabidopsis agp6 agp11 null mutants. Journal of Experimental Botany. 2009;60:3133–3142. doi: 10.1093/jxb/erp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coimbra S, Costa ML, Mendes MA, Pereira A, Pinto J, Pereira LG. Early germination of Arabidopsis pollen in a double null mutant for the arabinogalactan protein genes AGP6 and AGP11. Sexual Plant Reproduction. 2010;23:199–205. doi: 10.1007/s00497-010-0136-x. [DOI] [PubMed] [Google Scholar]
- Costa ML, Pereira LG, Coimbra S. Workshop on Molecular Mechanisms Controlling Flower Development. Maratea, Italy: 2011. Arabidopsis pollen specific AGPs are essential for pollen development and fitness; pp. 14–17. June 2011. [Google Scholar]
- Cooper JB, Varner JE. Selective inhibition of proline hydroxylation by 3,4-dehydroproline. Plant Physiology. 1983;73:324–328. doi: 10.1104/pp.73.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE. Oligosaccharins, brassinolides, and jasmonates: non traditional regulators of plant growth, development and gene expression. The Plant Cell. 1997;9:1211–1223. doi: 10.1105/tpc.9.7.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Garcia F, Hancock CN, Kim D, McClure BA. Stylar glycoproteins bind to S-RNase in vitro. The Plant Journal. 2005;42:295–304. doi: 10.1111/j.1365-313X.2005.02375.x. [DOI] [PubMed] [Google Scholar]
- Dardelle F, Lehner A, Ramdani Y, et al. Biochemical and immunocytological characterizations of Arabidopsis pollen tube cell wall. Plant Physiology. 2010;153:1563–1576. doi: 10.1104/pp.110.158881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Zhu JK. A role for arabinogalactan-proteins in root epidermal cell expansion. Planta. 1997;203:289–294. doi: 10.1007/s004250050194. [DOI] [PubMed] [Google Scholar]
- Dolan L, Roberts K. Secondary thickening in roots of Arabidopsis thaliana: anatomy and cell surface changes. New Phytologist. 1995;131:121–128. doi: 10.1111/j.1469-8137.1995.tb03061.x. [DOI] [PubMed] [Google Scholar]
- Dolan L, Linstead P, Roberts K. An AGP epitope distinguishes a central metaxylem initial from other vascular initials in the Arabidopsis roots. Protoplasma. 1995;189:145–155. [Google Scholar]
- Downie JA. The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legumes roots. FEMS Microbiology Reviews. 2010;34:150–170. doi: 10.1111/j.1574-6976.2009.00205.x. [DOI] [PubMed] [Google Scholar]
- Dresselhaus T, Márton ML. Micropylar pollen tube guidance and burst: adapted from defense mechanisms? Current Opinion in Plant Biology. 2009;12:773–780. doi: 10.1016/j.pbi.2009.09.015. [DOI] [PubMed] [Google Scholar]
- Driouich A, Baskin TI. Intercourse between cell wall and cytoplasm exemplified by arabinogalactan proteins and cortical microtubules. American Journal of Botany. 2008;95:1491–1497. doi: 10.3732/ajb.0800277. [DOI] [PubMed] [Google Scholar]
- Driouich A, Zhang GF, Staehelin LA. Effect of Brefeldin A on the structure of the Golgi apparatus and on the synthesis and secretion of proteins and polysaccharides in sycamore suspension culture cells. Plant Physiology. 1993;101:1363–1373. doi: 10.1104/pp.101.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driouich A, Durand C, Vicré-Gibouin M. Formation and separation of root border cells. Trends in Plant Science. 2007;12:14–19. doi: 10.1016/j.tplants.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Driouich A, Durand C, Cannesan MA, Percoco G, Vicré-Gibouin M. Border cells versus border-like cells: are they alike? Journal of Experimental Botany. 2010;61:3827–3831. doi: 10.1093/jxb/erq216. [DOI] [PubMed] [Google Scholar]
- Driouich A, Cannesan MA, Dardelle F, et al. Unity is strength: the power of border cells and border like cells in relation with plant defense. In: Vivanco JM, Baluska F, editors. Secretions and exudates in biological systems. Heidelberg: Springer; 2012. pp. 91–108. [Google Scholar]
- Du H, Simpson RJ, Moritz RL, Clarke AE, Bacic A. lsolation of the protein backbone of an arabinogalactan-protein from the styles of Nicotiana alata and characterization of a corresponding cDNA. The Plant Cell. 1994;6:1643–1653. doi: 10.1105/tpc.6.11.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Clarke AE, Bacic A. Arabinogalactan-proteins: a class of extracellular matrix proteoglycans involved in plant growth and development. Cell Biology. 1996;6:411–414. doi: 10.1016/s0962-8924(96)20036-4. [DOI] [PubMed] [Google Scholar]
- Durand C, Vicré-Gibouin M, Follet-Gueye ML, et al. The organization pattern of root border like cells of Arabidopsis is dependent on cell wall homogalacturonan. Plant Physiology. 2009;150:1411–1421. doi: 10.1104/pp.109.136382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M, Egelund J, Schultz CJ, Bacic A. Arabinogalactan proteins: key regulators at the cell surface. Plant Physiology. 2010;153:403–419. doi: 10.1104/pp.110.156000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquerré-Tugaye MT, Mazau D. Effect of a fungal disease on extension, the plant cell wall glycoprotein. Journal of Experimental Botany. 1974;25:509–513. [Google Scholar]
- Estévez J, Kieliszewski MJ, Khitrov N, Somerville C. Characterization of synthetic hydroxyproline-rich proteoglycans with arabinogalactan-protein and extensin motifs in Arabidopsis. Plant Physiology. 2006;142:458–470. doi: 10.1104/pp.106.084244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang K, Wang Y, Yu T, et al. Isolation of de-exined pollen and cytological studies of the pollen intines from Pinus bungeana Zucc. Ex Endl. and Picea wilsonii Mast. Flora. 2008;203:332–340. [Google Scholar]
- Feijó J, Malhó R, Obermeyer G. Ion dynamics and its possible role during in vitro pollen germination and tube growth. Protoplasma. 1995;187:155–167. [Google Scholar]
- Ferguson C, Bacic A, Anderson MA, Read SM. Subcellular distribution of arabinogalactan proteins in pollen grains and tubes as revealed with a monoclonal antibody raised against stylar arabinogalactan proteins. Protoplasma. 1999;206:105–117. [Google Scholar]
- Fernando DD, Quinn CR, Brenner E, Owens JN. Male gametophyte development and evolution in Gymnosperms. International Journal of Plant Developmental Biology. 2010;4:47–63. [Google Scholar]
- Fincher GB, Stone BA, Clarke AE. Arabinogalactan-proteins: structure, biosynthesis and function. Annual Review of Plant Physiology. 1983;34:47–70. [Google Scholar]
- Francis KE, Lam SY, Copenhaver GP. Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiology. 2006;142:1004–1013. doi: 10.1104/pp.106.085274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, Aldington S, Hetherington PR, Aitken J. Oligosaccharides as signals and substrates in the plant cell wall. Plant Physiology. 1993;103:1–5. doi: 10.1104/pp.103.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage DJ, Margolin W. Hanging by a thread: invasion of legume plants by rhizobia. Current Opinion in Microbiology. 2000;3:613–617. doi: 10.1016/s1369-5274(00)00149-1. [DOI] [PubMed] [Google Scholar]
- Gane AM, Weinhandl JA, Bacic A, Harris PJ. Histochemistry and composition of the cell walls of styles of Nicotiana alata Link et Otto. Planta. 1994;195:217–225. [Google Scholar]
- Gane AM, Craik D, Munro SLA, Howlett GJ, Clarke AE, Bacic A. Structural analysis of the carbohydrate moiety of arabinogalactan-proteins from stigmas and styles of Nicotiana alata. Carbohydrate Research. 1995;277:67–85. doi: 10.1016/0008-6215(95)00197-2. [DOI] [PubMed] [Google Scholar]
- Gao M, Showalter AM. Yariv reagent treatment induces programmed cell death in Arabidopsis cell cultures and implicated arabinogalactan proteins involvement. The Plant Journal. 1999;19:321–331. doi: 10.1046/j.1365-313x.1999.00544.x. [DOI] [PubMed] [Google Scholar]
- Gao M, Kieliszewski MJ, Lamport DTA, Showalter AM. Isolation, characterization and immunolocalization of a novel, modular tomato arabinogalactan-protein corresponding to the LeAGP1 gene. The Plant Journal. 1999;18:43–55. doi: 10.1046/j.1365-313x.1999.00428.x. [DOI] [PubMed] [Google Scholar]
- Gaspar YM, Nam J, Schultz CJ, et al. Characterization of the Arabidopsis lysine-rich arabinogalactan-protein AtAGP17 mutant (rat1) that results in a decreased efficiency of Agrobacterium transformation. Plant Physiology. 2004;135:2162–2171. doi: 10.1104/pp.104.045542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geitmann A, Steer M. The architecture and properties of the pollen tube cell wall. In: Malhó R, editor. The pollen tube: a cellular and molecular perspective (Plant Cell Monographs) Heidelberg: Springer; 2006. pp. 177–200. [Google Scholar]
- Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG. Arbuscular mycorrhizal fungi elicits a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. The Plant Cell. 2005;17:3489–3499. doi: 10.1105/tpc.105.035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gens JS, Fujiki M, Pickard BG. Arabinogalactan proteins and wall associated kinases in a plasmalemmal reticulum with specialized vesicles. Protoplasma. 2000;212:115–134. doi: 10.1007/BF01279353. [DOI] [PubMed] [Google Scholar]
- Gerster J, Allard S, Robert LS. Molecular characterization of two Brassica napus pollen-expressed genes encoding putative arabinogalactan proteins. Plant Physiology. 1996;110:1231–1237. doi: 10.1104/pp.110.4.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinazzi-Pearson V, Denarié J. Red carpet genetic programmes for root endosymbioses. Trends in Plant Science. 1997;10:371–372. [Google Scholar]
- Gilson P, Gaspar YM, Oxley D, Youl JJ, Bacic A. NaAGP4 is an arabinogalactan protein whose expression is suppressed by wounding and fungal infection in Nicotiana alata. Protoplasma. 2001;215:128–139. doi: 10.1007/BF01280309. [DOI] [PubMed] [Google Scholar]
- Goldraij A, Kondo K, Lee CB, et al. Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature. 2006;439:805–810. doi: 10.1038/nature04491. [DOI] [PubMed] [Google Scholar]
- de Graaf BHJ, Knuiman BA, Derksen J, Mariani C. Characterization and localization of the transmitting tissue-specific PELPIII proteins of Nicotiana tabacum. Journal of Experimental Botany. 2003;54:55–63. doi: 10.1093/jxb/erg002. [DOI] [PubMed] [Google Scholar]
- Grennan AK. Lipid raft in plants. Plant Physiology. 2007;143:1083–1085. doi: 10.1104/pp.104.900218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MG, Lerner DR, Fitter MS, Norman PM, Lamb CJ. Characterization of monoclonal antibodies to protoplast membranes of Nicotiana tabacum identified by an enzyme-linked immunosorbent assay. Planta. 1987;171:453–465. doi: 10.1007/BF00392292. [DOI] [PubMed] [Google Scholar]
- Hancock CN, Kent L, McClure BA. The stylar 120-kDa glycoprotein is required for S-specific pollen rejection in Nicotiana. The Plant Journal. 2005;43:716–723. doi: 10.1111/j.1365-313X.2005.02490.x. [DOI] [PubMed] [Google Scholar]
- Harris PJ, Ferguson LR, Roberton AM, Mckenzie RJ, White JB. Cell wall histochemistry and anatomy of taro (Colocasia esculenta) Australian Journal of Botany. 1992;40:207–222. [Google Scholar]
- Hasegawa Y, Nakamura S, Kakizoe S, Sato M, Nakamura N. Immunocytochemical and chemical analyses of Golgi vesicles isolated from the germinated pollen of Camellia japonica. Journal of Plant Research. 1998;111:421–429. [Google Scholar]
- Hawes MC, Gunawardena U, Miyasaka S, Zhao X. The role of border cells in plant defense. Trends in Plant Science. 2000;5:128–133. doi: 10.1016/s1360-1385(00)01556-9. [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K. Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis. The Plant Journal. 2002;32:105–113. doi: 10.1046/j.1365-313x.2002.01406.x. [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K. AtAGP30, an arabinogalactan-protein in the cell walls of the primary root, plays a role in root regeneration and seed germination. The Plant Journal. 2003;36:256–270. doi: 10.1046/j.1365-313x.2003.01874.x. [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, van Kammen A, de Vries SC. A relationship between seeds development, arabinogalactan proteins (AGPs) and the AGP mediated promotion of somatic embryogenesis. Physiologia Plantarum. 2002;114:637–644. doi: 10.1034/j.1399-3054.2002.1140418.x. [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Barber C, Roberts K. The expression patterns of arabinogalactan-protein AtAGP30 and GLABRA2 reveal a role for abscisic acid in the early stages of root epidermal patterning. The Plant Journal. 2004;39:70–83. doi: 10.1111/j.1365-313X.2004.02104.x. [DOI] [PubMed] [Google Scholar]
- Hepler PK, Vidali L, Cheung AY. Polarized cell growth in higher plants. Annual Review of Cell and Developmental Biology. 2001;17:159–187. doi: 10.1146/annurev.cellbio.17.1.159. [DOI] [PubMed] [Google Scholar]
- Hervé C, Marcus SE, Knox JP. Monoclonal antibodies, carbohydrate-binding modules, and the detection of polysaccharides in plant cell walls. In: Popper Z, editor. The plant cell wall: methods and protocols. New York, NY: Humana Press; 2011. pp. 103–113. [DOI] [PubMed] [Google Scholar]
- Hird DL, Worrall D, Hodge R, Smartt S, Paul W, Scott R. The anther-specific protein encoded by the Brassica napus and Arabidopsis thaliana A6 gene displays similarity to β-1,3-glucanases. The Plant Journal. 1993;4:1023–1033. doi: 10.1046/j.1365-313x.1993.04061023.x. [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biology. 2004;5:R85. doi: 10.1186/gb-2004-5-11-r85. doi:10.1186/gb-2004-5-11-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Qin Y, Zhao J. Localization of an arabinogalactan protein epitope and the effects of Yariv phenylglycoside during zygotic embryo development of Arabidopsis thaliana. Protoplasma. 2006;229:21–31. doi: 10.1007/s00709-006-0185-z. [DOI] [PubMed] [Google Scholar]
- Huang L, Cao J-S, Zhang A-H, Ye YQ. Characterization of a putative pollen-specific arabinogalactan protein gene, BcMF8, from Brassica campestris ssp. chinensis. Molecular Biology Reports. 2008;35:631–639. doi: 10.1007/s11033-007-9133-z. [DOI] [PubMed] [Google Scholar]
- Jauh GY, Lord EM. Localization of pectins and arabinogalactan-proteins in lily (Lilium longiflorum L.) pollen tube and style, and their possible roles in pollination. Planta. 1996;199:251–261. [Google Scholar]
- Johnson KL, Jones BJ, Bacic A, Schultz CJ. The fasciclin arabinogalactan protein-like arabinogalactan proteins of Arabidopsis: a multigene family of putative cell adhesion molecules. Plant Physiology. 2003;133:1911–1925. doi: 10.1104/pp.103.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KL, Kibble NAJ, Bacic A, Schultz CJ. A fasciclin like arabinogalactan protein (FLA) mutant of Arabidopsis thaliana, fla1, shows defects in shoot regeneration. PLoS One. 2011;6(9):e25154. doi: 10.1371/journal.pone.0025154. doi:10.1371/journal.pone.0025154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Lord EM. Extracellular guidance cues and intracellular signaling pathways that direct pollen tube growth. In: Malho R, editor. The pollen tube: a cellular and molecular perspective (Plant Cell Monographs) Heidelberg: Springer; 2006. pp. 223–242. [Google Scholar]
- Journet E-P, El-Gachtouli N, Vernoud V, et al. Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Molecular Plant–Microbe Interactions. 2001;14:737–748. doi: 10.1094/MPMI.2001.14.6.737. [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Noshiro M, Shen M, et al. Structural and phylogenetic analyses of RGD-CAP/beta ig-h3, a fasciclin-like adhesion protein expressed in chick chondrocytes. Biochimica et Biophysica Acta. 1998;1395:288–292. doi: 10.1016/s0167-4781(97)00172-3. [DOI] [PubMed] [Google Scholar]
- Kessler S, Shimosato-Asano H, Friderike Keinath N, et al. Conserved molecular components for pollen tube reception and fungal invasion. Science. 2010;330:968–971. doi: 10.1126/science.1195211. [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ, Shpak E. Synthetic genes for the elucidation of glycosylation codes for arabinogalactan-proteins and other hydroxyproline-rich glycoproteins. Cell and Molecular Life Sciences. 2001;58:1386–1398. doi: 10.1007/PL00000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Ohinata A, Tsumuraya Y, Hashimoto Y, Kaneko Y, Matsushima H. Production and characterization of antibodies to the beta-1,6-galactotetraosyl group and their interaction with arabinogalactan proteins. Planta. 1993;190:525–535. doi: 10.1007/BF00224792. [DOI] [PubMed] [Google Scholar]
- Kim JE, Jeong HW, Nam JO, et al. Identification of motifs in the fasciclin domains of the transforming growth factor-b-induced matrix protein big-h3 that interact with the avb5 integrin. Journal of Biological Chemistry. 2002;277:46159–46165. doi: 10.1074/jbc.M207055200. [DOI] [PubMed] [Google Scholar]
- Kjellbom P, Snogerup L, Stohr C, Reuzeau C, McCabe PF, Pennell RI. Oxidative crosslinking of plasma membrane arabinogalactan proteins. The Plant Journal. 1997;12:1189–1196. doi: 10.1046/j.1365-313x.1997.12051189.x. [DOI] [PubMed] [Google Scholar]
- Knee EM, Gong F-C, Gao M, et al. Root mucilage from pea and its utilization by rhizosphere bacteria as a sole carbon source. Molecular Plant–Microbe Interactions. 2001;14:775–784. doi: 10.1094/MPMI.2001.14.6.775. [DOI] [PubMed] [Google Scholar]
- Knox JP. Developmental regulated proteoglycans and glycoproteins of the plant cell surface. Federation of American Societies for Experimental Biology Letters. 1995;9:1004–1012. doi: 10.1096/fasebj.9.11.7544308. [DOI] [PubMed] [Google Scholar]
- Knox JP. The use of antibodies to study the architecture and developmental regulation of plant cell walls. International Review of Cytology. 1997;171:79–120. doi: 10.1016/s0074-7696(08)62586-3. [DOI] [PubMed] [Google Scholar]
- Knox JP, Day S, Roberts K. A set of cell surface glycoproteins forms an early marker of cell position, bu not cell type, in the root apical meristem of Daucus carota L. Development. 1989;106:47–56. [Google Scholar]
- Knox JP, Linstead PJ, Peart J, Cooper C, Roberts K. Developmentally regulated epitopes of cell surface arabinogalactan proteins and their relation to root tissue pattern formation. The Plant Journal. 1991;1:317–326. doi: 10.1046/j.1365-313X.1991.t01-9-00999.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Motose H, Iwamoto K, Fukuda H. Expression and genome-wide analysis of the xylogen-type gene family. Plant and Cell Physiology. 2011;52:1095–1106. doi: 10.1093/pcp/pcr060. [DOI] [PubMed] [Google Scholar]
- Kumar A, McClure B. Pollen–pistil interactions and the endomembrane system. Journal of Experimental Botany. 2010;61:2001–2013. doi: 10.1093/jxb/erq065. [DOI] [PubMed] [Google Scholar]
- Lalanne E, Honys D, Johnson A, et al. SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. The Plant Cell. 2004;16:229–240. doi: 10.1105/tpc.014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport DT, Kieliszewski MJ, Showalter AM. Salt-stress upregulates arabinogalactan-proteins: using salt-stress to analyse AGP function. New Phytologist. 2006;169:479–492. doi: 10.1111/j.1469-8137.2005.01591.x. [DOI] [PubMed] [Google Scholar]
- Laus MC, Logman TJ, Lamers GE, Van Brussel AAN, Carlson RW, Kijne JW. A novel polar surface polysaccharide from Rhizobium leguminosarum binds host plant lectin. Molecular Microbiology. 2006;59:1704–1713. doi: 10.1111/j.1365-2958.2006.05057.x. [DOI] [PubMed] [Google Scholar]
- Lee CB, Swatek KN, McClure B. Pollen proteins bind to the C-terminal domain of Nicotiana alata pistil arabinogalactan proteins. Journal of Biological Chemistry. 2008;283:26965–26973. doi: 10.1074/jbc.M804410200. [DOI] [PubMed] [Google Scholar]
- Lee CB, Kim S, McClure B. A pollen protein, NaPCCP, that binds pistil arabinogalactan proteins also binds phosphatidylinositol 3-phophate and associates with the pollen tube endomembrane system. Plant Physiology. 2009;149:791–802. doi: 10.1104/pp.108.127936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KJD, Sakata Y, Mau S-L, et al. Arabinogalactan proteins are required for apical cell extension in the moss Physcomitrella patens. The Plant Cell. 2005;17:3051–3065. doi: 10.1105/tpc.105.034413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon KA, Lord EM. In vivo pollen tube cell of Arabidopsis thaliana. I. Tube cell cytoplasm and wall. Protoplasma. 2000;214:45–56. [Google Scholar]
- Lennon KA, Roy S, Hepler PK, Lord E. The structure of the transmitting tissue of Arabidopsis thaliana (L.) and the path of pollen tube growth. Sexual Plant Reproduction. 1998;11:49–59. [Google Scholar]
- Lerouxel O, Choo TS, Séveno M, et al. Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiology. 2002;130:1754–1763. doi: 10.1104/pp.011965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitin B, Richter D, Markovich I, Zik M. Arabinogalactan proteins 6 and 11 are required for stamen and pollen function in Arabidopsis. The Plant Journal. 2008;56:351–363. doi: 10.1111/j.1365-313X.2008.03607.x. [DOI] [PubMed] [Google Scholar]
- Li J, Yu M, Geng L-L, Zhao J. The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. The Plant Journal. 2010;64:482–497. doi: 10.1111/j.1365-313X.2010.04344.x. [DOI] [PubMed] [Google Scholar]
- Li S, Showalter AM. Cloning and developmental/stress-regulated expression of a gene encoding a tomato arabinogalactan protein. Plant Molecular Biology. 1996;32:641–652. doi: 10.1007/BF00020205. [DOI] [PubMed] [Google Scholar]
- Li YQ, Bruun L, Pierson ES, Cresti M. Periodic deposition of arabinogalactan epitopes in the cell wall of pollen tubes of Nicotiana tabacum L. Planta. 1992;158:422–427. doi: 10.1007/BF00197045. [DOI] [PubMed] [Google Scholar]
- Li YQ, Faleri C, Geitmann A, Zhang H-Q, Cresti M. Immunogold localization of arabinogalactan proteins, unesterified and esterified pectins in pollen grains and pollen tubes of Nicotiana tabacum L. Protoplasma. 1995;189:26–36. [Google Scholar]
- Lind JL, Bacic A, Clarke A, Anderson MA. A style specific hydroxyproline-rich glycoprotein with properties of both extensins and arabinogalactan proteins. The Plant Journal. 1994;6:491–502. doi: 10.1046/j.1365-313x.1994.6040491.x. [DOI] [PubMed] [Google Scholar]
- Lind JL, Bönig I, Clarke AE, Anderson MA. A style-specific 120-kDa glycoprotein enters pollen tubes of Nicotiana alata in vivo. Sexual Plant Reproduction. 1996;9:75–86. [Google Scholar]
- Liu C, Mehdy MC. A nonclassical arabinogalactan protein gene highly expressed in vascular tissues, AGP31, is transcriptionally repressed by methyl jasmonic acid in Arabidopsis. Plant Physiology. 2007;145:863–874. doi: 10.1104/pp.107.102657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe PF, Valentine TA, Forsberg LS, Pennell RI. Soluble signals from cells identified at the cell wall establish a developmental pathway in carrot. The Plant Cell. 1997;9:2225–2241. doi: 10.1105/tpc.9.12.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubbin AG, Kao TH. Molecular recognition and response in pollen and pistil interactions. Annual Review of Cell and Developmental Biology. 2000;16:333–364. doi: 10.1146/annurev.cellbio.16.1.333. [DOI] [PubMed] [Google Scholar]
- Ma H, Zhao J. Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.) Journal of Experimental Botany. 2010;61:2647–2668. doi: 10.1093/jxb/erq104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Muthreich N, Liao C, et al. The mucilage proteome of maize (Zea mays L.) primary roots. Journal of Proteome Research. 2010;9:2968–2976. doi: 10.1021/pr901168v. [DOI] [PubMed] [Google Scholar]
- Majeswska-Sawka A, Rodriguez-Garcia I. Immunodetection of pectin and arabinogalactan protein epitopes during pollen exine formation of Beta vulgaris L. Protoplasma. 2006;228:41–47. doi: 10.1007/s00709-006-0160-8. [DOI] [PubMed] [Google Scholar]
- Malhó R, Read N, Trewavas A, Pais MS. Calcium channel activity during pollen tube growth and reorientation. The Plant Cell. 1995;7:1173–1184. doi: 10.1105/tpc.7.8.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, Asami T, Suzuki Y. Genome-wide identification, structure and expression studies, and mutant collection of 22 early nodulin-like protein genes in Arabidopsis. Bioscience Biotechnology Biochemistry. 2009;73:2452–2459. doi: 10.1271/bbb.90407. [DOI] [PubMed] [Google Scholar]
- Matthysse AG, Kijne JW. Attachment of Rhizobiaceae to plant cells. In: Spaink HP, Kondorosi A, Hooykaas PJJ, editors. The rhizobiaceae. Dordrecht: Kluwer Academic; 1998. pp. 135–249. [Google Scholar]
- Mogami N, Nakamura S, Nakamura N. Immunolocalization of the cell wall components in Pinus densiflora pollen. Protoplasma. 1999;206:1–10. [Google Scholar]
- Moller I, Marcus SE, Haeger A, et al. High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchical clustering of their carbohydrate microarray binding profiles. Glycoconjugate Journal. 2008;25:37–48. doi: 10.1007/s10719-007-9059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet JC, Kim S, Jauh GY, Lord EM. Arabinogalactan proteins, pollen tube growth, and the reversible effects of Yariv phenylglycoside. Protoplasma. 2002;219:89–98. doi: 10.1007/s007090200009. [DOI] [PubMed] [Google Scholar]
- Mollet JC, Faugeron C, Morvan H. Cell adhesion, separation and guidance in compatible plant reproduction. Annual Plant Reviews. 2007;25:69–90. [Google Scholar]
- Mongrand S, Morel J, Laroche J, et al. Lipid raft in higher plant cells: purification and characterization of of triton X-100-insoluble microdomains from tobacco plasma membrane. Journal of Biological Chemistry. 2004;279:36277–36286. doi: 10.1074/jbc.M403440200. [DOI] [PubMed] [Google Scholar]
- Moody SF, Clarke AE, Bacic A. Structural analysis of secreted lime from wheat and cowpea roots. Phytochemistry. 1988;27:2857–2861. [Google Scholar]
- Motose H, Sugiyama M, Fukuda H. A proteoglycan mediates inductive interaction during plant vascular development. Nature. 2004;429:873–878. doi: 10.1038/nature02613. [DOI] [PubMed] [Google Scholar]
- Nam J, Mysore KS, Zheng C, Knue MK, Mathysse AG, Gelvin SB. Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Molecular and General Genetics. 1999;261:429–438. doi: 10.1007/s004380050985. [DOI] [PubMed] [Google Scholar]
- Nguema-Ona E, Andeme-Onzighi C, Aboughe-Angone S, et al. The reb1-1 mutation of Arabidopsis: effect on the structure and localization of galactose-containing cell wall polysaccharides. Plant Physiology. 2006;140:1406–1417. doi: 10.1104/pp.105.074997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguema-Ona E, Bannigan A, Chevalier L, Baskin TI, Driouich A. Disruption of arabinogalactan-proteins disorganizes cortical microtubules in the root of Arabidopsis thaliana. The Plant Journal. 2007;52:240–251. doi: 10.1111/j.1365-313X.2007.03224.x. [DOI] [PubMed] [Google Scholar]
- Nguema-Ona E, Moore JP, Fagerström A, et al. Profiling the main cell wall polysaccharides of tobacco leaves using high throughput techniques. Carbohydrate Polymers. 2012;88:939–949. doi: 10.1016/j.carbpol.2013.08.013. [DOI] [PubMed] [Google Scholar]
- Norman PM, Wingate VPM, Fitter MS, Lamb CJ. Monoclonal antibodies to plant plasma-membrane antigens. Planta. 1986;167:452–459. doi: 10.1007/BF00391220. [DOI] [PubMed] [Google Scholar]
- Nothnagel E. Proteoglycans and related components in plant cells. International Review of Cytology. 1997;174 doi: 10.1016/s0074-7696(08)62118-x. 195–191. [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Johnson MA. Functional genomics of pollen tube–pistil interactions in Arabidopsis. Biochemical Society Transactions. 2010;38:593–597. doi: 10.1042/BST0380593. [DOI] [PubMed] [Google Scholar]
- Park BS, Kim JS, Kim SH, Park YD. Characterization of a pollen preferential gene, BAN102, from chinese cabbage. The Plant Cell. 2005;14:1–8. doi: 10.1007/s00299-005-0007-3. [DOI] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Baldwin D, et al. A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiology. 2010;153:514–525. doi: 10.1104/pp.109.151985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell RI, Knox JP, Scofield GN, Selvendran RR, Roberts K. A family of abundant plasma membrane-associated glycoproteins related to the arabinogalactan proteins is unique to flowering plants. Journal of Cell Biology. 1989;108:1967–1977. doi: 10.1083/jcb.108.5.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell RI, Janniche L, Kjellbom P, Scofield GN, Peart JM, Roberts K. Developmental regulation of a plasma membrane arabinogalactan protein epitope in oilseed rape flowers. The Plant Cell. 1991;3:1317–1326. doi: 10.1105/tpc.3.12.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira LG, Coimbra S, Oliveira H, Monteiro L, Sottomayor M. Expression of arabinogalactan protein genes in pollen tubes of Arabidopsis thaliana. Planta. 2005;223:374–380. doi: 10.1007/s00425-005-0137-4. [DOI] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijó JA, Becker JD. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiology. 2005;138:744–756. doi: 10.1104/pp.104.057935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A, Hirt H. New insights into an old story: Agrobacterium induced tumour formation in plants by plant transformation. EMBO Journal. 2010;29:1021–1032. doi: 10.1038/emboj.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Davies C. Characterization of a cDNA encoding the protein moiety of a putative arabinogalactan protein from Lycopersicon esculentum. Plant Molecular Biology. 1995;28:347–352. doi: 10.1007/BF00020254. [DOI] [PubMed] [Google Scholar]
- Preuss D. Sexual signaling on a cellular level: lesson from plant reproduction. Molecular Biology of the Cell. 2002;13:1803–1805. doi: 10.1091/mbc.ES-01-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhlmann J, Bucheli E, Swain MJ, et al. Generation of monoclonal antibodies against plant cell wall polysaccharides. I. Characterization of a monoclonal antibody to a terminal α-(1→2)-linked fucosyl-containing epitope. Plant Physiology. 1994;104:699–710. doi: 10.1104/pp.104.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Fong C, Lamport DTA. Gum arabic glycoprotein is a twisted hairy rope. Plant Physiology. 1991;96:848–855. doi: 10.1104/pp.96.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Zhao J. Localization of arabinogalactan proteins in egg cells, zygotes, and two-celled proembryos and effects of beta-d-glucosyl Yariv reagent on egg cell fertilization and zygote division in Nicotiana tabacum. Journal of Experimental Botany. 2006;57:2061–2074. doi: 10.1093/jxb/erj159. [DOI] [PubMed] [Google Scholar]
- Qin Y, Chen D, Zhao J. Localization of arabinogalactan proteins in anther, pollen, and pollen tube of Nicotiana tabacum L. Protoplasma. 2007;231:43–53. doi: 10.1007/s00709-007-0245-z. [DOI] [PubMed] [Google Scholar]
- Qin Y, Leydon AR, Manziello A, et al. Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genetics. 2009;5((8): e1000621. doi 10·1371/journal.pgen.1000621) doi: 10.1371/journal.pgen.1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Wu Y Z, Du S, Erickson L. A new arabinogalactan protein-like gene expressed in the pollen of alfalfa. Plant Science. 1997;124:41–47. [Google Scholar]
- Qu Y, Egelund J, Gilson PR, et al. Identification of a novel group of putative Arabidopsis thaliana β-(1,3)-galactosyltransferases. Plant Molecular Biology. 2008;68:43–59. doi: 10.1007/s11103-008-9351-3. [DOI] [PubMed] [Google Scholar]
- Rae AL, Harris PJ, Bacic A, Clarke AE. Composition of the cell walls of Nicotiana alata Link et Otto pollen tubes. Planta. 1985;166:128–133. doi: 10.1007/BF00397395. [DOI] [PubMed] [Google Scholar]
- Raghavan V. In: Molecular embryology of flowering plants. Cambridge: Cambridge University Press; 1997. Gene expression during pollen development; pp. 90–119. [Google Scholar]
- Rathbun EA, Naldrett MJ, Brewin NJ. Identification of a family of extensin-like glycoproteins in the lumen of Rhizobium-induced infection threads in pea root nodules. Molecular Plant–Microbe Interactions. 2002;15:350–359. doi: 10.1094/MPMI.2002.15.4.350. [DOI] [PubMed] [Google Scholar]
- Rhee SY, Osborne E, Poindexter PD, Somerville CR. Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiology. 2003;133:1170–1180. doi: 10.1104/pp.103.028266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli C. Monitoring the outside: cell wall-sensing mechanisms. Plant Physiology. 2010;153:1445–1452. doi: 10.1104/pp.110.154518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Fernandez AG, Fujita M, et al. COBRA, an Arabidopsis extracellular glycosylphosphatidylinositol anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. The Plant Cell. 2005;17:1749–1763. doi: 10.1105/tpc.105.031732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Eckard KJ, Lancelle S, Kepler PK, Lord EM. High-pressure freezing improves the ultrastructural preservation of in vivo grown lily pollen tubes. Protoplasma. 1997;200:87–98. [Google Scholar]
- Roy S, Jauh GY, Hepler PK, Lord EM. Effects of Yariv phenylglycoside on cell wall assembly in the lily pollen tube. Planta. 1998;204:450–458. doi: 10.1007/s004250050279. [DOI] [PubMed] [Google Scholar]
- Samaj J, Braun M, Baluska F, Ensikat H-J, Tsumuraya Y, Volkmann D. Specific localization of arabinogalactan protein epitopes at the surface of maize root hairs. Plant and Cell Physiology. 1999;40:874–883. [Google Scholar]
- Sardar HS, Yang J, Showalter AM. Molecular interactions of arabinogalactan-proteins with cortical microtubules and F-actin in Bright-Yellow-2 tobacco cultured cells. Plant Physiology. 2006;142:1469–1479. doi: 10.1104/pp.106.088716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Harrison MJ. Novel plant and fungal AGP like proteins in the Medicago truncatula–Glomus intraradices arbuscular mycorrhizal symbiosis. Mycorrhiza. 2008;18:403–412. doi: 10.1007/s00572-008-0194-1. [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Hauser K, Lind JL, et al. Molecular characterisation of a cDNA sequence encoding the backbone of a style-specific 120-kDa glycoprotein which has features of both extensins and arabinogalactan-proteins. Plant Molecular Biology. 1997;35:833–845. doi: 10.1023/a:1005816520060. [DOI] [PubMed] [Google Scholar]
- Schultz C, Gilson P, Oxley D, Youl J, Bacic A. GPI-anchors on arabinogalactanproteins: implications for signalling in plants. Trends in Plant Science. 1998;3:426–431. [Google Scholar]
- Schultz CJ, Johnson KL, Currie G, Bacic A. The classical arabinogalactan protein gene family of Arabidopsis. The Plant Cell. 2000;12:1751–1768. doi: 10.1105/tpc.12.9.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Rumsewicz MP, Johnson KL, Jones BJ, Gaspar YM, Bacic A. Using genomic resources to guide research directions: the arabinogalactan protein gene family as a test case. Plant Physiology. 2002;129:1448–1463. doi: 10.1104/pp.003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ, Roberts K. The biology of arabinogalactan proteins. Annual Review in Plant Biology. 2007;58:137–161. doi: 10.1146/annurev.arplant.58.032806.103801. [DOI] [PubMed] [Google Scholar]
- Seifert GJ, Barber C, Wells B, Dolan L, Roberts K. Galactose biosynthesis in Arabidopsis: genetic evidence for substrate channeling from UDP-D-galactose into cell wall polymers. Current Biology. 2002;12:1840–1845. doi: 10.1016/s0960-9822(02)01260-5. [DOI] [PubMed] [Google Scholar]
- Serpe MD, Nothnagel EA. Effects of Yariv phenyl glycosides on rose cell-suspensions: evidence for the involvement of arabinogalactan proteins in cell proliferation. Planta. 1994;193:542–550. [Google Scholar]
- Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK. The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. The Plant Cell. 2003;15:19–32. doi: 10.1105/tpc.007872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM. Arabinogalactan proteins: structure, expression and function. Cellular and Molecular Life Sciences. 2001;58:1399–1417. doi: 10.1007/PL00000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM, Keppler B, Lichtenberg J, Gu D, Welch LR. A bioinformatic approach to the identification, classification and analysis of hydroxyproline rich glycoproteins. Plant Physiology. 2010;153:485–513. doi: 10.1104/pp.110.156554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E, Barbar E, Leykam JF, Kieliszewski MJ. Contiguous hydroxyproline residues direct hydroxyproline arabinosylation in Nicotiana tabacum. Journal of Biological Chemistry. 2001;276:11272–11278. doi: 10.1074/jbc.M011323200. [DOI] [PubMed] [Google Scholar]
- Simon-Plas F, Perraki A, Bayer E, Gerbeau-Pissot P, Mongrand S. An update on plant membrane raft. Current Opinion in Plant Biology. 2011;14:642–649. doi: 10.1016/j.pbi.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Smallwood M, Beven A, Donovan N, et al. Localization of cell wall protein in relation to the developmental anatomy of the carrot root apex. The Plant Journal. 1994;5:237–246. [Google Scholar]
- Smallwood M, Yates EA, Willats WGT, Martin H, Knox JP. Immunochemical comparison of membrane-associated and secreted arabinogalactan proteins in rice and carrots. Planta. 1996;198:452–459. [Google Scholar]
- Sommer-Knudsen J, Clarke AE, Bacic A. A galactose-rich, cell-wall glycoprotein from styles of Nicotiana alata. The Plant Journal. 1996;9:71–83. doi: 10.1046/j.1365-313x.1996.09010071.x. [DOI] [PubMed] [Google Scholar]
- Sørensen I, Domozych D, Willats WGT. How have plant cell walls evolved? Plant Physiology. 2010;153:366–372. doi: 10.1104/pp.110.154427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speranza A, Taddei AR, Gambellini G, Ovidi E, Scoccianti V. The cell wall of kiwifruit pollen tubes is a target for chromium toxicity: alterations to morphology, callose pattern and arabinogalactan protein distribution. Plant Biology. 2009;11:179–193. doi: 10.1111/j.1438-8677.2008.00129.x. [DOI] [PubMed] [Google Scholar]
- Steffan W, Kova P, Albersheim P, Darvill AG, Hahn MG. Characterization of a monoclonal antibody that recognizes an arabinosylated beta-1,6-d-galactan epitope in plant complex carbohydrates. Carbohydrate Research. 1995;275:295–307. doi: 10.1016/0008-6215(95)00174-r. [DOI] [PubMed] [Google Scholar]
- Sun W, Zhao ZD, Hare MC, Kieliszewski MJ, Showalter AM. Tomato LeAGP-1 is a plasma membrane-bound, glycosylphosphatidylinositol-anchored arabinogalactan-protein. Physiologia Plantarum. 2004;120:319–327. doi: 10.1111/j.0031-9317.2004.0236.x. [DOI] [PubMed] [Google Scholar]
- Sun W, Xu J, Yang J, Kieliszewski MJ, Showalter AM. The lysine-rich arabinogalactan-protein subfamily in arabidopsis: gene expression, glycoprotein purification and biochemical characterization. Plant and Cell Physiology. 2005;46:975–984. doi: 10.1093/pcp/pci106. [DOI] [PubMed] [Google Scholar]
- Svetek J, Yadav MP, Nothnagel EA. Presence of a glycosylphosphatidylinositol lipid anchor on rose arabinogalactan proteins. Journal of Biological Chemistry. 1999;274:14724–14733. doi: 10.1074/jbc.274.21.14724. [DOI] [PubMed] [Google Scholar]
- Takata R, Tokita K, Mori S, et al. Degradation of carbohydrate moieties of arabinogalactan-proteins by glycoside hydrolases from Neurospora crassa. Carbohydrate Research. 2010;345:2516–2522. doi: 10.1016/j.carres.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Tan L, Leykam JF, Kieliszewski MJ. Glycosylation motifs that direct arabinogalactan addition to arabinogalactan-proteins. Plant Physiology. 2003;132:1362–1369. doi: 10.1104/pp.103.021766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Qiu F, Lamport DTA, Kieliszewski MJ. Structure of a hydroxyproline (Hyp)-arabinogalactan polysaccharide from repetitive Ala-Hyp expressed in transgenic Nicotiana tabacum. Journal of Biological Chemistry. 2004;279:13156–13165. doi: 10.1074/jbc.M311864200. [DOI] [PubMed] [Google Scholar]
- Tan L, Varnai P, Lamport DTA, et al. Plant O-glycosylation arabinogalactans are composed of repeating trigalactosyl subunits with short bifurcated side chains. Journal of Biological Chemistry. 2010;285:24575–24583. doi: 10.1074/jbc.M109.100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Ezcurra I, Muschietti J, McCormick S. A cysteine-rich extracellular protein, LAT52, interacts with the extracellular domain of the pollen receptor kinase LePRK2. The Plant Cell. 2002;14:2277–2287. doi: 10.1105/tpc.003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X-C, He Y-Q, Wang Y, Sun M-X. The role of arabinogalactan proteins binding to Yariv reagents in the initiation, cell developmental fate, and maintenance of microspore embryogenesis in Brassica napus L. cv. Topas. Journal of Experimental Botany. 2006;57:2639–2650. doi: 10.1093/jxb/erl027. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the Flowering plant Arabidopsis thaliana. Nature. 2000;408:796–814. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Tryfona T, Liang H-C, Kotake T, et al. Carbohydrate structural analysis of wheat flour arabinogalactan protein. Carbohydrate Research. 2010;345:2648–2656. doi: 10.1016/j.carres.2010.09.018. [DOI] [PubMed] [Google Scholar]
- Tsumuraya Y, Ogura K, Hashimoto Y, Mukoyama H, Yamamoto S. Arabinogalactan-proteins from primary and mature roots of radish (Raphanus sativus L.) Plant Physiology. 1988;86:155–160. doi: 10.1104/pp.86.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsyganova AV, Tsyganov VE, Findlay KC, Borisov AY, Tikhonovich IA, Brewin NJ. Distribution of legume arabinogalactan proteins-extensin (AGPE) glycoproteins in symbiotically defective pea mutants with abnormal infection threads. Cell and Tissue Biology. 2009;3:93–102. [PubMed] [Google Scholar]
- Velasquez SM, Ricardi MM, Dorosz JG, et al. O-Glycosylated cell wall proteins are essential in root hair growth. Science. 2011;332:1401–1403. doi: 10.1126/science.1206657. [DOI] [PubMed] [Google Scholar]
- Vicré M, Santaella C, Blanchet S, Gateau A, Driouich A. Root border-like cells of Arabidopsis: microscopical characterization and role in the interaction with rhizobacteria. Plant Physiology. 2005;138:998–1008. doi: 10.1104/pp.104.051813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Spano T, Vlad D, Boudaher F, Ouelhadj A, Kalaitzis P. Arabidopsis prolyl-4 hydroxylases are differentially expressed in response to hypoxia, anoxia and mechanical wounding. Physiologia Plantarum. 2007;130:471–483. [Google Scholar]
- Wagner TA, Kohorn BD. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. The Plant Cell. 2001;13:303–318. doi: 10.1105/tpc.13.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker TS, Bais HP, Grotewold E, Vivanco JM. Root exudation and rhizosphere biology. Plant Physiology. 2003;132:44–51. doi: 10.1104/pp.102.019661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wu HM, Cheung AY. Developmental and pollination regulation of the accumulation and glycosylation of a transmitting tissue-specific proline-rich glycoprotein. The Plant Cell. 1993;5:1639–1650. doi: 10.1105/tpc.5.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HJ, Huang JC, Jauh GY. Pollen germination and tube growth. Advances in Botanical Research. 2010;54:1–52. [Google Scholar]
- Wang Y, Zhang WZ, Song LF, Zou JJ, Su Z, Wu WH. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiology. 2008;148:1201–1211. doi: 10.1104/pp.108.126375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F, van Etten HD, Tsaprailis G, Hawes MC. Extracellular proteins in pea roots tip and border cell exudates. Plant Physiology. 2007;143:773–783. doi: 10.1104/pp.106.091637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats GTW, Knox JP. A role for arabinogalactan-proteins in plant cell expansion: evidence from studies on the interaction of β-glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. The Plant Journal. 1996;9:919–925. doi: 10.1046/j.1365-313x.1996.9060919.x. [DOI] [PubMed] [Google Scholar]
- Williams A, Wilkinson A, Krehenbrink M, Russo DM, Zorreguieta A, Downie JA. Glucomannan-mediated attachment of Rhizobium leguminosarum to pea root hairs is required for competitive nodule infection. Journal of Bacteriology. 2008;190:4706–4715. doi: 10.1128/JB.01694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewska E, Majewska-Sawka A. Cell wall polysaccharides in differentiating anthers and pistils of Lolium perenne. Protoplasma. 2006;228:65–71. doi: 10.1007/s00709-006-0175-1. [DOI] [PubMed] [Google Scholar]
- Wu HM, Wang H, Cheung AY. A pollen tube growth stimulatory glycoprotein is deglycosylated by pollent tubes and displays a glycosylation gradient in the flower. Cell. 1995;82:395–403. doi: 10.1016/0092-8674(95)90428-x. [DOI] [PubMed] [Google Scholar]
- Wu HM, Wong E, Ogdahl J, Cheung AY. A pollen tube growth-promoting arabinogalactan protein from Nicotiana alata is similar to the tobacco TTS protein. The Plant Journal. 2000;22:167–176. doi: 10.1046/j.1365-313x.2000.00731.x. [DOI] [PubMed] [Google Scholar]
- Wu Y, Williams M, Bernard S, Driouich A, Showalter AM, Faik A. Functional identification of two nonredundant Arabidopsis (1,2)fucosyltransferases specific to arabinogalactan proteins. Journal of Biological Chemistry. 2010;285:13638–13645. doi: 10.1074/jbc.M110.102715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Ma L, Samaj J, Xu C. Immunohistochemical analysis of cell wall hydroxyproline-rich glycoproteins in the roots of resistant and susceptible wax gourd cultivars in response to Fusarium oxysporum f. sp. Benincasae infection and fusaric acid treatment. Plant Cell Report. 2011;30:1555–1569. doi: 10.1007/s00299-011-1069-z. [DOI] [PubMed] [Google Scholar]
- Xie F, Williams A, Edwards A, Downie JA. A plant arabinogalactan-like glycoprotein promotes a novel type of polar surface attachment by Rhizobium leguminosarum. Molecular Plant–Microbe Interactions. 2012;25:250–258. doi: 10.1094/MPMI-08-11-0211. [DOI] [PubMed] [Google Scholar]
- Xu J, Tan L, Lamport DTA, Showalter AM, Kieliszewski MJ. The O-Hyp glycosylation code in tobacco and Arabidopsis and a proposed role of Hyp-glycans in secretion. Phytochemistry. 2008;69:1631–1640. doi: 10.1016/j.phytochem.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Yang J, Kieliszewski MJ, Showalter AM. The lysine-rich arabinogalactan-protein subfamily in Arabidopsis: gene expression, glycoprotein purification and biochemical characterization. Plant and Cell Physiology. 2005;46:975–984. doi: 10.1093/pcp/pci106. [DOI] [PubMed] [Google Scholar]
- Yariv J, Rapport MM, Graf L. The interaction of glycosides and saccharides with antibody to the corresponding phenylazo glycosides. Biochemical Journal. 1962;85:383–388. doi: 10.1042/bj0850383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yariv J, Lis H, Katchalski E. Precipitation of arabic acid and some seed polysaccharides by glycosylphenylazo dyes. Biochemical Journal. 1967;105 doi: 10.1042/bj1050001c. p1C–2C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates EA, Knox JP. Investigations into the occurrence of plant cell surface epitopes in exudate gums. Carbohydrate Polymers. 1994;24:281–286. [Google Scholar]
- Yates EA, Valdor J-F, Haslam SM, et al. Characterization of carbohydrate structural features recognized by anti arabinogalactan-protein monoclonal antibodies. Glycobiology. 1996;6:131–139. doi: 10.1093/glycob/6.2.131. [DOI] [PubMed] [Google Scholar]
- Yatomi R, Nakamura S, Nakamura N. Immunochemical and cytochemical detection of wall components of germinated pollen of Gymnosperms. Grana. 2002;41:21–28. [Google Scholar]
- Youl JJ, Bacic A, Oxley D. Arabinogalactan-proteins from Nicotiana alata and Pyrus communis contain glycosyl-phosphatidylinositol membrane anchors. Proceedings of the National Academy of Sciences of the USA. 1998;95:7921–7926. doi: 10.1073/pnas.95.14.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, et al. The genomes of Oryza sativa: a history of duplications. PLoS Biology. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. doi:10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Y, Yang J, Showalter A. AtAGP18, a lysine-rich arabinogalactan protein in Arabidopsis thaliana, functions in plant growth and development as a putative co-receptor for signal transduction. Plant Signaling and Behaviour. 2011;6:855–857. doi: 10.4161/psb.6.6.15204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Mollet JC, Lord EM. Lily (Lilium longiflorum L.) pollen protoplast adhesion is increased in the presence of the peptide SCA. Sexual Plant Reproduction. 2004;16:227–233. [Google Scholar]
- Zhao ZD, Tan L, Showalter AM, Lamport DTA, Kieliszewski MJ. Tomato LeAGP-1 arabinogalactan-protein purified from transgenic tobacco corroborates the Hyp contiguity hypothesis. The Plant Journal. 2002;31:431–444. doi: 10.1046/j.1365-313x.2002.01365.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. Genevestigator. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]