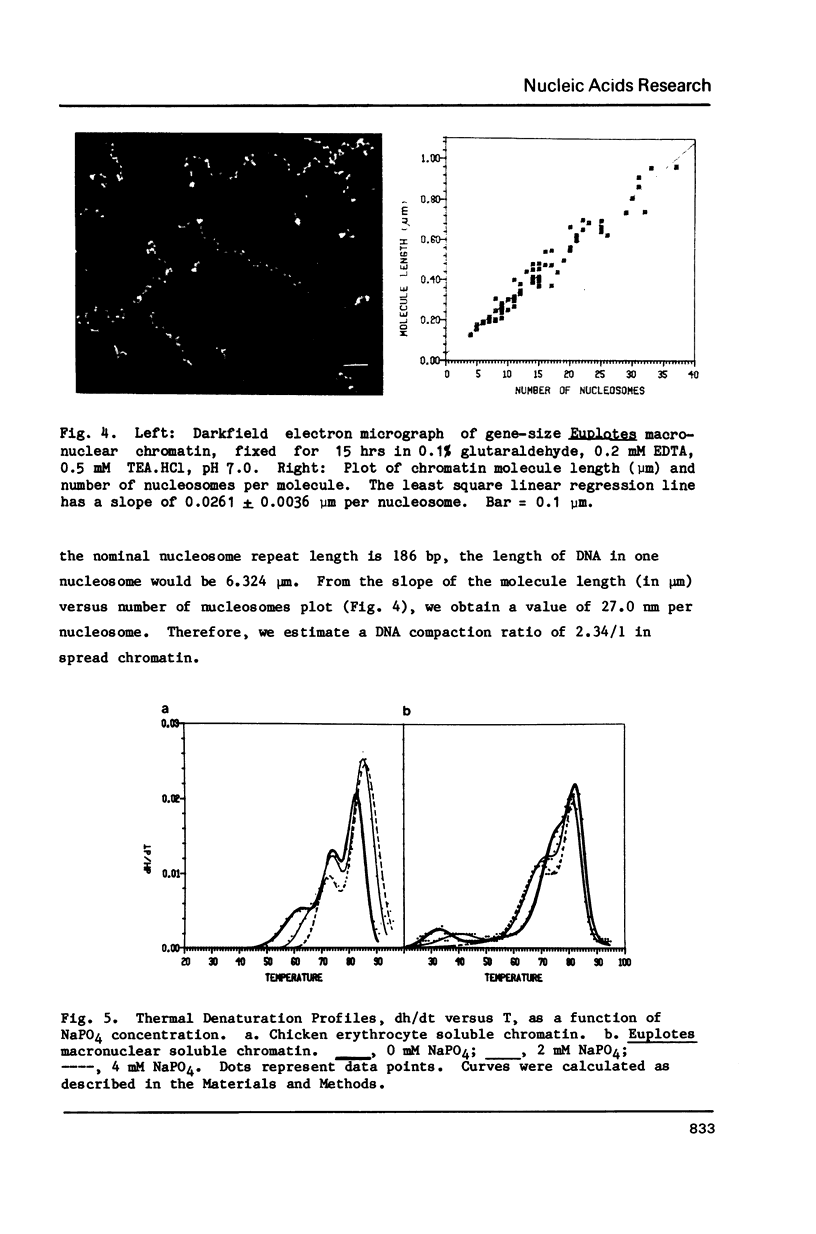
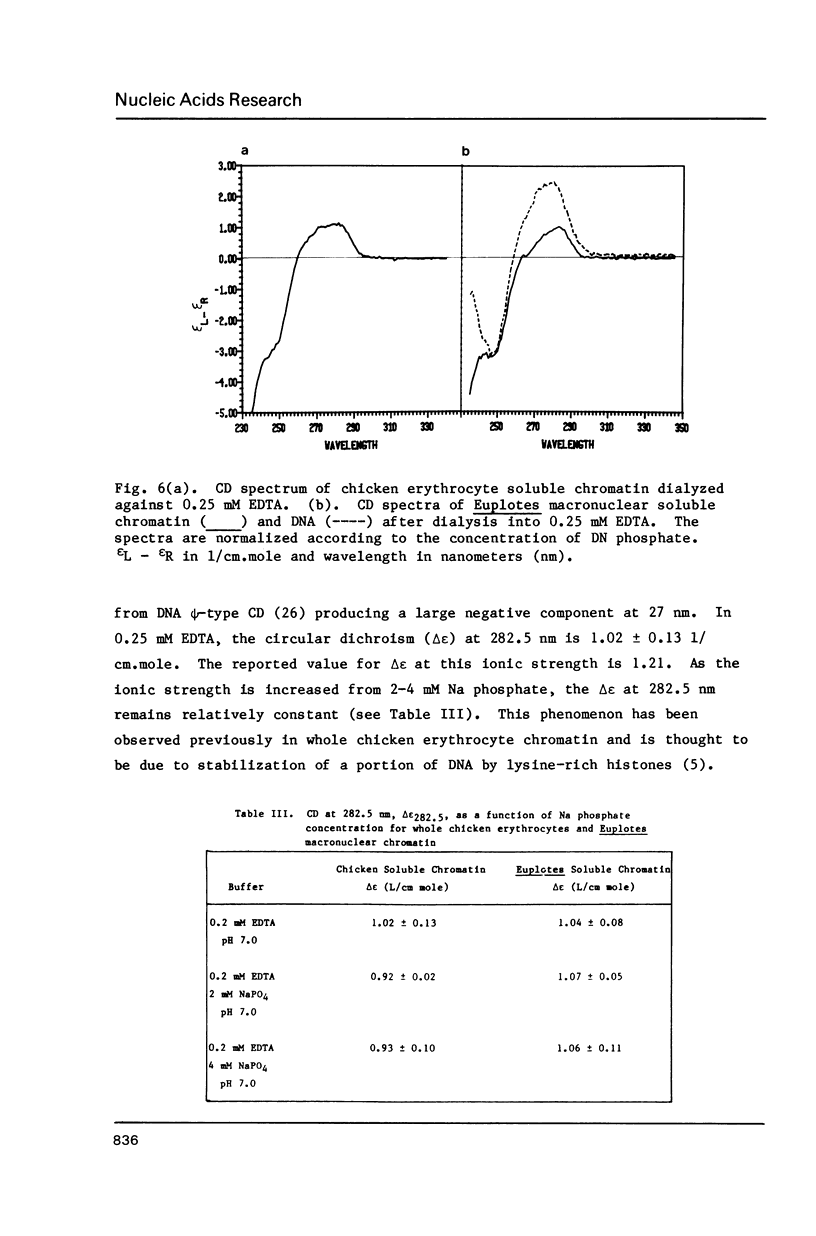
Abstract
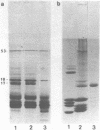
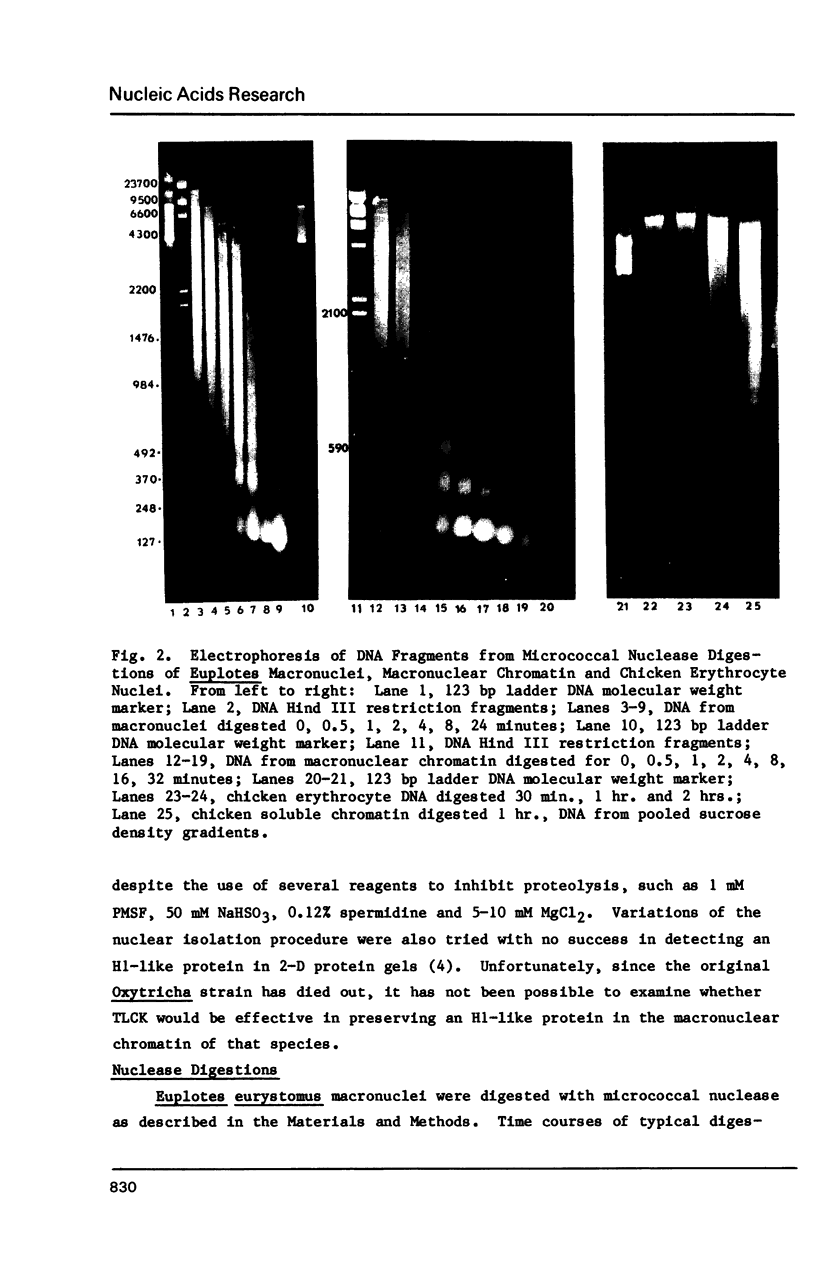
Euplotes eurystomus is a hypotrichous ciliate containing a transcriptionally active macronucleus (MAC) and a transcriptionally inactive micronucleus. Soluble MAC chromatin contains a normal complement of inner histones, an H1-like protein which is very sensitive to proteolysis, and a considerable proportion of non-histone proteins. A combination of N-Tosyl lysine chloromethyl ketone (TLCK) and PMSF was found to be most effective in preventing proteolysis. Microccocal nuclease digestion yielded an average nucleosome repeat length of 187 +/- 25 bp for soluble chromatin; and 190 +/- 9 bp for isolated macronuclei. Thermal denaturation profiles of MAC chromatin in 0.25 mM EDTA display two main transitions at about 76 and 83 degrees C, resembling the melting of soluble chicken erythrocyte chromatin. Circular dichroic spectra of MAC chromatin were compared to soluble chicken erythrocyte chromatin under the same ionic strength conditions and were found to be very similar. 2D chromatin/DNA agarose gel electrophoresis resulted in a diagonal line of DNA staining, which establishes a strict correlation between DNA and chromatin electrophoretic mobility.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ammermann D., Muenz A. DNA and protein content of different hypotrich ciliates. Eur J Cell Biol. 1982 Apr;27(1):22–24. [PubMed] [Google Scholar]
- Bryan P. N., Wright E. B., Hsie M. H., Olins A. L., Olins D. E. Physical properties of inner histone-DNA complexes. Nucleic Acids Res. 1978 Oct;5(10):3603–3617. doi: 10.1093/nar/5.10.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A. P., Laughlin T. J., Cadilla C. L., Henry J. M., Olins D. E. Physical structure of gene-sized chromatin from the protozoan Oxytricha. Nucleic Acids Res. 1984 Apr 11;12(7):3201–3217. doi: 10.1093/nar/12.7.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulmer A. W., Fasman G. D. Analysis of chromatin reconstitutiion. Biochemistry. 1979 Feb 20;18(4):659–668. doi: 10.1021/bi00571a017. [DOI] [PubMed] [Google Scholar]
- Fulmer A. W., Fasman G. D. Ionic strength-dependent conformational transitions of chromatin. Circular dichroism and thermal denaturation studies. Biopolymers. 1979 Nov;18(11):2875–2891. doi: 10.1002/bip.1979.360181115. [DOI] [PubMed] [Google Scholar]
- Gottschling D. E., Cech T. R. Chromatin structure of the molecular ends of Oxytricha macronuclear DNA: phased nucleosomes and a telomeric complex. Cell. 1984 Sep;38(2):501–510. doi: 10.1016/0092-8674(84)90505-1. [DOI] [PubMed] [Google Scholar]
- Kaplan L. J., Bauer R., Morrison E., Langan T. A., Fasman G. D. The structure of chromatin reconstituted with phosphorylated H1. Circular dichroism and thermal denaturation studies. J Biol Chem. 1984 Jul 25;259(14):8777–8785. [PubMed] [Google Scholar]
- Klobutcher L. A., Swanton M. T., Donini P., Prescott D. M. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3' terminus. Proc Natl Acad Sci U S A. 1981 May;78(5):3015–3019. doi: 10.1073/pnas.78.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger R. C. The inhibition of chromatin solubilization by proteolytic inhibitors. Biochem Biophys Res Commun. 1983 Jan 14;110(1):216–219. doi: 10.1016/0006-291x(83)91282-2. [DOI] [PubMed] [Google Scholar]
- Laughlin T. J., Herrmann A. L., Olins D. E. Fractionation of the gene-size macronuclear chromatin fragments of the binucleated eukaryote Oxytricha. Mol Cell Biochem. 1984 Jun;62(2):157–163. doi: 10.1007/BF00223306. [DOI] [PubMed] [Google Scholar]
- Levinger L., Barsoum J., Varshavsky A. Two-dimensional hybridization mapping of nucleosomes. comparison of DNA and protein patterns. J Mol Biol. 1981 Mar 5;146(3):287–304. doi: 10.1016/0022-2836(81)90389-2. [DOI] [PubMed] [Google Scholar]
- Lipps H. J., Gruissem W., Prescott D. M. Higher order DNA structure in macronuclear chromatin of the hypotrichous ciliate Oxytricha nova. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2495–2499. doi: 10.1073/pnas.79.8.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipps H. J., Nock A., Riewe M., Steinbrück G. Chromatin structure in the macronucleus of the ciliate Stylonychia mytilus. Nucleic Acids Res. 1978 Dec;5(12):4699–4709. doi: 10.1093/nar/5.12.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P. T., Burgess D. R. SDS microslab linear gradient polyacrylamide gel electrophoresis. Anal Biochem. 1978 Jul 1;87(2):386–396. doi: 10.1016/0003-2697(78)90688-7. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Carlson R. D., Wright E. B., Olins D. E. Chromatin nu bodies: isolation, subfractionation and physical characterization. Nucleic Acids Res. 1976 Dec;3(12):3271–3291. doi: 10.1093/nar/3.12.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottensmeyer F. P., Whiting R. F., Schmidt E. E., Clemens R. S. Electron microtephroscopy of proteins. A close look at the ashes of myokinase and protamine. J Ultrastruct Res. 1975 Aug;52(2):193–201. doi: 10.1016/s0022-5320(75)80111-0. [DOI] [PubMed] [Google Scholar]
- Ralph-Edwards A., Silver J. C. Nucleosome DNA repeat length and histone complement in a fungus exhibiting condensed chromatin. Exp Cell Res. 1983 Oct 15;148(2):363–376. doi: 10.1016/0014-4827(83)90159-3. [DOI] [PubMed] [Google Scholar]
- Sasi R., Fasman G. D. The effect of a high mobility group protein (HMG 17) on the structure of acetylated and control core HeLa cell chromatin. Biochim Biophys Acta. 1984 May 15;782(1):55–66. doi: 10.1016/0167-4781(84)90106-4. [DOI] [PubMed] [Google Scholar]
- Shin Y. A., Eichhorn G. L. Formation of psi (+) and psi (-) DNA. Biopolymers. 1984 Feb;23(2):325–335. doi: 10.1002/bip.360230211. [DOI] [PubMed] [Google Scholar]
- Swanton M. T., McCarroll R. M., Spear B. B. The organization of macronuclear rDNA molecules of four hypotrichous ciliated protozoans. Chromosoma. 1982;85(1):1–9. doi: 10.1007/BF00344590. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T. Influence of histone H1 on chromatin structure. Cell. 1977 Sep;12(1):101–107. doi: 10.1016/0092-8674(77)90188-x. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]