Abstract
The increasing demand for antibody-based therapeutics has emphasized the need for technologies to improve recombinant antibody titers from mammalian cell lines. Moreover, as antibody therapeutics address an increasing spectrum of indications, interest has increased in antibody engineering to improve affinity and biological activity. However, the cellular mechanisms that dictate expression and the relationships between antibody sequence and expression level remain poorly understood. Fundamental understanding of how mammalian cells handle high levels of transgene expression and of the relationship between sequence and expression are vital to the development of new antibodies and for increasing recombinant antibody titers. In this work, we analyzed a pair of mutants that vary by a single amino acid at Kabat position 49 (heavy chain framework), resulting in differential transient and stable titers with no apparent loss of antigen affinity. Through analysis of mRNA, gene copy number, intracellular antibody content, and secreted antibody, we found that while translational/post-translational mechanisms are limiting in transient systems, it appears that the amount of available transgenic mRNA becomes the limiting event upon stable integration of the recombinant genes. We also show that amino acid substitution at residue 49 results in production of a non-secreted HC variant and postulate that stable antibody expression is maintained at a level which prevents toxic accumulation of this HC-related protein. This study highlights the need for proper sequence engineering strategies when developing therapeutic antibodies and alludes to the early analysis of transient expression systems to identify the potential for aberrant stable expression behavior.
Introduction
Chinese hamster ovary (CHO) cells are a commonly employed system for the expression of recombinant therapeutic proteins due to their ability to secrete glycosylated, correctly folded proteins.1 Recombinant monoclonal antibodies (mAbs) constitute a significant fraction of the biopharmaceutical market share2 and the drive to produce large-scale, high-quality product at low cost has led to a significant research effort towards maximizing antibody titers. Strategies for rational engineering of antibody sequences are employed to improve function, minimize heterogeneity, and control pharmacodynamic behavior of therapeutic candidates, but the relationship between antibody sequence and recombinant expression levels are still poorly understood.
Due to the complexity of protein transcription, mRNA turnover, translation, post-translational processing, and secretion in mammalian cells, there are many stages at which observed (i.e. secreted) antibody expression can be affected. Several studies over the past several years have analyzed clones that produce identical proteins at different specific productivities to identify the relationship between transgene copy number, mRNA levels, and specific productivity, in an attempt to create rigorous selection criteria for development of high-producing stable cell lines.3–10 The results of these studies were inconsistent due to variation in mammalian cell lines (NS0, CHO), selection systems (DFHR, GS), and protein selected for expression (mAbs, luciferase). However, many studies observed positive correlations between heavy chain (HC) mRNA levels and secreted mAb titer and/or specific productivities. Correlations between mRNA levels and protein expression often broke down in highly-amplified expression systems or in very high-producing cell lines. Other studies, focused specifically on mAb production, implicated protein folding and assembly as the limiting mechanisms.11, 12
Antibody folding and assembly is a complex process which has to pass several checkpoints and quality control mechanisms in the cell.13 This is to prevent secretion of partially folded or mis-folded antibodies which would not elicit the desired immune response. In general, antibody folding begins co-translationally in the endoplasmic reticulum (ER), then HC dimers are formed through Fc association, and finally the light chains (LCs) are added through inter-domain disulfide formation between the CH1 and CL domains. Immunoglobulin heavy-chain binding protein (BiP) is transiently associated with the HC in antibody intermediates to prevent aggregation. All domains posses an intra-domain disulfide bridge for stability and the constant domains have to undergo a peptidylprolyl isomerization reaction to convert the Pro residue from the trans to the cis configuration. These reactions are rate-limiting and can take several minutes to reach completion at room temperature. Antibodies that fail to fold or assemble correctly may engage the unfolded protein response (UPR) which attempts to alleviate the stress through increasing the ER folding capacity, downregulating transcription/translation, or directing the aberrant protein for degradation.14
In this study, we examined an antibody pair whereby a single amino acid substitution impacts antibody expression without altering the innate antibody function (e.g. antigen binding). By elucidating the cellular mechanisms responsible for differential expression of the mutant pair, we hope to gain understanding of how CHO cells regulate exogenous antibody expression. These studies may provide broader insight to the potential repercussions of antibody sequence engineering, leading to strategies for optimizing antibodies for compatibility with the biomanufacturing process.
Materials and Methods
Cell Culture
CHO-S (Invitrogen, Paisley, UK) and CHOK1-SV (Lonza Biologics, Slough, Berkshire, UK) cells were cultured in CDCHO medium (Invitrogen) supplemented with 4 mM GlutaMax; all stable, recombinant IgG-producing clones using the glutamine synthetase (GS) selection system were cultured in CDCHO medium containing 50 μM methionine sulfoximine (MSX). All cells were subcultured every 3–4 days in polycarbonate Erlenmeyer shake flasks with an inoculum viable cell density (VCD) of 3×105 cells/mL. Flasks were incubated at 37°C in 8% CO2 and shaken at 140 rpm. All cell counts were performed using a Cedex A520 automated cell counter (Innovatis, Bielefeld, Germany).
Specific productivity, Qp (pg/cell/day), was calculated during exponential growth phase using the following equation:
where t is the elapsed culture time in days, n0 and nt are the viable cell densities (106 cells/mL) at the start and end of culture, respectively, and ΔP is the change in antibody titer (μg/mL) between the start and end of the culture.
Plasmids
Single and double gene vectors containing the heavy and/or light chain genes under individual control of the human CMV promoter were used for transient experiments. The bicistronic vectors were modified to include the GS gene under control of the SV-40 promoter for all stable transfections. All plasmids were prepared using MiniPrep or MaxiPrep kits (Qiagen, West Sussex, UK). DNA concentrations were measured using a Nanodrop 1000 spectrophotometer; DNA was considered pure if A260/A280 ratio ≥ 1.8.
Site-specific mutagensis was performed using a Stratagene QuikChange Multi SDM Kit (Agilent, Stockport, Cheshire, UK). Primers were designed using Stratagene's QuikChange Primer Design Program (http://www.stratagene.com/qcprimerdesign). Samples were sequenced on a Genetic Analyzer system (Applied Biosystems, Foster City, CA) followed by analysis using Sequencher v4.7 software (Gene Codes).
Transient Transfections
For initial shake-flask studies, CHO-S cells in log phase were counted, and 4–5×107 cells per transfection were centrifuged at 1200 rpm for 5 min and resuspended in 15 mL of fresh media. In a polypropylene tube, 150 μL of Lipofectamine 2000 (Invitrogen) was diluted with 1.5 mL of Opti-MEM I reduced serum medium and incubated for 5 min at room temperature. In a separate tube, 60 μg of DNA was diluted with 1.5 mL of Opti-MEM. The Lipofectamine and DNA mixtures were combined and incubated for 20 min at room temperature. After formation of the DNA complexes, the 3-mL Lipofectamine/DNA mixture was added to the 15 mL of cell suspension in a 125-mL Erlenmeyer flask and incubated at 37°C with shaking (125 rpm) for 4 h. At the end of the incubation period, 27 mL of fresh media was added to the flask. Gene expression was assayed 2–9 days post-transfection.
For the synonymous codon variants, the transient transfections were performed with CHO-S cells using an Amaxa Nucleofector II (Nucleofector Kit V). Kits were used according to the manufacturers' instructions. For each transfection, ~5×106 cells were centrifuged and resuspended in 100 μL of Nucleofector solution. The cells were transferred to a supplied cuvette and 2 μg of DNA was added. Program U-024 was applied to the cell/DNA mixture, 0.5 mL of media was added to the cuvette, and then the cells were immediately transferred to 5 mL of prewarmed media in a 50 mL spin tube. The tubes were incubated at 37°C in 8% CO2 and shaken at 250 rpm.
Stable Cell Line Generation
For transfection of 1×107 CHOK1SV cells, 40 μg of linearized DNA was diluted with ultrapure water to a final volume of 100 μL. CHO cells in log phase were centrifuged and resuspended in 700 μL of CDCHO. The cells and DNA were transferred to a Biorad cuvette and electroporated using a Bio-Rad GenePulser (Bio-Rad, Hemel Hempsted, UK) instrument set at 0.3kV and 960μF. The cell suspension was carefully transferred into a T-175 flask containing 50 mL of prewarmed media. The transfected cells were allowed to recover for 24 hours. Cells were then subcultured into T-25 tissue culture flasks at a density of ~1×106 cells/flask (10 mL working volume) and 50 μM MSX was added. The transfectants were incubated under static culture conditions at 37°C and 8% CO2 until nine days post-transfection when a media change was performed, and the cells were returned to T-25 flasks. After ~21 days, cells were counted for each pool, antibody titers were measured by ELISA, and if the VCD was acceptable (>3×105 cells/mL and >90% viability), cells were expanded into shake-flask cultures or clonal selection was performed.
For clonal selection, semi-solid CDCHO GS CloneMedia (Genetix, New Milton, UK) was supplemented with 50 μM MSX and divided into 45 mL aliquots. Cells from the polyclonal pools were added to the aliquots for a final density of 1000 cells/mL and the mixture was transferred to PetriWell-6 plates (Genetix) at ~2 mL/well. Colonies were allowed to grow for 10–14 days and then plates were analyzed using a ClonePix FL automated colony picker. Approximately 500 colonies from each bulk pool were identified. The top 196 colonies based on size (total area) were selected by white light analysis and plated by colony size into two 96-well sterile tissue culture plates containing selection-supplemented CDCHO medium. Colony expansion into larger volumes was performed for clones that exhibited the desired growth characteristics and antibody production.
Antibody Assays
For antibody concentrations over 0.5 μg/mL, titers were measured using an Octet QK (ForteBio, Menlo Park, CA) with Protein A Biosensors according to the manufacturer's instructions. The appropriate number of biosensors were prewet in 200 μL media or lysis buffer for a minimum of 5 min prior to use in a 96-well plate. The samples for analysis were prepared by adding 200 μL to a black, flat-bottomed 96-well plate. The analysis was performed at 30°C with a 2-min run time and 200 rpm plate agitation. Final antibody titers were compared to a standard IgG4 curve over a working range of 1 to 700 μg/mL.
Quantitative real-time PCR
Total RNA and genomic DNA (gDNA) were isolated from 1×106 cells in mid-exponential phase (~day 4) using a Qiagen AllPrep kit. The concentration and purity of the extracted RNA was determined using the Nanodrop 1000 spectrophotometer and was considered pure if the A260/A280 ratio was ≥ 2.0. Up to 2 μg of total RNA was used to generate cDNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The cDNA was diluted 1:100 with ultrapure DNase, RNase-free water. TaqMan master mix solutions were prepared by mixing 5 μL of TaqMan Gene Expression Master Mix (Applied Biosystems) with 0.5 μL of 20× TaqMan custom gene expression assay per sample. The TaqMan primer and probe sequences are listed in Table 1. Triplicate qPCR reactions were loaded onto a 384-well plate containing 4.5 μL of sample and 5.5 μL of TaqMan master mix. Plates were analyzed on an ABI 7900HT Fast Real-Time PCR system using an initial 10 min activation step at 95°C followed by 40 cycles of 15 seconds denaturation at 95° and 1 min anneal/extend at 60°C. Relative quantitation data analysis was performed using the comparative quantification method, ΔΔCt, with GAPDH as the endogenous reference. The amplification efficiencies of the genes of interest were comparable over the input cDNA concentrations used in this study. The calibrator used is specified in the appropriate results section.
Table 1.
TaqMan primer/probe sequences used in qPCR reactions
| Target | Primer | Sequence (5'→3') |
|---|---|---|
| Human constant κ (Light Chain) | Forward | AAAGTACAGTGGAAGGTGGATAACG |
| Reverse | CTTGCTGTCCTGCTCTGTGA | |
| Probe | CCAATCGGGTAACTCC | |
| Human γ4 (Heavy Chain) | Forward | CCCAAGGACACTCTCATGATCTC |
| Reverse | CCATCCACGTACCAGTTGAACT | |
| Probe | ACGCACGTGACCTCAG | |
| Glutamine Synthetase | Forward | TGCCCAGTGGGAATTCCAAATAG |
| Reverse | GGGCCACCCAGAGATGATC | |
| Probe | CCATGCGGATTCCTT | |
| Hamster GAPDH | Forward | CTGCCACCCAGAAGACTGT |
| Reverse | GTGGATGCAGGGATGATGTTCT | |
| Probe | ATCACGCCACAGCTTT |
Gene copy number was determined by adding 10 ng of purified gDNA, 5.5 μL of TaqMan master mix containing the appropriate probe, and water to a 384-well plate for a final volume of 10 μL/well. The results were compared to a standard curve, which was constructed by mixing known amounts of plasmid DNA containing the probe gene with 10 ng of gDNA purified from the untransfected host CHOK1SV cell line. gDNA was purified from CHOK1SV cells using a Promega Wizard Genomic DNA Purification Kit. The plates were analyzed as described above. All qPCR reactions were performed in triplicate. For analysis of gene copy number, it was assumed that CHO gDNA weighs 6.6 pg/cell (internal communication), and therefore 10 ng of gDNA contains ~1568 copies of the genome.
SDS-PAGE and Immunoblots
Cells were washed twice with cold PBS and lysed on ice with cold RIPA buffer (150 mM NaCl, 25 mM Tris, 1% (w/v) NP-40, 1% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, pH 7.6) containing protease inhibitors. The lysates were agitated for 30 minutes at 4°C followed by centrifugation in a microcentrifuge at maximum speed (> 14,000 × g) for 15 min to pellet cellular debris. The supernatants were removed and transferred to a clean microcentrifuge tube. Total protein concentration was determined by bicinchoninic acid (BCA) assay (Thermo Scientific, Rockford, IL) according to manufacturers' instructions. A total of 20 μg/well of protein was mixed with the appropriate amount of 4× NuPage LDS Sample Buffer (Invitrogen) and 10× NuPage Sample Reducing Agent (Invitrogen) for reducing conditions or 100 mM N-ethylmaleimide (NEM) for non-reducing conditions. Samples were boiled at 100°C for 10 min before adding to a 4–12% Bis-Tris gel (Invitrogen). Electrophoresis was performed at 22.5 V/cm for 55 min. Proteins were transferred from the gel onto nitrocellulose membranes using the semi-dry iBlot system (Invitrogen). Gels were post-stained with Coomassie blue to ensure even loading and transfer onto the membrane. Membranes were blocked with Superblock T20 PBS blocking buffer (Thermo Scientific) for an hour at room temperature followed by incubation with an HRP-conjugated anti-human Fcγ antibody (Stratech, Newmarket Suffolk, UK) diluted 1:10,000 in blocking buffer for at least 4 hours at 4°C. The membrane was washed a minimum of three times for 5 min with PBS containing 0.1% (v/v) Tween-20. Blots were revealed using ECL chemiluminescent reagent (Invitrogen) and exposed to Kodak x-ray film. The resulting films were visualized using an XOgraph automatic developer.
Protein Purification
Supernatants containing antibody product from stable cell lines were purified on a Mab Select SuRe Protein A column using an AKTA Prime system (GE Healthcare, Little Chalfont, UK). The column was equilibrated with 5 column volumes (CV's) of PBS, followed by loading of the supernatant or cell lysate. Column loading was monitored by measuring the absorbance at 280 nm. The loaded column was rinsed with 5 CV's of PBS or until the A280 baseline returned to zero. The bound antibody was then eluted with 0.1 M sodium citrate buffer, pH 3. Eluted fractions were immediately neutralized with 1/4 the fraction volume of 2M Tris·HCl, pH 8. The purified mAbs were buffer exchanged into phosphate buffered saline (PBS), pH 7.4 or 20mM sodium phosphate, pH 8 using PD-10 desalting columns (GE Healthcare). The mAb concentration was measured by OD280 using a calculated extinction coefficient based on the protein sequence.
Biomolecular Interaction Analysis
Samples were analyzed using a Biacore 3000 instrument (GE Healthcare). All experiments were performed at 25°C. Affinipure F(ab')2 fragment goat anti-human IgG, Fc fragment specific (Stratech) was immobilized on a CM5 Sensor Chip (GE Healthcare) via amine coupling chemistry to a capture level of ~7000 response units (RUs). HBSEP buffer (10mM HEPES pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.005 % Surfactant P20, GE Healthcare) was used as the running buffer with a flow rate of 10 μL/min. A 10 μL injection of 277 mAb at 1.5 μg/ml was captured by the immobilized anti-human IgG, Fc. Antigen was titrated over the captured mAb 277 at doubling dilutions from 10 nM to sub nM concentrations at a flow rate of 30 μL/min. The surface was regenerated at a flow rate of 10 μL/min with 2 × 10 μL injections of 40 mM HCl, followed by a 5 μL injection of 5 mM NaOH. The background signal was removed by subtracting duplicate buffer blank runs from the sample signal. The resulting binding curves were analyzed using the BIAevaluation software (version 3.2) following standard procedures. Kinetic parameters were determined from the fitting algorithm.
Differential Scanning Fluorimetry
Purified mAb was diluted to 0.1 mg/mL with PBS. A 30× fluorescent dye solution was made by mixing 6 μL 5000× Sypro Orange dye (Invitrogen) with 994 μL PBS. Samples were prepared by mixing 45 μL diluted mAb with 5 μL of 30× dye and adding 10 μL of the mixture to a 384-well plate in quadruplicate. The plate was sealed with optical film and analyzed on the ABI 7900HT Fast system using an initial 5 min hold at 20°C, ramp to 99°C at 1°C/min, and a final hold for 20 min at 99°C. Data was analyzed using an in-house Excel macro.
Circular Dichroism
Circular dichrosim (CD) spectroscopy was performed using a Chirascan Spectrometer (Applied Photophysics, Leatherhead, UK). For measurements in the far UV region (190–260 nm) samples were prepared at 0.5 mg/mL in 20 mM sodium phosphate buffer, pH 8, and read in a 0.5 mm pathlength rectangular quartz cuvette. For measurements in the near UV region (250–320 nm), samples were prepared at 1 mg/mL in phosphate buffer and added to a 1 cm pathlength quartz cuvette. Scans were performed in triplicate at 10°C using a 1 s acquisition time and 0.5 nm step size. Any background signal was removed by subtracting the phosphate buffer spectrum. The far UV data was converted from the machine units of ellipticity (θ, mdeg) to per residue delta epsilon (Δε, deg cm2 dmol−1) using:
where C is the protein concentration in mg/mL, MRW is the mean residue molecular weight (protein molecular weight/number amino acids (N) in the sequence), and l is the cuvette pathlength in cm. The near UV data was converted to molar ellipticity ([θ], deg cm2 dmol−1). Temperature-ramped far UV CD measurements were acquired between 200–260 nm using a 1°C/min ramp rate. To keep spectrum acquisition less than 1 min, the time-per-point was reduced to 0.4 s and a 1 nm step size was used. Secondary structure analysis was performed using the CDPro software package.
Results and Discussion
Transient Expression
A mutation from Ala to Gly at Kabat position 49 (framework 2 of the VH domain) in the 277 IgG4 mAb selected for this study had previously been observed to result in an average four-fold decrease in transient expression without an apparent effect on antigen affinity (unpublished results). The objective of the current studies was to assess the reproducibility of sequence-dependent expression and to identify the cellular mechanisms responsible for differential expression in transient systems. Initial assessment of the mAb 277 Ala and Gly expression in small-scale shake flask culture using various transfection parameters routinely resulted in a two to six-fold difference in secreted mAb titer four days after transfection (data not shown). This observed difference in antibody expression between the Ala and Gly variants remained constant when measured anywhere between days four and nine days post-transfection.
To determine the processes responsible for differential expression, intracellular mRNA, intracellular mAb, and secreted mAb levels were compared between Ala and Gly variants. The synonymous codon variants for Ala and Gly were also included in the study to determine if codon choice at this position has an impact on secreted mAb titer. The mRNA levels for HC and LC were measured by RT-qPCR four days post-transfection and are shown in Figure 1a. Message levels for all probes were calibrated to the amount of corresponding message from the Ala-GCC construct. In all samples, the amount of transgenic HC and LC mRNA varied less than two-fold between Ala and Gly constructs. The XXT codons did exhibit slightly lower HC and LC mRNA levels. A possible explanation for this decrease in mRNA levels is that introduction of a nucleotide motif that is avoided in mammalian genes, the U|A codon junction, may result in endoribonuclease cleavage.15
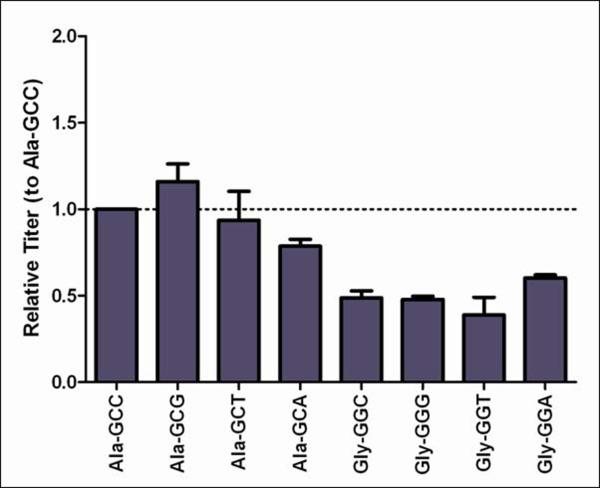
Figure 1.
Transient protein and mRNA expression from Ala and Gly synonymous codon variants.
In (A) the message levels for HC and LC are normalized to Ala-GCC, and the error bars represent ±1 standard deviation from experimental replicates (n=3). In (B) the corresponding antibody titers are the average of biological replicates (n=2) and have also been normalized to Ala-GCC.
The amount of secreted mAb present in the supernatant on day four was ~two-fold higher in the Ala constructs than in the Gly constructs (Figure 1b). While mAb titer appeared to have some dependence on the selected codon, repeat transfections showed codon-dependent expression varied (i.e. Ala-GCA was not always the highest expressing codon); the average of all Ala codon variants was always a minimum two-fold higher than the average of all Gly variants (data not shown). This variation in preferred codon for expression may be a reflection of the relative tRNA levels available in the cell at the time of translation. When the secreted mAb titer was compared to HC and LC mRNA levels, no apparent relationship was observed. Assuming the Ala and Gly variant mRNA was degraded at a similar rate (preliminary mRNA half-life experiments support this) and relative mAb accumulation remains constant over the culture lifetime, then transcription-level events were unlikely to be responsible for the observed differential transient expression.
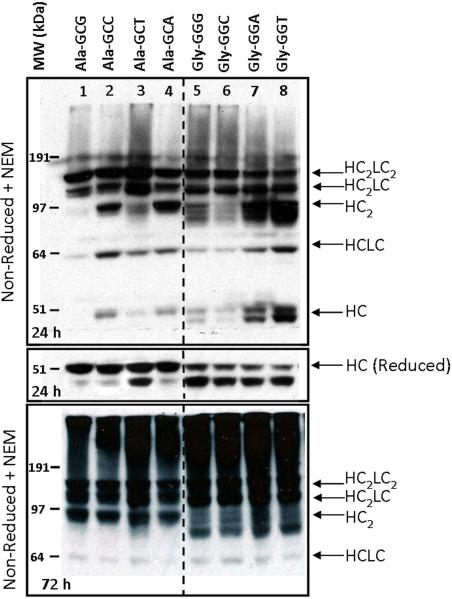
For the full-length mAb to be secreted from the cell, first HC dimers must form followed by association with two folded LCs.13 If this fails to happen, the UPR may be engaged to assist in the folding/assembly of the mAb or the aberrant protein will be targeted for degradation. Accumulation of intracellular mAb intermediates may identify sequence-related complications with protein folding/assembly which lead to failure of the full-length mAb to be secreted from the cell. Intracellular mAb formation was qualitatively analyzed by comparing the relative fractions of HC-related mAb intermediates in equivalent loading of lysate from the Ala and Gly transfectants. Cell lysates were probed with an anti-human Fcγ antibody 24 and 72 h after transfection with the 277 Ala or Gly codon variants (Figure 2). The 24 h non-reduced blot (Figure 2, top panel) shows that a fraction of intracellular HC was present as mAb HCLC, HC2, and HC2LC intermediates. For all Ala codon variants (lanes 1–4), the majority of the HC appeared to be involved in late-stage intermediates. The Gly codon variants (lanes 5–8) showed different intracellular HC behavior, including triplet banding in the HC dimer region and a doublet around the free HC. Reduction of all samples resulted in all bands converging to two distinct HC-related bands around 45–50 kDa (Figure 2, middle panel). The Ala variants were enriched in the top band, whereas the Gly variants had a stronger lower band. A comparison of the reduced intracellular lysate to reduced secreted mAb showed only the upper HC band was secreted in association with the LC (data not shown). Treatment with endoglycosidases PNGase F and EndoH resulted in a slight downward shift of both bands (data not shown) indicating that all the intracellular HC was glycosylated, and the smaller HC-related band was not simply a differentially or aglycosylated form. N-terminal sequencing was attempted on both HC bands, but was only successful with the larger, full-length HC band. Sequencing of the smaller band failed due to rapid aggregation.
Figure 2.
Western blot of HC-related proteins in lysates from transient transfectants expressing mAb 277 Ala and Gly variants.
HC-related proteins were probed with an HRP-conjugated α-human Fcγ F(ab')2. Top panel: Non-reduced NEM-treated transient lysates 24 h after transfection. Middle panel: Reduced transient lysates 24 h after transfection. Bottom Panel: Non-reduced transient lysates 72 h after transfection. The bands corresponding to HC (50 kDa), HCLC (75 kDa), HC2 (100 kDa), HC2LC (125 kDa), and HC2LC2 (150 kDa) are labeled.
A repeat of the western blot analysis 72 h after transfection indicated little to no free HC and an increase in the relative amounts of late-stage intermediates and high-order aggregates in all samples (Figure 2, bottom panel). The relative amounts of non-secreted intermediates, high-order aggregates, and further degradation of the HC2 bands were markedly increased in the Gly codon variants compared to the Ala variants. The decrease in secreted transient mAb titers is likely attributed to the increased levels of HC dimer degradation and prevalence of the non-secreted HC-related band in the Gly variant western blots.
Stable Expression
Transient expression is often used to compare relative secreted titers of various antibody grafts before engaging in the time consuming process of generating a stable cell line. However, it is not clear if the cellular mechanisms responsible for differential expression in both transient and stable systems are the same. If the limiting mechanisms stem from the same processes, then this would imply transient behavior can be predictive of stable behavior. To address this issue, stable cell lines were established for the 277 Ala (GCC) and Gly (GGC) sequence variants and their differential expression behavior was evaluated.
The stable expression of clones capable of growing in selective media was widely varied and reached ~160 mg/L by day 6 for the highest expressing Ala clones. In contrast, the highest Gly clones only achieved maximum titers of ~1.7 mg/L. This 80-fold difference in final titer was also reflected in the specific productivities of the best Ala and Gly clones (see Table 2). Therefore the differential expression of the Ala and Gly sequence variants observed in transient expression was also observed in stable cell lines, but the difference was enhanced nearly 20-fold.
Table 2.
Specific productivities of stable Ala and Gly clones isolated by the ClonePix system
| Bulk Construct | mAb Titers (μg/mL)† | qmAb (pg/cell/day) |
|---|---|---|
| Ala-6 | 56.383 | 9.53 |
| Ala-65 | 42.535 | 5.62 |
| Ala-115 | 35.868 | 4.72 |
| Ala-123 | 15.924 | 3.45 |
| Ala-138* | 158.555 | 23.92 |
| Ala-187* | 132.995 | 23.23 |
|
| ||
| Gly-26* | 1.723 | 0.36 |
| Gly-55 | 0.974 | 0.22 |
| Gly-71 | 1.027 | 0.28 |
| Gly-75* | 0.876 | 0.27 |
| Gly-80 | 1.026 | 0.22 |
| Gly-81 | 0.915 | 0.31 |
Antibody expression measured on day 6.
Highest expressing clones for each variant. These clones were further analyzed as described in the text.
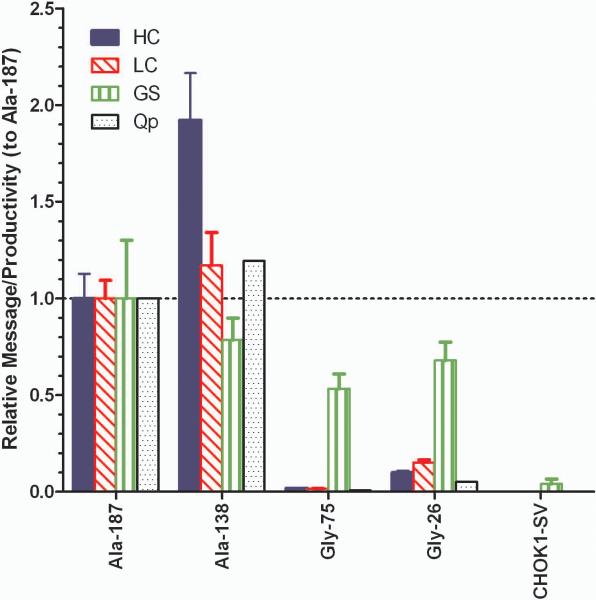
The relationship between antibody expression and message level was determined through analysis of samples in mid-exponential growth phase. Figure 3 shows the message levels of the two highest expressing Ala (Ala-187, Ala-138) and Gly clones (Gly-75, Gly-26) and the parental CHOK1-SV control relative to Ala-187. The highest expressing cell line, Ala-138, appeared to have slightly more HC message than Ala-187, but the difference was less than 2-fold. The Gly samples contain much less HC and LC message; Gly-75 message expression was two orders of magnitude less than the Ala constructs, which mimics the observed difference in secreted antibody titer. The GS message exhibited less than 2-fold variation between all mAb constructs, but was an order of magnitude higher than the endogenous GS expression exhibited by the CHOK1-SV control. Endogenous GS in CHO cells is thought to be inhibited by 3 μM MSX.16 The adaptation of the stable clones to 50 μM MSX appears to be reflected in the order of magnitude increase in GS mRNA and is independent of HC and LC gene expression despite the GS, HC, and LC genes being located on the same vector. However, GS mRNA in the host cell line may be underestimated due to lack of MSX in the culture media which may induce endogenous GS expression. If HC and LC message are compared to specific productivity (Qp) rather than titer (thus accounting for the difference in growth rate), the productivity tracks the mRNA levels well (Figure 3), suggesting that the amount of available translatable mRNA was primarily responsible for differential expression in mAb 277 stable cell lines.
Figure 3.
Message expression and specific productivity (Qp) in mAb 277 Ala and Gly stable cell lines. Message levels for HC, LC, and glutamine synthetase (GS) were measured by qPCR from batch cultures on day 4. The message levels of each gene were normalized to the amount present in the Ala-187 sample and error bars are ±1 standard deviation (n=3).
To ensure that the same number of copies of each gene was successfully integrated into the host genome, the approximate gene copy number (GCN) was measured by qPCR. The samples were compared to a standard curve containing known amounts of linearized plasmid diluted in CHOK1-SV genomic DNA to eliminate matrix effects. The Ala clones contained a single copy and Gly clones contained 3–4 copies of the HC and LC (data not shown). All clones contained a single GS copy. No significant gene amplification (<10 copies), which can lead to genetic instability, was observed in any clones. When the relative GCN data was compared to the relative message expression, no correlation was observed. Assuming mRNA degradation rates were similar in all cell lines (preliminary mRNA half-life results suggest this is true), then transcription, or more accurately the amount of mRNA available for translation, still appears to be the limiting event in stable cell line expression.
In addition to correlating intracellular HC and LC message to secreted mAb, the intracellular protein behavior needs to be compared between the Ala and Gly constructs. The highest expressing Ala and Gly cell lines (Ala-138 and Gly-26) were again used for comparison. Supernatants and lysates were sampled from triplicate cultures in exponential growth phase (day four) and the resulting protein levels are compared in Table 3. The crude lysates were analyzed using the Octet instrument fitted with protein A biosensors to measure the amount of intracellular IgG in each cell line. Lysate from the non-expressing parental CHOK1-SV cell line was also included as a control to subtract out any background binding. The total protein concentration was measured by BCA assay which can be used to calculate the total protein content per cell (assuming complete cell lysis). The protein content of mammalian cells is cell line dependent, but is on average about 18–20% of the total cell weight (~300–500 pg/cell).17 After subtracting the background signal from host cell protein (HCP) binding in the parental CHOK1-SV sample, 2.7% of the total protein content in Ala-138 was IgG. By contrast only 0.2% of the Gly-26 protein was IgG. This equates to a 14-fold difference in intracellular protein expression and is comparable to the 20-fold difference in mRNA expression shown in Figure 3a. While it is possible that translation is slightly slower in the Gly-26 construct, it does not appear to be the main contributor to the observed differential stable expression. However, when the supernatant IgG concentrations were compared, there was a 48-fold difference in secreted mAb titer between the Ala-138 and Gly-26 constructs. This suggests there is a greater proportion of non-secreted intracellular mAb in the Gly-26 construct than in the Ala-138 construct.
Table 3.
Intracellular versus extracellular protein levels in the highest expressing Ala and Gly stable cell lines
| Construct | Total Protein/Cell (pg/cell) | IgG/Cell (pg/cell)a | % IgG/Cell | Supernatant IgG (μg/mL)b |
|---|---|---|---|---|
| Ala-138 | 263.2 | 7.1 | 2.7% | 181.0 ± 4.6 |
| Gly-26 | 293.1 | 0.6 | 0.2% | 3.8 ± 0.07 |
|
| ||||
| Ala/Gly | 0.9 | 11.8 | 13.5 | 47.6 |
Error is ±1 standard deviation of biological replicates (n=3)
Value is corrected for background binding onto the Protein A Biosensors and was subtracted before determining the % intracellular IgG per cell.
IgG titer in supernatant on day of sampling (day 4)
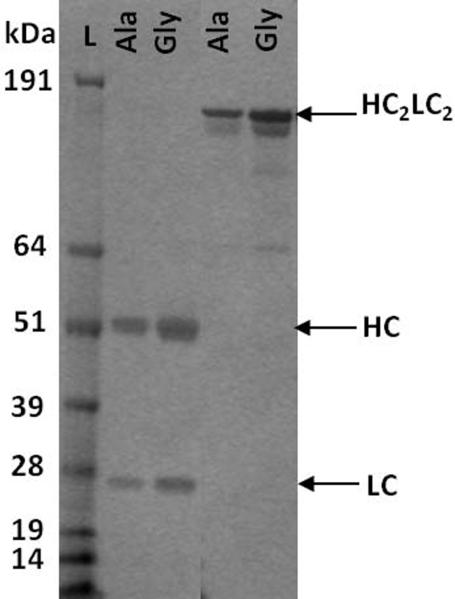
To address the amount and nature of the non-secreted intracellular mAbs, western blots were performed on non-reduced lysates obtained from the stable Ala-187, Ala-138, Gly-75, and Gly-26 clones in mid-exponential growth, using the same method as the transient lysates. Identical banding patterns to the transient transfections, including the doublet HC bands, were observed in all samples (data not shown). PNGase F and Endo H treatment of the reduced lysates gave identical results to the transient samples with both HC-related bands migrating further down the gel. In addition, antibodies and other HC containing species were purified from cell lysates using protein A chromatography. Western blots performed on the eluted fractions (reduced) from the Ala and Gly lysate purification (Figure 4a) show that both HC-related bands can be isolated during protein A purification. This implies that the lower HC-related band contained an intact protein A binding site within the Fc domain in addition to the binding sites for the α-human Fcγ antibody used for the western blots. This supports our hypothesis that the multiple banding of HC-related product in the Gly samples, and to a lesser extent in the Ala samples, may be indicative of a cleavage or premature truncation process resulting in differential migration in the gel. When the intracellular western blots were compared to the secreted supernatants from the stable cell lines, it was again observed that the lower-HC related band was not secreted outside the cell (Figure 4b). The ability of this band to bind to protein A but not engage in full-length mAb assembly may account for the greater proportion of non-secreted mAb in the Gly-26 lysates (Table 3).
Figure 4.
Analysis of purified mAb 277 Ala and Gly stable lysates and supernatants
(A) The eluted fractions (lanes 1–6, in the order they were eluted from the column from first (1) to last (6)) from protein A purified Ala-138 (top panel) and Gly-26 (bottom panel) lysates were probed using the same α-human Fcγ antibody under reducing conditions. The top band for the Ala-138 and Gly-26 panels is ~50 kDa. Lane 7 is the crude lysate. (B) SDS-PAGE of the reduced (left) and non-reduced (right) purified supernatant shows that none of the low molecular weight HC band was secreted.
Biophysical Characterization
The purified mAb 277 variants were analyzed by biomolecular interaction analysis (BIA) to determine the binding kinetics with antigen (Table 4). Purified 277 wild-type (Ala variant) reference standard was included in the analysis as a control. The ka values indicated strong binding which were consistent between samples and within the working range of the instrument. This was complemented by small kd values, indicating very slow dissociation. The equilibrium dissociation rate, KD, showed all the samples exhibit low pM binding and were in good agreement with the 277 Ala control samples. The close agreement of the Ala and Gly affinity confirms the initial observations that the variants exhibit equivalent antigen-binding properties despite exhibiting different expression properties.
Table 4.
Antigen affinities of the mAb 277 Ala and Gly variants determined by BIA analysis
| Sample | ka (1/Ms) | kd (1/s) | KD (pM) |
|---|---|---|---|
| 277 Ala Std | 1.93E+06 | 1.42E−05 | 7.35 |
| Ala-138 | 1.88E+06 | 1.60E−05 | 8.49 |
| Gly-26 | 2.42E+06 | 1.81E−05 | 7.47 |
| 277 Ala Std | 1.85E+06 | 1.70E−05 | 9.22 |
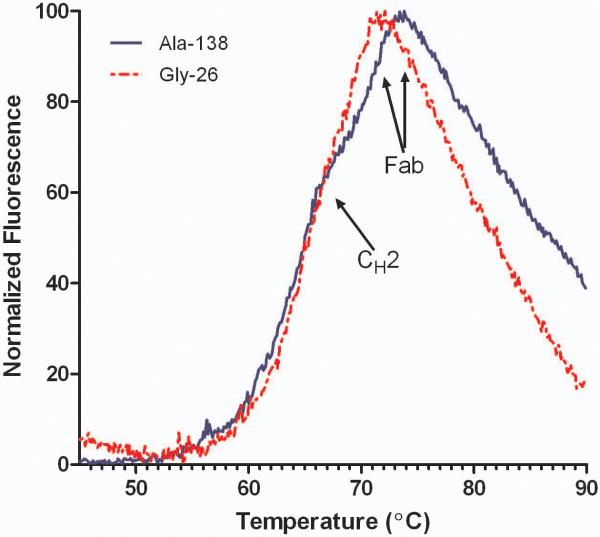
Antibody stability was assessed using differential scanning fluorimetry (DSF). This technique can be used to monitor protein unfolding as a function of temperature. Instead of measuring the heat flux as in traditional calorimetric methods, DSF measures the fluorescence of an environmentally sensitive dye (SYPRO Orange) that binds to hydrophobic areas of protein.18 For IgG4 antibodies, 65°C is the expected melting temperature (Tm) for a glycosylated Fc region. Only one transition is observed due to cooperative unfolding of the CH2 and CH3 domains.19 Decoupling of the Fc is achieved by deglycosylation which results in an approximate Tm of 61°C for the aglycosylated CH2 domain and 68°C for the CH3 domain. Humanized Fab domains exhibit a wide range of Tms, typically between 60–80°C.20 The differences in Fab stability are attributed to the varied intrinsic stability of the parental germline sequence and interactions within the HC and LC interface.21 As seen in Figure 5, the DSF traces showed two transitions; the shoulder on the peak is the unfolding of the CH2/CH3 domains around 65°C while the more intense Fab unfolding occurred at ~70°C. The Ala and Gly variants exhibited similar CH2/CH3 melting, but the Fab Tm's differed by ~2°C. This suggests that the substitution of the Gly residue at position 49 had a destabilizing effect on the Fab domain.
Figure 5.
Stability of purified mAb 277 Ala and Gly variants
Differential scanning fluorimetry (DSF) traces of purified Ala-138 and Gly-26. Each trace is the mean of four technical replicates. The transitions of the CH2 and Fab unfolding are labeled.
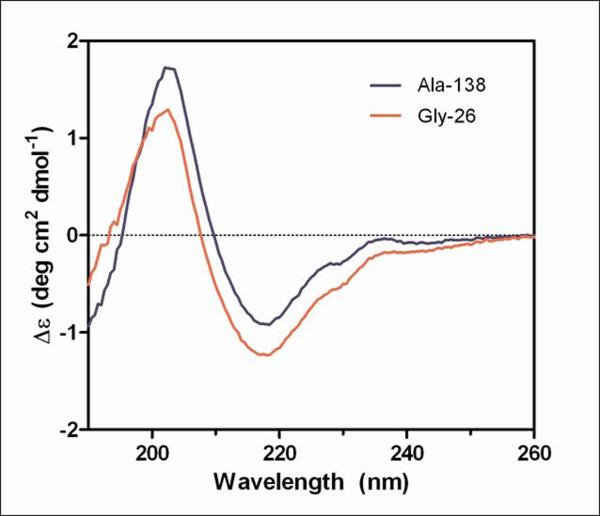
The secondary structure of the purified Ala-138 and Gly-26 antibodies was compared using far-UV CD. All spectra exhibited characteristic profiles typical of high β-sheet content which consist of a negative band at ~217 nm and a positive band at~200 nm (Figure 6a).22–24 A small shoulder at 230 nm was also present in all spectra and was attributed to contributions from aromatic side chain asymmetry. The downward shift in the Gly spectrum compared to the Ala spectrum can be ascribed to an increased fraction of α-helical content in the Gly sample. The signal from helical structures is much stronger than signal from β-sheets, and therefore, subtle increases in helical content can dominate the spectrum, resulting in difficulty estimating relative proportions of secondary structure.24 Thus, the differences in the secondary structure may be subtle and further structural characterization would be required to determine if the Gly mutation at position 49 induces formation of α-helix in the VH domain.
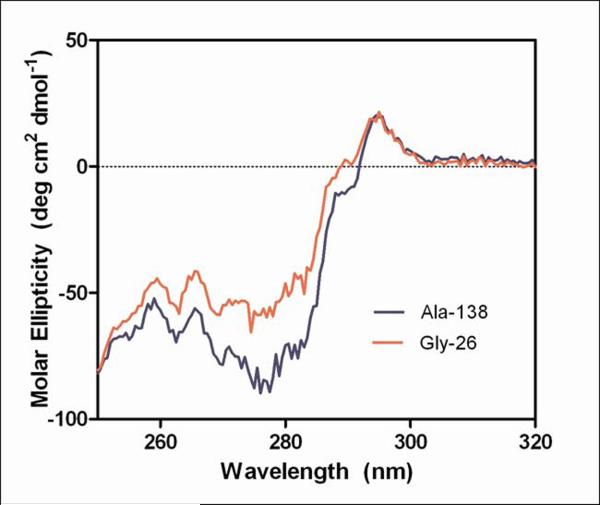
Figure 6.
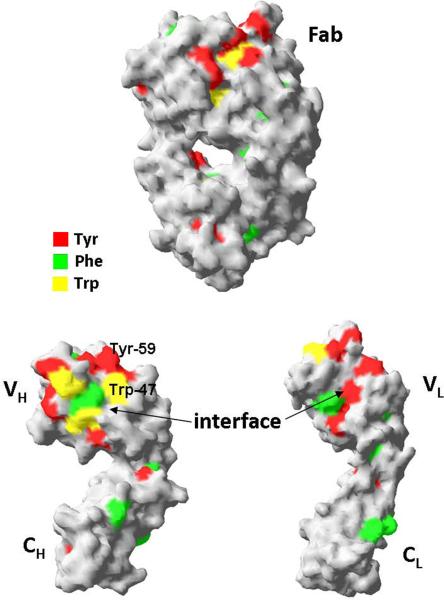
Circular dichroism measurements of purified mAb 277 Ala and Gly variants.
Far-UV (A) and near-UV (B) CD spectra of purified mAb from Ala-138 and Gly-26 stable cell lines. Each trace is the average of three acquisitions and has had the 20 mM phosphate buffer, pH 8 buffer signal subtracted. The Tyr, Phe, and Trp residues in the 277 Fab which contribute to the near-UV spectrum are highlighted in the crystal structure shown in (C).
The near UV CD spectra, which were used to compare the tertiary conformation of the samples, indicated that the Ala and Gly mAbs differ from each other (Figure 6b). The most noticeable deviations in the spectra occurred between 270–295 nm, which is mainly comprised of Tyr (270–290 nm) and Trp (280–300 nm) signals.23 Preliminary analysis of the Fab domain crystal structure (unpublished results) indicated the majority of these residues are either buried or present at the HC/LC interface (Figure 6c). The amino acid occupying Kabat position 49 potentially interacts with, or impacts the conformation of, two Trp and two Tyr residues on the VH interface. The side chain of Ala interacts with the buried Trp-36 and Tyr-60 residues whereas the Trp-47 and Tyr-59 reside on the surface. Trp-47 is a highly conserved residue that directly interacts with the VL interface.25 Replacing Ala with Gly may eliminate several interactions within the hydrophobic core of the VH domain which could result in the observed alternate conformation and the decreased stability of the Fab domain.
Taking all the biophysical data into consideration, it is apparent that the Ala and Gly mAb secreted from the cell are correctly assembled and fully functional, suggesting the secreted mAbs passed the “quality control” checkpoints in the cell.13 The differences in stability and conformation imply position 49 may have an important role in folding and assembly of the VH domain. The sequence analysis of mAb 277 shows that position 49 in the VH domain is typically occupied by small amino acids and is surrounded by hydrophobic amino acids which are critical to both core and interface stability. Previous studies on the stability of various VH domains identified position 49 as a key residue that participates in formation of the hydrophobic core as well as influences the integrity of the VH/VL, interface.25,26 The crystal structure and homology models of the VH domains showed that the Ala-49 side chain participates in several hydrophobic interactions within the β-barrel core which are lost when a Gly is substituted at this position. One could envision two scenarios which arise from the loss of these hydrophobic interactions. In the first scenario, the Gly VH domain may compensate for the loss in stability by adopting an alternate conformation which “rescues” the stability of the hydrophobic core. However, this may alter the orientation of the key VH interface residues and prove detrimental to formation of the VH/VL dimer. This would be damaging to expression because the HC and LC must correctly assemble for mAb to secrete from the cell. In the second scenario, the Gly variant adopts a conformation similar to the Ala variant, which maintains the VH/VL contacts, but at the expense of VH stability. The competition between these conformations may explain the reduced expression of the Gly which coincides with the increased presence of the non-secreted HC-related band in the intracellular western blots. If the VH domain folding exhibits a degree of dependence on VL association, then any disruption to the interface could be detrimental to expression due to intracellular mAb components being retained in the cell's quality control systems.
Conclusions
Understanding the effect of sequence changes on antibody folding, stability, and expression is vital to developing effective, safe, and affordable therapeutic candidates. Antibodies typically undergo the highest degree of sequence engineering during the humanization process in which immunogenic murine residues are replaced with human equivalents. Published grafting strategies suggest that antigen-contacting, conformationally important (Pro, Gly), upper-core, and highly conserved VL/VH residues should be maintained in order to retain antigen affinity.27 The Ala to Gly mutation at Kabat position 49 is present in several germline sequences within the VH3 family and is present in >50% of all germline sequences. Therefore the traditional grafting strategies would suggest that Gly be introduced at position 49, but this resulted decreased expression and stability of mAb 277. In this study, we observed that the mutation from an Ala to a Gly residue at Kabat position 49 in the VH domain resulted in decreased expression but no loss of antigen affinity. The decreased expression was observed in both transient and stable expression systems. We further observed that antibody from the Gly variant was less stable than antibody containing the Ala residue. It was proposed that protein folding and assembly issues were responsible for the observed differential expression. More so, protein folding/assembly limitations presented at the transient level appear to be related to the transcriptional limitations in stable cell lines. In the case of the Gly constructs, transient and stable lysates exhibit a larger fraction of non-secreted HC-related protein than the correct HC form. It is not clear how this truncated version arises and whether the presence of this truncated version is related to the decrease in stability of the secreted antibody, though one could envision mechanisms by which they could be related (e.g. intracellular clipping of slowly folding HC). Accumulation of the misfolded or truncated version of the HC could be cytotoxic and lead to apoptosis. Upon stable transgene integration, only Gly cell lines which exhibit decreased HC expression, reduced to a level where the cell can accommodate the misfolded protein load, are able to survive. One could postulate that the Ala variant, which contains only a small proportion of the non-secreted HC product, can express higher levels of nascent HC before potentially toxic accumulation of the non-secreted HC component. This suggests that both transient and stable systems are limited by antibody folding and assembly, but the comparatively long evolutionary time lines and selective pressures used in stable systems allow only those Gly cell lines with reduced transgene mRNA levels to propagate. Further work is needed to evaluate if position 49 is critical to expression in other HC/LC antibody pairings and to identify if there are other potential sites where non-germline preferred residues result in enhanced expression.
Acknowledgements
The authors would like to thank Lonza Biologics for permission to use the GS Expression System. Funding for this project was provided by a NSF GOALI grant (CBET-0854099); Megan Mason was supported in part by a National Institutes of Health Training Grant (T32GM067545). This paper is dedicated to Howard P. Isermann on the occasion of his 90th birthday in appreciation for his support for undergraduate and graduate education in Chemical and Biological Engineering at RPI.
References
- 1.Jayapal KR, Wlaschin KF, Hu WS, Yap MGS. Recombinant protein therapeutics from CHO cells - 20 years and counting. Chem Eng Prog. 2007;103:40–47. [Google Scholar]
- 2.Ziegelbauer K, Light DR. Monoclonal antibody therapeutics: Leading companies to maximise sales and market share. J Commer Biotechnol. 2007;14:65–72. [Google Scholar]
- 3.Barnes LM, Bentley CM, Dickson AJ. Molecular definition of predictive indicators of stable protein expression in recombinant NS0 myeloma cells. Biotechnol Bioeng. 2004;85:115–21. doi: 10.1002/bit.10893. [DOI] [PubMed] [Google Scholar]
- 4.Barnes LM, Bentley CM, Moy N, Dickson AJ. Molecular analysis of successful cell line selection in transfected GS-NS0 myeloma cells. Biotechnol Bioeng. 2007;96:337–48. doi: 10.1002/bit.21119. [DOI] [PubMed] [Google Scholar]
- 5.Chusainow J, Yang YS, Yeo JH, Toh PC, Asvadi P, Wong NS, Yap MG. A study of monoclonal antibody-producing CHO cell lines: what makes a stable high producer? Biotechnol Bioeng. 2009;102:1182–96. doi: 10.1002/bit.22158. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Z, Huang Y, Sharfstein ST. Regulation of recombinant monoclonal antibody production in Chinese hamster ovary cells: a comparative study of gene copy number, mRNA level, and protein expression. Biotechnology Progress. 2006;22:313–8. doi: 10.1021/bp0501524. [DOI] [PubMed] [Google Scholar]
- 7.Lee CJ, Seth G, Tsukuda J, Hamilton RW. A Clone Screening Method Using mRNA Levels to Determine Specific Productivity and Product Quality for Monoclonal Antibodies. Biotechnol Bioeng. 2009;102:1107–1118. doi: 10.1002/bit.22126. [DOI] [PubMed] [Google Scholar]
- 8.Mead EJ, Chiverton LM, Smales CM, von der Haar T. Identification of the Limitations on Recombinant Gene Expression in CHO Cell Lines With Varying Luciferase Production Rates. Biotechnol Bioeng. 2009;102:1593–1602. doi: 10.1002/bit.22201. [DOI] [PubMed] [Google Scholar]
- 9.O'Callaghan PM, McLeod J, Pybus LP, Lovelady CS, Wilkinson SJ, Racher AJ, Porter A, James DC. Cell Line-Specific Control of Recombinant Monoclonal Antibody Production by CHO Cells. Biotechnol Bioeng. 2010;106:938–951. doi: 10.1002/bit.22769. [DOI] [PubMed] [Google Scholar]
- 10.Reisinger H, Steinfellner W, Stern B, Katinger H, Kunert R. The absence of effect of gene copy number and mRNA level on the amount of mAb secretion from mammalian cells. Appl Microbiol Biotechnol. 2008;81:701–710. doi: 10.1007/s00253-008-1701-1. [DOI] [PubMed] [Google Scholar]
- 11.Barnes LM, Dickson AJ. Mammalian cell factories for efficient and stable protein expression. Curr Opin Biotechnol. 2006;17:381–386. doi: 10.1016/j.copbio.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Dinnis DM, James DC. Engineering mammalian cell factories for improved recombinant monoclonal antibody production: lessons from nature? Biotechnol Bioeng. 2005;91:180–9. doi: 10.1002/bit.20499. [DOI] [PubMed] [Google Scholar]
- 13.Feige MJ, Hendershot LM, Buchner J. How antibodies fold. Trends Biochem Sci. 2010;35:189–198. doi: 10.1016/j.tibs.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–89. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 15.Duan J, Antezana M. Mammalian Mutation Pressure, Synonymous Codon Choice, and mRNA Degradation. J Mol Evol. 2003;57:694–701. doi: 10.1007/s00239-003-2519-1. [DOI] [PubMed] [Google Scholar]
- 16.Birch JR, Racher AJ. Antibody production. Adv Drug Del Rev. 2006;58:671–85. doi: 10.1016/j.addr.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Brown TA. Genomes 3. Garland Science Publishing; New York, NY: 2007. [Google Scholar]
- 18.Senisterra GA, Finerty PJ., Jr. High throughput methods of assessing protein stability and aggregation. Mol Biosyst. 2009;5:217–23. doi: 10.1039/b814377c. [DOI] [PubMed] [Google Scholar]
- 19.Ghirlando R, Lund J, Goodall M, Jefferis R. Glycosylation of human IgG-Fc: influences on structure revealed by differential scanning micro-calorimetry. Immunol Lett. 1999;68:47–52. doi: 10.1016/s0165-2478(99)00029-2. [DOI] [PubMed] [Google Scholar]
- 20.Garber E, Demarest SJ. A broad range of Fab stabilities within a host of therapeutic IgGs. Biochem Biophys Res Commun. 2007;355:751–7. doi: 10.1016/j.bbrc.2007.02.042. [DOI] [PubMed] [Google Scholar]
- 21.Rothlisberger D, Honegger A, Pluckthun A. Domain interactions in the Fab fragment: A comparative evaluation of the single-chain Fv and Fab format engineered with variable domains of different stability. J Mol Biol. 2005;347:773–789. doi: 10.1016/j.jmb.2005.01.053. [DOI] [PubMed] [Google Scholar]
- 22.Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nat Protoc. 2006;1:2876–90. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Bba-Proteins Proteom. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Sreerama N, Woody RW. Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 2004;383:318–51. doi: 10.1016/S0076-6879(04)83013-1. [DOI] [PubMed] [Google Scholar]
- 25.Wang N, Smith WF, Miller BR, Aivazian D, Lugovskoy AA, Reff ME, Glaser SM, Croner LJ, Demarest SJ. Conserved amino acid networks involved in antibody variable domain interactions. Proteins. 2009;76:99–114. doi: 10.1002/prot.22319. [DOI] [PubMed] [Google Scholar]
- 26.Honegger A, Malebranche AD, Rothlisberger D, Pluckthun A. The influence of the framework core residues on the biophysical properties of immunoglobulin heavy chain variable domains. Protein Eng Des Sel. 2009;22:121–134. doi: 10.1093/protein/gzn077. [DOI] [PubMed] [Google Scholar]
- 27.Honegger A. Engineering antibodies for stability and efficient folding. In: Chernajovsky Y, Nissim A, editors. Therapeutic Antibodies. Vol. 181. Springer; 2008. pp. 47–68. [DOI] [PubMed] [Google Scholar]