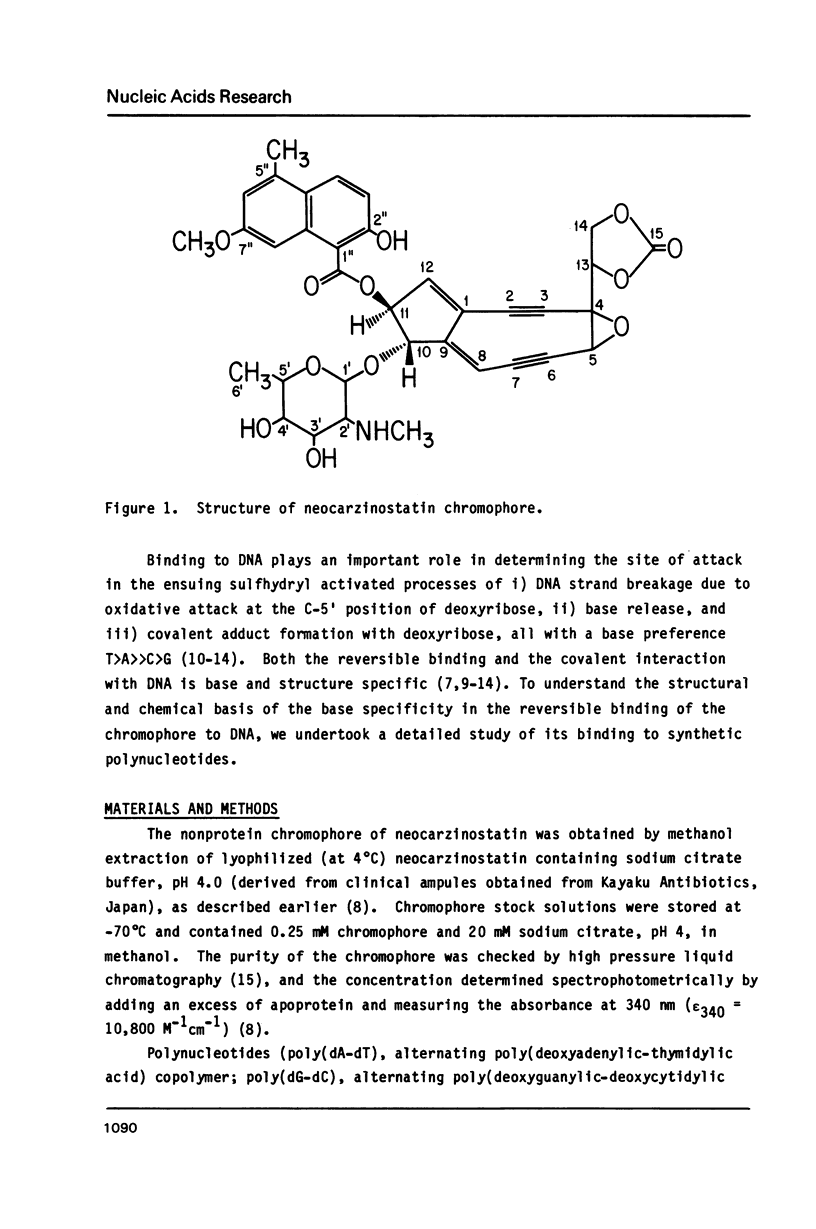
Abstract
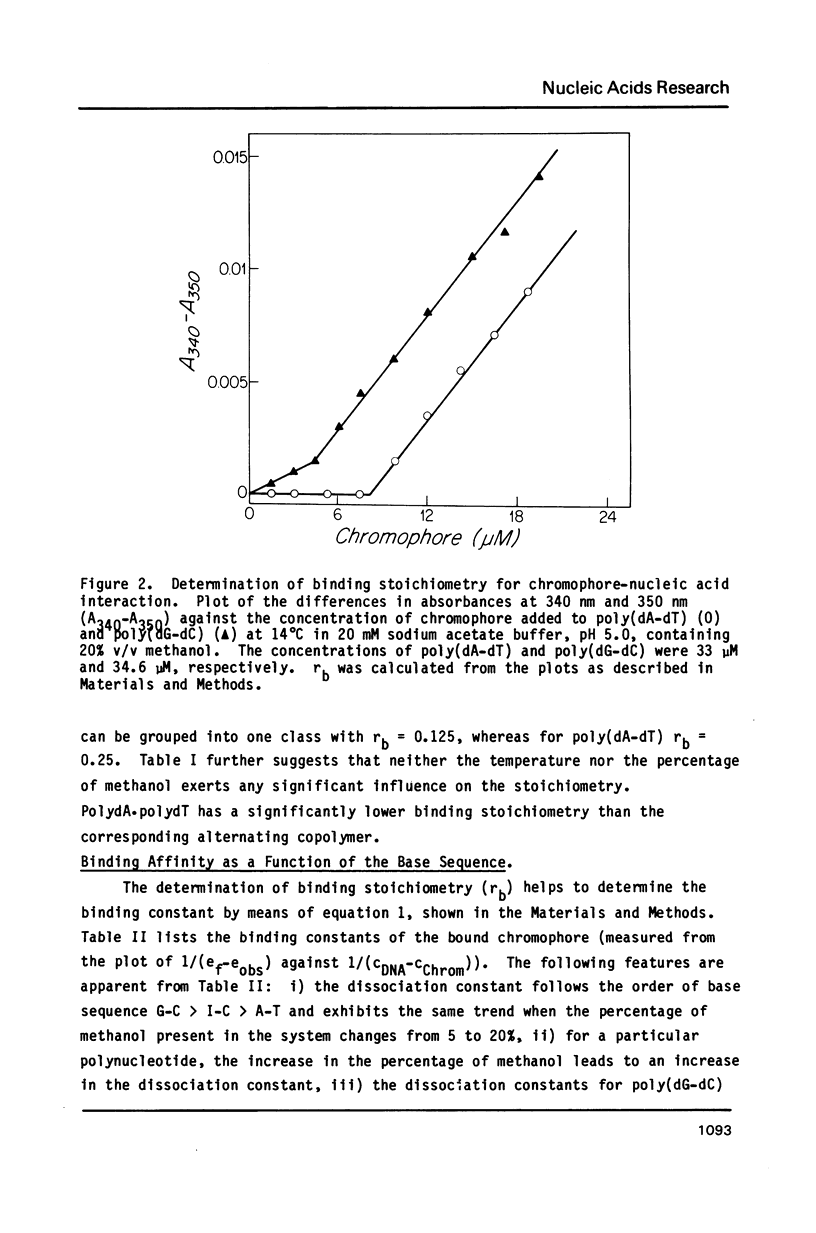
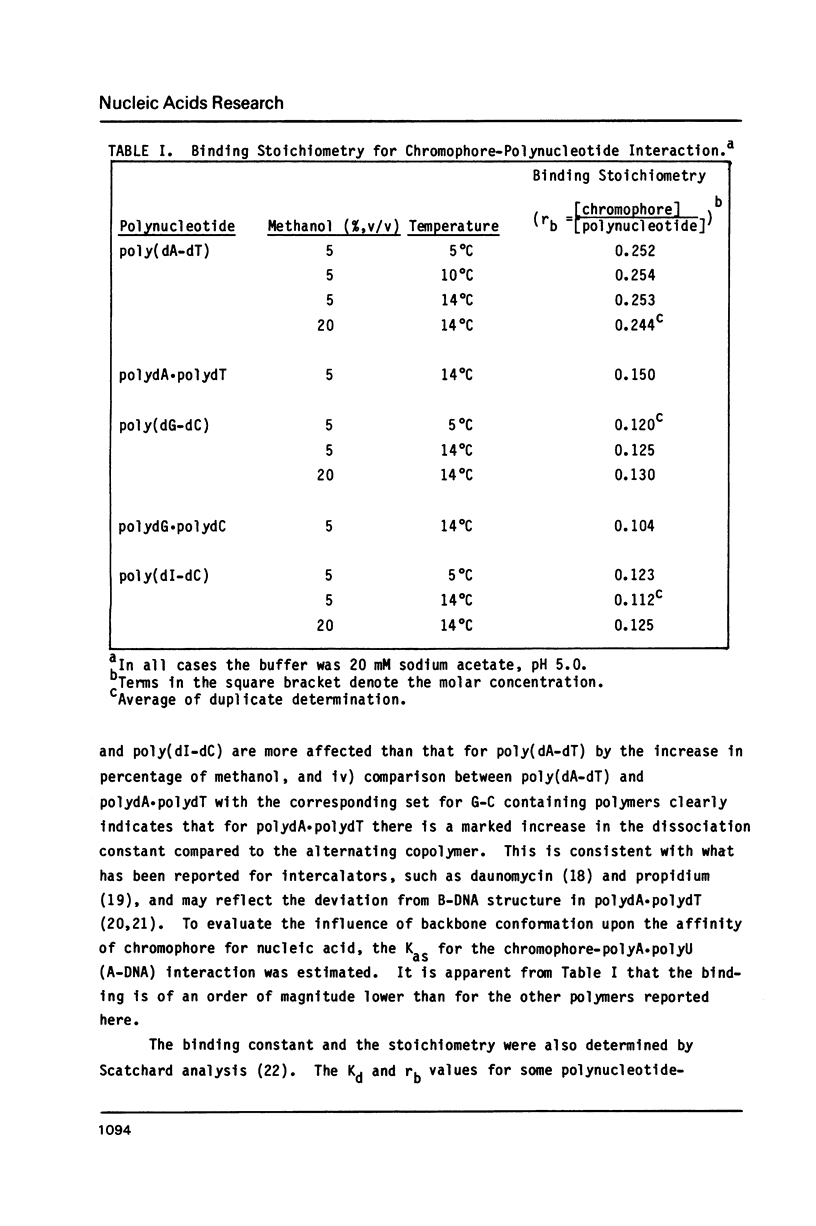
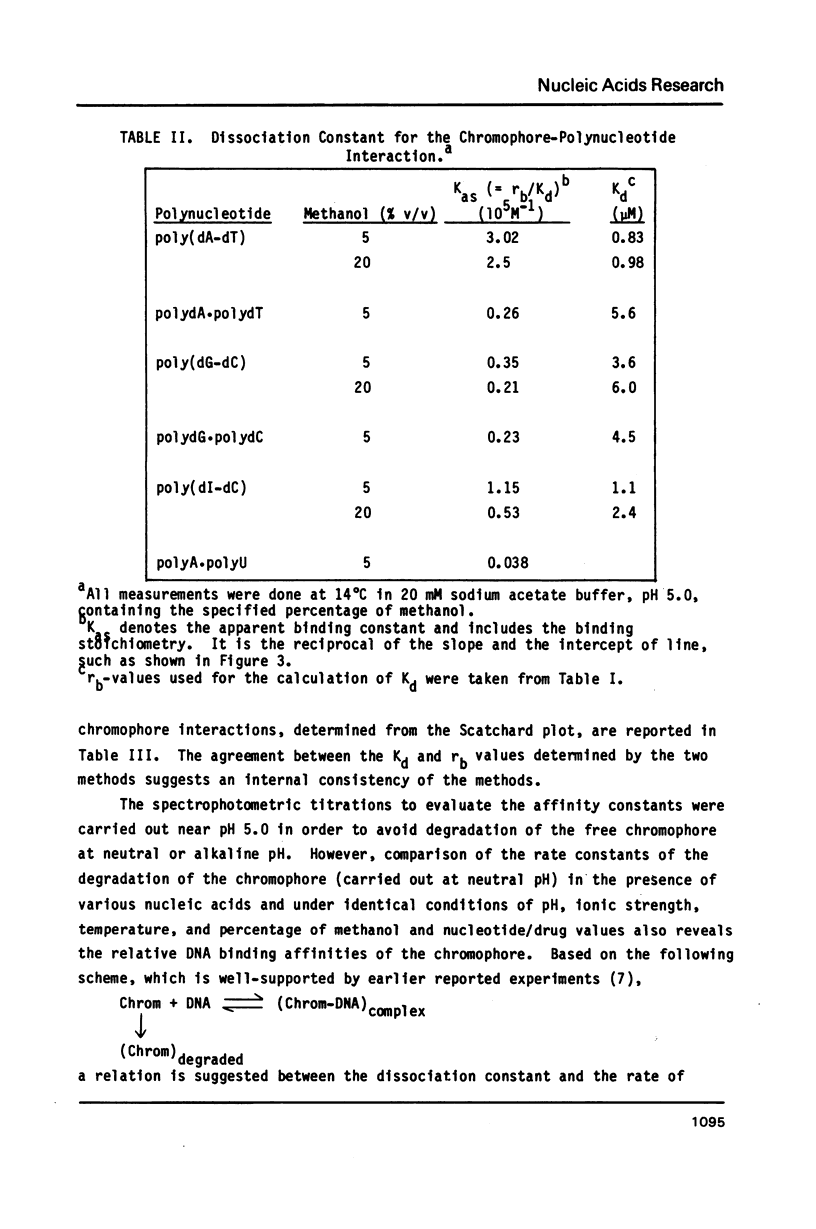
The reversible binding of neocarzinostatin chromophore to polynucleotides was studied in order to understand the molecular basis of its base sequence-specificity in DNA damage production. Studies of the spectroscopic and thermodynamic properties of chromophore-polynucleotide interactions reveal that the binding of the chromophore to poly(dA-dT) is qualitatively and quantitatively different from that to poly(dG-dC) (and poly(dI-dC]. From these and other experiments using double-stranded mixtures of homopolynucleotides, it is proposed that the observed A T specific intercalation might result from differential recognition of minor variations in the B-DNA type structure by the chromophore.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aktipis S., Martz W. W. Circular dichroism and temperature--optical density studies on the conformation of polynucleotide--ethidium bromide complexes. Biochemistry. 1974 Jan 1;13(1):112–118. doi: 10.1021/bi00698a018. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Hall I. H., Puigjaner L. C. Heteronomous DNA. Nucleic Acids Res. 1983 Jun 25;11(12):4141–4155. doi: 10.1093/nar/11.12.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assa-Munt N., Granot J., Behling R. W., Kearns D. R. 1H NMR relaxation studies of the hydrogen-bonded imino protons of poly(dA-dT). Biochemistry. 1984 Feb 28;23(5):944–955. doi: 10.1021/bi00300a023. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Elgin S. C. Analysis of chromatin structure and DNA sequence organization: use of the 1,10-phenanthroline-cuprous complex. Nucleic Acids Res. 1982 Oct 11;10(19):5835–5852. doi: 10.1093/nar/10.19.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaires J. B. Equilibrium studies on the interaction of daunomycin with deoxypolynucleotides. Biochemistry. 1983 Aug 30;22(18):4204–4211. doi: 10.1021/bi00287a007. [DOI] [PubMed] [Google Scholar]
- Dahl K. S., Pardi A., Tinoco I., Jr Structural effects on the circular dichroism of ethidium ion-nucleic acid complexes. Biochemistry. 1982 May 25;21(11):2730–2737. doi: 10.1021/bi00540a024. [DOI] [PubMed] [Google Scholar]
- Dalgleish D. G., Peacocke A. R. The circular dichroism in the ultraviolet of aminoacridines and ethidium bromide bound to DNA. Biopolymers. 1971 Oct;10(10):1853–1863. doi: 10.1002/bip.360101008. [DOI] [PubMed] [Google Scholar]
- Dasgupta D., Goldberg I. H. Mode of reversible binding of neocarzinostatin chromophore to DNA: evidence for binding via the minor groove. Biochemistry. 1985 Nov 19;24(24):6913–6920. doi: 10.1021/bi00345a025. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R., Conner B. N., Wing R. M., Fratini A. V., Kopka M. L. The anatomy of A-, B-, and Z-DNA. Science. 1982 Apr 30;216(4545):475–485. doi: 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Lomonossoff G. P., Laskey R. A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Wing R. M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G., Sarma M. H., Sarma R. H. Structure and dynamics of netropsin-poly(dA-dT).poly(dA-dT) complex: 500 MHz 1H NMR studies. J Biomol Struct Dyn. 1984 Jun;1(6):1457–1472. doi: 10.1080/07391102.1984.10507530. [DOI] [PubMed] [Google Scholar]
- Hatayama T., Goldberg I. H., Takeshita M., Grollman A. P. Nucleotide specificity in DNA scission by neocarzinostatin. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3603–3607. doi: 10.1073/pnas.75.8.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensens O. D., Dewey R. S., Liesch J. M., Napier M. A., Reamer R. A., Smith J. L., Albers-Schönberg G., Goldberg I. H. Neocarzinostatin chromophore: presence of a highly strained ether ring and its reaction with mercaptan and sodium borohydride. Biochem Biophys Res Commun. 1983 Jun 15;113(2):538–547. doi: 10.1016/0006-291x(83)91759-x. [DOI] [PubMed] [Google Scholar]
- Jollès B., Laigle A., Chinsky L., Turpin P. Y. The poly dA strand of poly dA.poly dT adopts an A-form in solution: a UV resonance Raman study. Nucleic Acids Res. 1985 Mar 25;13(6):2075–2085. doi: 10.1093/nar/13.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappen L. S., Goldberg I. H. Activation of neocarzinostatin chromophore and formation of nascent DNA damage do not require molecular oxygen. Nucleic Acids Res. 1985 Mar 11;13(5):1637–1648. doi: 10.1093/nar/13.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappen L. S., Goldberg I. H. Stabilization of neocarzinostatin nonprotein chromophore activity by interaction with apoprotein and with HeLa cells. Biochemistry. 1980 Oct 14;19(21):4786–4790. doi: 10.1021/bi00562a011. [DOI] [PubMed] [Google Scholar]
- Kappen L. S., Napier M. A., Goldberg I. H. Roles of chromophore and apo-protein in neocarzinostatin action. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1970–1974. doi: 10.1073/pnas.77.4.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A., Rhodes D., Smith J., Finch J. T., Thomas J. O. A low resolution structure for the histone core of the nucleosome. Nature. 1980 Oct 9;287(5782):509–516. doi: 10.1038/287509a0. [DOI] [PubMed] [Google Scholar]
- Napier M. A., Holmquist B., Strydom D. J., Goldberg I. H. Neocarzinostatin chromophore: purification of the major active form and characterization of its spectral and biological properties. Biochemistry. 1981 Sep 15;20(19):5602–5608. doi: 10.1021/bi00522a038. [DOI] [PubMed] [Google Scholar]
- Napier M. A., Holmquist B., Strydom D. J., Goldberg I. H. Neocarzinostatin: spectral characterization and separation of a non-protein chromophore. Biochem Biophys Res Commun. 1979 Jul 27;89(2):635–642. doi: 10.1016/0006-291x(79)90677-6. [DOI] [PubMed] [Google Scholar]
- Poon R., Beerman T. A., Goldberg I. H. Characterization of DNA strand breakage in vitro by the antitumor protein neocarzinostatin. Biochemistry. 1977 Feb 8;16(3):486–493. doi: 10.1021/bi00622a023. [DOI] [PubMed] [Google Scholar]
- Povirk L. F., Dattagupta N., Warf B. C., Goldberg I. H. Neocarzinostatin chromophore binds to deoxyribonucleic acid by intercalation. Biochemistry. 1981 Jul 7;20(14):4007–4014. doi: 10.1021/bi00517a009. [DOI] [PubMed] [Google Scholar]
- Povirk L. F., Goldberg I. H. Binding of the nonprotein chromophore of neocarzinostatin to deoxyribonucleic acid. Biochemistry. 1980 Oct 14;19(21):4773–4780. doi: 10.1021/bi00562a009. [DOI] [PubMed] [Google Scholar]
- Povirk L. F., Goldberg I. H. Detection of neocarzinostatin chromophore-deoxyribose adducts as exonuclease-resistant sites in defined-sequence DNA. Biochemistry. 1985 Jul 16;24(15):4035–4040. doi: 10.1021/bi00336a035. [DOI] [PubMed] [Google Scholar]
- Takeshita M., Kappen L. S., Grollman A. P., Eisenberg M., Goldberg I. H. Strand scission of deoxyribonucleic acid by neocarzinostatin, auromomycin, and bleomycin: studies on base release and nucleotide sequence specificity. Biochemistry. 1981 Dec 22;20(26):7599–7606. doi: 10.1021/bi00529a039. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Larson J. E., Grant R. C., Shortle B. E., Cantor C. R. Physicochemical studies on polydeoxyribonucleotides containing defined repeating nucleotide sequences. J Mol Biol. 1970 Dec 28;54(3):465–497. doi: 10.1016/0022-2836(70)90121-x. [DOI] [PubMed] [Google Scholar]
- Wilson W. D., Jones R. L. Interaction of actinomycin D, ethidium, quinacrine, daunorubicin, and tetralysine with DNA: 31P NMR chemical shift and relaxation investigation. Nucleic Acids Res. 1982 Feb 25;10(4):1399–1410. doi: 10.1093/nar/10.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W. D., Wang Y. H., Krishnamoorthy C. R., Smith J. C. Poly(dA).poly(dT) exists in an unusual conformation under physiological conditions: propidium binding to poly(dA).poly(dT) and poly[d(A-T)].poly[d(A-T)]. Biochemistry. 1985 Jul 16;24(15):3991–3999. doi: 10.1021/bi00336a029. [DOI] [PubMed] [Google Scholar]


