Abstract
Benzyl isothiocyanate (BITC) is a promising anticancer constituent of edible cruciferous vegetables with in vivo efficacy against chemically-induced as well as oncogene-driven breast cancer in experimental rodents. However, the mechanism underlying anticancer effect of BITC is not fully understood. The present study was undertaken to determine the role of Notch signaling in anticancer responses to BITC as this pathway is often hyperactive in human breast cancer. Exposure of MCF-7, MDA-MB-231, and SUM159 human breast cancer cells to pharmacologic concentrations of BITC (2.5 and 5 μM) resulted in cleavage (activation) of Notch1, Notch2, and Notch4, which was accompanied by induction of γ-secretase complex components Presenilin1 and/or Nicastrin. The BITC-mediated cleavage of Notch was associated with its transcriptional activation as revealed by RBP-Jk and Hes-1A/B luciferase reporter assays. Inhibition of cell migration or cell viability resulting from BITC exposure was not influenced by pharmacological suppression of Notch1 using a γ-secretase inhibitor or RNA interference of Notch 1 as well as Notch4. On the other hand, the BITC-mediated inhibition of cell migration, but not cell viability, was significantly augmented by siRNA and shRNA knockdown of Notch2 protein. Furthermore, the BITC-mediated inhibition of MDA-MB-231 xenograft growth in vivo was associated with a significant increase in nuclear levels of cleaved Notch2 and Hes-1 proteins. In conclusion, the results of the present study indicate that (a) BITC treatment activates Notch2 in cultured and xenografted human breast cancer cells, and (b) Notch2 activation impedes inhibitory effect of BITC on cell migration.
Keywords: benzyl isothiocyanate, Notch2, breast cancer, cell migration
Introduction
Population-based case-control studies continue to provide compelling evidence for an inverse association between dietary intake of cruciferous vegetables and the risk of cancer, and this association is quite persuasive for breast cancer [1-4]. Anticancer effect of cruciferous vegetables is partly attributed to chemicals with an isothiocyanate (−N=C=S) functional group [5]. Benzyl isothiocyanate (BITC) is one such naturally-occurring constituent of edible cruciferous vegetables (e.g., garden cress) with in vivo efficacy against breast cancer in experimental animals [6-9]. Cancer protective effect of BITC was first recognized by Wattenberg [6], who demonstrated inhibition of 7,12-dimethylbenz[a]anthracene-induced mammary tumor formation in female Sprague-Dawley rats. Studies from our own laboratory have shown that BITC administration in the diet (3 μmol BITC/g diet) confers significant protection against mammary hyperplasia and carcinoma incidence and/or burden in a clinically-relevant transgenic mouse model [7]. The BITC administration was also shown to inhibit in vivo growth of transplanted breast cancer cells in mice [8,9].
The mechanism by which BITC inhibits growth of breast cancer cells is still not fully understood, but known pharmacological effects potentially contributing to its anticancer response include growth arrest [10], p53-independent apoptosis induction facilitated by downregulation of X-linked inhibitor of apoptosis protein [11-13], suppression of estrogen receptor-α expression [14], inhibition of signal transducer and activator of transcription 3 [15], and tumor infiltration of T cells [7]. Because pathogenesis of breast cancer is complex often involving abnormalities in various checkpoints and activation of different oncogenes, ability to target multiple pathways is desirable for preventive agents. Agents targeting a single pathway may have limited clinical utility as exemplified by selective estrogen receptor modulators [16].
More recent studies from our laboratory have revealed that BITC is a potent inhibitor of epithelial-mesenchymal transition (EMT) in cultured and xenografted human breast cancer cells [17]. However, the molecular mechanism by which BITC inhibits EMT is still elusive. EMT is a normal physiological process essential for embryonic development, tissue remodeling, and wound healing. At the same time, EMT is one of the key mechanisms contributing to tumor invasion and metastasis [18-20]. Mechanistic understanding of the EMT induction in cancer cells continues to evolve, but several pathways have been implicated in regulation of this process including Notch signaling [18-21]. The Notch pathway regulates expression of genes involved in cell fate determination including proliferation and differentiation [22-24]. Moreover, Notch pathway is implicated in mammary carcinogenesis [25-28]. The present study was undertaken to explore the possibility of whether BITC inhibits Notch activation using a panel of human breast cancer cell lines (MCF-7, MDA-MB-231, and SUM159) and MDA-MB-231 xenografts from control and BITC-treated mice.
Materials and methods
Ethics statement
Archived tumor sections from our previously published in vivo study [8] were used to determine the effect of BITC administration on expression of cleaved Notch2 and Hes-1. Use of mice and their care [8] was in accordance with the University of Pittsburgh Institutional Animal Care and Use Committee guidelines (protocol number 0704557).
Reagents
The BITC (purity >98 %) was purchased from the LKT Laboratories. Cell culture reagents including fetal bovine serum, antibiotics, and Oligofectamine were purchased from Invitrogen-Life Technologies (Carlsbad, CA). Antibodies specific for detection of cleaved Notch1, transmembrane (uncleaved) Notch1, transmembrane (uncleaved) Notch2, Jagged1, Jagged2, Presenilin1, and Nicastrin were from Cell Signaling Technology (Beverly, MA); an antibody specific for detection of cleaved Notch2 was from EMD Millipore (Billerica, MA); an antibody against transmembrane (uncleaved) Notch4 was from Santa Cruz Biotechnology (Santa Cruz, CA); antibody against Hes-1 was from Novus Biologicals (Littleton, CO); and antibodies against actin and cleaved Notch4 were from Sigma-Aldrich (St. Louis, MO). A γ-secretase inhibitor {N-[N-(3,5-difluorophenacetyl-L-alanyl)]-S-phenylglycine-t-butyl ester; hereafter abbreviated as DAPT} was purchased from Calbiochem (Billerica, MA). The Notch1, Notch2 and Notch4-targeted small interfering RNA (siRNA) were purchased from Santa Cruz Biotechnology. A nonspecific control siRNA was purchased from Qiagen (Germantown, MD). The Notch2-targeted small hairpin RNA (shRNA) and control shRNA were purchased from Santa Cruz Biotechnology.
Cell lines
The MCF-7 and MDA-MB-231 cell lines were purchased from American Type Culture Collection (Manassas, VA) and maintained as described by us previously [10, 12]. The SUM159 cell line was purchased from Asterand (Detroit, MI) and cultured in Ham’s F-12 media supplemented with 5% fetal bovine serum, 1 μg/mL hydrocortisone, 5 μg/mL insulin, and 10 mM HEPES.
Western blotting
Stock solution of BITC was prepared in dimethyl sulfoxide (DMSO), and an equal volume of DMSO (final concentration 0.1%) was added to controls. After treatment, the cells were collected and processed for immunoblotting as described by us previously [29, 30]. Immunoreactive bands were visualized using enhanced Chemiluminescence reagent.
Luciferase reporter assay
Cignal RBP-Jk luciferase reporter kit (SABiosciences-Qiagen) was used to determine the effect of BITC treatment on transcriptional activity of Notch. The RBP-Jk [C protein binding factor 1/Suppressor of Hairless/Lag1 (CBF1/Su(H)/Lag1)] is a direct downstream modulator of Notch signaling [23, 24]. Notch intracellular domain (cleaved Notch) binds to CSL/CBF1/Su(H)/Lag1 and converts it from a transcriptional repressor to a transcriptional activator [23, 24]. The Hes-1A/B reporter construct was a generous gift from Dr Kimberly E. Foreman (Loyola University Chicago, IL). The Hes-1A/B luciferase reporter construct contains the −194 to +160 promoter fragment of the Hes-1 gene inserted upstream of the luciferase gene in pGL2 [31]. The cells were transfected with the RBP-Jk or Hes-1A/B reporter construct and pRL-CMV using Fugene6. Twenty-four hours after transfection, the cells were treated with DMSO (control) or BITC (2.5 and 5 μM) for 24 hours. Luciferase activity was determined using Dual-Luciferase Reporter Assay kit from Promega (Madison, WI) as described by us previously [17].
RNA interference of Notch
The cells were transfected at ~50% confluency with a control nonspecific siRNA or Notch1-targeted siRNA (100 nM), Notch2-targeted siRNA (100 nM), or Notch4-targeted siRNA (200 nM) using Oligofectamine. Twenty-four hours after transfection, the cells were treated with DMSO (control) or BITC (5 μM) for 24 hours. Subsequently, the cells were collected and processed for western blotting and cell proliferation and cell migration assays.
Stable transfection with Notch2-targeted shRNA
The MDA-MB-231 cells were transfected with 2 μg of a control shRNA or 2 μg of a Notch2-targeted shRNA using shRNA transfection medium and reagent from Santa Cruz Biotechnology, and selected by culture in medium supplemented with 10 μg/mL puromycin over 4 weeks.
Cell proliferation assay
Cell proliferation was determined using MTS reagent (Promega) according to the supplier’s instructions. The number of living cells is directly proportional to the absorbance at 490 nm of a formazan product.
Cell migration assay
Transwell Boyden chamber containing 8 μm polycarbonate filter was used to determine cell migration. Cell migration was determined as described by us previously [17].
Immunohistochemistry for cleaved Notch2 and Hes-1 in MDA-MB-231 xenografts
Immunohistochemistry was performed to determine the effect of BITC administration on expression of cleaved Notch2 and Hes-1 in vivo as described by us previously for other proteins [30]. Expression of cleaved Notch2 and Hes-1 in the nucleus was determined using Nuclear v9.1 algorithm of Aperio Image Scope software which automatically counts blue-negative and brown-positive stained nuclei and categorizes them according to intensity (0, 1+, 2+ or 3+). Results are computed as percent positive nuclei accounting for both count and intensity.
Results
BITC treatment increased levels of cleaved Notch1, cleaved Notch2, and cleaved Notch4 in human breast cancer cells
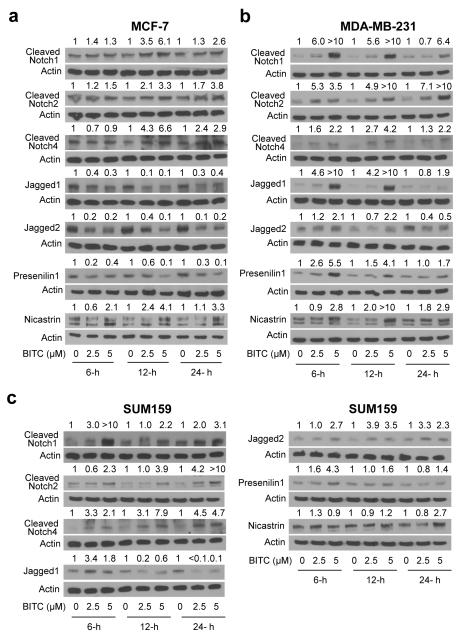
Activation of notch involves its binding to adjoining ligand (e.g., Jagged1 and Jagged2) leading to a conformational change within the receptor followed by another cleavage mediated by the γ-secretase complex at a site located within the Notch transmembrane domain [24]. The net outcome of the second cleavage is release of the Notch intracellular domain into the cytoplasm, which then translocates to the nucleus to regulate target gene expression [23, 24]. Levels of cleaved Notch1, cleaved Notch2, and cleaved Notch4 proteins were increased markedly upon 12- and 24-hour treatment with BITC in MCF-7 cells (Fig.1a), which are estrogen-responsive and express wild-type p53. The BITC-mediated cleavage of Notch1, Notch2, and Notch4 was also observed in the MDA-MB-231 cell line, which is an estrogen-independent cell line with mutant p53 (Fig. 1b). However, the MDA-MB-231 cell line was relatively more sensitive to BITC-mediated cleavage of Notch1 and Notch2 compared with MCF-7 cells (Fig. 1a,b). The BITC-mediated increase in levels of cleaved Notch1, cleaved Notch2, and cleaved Notch4 was also evident in SUM159 (Fig. 1c), which is a triple-negative human breast cancer cell line. Effect of the BITC treatment on Jagged1 and Jagged2 protein expression was different between MCF-7 and MDA-MB-231 cells. The BITC-treated MCF-7 cells exhibited a marked decrease in expression of both Jagged1 and Jagged2 proteins, which was evident as early as 6 hours post-exposure (Fig. 1a). On the other hand, the BITC treatment resulted in a marked induction of Jagged1 and Jagged2 proteins in MDA-MB-231 cells at 6- and 12-hour time points, but this effect was diminished at 24-hour time point (Fig. 1b). In SUM159 cells, the BITC treatment caused transient induction of Jagged1 (6-hours) followed by decline in its expression (12- and 24-hours) but sustained Jagged2 induction when compared with DMSO-treated control (Fig. 1c). Finally, the expression of γ-secretase complex component Presenilin1 was decreased after treatment with BITC at each time point in MCF-7 cells (Fig. 1a). On the other hand, the BITC treatment resulted in induction of Nicastrin in MCF-7 cells, which was sustained for the duration of the study at 5 μM concentration (Fig. 1a). Presenilin1 and Nicastrin proteins were increased robustly upon BITC exposure in MDA-MB-231 cells (Fig. 1b). Induction of Presenilin1 and Nicastrin was also observed in SUM159 cells, albeit with different kinetics (Fig. 1c). Specifically, induction of Presenilin1 expression in SUM159 cells peaked between 6- and 12-hours, whereas the BITC-mediated increase in Nicastrin expression was maximal at 24-hour time point at least with the 5 μM dose (Fig. 1c). Collectively, these results indicated that BITC treatment resulted in cleavage of Notch1, Notch2, and Notch4 in breast cancer cells.
Fig. 1.
The BITC treatment increases levels of cleaved Notch1, cleaved Notch2, and cleaved Notch4 in breast cancer cells. Immunoblotting for cleaved Notch1, cleaved Notch2, cleaved Notch4, Jagged1, Jagged2, Presenilin1, and Nicastrin proteins using lysates from (a) MCF-7 cells, (b) MDA-MB-231 cells, and (c) SUM159 cells after 6-, 12-, or 24-hour treatment with DMSO or BITC (2.5 or 5 μM). The number on top of the immunoreactive band represents change in protein level relative to corresponding DMSO-treated control. Western blotting for each protein was performed at least twice using independently prepared lysates and the results were comparable.
Effect of BITC treatment on transcriptional activity of Notch
Next, we determined the effect of BITC treatment on transcriptional activity of Notch. Exposure of MCF-7, MDA-MB-231, and SUM159 cells to BITC for 24 hours resulted in a statistically significant increase in RBP-Jk luciferase reporter activity especially at the 5 μM concentration (Fig. 2a). The Hes1A/B-associated luciferase activity was also increased significantly upon 24-hour treatment with 5 μM BITC in each cell line (Fig. 2b).
Fig. 2.
The BITC treatment increases RBP-Jk and Hes-1A/B luciferase reporter activity in breast cancer cells. a RBP-Jk luciferase reporter activity (a measure of transcriptional activity of Notch) in MCF-7, MDA-MB-231, and SUM159 cells after 24-hour treatment with DMSO or the indicated concentrations of BITC. b Hes-1A/B luciferase activity in MCF-7, MDA-MB-231, and SUM159 cells after 24-hour treatment with DMSO or the indicated concentrations of BITC. Results shown are mean ± SD (n = 3). *Significantly different (P<0.05) compared with DMSO-treated control by one-way ANOVA followed by Dunnett’s test. Each experiment was repeated at least twice with comparable results.
Effect of pharmacological and genetic suppression of Notch1 cleavage on BITC-mediated inhibition of cell migration
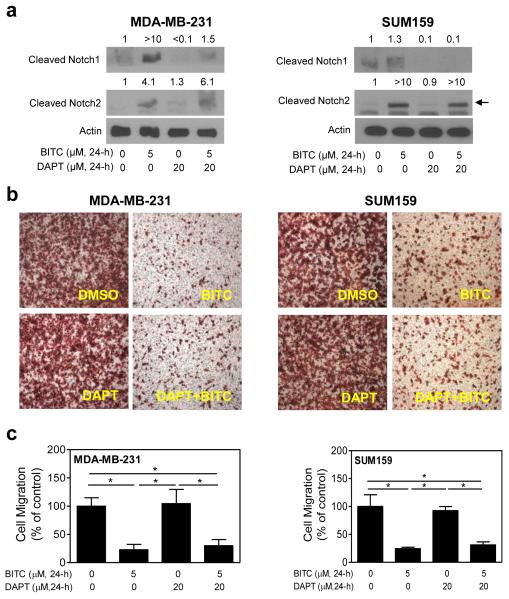
Previous studies have shown that knockdown of Notch1 decreases cell migration in cancer cells [32]. We designed experiments using MDA-MB-231 and SUM159 cells (MCF-7 cells were not used because of weak migratory potential) and a pharmacological inhibitor of γ-secretase (DAPT) to test the consequences of Notch1 activation in BITC-mediated inhibition of cell migration. The BITC-mediated increase in levels of cleaved Notch1 protein (Fig. 3a), but not cleaved Notch2, was markedly suppressed in the presence of 20 μM DAPT in MDA-MB-231 and SUM159 cells. These results indicated that DAPT preferentially inhibited the BITC-mediated cleavage of Notch1. These observations are intriguing but not unique to our study. For example, Harrison et al [33] have shown previously that treatment with 10 μM DAPT decreases levels of cleaved Notch1 but not cleaved Notch4 in MCF-7, MDA-MB-231, and BT474 human breast cancer cells. We tend to agree with the conclusions of the Harrison et al [33] that DAPT preferentially affects Notch1 activity. At the same time, the possibility that BITC-mediated cleavage of Notch2 involves other mechanism(s) can’t be discarded. The BITC-mediated inhibition of MDA-MB-231 or SUM159 cell migration was not affected by DAPT in either cell line (Fig. 3c). These results suggested that Notch1 activation did not impart a significant effect on the BITC-mediated inhibition of cell migration.
Fig. 3.
The BITC-mediated inhibition of breast cancer cell migration is not affected by co-treatment with a γ-secretase inhibitor. a Immunoblotting for cleaved Notch1 and cleaved Notch2 proteins using lysates from the MDA-MB-231 cells and the SUM159 cells treated for 24 hours with DMSO (control) or 5 μM BITC in the absence or presence of 20 μM DAPT. Number above band represents change in protein level relative to DMSO-treated control. b Representative images (Boyden chamber assay) depicting migration by the MDA-MB-231 cells and the SUM159 cells after 24-hour treatment with DMSO or 5 μM BITC in the absence or presence of 20 μM DAPT (100× magnification). c Quantitation of cell migration after 24-hour treatment with DMSO or 5 μM BITC in the absence or presence of 20 μM DAPT. Results shown are mean ± SD (n = 3). *Significantly different (P<0.05) between the indicated groups by one-way ANOVA followed by Bonferroni’s multiple comparison test. Each experiment was repeated twice and representative data from one such experiment are shown.
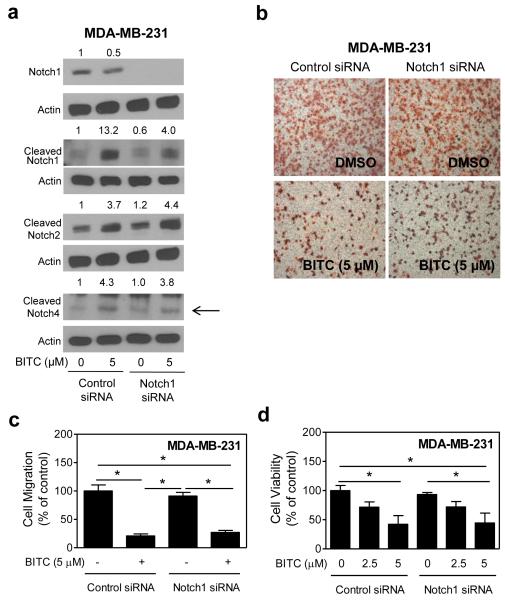
We utilized siRNA targeted against Notch1 to confirm the results with DAPT. The MDA-MB-231 cells transiently transfected with the Notch1-targeted siRNA exhibited a near complete loss in the level of transmembrane (uncleaved) Notch1 when compared with the cells transfected with control siRNA (Fig. 4a). Consistent with the results using DAPT, RNA interference of Notch1 had minimal effect on BITC-mediated inhibition of MDA-MB-231 cell migration (Fig. 4b,c). Moreover, the BITC-mediated inhibition of MDA-MB-231 cell viability was not affected by RNA interference of Notch1 (Fig. 4d). Collectively, these results indicated that Notch1 activation had no impact on BITC’s ability to inhibit cell migration or cell viability.
Fig. 4.
RNA interference of Notch1 fails to confer protection against BITC-mediated inhibition of cell migration. a Immunoblotting for transmembrane (uncleaved) Notch1, cleaved Notch1, cleaved Notch2, and cleaved Notch4 proteins using lysates from the MDA-MB-231 cells transiently transfected with a control (nonspecific) siRNA or a Notch1-targeted siRNA and treated for 24 hours with DMSO or 5 μM BITC. The number above band represents change in protein level relative to control siRNA-transfected cells treated with DMSO. b Microscopic images depicting MDA-MB-231 cell migration in control siRNA transfected or Notch1-targeted siRNA cells after 24-hour treatment with DMSO or 5 μM BITC (100× magnification). c Quantitation of migrated MDA-MB-231 cells from experimental data shown in (b). d Viability of MDA-MB-231 cells transiently transfected with a control siRNA or a Notch1-targeted siRNA and treated for 24 hours with DMSO (control) or BITC (2.5 or 5 μM). Results are expressed as mean ± SD (n = 3). *Significantly different (P<0.05) between the indicated groups by one-way ANOVA followed by Bonferroni’s multiple comparison test. Each experiment was repeated twice, and representative data from one such experiment are shown.
RNA interference of Notch2 augments BITC-mediated inhibition of cell migration
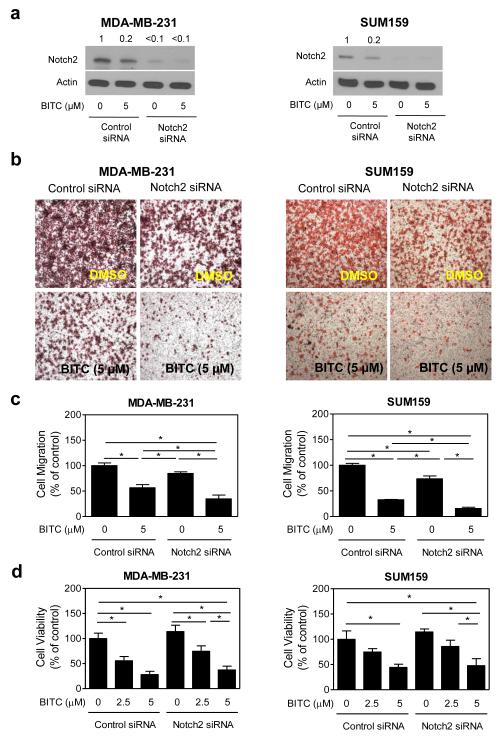
Levels of transmembrane (uncleaved) Notch2 were decreased by >90% upon transfection of the MDA-MB-231 and SUM159 cells with the Notch2-trageted siRNA when compared with the cells transfected with the control siRNA (Fig. 5a). The increase in levels of cleaved Notch2, but not cleaved Notch1 or cleaved Notch4, upon treatment with BITC was markedly suppressed by RNA interference of Notch2 in both MDA-MB-231 and SUM159 cells (Fig 6). Transient transfection with the Notch2-targeted siRNA alone resulted in a decrease in MDA-MB-231 and SUM159 cell migration compared with the control siRNA-transfected cells (Fig. 5b). The BITC-mediated inhibition of cell migration was significantly augmented by knockdown of the Notch2 protein in both cell lines (Fig. 5c). On the other hand, inhibition of cell viability resulting from BITC exposure was not affected by Notch2 protein knockdown in either MDA-MB-231 cells or SUM159 cells (Fig. 5d).
Fig. 5.
Notch2 knockdown augments BITC-mediated inhibition of cell migration. a Immunoblotting for transmembrane (uncleaved) Notch2 protein using lysates from the MDA-MB-231 cells and the SUM159 cells transiently transfected with a control siRNA or a Notch2-targeted siRNA and treated for 24 hours with DMSO or 5 μM BITC. The number above band represents change in protein level relative to control siRNA-transfected cells treated with DMSO. b Representative microscopic images depicting cell migration in cells transiently transfected with a control siRNA or a Notch2-targeted siRNA and treated for 24 hours with DMSO or 5 μM BITC (100× magnification). c Quantitation of migrated MDA-MB-231 and SUM159 cells from the experimental data shown in panel (b). d Viability of the MDA-MB-231 cells and the SUM159 cells transiently transfected with a control siRNA or a Notch2-targeted siRNA and treated for 24 hours with DMSO (control) or BITC (2.5 or 5 μM). Results shown are mean ± S.D (n = 3). *Significantly different (P<0.05) between the indicated groups by one-way ANOVA followed by Bonferroni’s multiple comparison test. Similar results were observed in two independent experiments in each cell line. Representative data from a single experiment are shown.
Fig.6.
Specificity of the Notch2-targeted siNRA. Immunoblotting for cleaved Notch2, cleaved Notch1, and cleaved Notch4 proteins using lysates from the MDA-MB-231 cells and the SUM159 cells transiently transfected with a control (nonspecific) siRNA or a Notch2-targeted siRNA and treated for 24 hours with DMSO or 5 μM BITC. The number above band represents change in protein relative to control siRNA-transfected cells treated with DMSO.
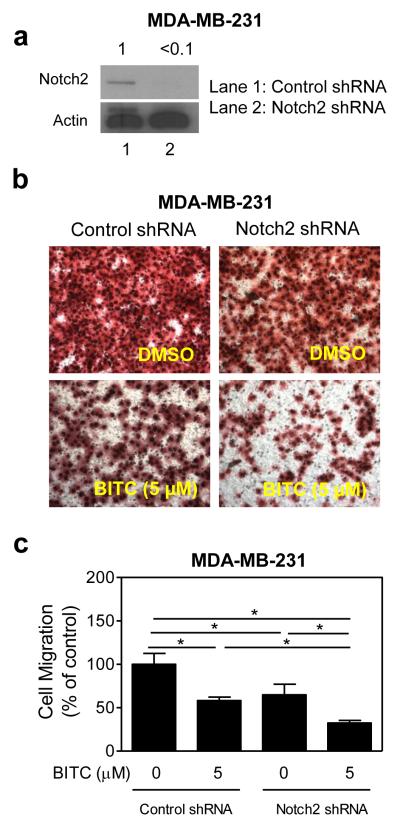
We created a variant of MDA-MB-231 cell line with stable knockdown of Notch2 using a shRNA to further test its role in BITC-mediated inhibition of cell migration. Level of Notch2 protein was decreased by >90% in MDA-MB-231 cells stably transfected with the Notch2-targeted shRNA compared with control shRNA-transfected cells (Fig. 7a). Similar to siRNA transfection experiments, the BITC-mediated inhibition of MDA-MB-231 cell migration was augmented by stable knockdown of Notch2 protein (Fig. 7b, c). Stable knockdown of Notch2 protein had no meaningful impact on BITC’s ability to inhibit cell proliferation (results not shown). Collectively, these results indicated that Notch2 activation by BITC impeded its inhibitory effect on cell migration.
Fig. 7.
The effect of stable knockdown of Notch2 protein on BITC-mediated inhibition of cell migration. a Immunoblotting for transmembrane (uncleaved) Notch2 protein using lysate from MDA-MB-231 cells stably transfected with a control shRNA or a Notch2-targeted shRNA. b Representative images depicting migration of MDA-MB-231 cells stably transfected with a control shRNA or a Notch2-targeted shRNA and treated for 24 hours with DMSO (control) or 5 μM BITC (200× magnification). c Quantitation of cell migration in the cells stably transfected with a control shRNA or a Notch2-targeted shRNA and treated for 24 hours with DMSO (control) or 5 μM BITC. Result shown are mean ± SD (n= 3) *Significantly different (P<0.05) between the indicated groups by one-way ANOVA followed by Bonferroni’s multiple comparison test.
Notch4 activation is dispensable for BITC-mediated inhibition of cell migration
Next, we proceeded to determine the role of Notch4 activation in BITC-mediated inhibition of cell migration using MDA-MB-231 cells. Level of transmembrane (uncleaved) Notch4 protein was decreased by 60% upon transient transfection of MDA-MB-231 cells with the Notch4-targeted siRNA (Fig. 8a). Similar to Notch1, RNA interference of Notch4 alone had minimal effect on MDA-MB-231 cell migration (Fig. 8b). Moreover, inhibition of MDA-MB-231 cell migration resulting from BITC exposure was not influenced by RNA interference of Notch4 (Fig. 8c).
Fig. 8.
The BITC-mediated inhibition of cell migration is not affected by Notch4 protein knockdown in the MDA-MB-231 cell line. a Immunoblotting for transmembrane (uncleaved) Notch4, cleaved Notch4, cleaved Notch1, and cleaved Notch2 proteins using lysate from the MDA-MB-231 cells transiently transfected with a control siRNA or a Notch4-targeted siRNA and treated for 24 hours with DMSO or 5 μM BITC. The number above band represents change in protein level relative to control siRNA-transfected cells treated with DMSO. b Representative images depicting migration of MDA-MB-231 cells transiently transfected with a control siRNA or a Notch4-targeted siRNA and treated for 24 hours with DMSO or 5 μM BITC (100× magnification). c Quantitation of cell migration in cells transfected with a control siRNA or a Notch4-targeted siRNA and treated for 24 hours with DMSO or 5 μM BITC. Results shown are mean ± S.D (n = 3). *Significantly different (P<0.05) between the indicated groups by one-way ANOVA followed by Bonferroni’s multiple comparison test. Each experiment was repeated twice, and representative data from one such experiment are shown.
Immunohistochemical analysis for cleaved Notch2 and Hes-1 proteins in MDA-MB-231 xenografts
We used archived MDA-MB-231 tumor xenografts from our previously completed study [8] to determine the effect of BITC administration on levels of cleaved Notch2 and Hes-1. Representative images for cleaved Notch2 expression in MDA-MB-231 tumor xenografts from three mice each of the control group and the BITC treatment groups are shown in Fig. 9a. Nuclear levels of cleaved Notch2 was significantly higher in the MDA-MB-231 tumor xenografts from BITC-treated mice compared with control mice (Fig. 9b). Similarly, BITC administration resulted in a modest yet significant increase in nuclear levels of Hes-1 protein compared with control (Fig. 9c, d). These results provided in vivo evidence for BITC-mediated activation of Notch2 in xenografted MDA-MB-231 cells.
Fig. 9.
The BITC administration to tumor bearing athymic mice increases in vivo expression of cleaved Notch2 and Hes-1 proteins in the MDA-MB-231 xenografts. a Immunohistochemical analysis for cleaved Notch2 protein in representative tumor sections from the control mice and those treated with 7.5 μmol BITC (scale bar, 50 μm; 400× magnification) [8]. Amplification of selected area is shown in the inset. b Quantitation of nuclear level of cleaved Notch2 protein in the tumors from the control mice and those treated with 7.5 μmol BITC. c Immunohistochemical analysis for Hes-1 expression in the tumor sections from the control mice and those treated with 7.5 μmol BITC (scale bar, 50 μm; 400× magnification) [8]. Amplification of selected area is shown in the inset. d Quantitation of nuclear levels of Hes-1 protein expression in the tumors from the control mice and those treated with 7.5 μmol BITC. At least four randomly selected fields on each tumor section were scored for cleaved Notch2 and Hes-1 protein expression. Results shown are mean percent positive nuclei with corresponding SD (n = 5 for the control group, n=3 for the BITC treatment group).
Discussion
Accumulating evidence implies critical role for Notch signaling in mammary carcinogenesis [22-28]. Overexpression of activated Notch1 and Notch3 in transgenic mice induces mammary tumorigenesis [25] and high level expression of Notch1 and its ligands is associated with poor outcome in breast cancer patients [26-28]. Induction of Notch1 activity and function by ErbB2 and cross-talk between Notch and estrogen receptor has also been documented in breast cancer cells [34, 35]. Impetus to determine the effect of BITC on Notch signaling stemmed from the following observations: (a) BITC is a potent inhibitor of EMT in breast cancer cells [17], (b) Jagged1-mediated Notch activation induces EMT through Slug-induced repression of epithelial marker E-cadherin [36], and anticancer effect of BITC is characterized by suppression of various oncogenic pathways including nuclear factor-κB [37], signal transducer and activator of transcription 3 [15], and Akt [38]. The present study reveals that BITC treatment increases levels of cleaved Notch1, cleaved Notch2, and cleaved Notch4 leading to transcriptional activation of Notch in human breast cancer cells, but only Notch2 activation has functional relevance in the context of BITC’s effect on cell migration. The BITC-mediated activation of Notch in breast cancer cells is evident at concentrations (2.5 to 5 μM) below peak plasma achievable dose of 6.5 μM [38]. The BITC administration to the tumor bearing athymic mice causes significant increase in nuclear levels of cleaved Notch2 in MDA-MB-231 xenografts. It is reasonable to propose that BITC-mediated inhibition of EMT is not caused by suppression of Notch.
We have consistently observed cell line-specific differences in effect of BITC treatment on expression of Jagged1 and Jagged2 proteins. In MCF-7 cells, BITC treatment consistently decreased levels of both Jagged1 and Jagged2 proteins (Fig. 1a). Surprisingly, the MDA-MB-231 cells consistently exhibited a transient increase followed by decline in expression of Jagged1 and Jagged2 proteins upon treatment with BITC (Fig. 1b). The molecular basis for differential effect of BITC on Jagged1 and Jagged2 protein expression in MCF-7 versus MDA-MB-231 cells remains to be determined, but may be related, for example, to differences in Notch3 and/or Wnt/β-catenin [39]. Both Notch3 and Wnt/β-catenin have been shown to be positive regulators of Jagged1 expression in ovarian cancer cells [39], and a similar association in breast cancer cells is highly plausible. Interestingly, the Wnt/β-catenin is constitutively active in the MCF-7 cells but not in the MDA-MB-231 cell line [40]. It is possible that the BITC-mediated decrease in Jagged1 expression in the MCF-7 cell line is due to inactivation of the Wnt/β-catenin pathway. Similarly, the possibility that cell line-specific differences in effect of BITC treatment on Jagged1 and Jagged2 protein expression in MCF-7 and MDA-MB-231 cells are due to differential Notch3 expression and/or activation status can’t be overlooked. Additional work is necessary to explore these possibilities. Nevertheless, the BITC-mediated decrease in levels of Jagged1 and Jagged2 proteins in MDA-MB-231 cells at the 24-hour time points is not due to their proteasomal degradation. This conclusion is based on our unpublished observations that proteasomal inhibitor MG132 (1 μM) is unable to rescue suppression of Jagged 1 and Jagged2 protein expression upon exposure to BITC (results not shown).
The mechanism by which Notch2 protein knockdown augments BITC-mediated inhibition of cell migration is not yet clear. Modulation of urokinase-type plasminogen activator (uPA) expression is one such possible mechanism because uPA is a direct transcriptional target of Jagged1-Notch signaling in breast cancer cells [41]. Moreover, high uPA expression correlates with breast cancer recurrence and metastasis [42]. However, our preliminary unpublished studies indicate that BITC treatment decreases uPA expression in breast cancer cells (Sehrawat A, Kim SH, Singh SV, unpublished observations). Our unpublished results are consistent with the BITC-mediated decrease in uPA levels observed in the HT29 colon cancer cells [43]. Further studies are needed to experimentally address this question.
Abbreviations
- BITC
benzyl isothiocyanate
- EMT
epithelial mesenchymal transition
- DAPT
N-[N-(3,5-difluorophenacetyl-L-alanyl)]-S-phenylglycine-t-butyl ester
- siRNA
small interfering RNA
- shRNA
small hairpin RNA
- DMSO
dimethyl sulfoxide
- uPA
urokinase-type plasminogen activator
Footnotes
Conflict of interest: None of the authors has any conflict of interest.
Financial Disclosure: The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fowke JH, Chung FL, Jin F, Qi D, Cai Q, Conaway C, Cheng JR, Shu XO, Gao YT, Zheng W. Urinary isothiocyanate levels, brassica, and human breast cancer. Cancer Res. 2003;63:3980–3986. [PubMed] [Google Scholar]
- 2.Ambrosone CB, McCann SE, Freudenheim JL, Marshall JR, Zhang Y, Shields PG. Breast cancer risk in premenopausal women is inversely associated with consumption of broccoli, a source of isothiocyanates, but is not modified by GST genotype. J Nutr. 2004;134:1134–1138. doi: 10.1093/jn/134.5.1134. [DOI] [PubMed] [Google Scholar]
- 3.Zhao H, Lin J, Grossman HB, Hernandez LM, Dinney CP, Wu X. Dietary isothiocyanates, GSTM1, GSTT1, NAT2 polymorphisms and bladder cancer risk. Int J Cancer. 2007;120:2208–2213. doi: 10.1002/ijc.22549. [DOI] [PubMed] [Google Scholar]
- 4.Moy KA, Yuan JM, Chung FL, van den Berg D, Wang R, Gao YT, Yu MC. Urinary total isothiocyanates and colorectal cancer: a prospective study of men in Shanghai, China. Cancer Epidemiol Biomarkers Prev. 2008;17:1354–1359. doi: 10.1158/1055-9965.EPI-07-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hecht SS. Inhibition of carcinogenesis by isothiocyanates. Drug Metab Rev. 2000;32:395–411. doi: 10.1081/dmr-100102342. [DOI] [PubMed] [Google Scholar]
- 6.Wattenberg LW. Inhibition of carcinogenic effects of polycyclic hydrocarbons by benzyl isothiocyanate and related compounds. J Natl Cancer Inst. 1977;58:395–398. doi: 10.1093/jnci/58.2.395. [DOI] [PubMed] [Google Scholar]
- 7.Warin R, Chambers WH, Potter DM, Singh SV. Prevention of mammary carcinogenesis in MMTV-neu mice by cruciferous vegetable constituent benzyl isothiocyanate. Cancer Res. 2009;69:9473–9480. doi: 10.1158/0008-5472.CAN-09-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warin R, Xiao D, Arlotti JA, Bommareddy A, Singh SV. Inhibition of human breast cancer xenograft growth by cruciferous vegetable constituent benzyl isothiocyanate. Mol Carcinogenesis. 2010;49:500–507. doi: 10.1002/mc.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim EJ, Hong JE, Eom SJ, Lee JY, Park JHY. Oral administration of benzyl-isothiocyanate inhibits solid tumor growth and lung metastasis of 4T1 murine mammary carcinoma cells in BALB/c mice. Breast Cancer Res Treat. 2011;130:61–71. doi: 10.1007/s10549-010-1299-8. [DOI] [PubMed] [Google Scholar]
- 10.Xiao D, Vogel V, Singh SV. Benzyl isothiocyanate-induced apoptosis in human breast cancer cells is initiated by reactive oxygen species and regulated by Bax and Bak. Mol Cancer Ther. 2006;5:2931–2945. doi: 10.1158/1535-7163.MCT-06-0396. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Tang L, Gonzalez V. Selected isothiocyanates rapidly induce growth inhibition of cancer cells. Mol Cancer Ther. 2003;2:1045–1052. [PubMed] [Google Scholar]
- 12.Xiao D, Powolny AA, Singh SV. Benzyl isothiocyanate targets mitochondrial respiratory chain to trigger reactive oxygen species-dependent apoptosis in human breast cancer cells. J Biol Chem. 2008;283:30151–30163. doi: 10.1074/jbc.M802529200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SH, Singh SV. p53-Independent apoptosis by benzyl isothiocyanate in human breast cancer cells is mediated by suppression of XIAP expression. Cancer Prev Res. 2010;3:718–726. doi: 10.1158/1940-6207.CAPR-10-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang L, Ding L, Wang ZY. Isothiocyanates repress estrogen receptor alpha expression in breast cancer cells. Oncol Rep. 2009;21:185–192. [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SH, Nagalingam A, Saxena NK, Singh SV, Sharma D. Benzyl isothiocyanate inhibits oncogenic actions of leptin in human breast cancer cells by suppressing activation of signal transducer and activator of transcription 3. Carcinogenesis. 2011;32:359–367. doi: 10.1093/carcin/bgq267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, Vogel V, Robidoux A, Dimitrov N, Atkins J, Daly M, Wieand S, Tan-Chiu E, Ford L, Wolmark N. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel project P-1 study. J Natl Cancer Inst. 1998;90:1371–1388. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 17.Sehrawat A, Singh SV. Benzyl isothiocyanate inhibits epithelial-mesenchymal transition in cultured and xenografted human breast cancer cells. Cancer Prev Res. 2011;4:1107–1117. doi: 10.1158/1940-6207.CAPR-10-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 19.Tomaskovic-Crook E, Thompson EW, Thiery JP. Epithelial to mesenchymal transition and breast cancer. Breast Cancer Res. 2009;11:213. doi: 10.1186/bcr2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollier BG, Evans K, Mani SA. The epithelial-to-mesenchymal transition and cancer stem cells: a coalition against cancer therapies. J Mammary Gland Biol Neoplasia. 2009;14:29–43. doi: 10.1007/s10911-009-9110-3. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Imanaka N, Chen J, Griffin JD. Hypoxia potentiates Notch signaling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. Br J Cancer. 2010;102:351–360. doi: 10.1038/sj.bjc.6605486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 2006;107:2223–2233. doi: 10.1182/blood-2005-08-3329. [DOI] [PubMed] [Google Scholar]
- 24.Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 25.Hu C, Dievart A, Lupien M, Calvo E, Tremblay G, Jolicoeur P. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol. 2006;168:973–990. doi: 10.2353/ajpath.2006.050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, Lockwood G, Egan SE. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 27.Dickson BC, Mulligan AM, Zhang H, Lockwood G, O’Malley FP, Egan SE, Reedijk M. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol. 2007;20:685–693. doi: 10.1038/modpathol.3800785. [DOI] [PubMed] [Google Scholar]
- 28.Reedijk M, Pinnaduwage D, Dickson BC, Mulligan AM, Zhang H, Bull SB, O’Malley FP, Egan SE, Andrulis IL. JAG1 expression is associated with a basal phenotype and reccurence in lymph node-negative breast cancer. Breast Cancer Res Treat. 2008;111:439–448. doi: 10.1007/s10549-007-9805-3. [DOI] [PubMed] [Google Scholar]
- 29.Xiao D, Srivastava SK, Lew KL, Zeng Y, Hershberger P, Johnson CS, Trump DL, Singh SV. Allyl isothiocyanate, a constituent of cruciferous vegetables, inhibits proliferation of human prostate cancer cells by causing G2/M arrest and inducing apoptosis. Carcinogenesis. 2003;24:891–897. doi: 10.1093/carcin/bgg023. [DOI] [PubMed] [Google Scholar]
- 30.Powolny AA, Bommareddy A, Hahm ER, Normolle DP, Beumer JH, Nelson JB, Singh SV. Chemopreventative potential of the cruciferous vegetable constituent phenethyl isothiocyanate in a mouse model of prostate cancer. J Natl Cancer Inst. 2011;103:571–584. doi: 10.1093/jnci/djr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curry CL, Reed LL, Nickoloff BJ, Miele L, Foreman KE. Notch-independent regulation of Hes-1 expression by c-Jun N-terminal kinase signaling in human endothelial cells. Lab Invest. 2006;86:842–852. doi: 10.1038/labinvest.3700442. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Fu L, Gu F, Ma Y. Notch1 is involved in migration and invasion of human breast cancer cells. Oncol Rep. 2011;26:1295–1303. doi: 10.3892/or.2011.1399. [DOI] [PubMed] [Google Scholar]
- 33.Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR, Bundred NJ, Clarke RB. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 2010;70:709–718. doi: 10.1158/0008-5472.CAN-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindsay J, Jiao X, Sakamaki T, Casimiro MC, Shirley LA, Tran TH, Ju X, Liu M, Li Z, Wang C, Katiyar S, Rao M, Allen KG, Glazer RI, Ge C, Stanley P, Lisanti MP, Rui H, Pestell RG. ErbB2 induces Notch1 activity and function in breast cancer cells. Clin Transl Sci. 2008;1:107–115. doi: 10.1111/j.1752-8062.2008.00041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rizzo P, Miao H, D’Souza G, Osipo C, Song LL, Yun J, Zhao H, Mascarenhas J, Wyatt D, Antico G, Hao L, Yao K, Rajan P, Hicks C, Siziopikou K, Selvaggi S, Bashir A, Bhandari D, Marchese A, Lendahl U, Qin JZ, Tonetti DA, Albain K, Nickoloff BJ, Miele L. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res. 2008;68:5226–5235. doi: 10.1158/0008-5472.CAN-07-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leong KG, Niessen K, Kulic I, Raouf A, Eaves C, Pollet I, Karsan A. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J Exp Med. 2007;204:2935–2948. doi: 10.1084/jem.20071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srivastava SK, Singh SV. Cell cycle arrest, apoptosis induction and inhibition of nuclear factor kappa B activation in anti-proliferative activity of benzyl isothiocyanate against human pancreatic cancer cells. Carcinogenesis. 2004;25:1701–1709. doi: 10.1093/carcin/bgh179. [DOI] [PubMed] [Google Scholar]
- 38.Boreddy SR, Pramanik KC, Srivastava SK. Pancreatic tumor suppression by benzyl isothiocyanate is associated with inhibition of PI3K/AKT/FOXO pathway. Clin Cancer Res. 2011;17:1784–1795. doi: 10.1158/1078-0432.CCR-10-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X, Stoeck A, Lee SJ, Shih IM, Wang MM, Wang TL. Jagged1 expression regulated by Notch3 and Wnt/β-catenin signaling pathway in ovarian cancer. Oncotarget. 2010;1:210–218. doi: 10.18632/oncotarget.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlange T, Matsuda Y, Lienhard S, Huber A, Hynes NE. Autocrine WNT signaling contributes to breast cancer cell proliferation via the canonical WNT pathway and EGFR transactivation. Breast Cancer Res. 2007;9:R63. doi: 10.1186/bcr1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu M, Cohen B, Goldvasser P, Berman H, Virtanen C, Reedijk M. Plasminogen activator uPA is a direct transcriptional target of the JAG1-Notch receptor signaling pathway in breast cancer. Cancer Res. 2011;71:277–286. doi: 10.1158/0008-5472.CAN-10-2523. [DOI] [PubMed] [Google Scholar]
- 42.Annecke K, Schmitt M, Euler U, Zerm M, Paepke D, Paepke S, von Minckwitz G, Thomssen C, Harbeck N. uPA and PAI-1 in breast cancer: review of their clinical utility and current validation in the prospective NNBC-3 trial. Adv Clin Chem. 2008;45:31–45. doi: 10.1016/s0065-2423(07)00002-9. [DOI] [PubMed] [Google Scholar]
- 43.Lai KC, Huang AC, Hsu SC, Kuo CL, Yang JS, Wu SH, Chung JG. Benzyl isothiocyanate (BITC) inhibits migration and invasion of human colon cancer HT29 cells by inhibiting matrix metalloproteinase-2/-9 and urokinase plasminogen (uPA) through PKC and MAPK signaling pathway. J Agric Food Chem. 2010;58:2935–2942. doi: 10.1021/jf9036694. [DOI] [PubMed] [Google Scholar]