Abstract
Background
In early-stage rectal cancer, the surgeon must decide between the high morbidity of radical surgery and the high recurrence rates of local excision. A prognostic marker could improve patient selection and lower recurrence rates. MicroRNAs (miRNAs), small RNAs that often inhibit tumor suppressors, have shown prognostic potential in colorectal cancer. We hypothesized that high miRNA levels in malignant tissue from early-stage rectal cancer patients could predict recurrence after local excision.
Materials & Methods
We identified 17 early-stage rectal cancer patients treated with local excision between 1990 and 2005, 4 of whom had recurred. Total RNA was extracted from benign and malignant tissue and used in quantitative real-time reverse transcriptase PCR (qRT-PCR) to probe for miR-20a, miR-21, miR-106a, miR-181b, and miR-203. MiRNA data was evaluated for association with recurrence using univariate analysis with Wilcoxon rank-sum test.
Results
Malignant tissue in both patients who recurred and patients who did not recur had equivalently high levels of miRNA. However, the benign tissue of patients who recurred contained significantly higher levels of all 5 miRNAs when compared to the benign tissue of non-recurrent patients despite having no histological differences.
Conclusions
This is the first study to show that high miRNA levels of histologically benign tissue obtained from the surgical margin of locally excised rectal cancers can predict recurrence. The malignant miRNA levels did not have predictive value. Further investigation of miRNAs is needed to explore their potential for a more accurate prognosis of rectal cancer.
Keywords: rectal cancer, adenocarcinoma, local excision, microRNA, prognosis, recurrence
INTRODUCTION
The American Cancer Society estimates that there will be nearly 40,000 new cases of rectal cancer diagnosed in the United States in 2011. While radiation and chemotherapy are important in the treatment of rectal cancer, surgery remains the most vital part of the treatment regimen. Currently, two basic surgical options exist for these patients: radical surgery and local excision. Radical surgery, involving complete or nearly complete removal the rectum, possesses a low recurrence rate (2–15%), but entails significant morbidity (20–30%) and possible loss of anorectal, bladder, and sexual function for patients [3, 10, 32]. In contrast, local excision allows for rapid recovery, low morbidity, and maintenance of bowel continence and rectal function. However, local excision has an unacceptably high rate (10–40%) of local recurrence [24]. Identification of a prognostic marker that could predict local recurrence in patients with rectal cancer would permit a more accurate assessment of patients’ candidacy for local excision. That is, patients identified as high risk could undergo radical surgery instead of local excision, and thereby lower their chance of local recurrence. Conversely, identification of patients at low risk of recurrence may allow local excision to be performed in patients who were previously thought to be poor candidates.
Exciting new research has shown that microRNA (miRNA) profiles have prognostic potential in cancer [5, 25, 34]. MiRNAs were first discovered in 1993, and were defined as non-coding, single-stranded RNAs that are approximately 22 nucleotides in length [1]. Investigations since their discovery have elucidated that they act by binding to the 3’ untranslated region of a messenger RNA which leads to deadenylation, translational repression, or cleavage of the target mRNA [12, 13]. Therefore, miRNAs negatively regulate gene expression at the posttranscriptional level. Numerous miRNAs are either up- or down-regulated in a variety of cancers [2, 5, 33]. Furthermore, many of their targets have been found to consist of oncogenes and tumor suppressor genes [8, 9, 11, 18, 33, 36]. This direct involvement in carcinogenesis makes miRNAs excellent candidates for prognostic markers. Recently, a large, multicenter study found that miR-20a, miR-21, miR-106a, miR-181b, and miR-203 were significantly upregulated in colon adenocarcinoma tumor tissue vs. paired normal tissue [25]. Moreover, all the aforementioned miRNAs were found to be associated with poor survival, and miR-21 was associated with poor therapeutic outcome [25]. Nevertheless, the molecular and genetic differences between colon and rectal cancer preclude a direct correlation of these results [30], and no studies to our knowledge have investigated the role of miRNAs in rectal cancer specifically, let alone examined their utility as prognostic indicators.
Considering the newly discovered prognostic significance of miRNAs and the high potential for improved outcomes in rectal cancer patients, this study aimed to evaluate the usefulness of the miRNA profile in predicting recurrence of rectal cancer after local resection. Specifically, we hypothesized that high miRNA levels in malignant tissue from early-stage rectal cancer patients could predict recurrence after local excision.
MATERIALS AND METHODS
Approval for this experiment was obtained from the University of Wisconsin Institutional Review Board. A colon and rectal cancer database that is maintained in the University of Wisconsin Department of Surgery was utilized to find all patients with early-stage (T1 and T2) rectal cancer who had undergone local excision from 1990 to 2005. 23 consecutive patients were identified using this search. Archived formalin-fixed paraffin embedded (FFPE) tissue samples were obtained from the Department of Pathology. Three patients were excluded because their tissue blocks were unable to be located in the tissue archive. An experienced department pathologist then identified and marked suitable benign and malignant tissue on FFPE tissue samples. Three additional patients were excluded from the study because the pathologist could not identify suitable tissue. Therefore, 17 patients were included in this study with clearly marked histologically normal and malignant tissue. This marked benign and malignant tissue was then macrodissected from associated stromal tissues. Notably, quantitative data regarding the margin status (mucosal and radial) was not available in the medical record, as it was not standard to record such data during the study enrollment period. Still, as part of our inclusion criteria, all patients had completely negative surgical margins in all directions according to their pathology reports. Furthermore, benign and malignant tissues were never macrodissected from the same tissue block, thereby decreasing the likelihood of cross contamination.
RNA was extracted from macrodissected tissue samples according to the RNeasy FFPE Kit protocol (Qiagen, Valencia, CA). The quality and quantity of the RNA was assured using the ND-1000 spectrophotometer (NanoDrop, Wilmington, DE). Using quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR), the normal and malignant tissue was assayed for the level of miR-20a, miR-21, miR-106a, miR-181b, and miR-203. qRT-PCR was done with the TaqMan MicroRNA Reverse Transcription Kit, TaqMan MicroRNA Assays, and the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol. Expression levels of small nuclear RNA U6B were used as the normalization control. All assays were performed separately in triplicate by two investigators who were blinded to the clinical data, yielding six quantifications per miRNA for each tissue sample.
Clinical information regarding age, gender, local and metastatic recurrence, survival, tumor grade, and clinical stage of disease was collected from the University of Wisconsin Department of Surgery colon and rectal cancer database. A Wilcoxon rank-sum test was used to determine differences between the benign and malignant levels of each miRNA. The same test was used to determine if patients with and without cancer recurrence showed differences in their miRNA profiles. For example, the benign tissue levels of a certain miRNA for patients who recurred were compared to the benign tissue levels of that miRNA for those who did not recur. Likewise, the malignant miRNA tissue levels were compared between patients who recurred and those who did not recur. All statistical analyses were done using MStat version 5.1 (McArdle Laboratory for Cancer Research, Madison, WI). Associations with clinical data were considered statistically significant if the P value was less than .05.
RESULTS
Patient and tumor characteristics are shown in Table 1. Patients who experienced cancer recurrence after local excision were not significantly different from recurrence-free patients with respect to age, length of follow up, and tumor characteristics (stage, size, and grade). No lymphovascular invasion, deep submucosal invasion, signet ring cells, or colloid histology was identified in the tissue samples. Patients had been clinically staged by a variety of methods including digital rectal exam, proctoscopy, flexible sigmoidoscopy, colonoscopy, biopsies, transrectal ultrasound, computed tomography, chest radiographs, or a combination thereof. Clinical T stage did not correlate with malignant or benign tissue miRNA expression (P > 0.05 for all miRNAs).
TABLE 1.
Patient and Tumor Characteristics
| Recurrent (n = 4) |
Non-recurrent (n = 13) |
P value | |
|---|---|---|---|
| Age | 68 | 69 | 0.89 |
| T-stage 1 | 2 | 6 | 0.91 |
| T-stage 2 | 2 | 7 | |
| Tumor size (cm) | 2.1 | 1.9 | 0.76 |
| Grade 1 | 2 | 4 | |
| Grade 2 | 2 | 6 | 0.78 |
| Grade not reported | 0 | 3 | |
| Average follow up (months) | 66 | 45 | 0.28 |
Malignant tissue miRNA levels were compared to benign tissue levels. MiRNA expression was significantly upregulated in malignant tissue (Fig. 1). The difference was highly significant (P < 0.00001) for all miRNAs studied (miR-20a, miR-21, miR-106a, miR-181b, and miR-203). Most notably, miR-21 levels showed approximately a 7-fold increase in malignant tissue.
FIGURE 1.
Relative miRNA expression in benign and malignant tissue. Error bars represent the standard error of the mean. **P < 0.00001.
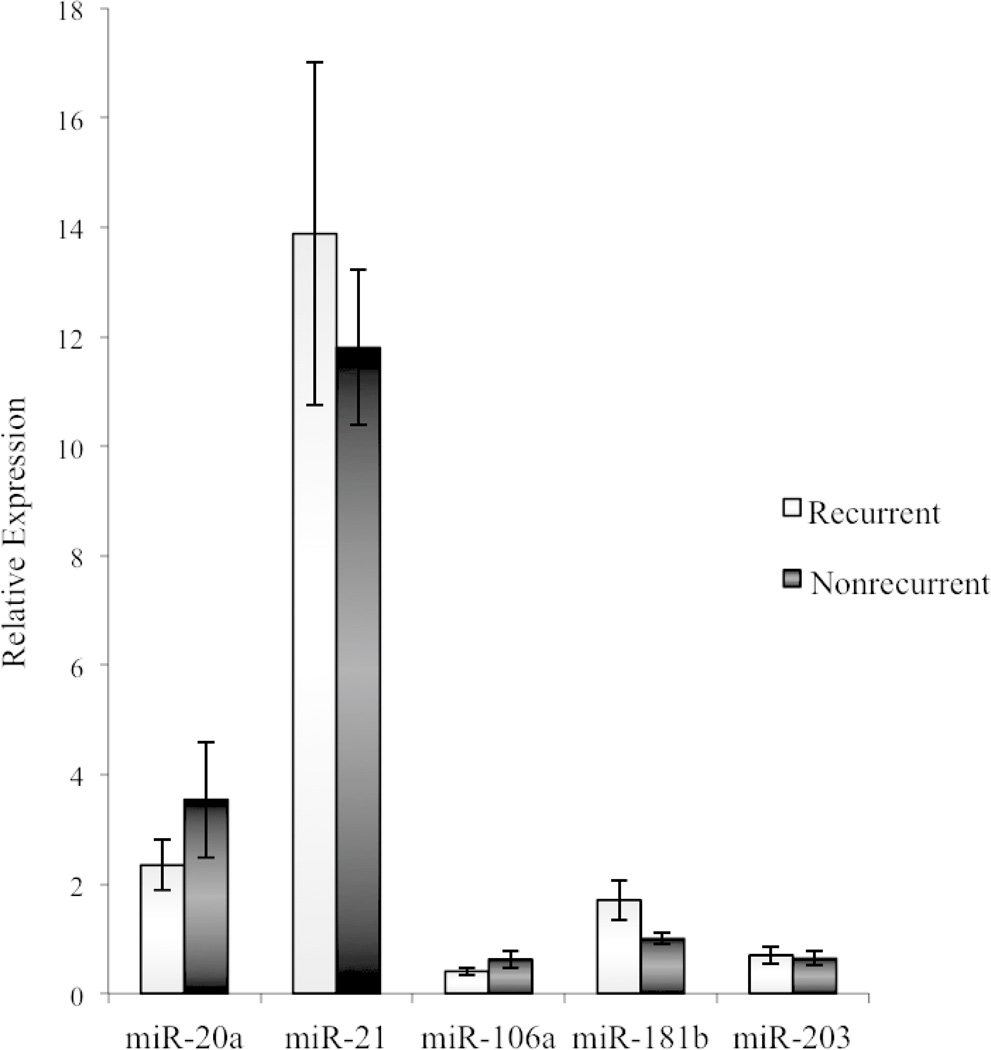
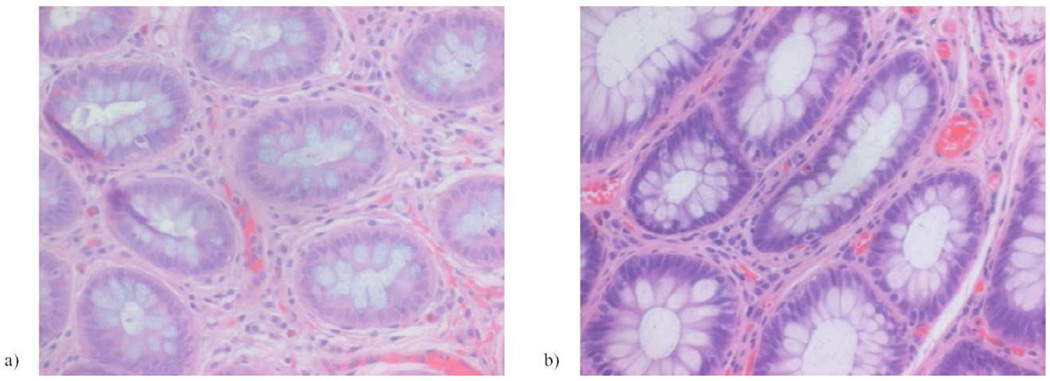
The miRNA profile of recurrent patients was compared to non-recurrent patients. In malignant tissue, miRNA expression in recurrent patients did not differ from non-recurrent patients (Fig. 2). That is, the miRNA expression in malignant tissue was equally elevated (relative to benign tissue) in recurrent and non-recurrent patients. However, benign tissue levels of all 5 miRNAs were significantly upregulated in patients who eventually experienced a cancer recurrence (P < 0.05) (Fig. 3). In other words, the benign tissue of recurrent patients showed molecular changes similar to malignant tissue (miRNA upregulation). This finding prompted reexamination of the pathology tissue blocks and slides to assess for any histologic signs of cancer at the tissue margins (labeled as benign tissue). After careful inspection by an experienced pathologist, all tissues from the cancer margin were found to contain only normal rectal mucosa(Fig. 4).
FIGURE 2.
Malignant tissue miRNA expression in recurrent and non-recurrent patients. The groups exhibited no differences in expression (P > 0.05). Error bars represent the standard error of the mean.
FIGURE 3.
Benign tissue miRNA expression in recurrent and non-recurrent patients. Error bars represent the standard error of the mean. *P < 0.05.
FIGURE 4.
Representative hematoxylin and eosin stained slides of tissue at the tumor margin are shown for a) non-recurrent and b) recurrent patients. All patients, both recurrent and non-recurrent, had histologically normal rectal mucosa at the tumor margin.
DISCUSSION
This study confirms the finding that miR-20a, miR-21, miR-106a, miR-181b, and miR-203 are upregulated in colorectal cancer [25]. Keeping in mind that colon and rectal cancers differ in their miRNA expression profiles [30], it is important to note that this is the first study showing that these miRNAs are upregulated specifically in rectal cancer tissue. This finding is consistent with the fact that these 5 miRNAs are all thought to act as oncogenes, largely by inhibiting tumor suppressor gene expression [15, 17, 35].
Given the proposed oncogenic activity of these miRNAs and their overexpression in rectal cancer tissue, we speculated that high tumor levels of miRNA would suggest a more aggressive tumor phenotype, and thereby predict cancer recurrence. Therefore, it was surprising to find that there was no difference in the tumor miRNA expression profiles of those patients who recurred versus those who did not. That is, the malignant tissue from recurrent and non-recurrent patients was molecularly similar. This result differed from other studies’ findings that the miRNA expression of colorectal tumor tissue could predict prognosis [20, 25, 28]. The discrepancy in results could be explained by differential expression of miRNAs between colon and rectal cancers [30].
Another unanticipated and intriguing finding was discovering miRNA upregulation in the benign tissue of patients who experienced cancer recurrence. These results show that even though recurrent and non-recurrent patients have normal histology at the tumor margin, the tissues’ molecular makeup is different. Such a finding suggests that miRNA expression has the ability to indicate a sort of precancerous state at the tumor margin. This conclusion is consistent with the concept of field cancerization, originally described by Slaughter in 1953 and supported by more recent molecular studies [4, 16, 31]. Interestingly, our finding is similar to a recent study demonstrating that miR-21 upregulation in the stroma of colon cancers can predict poor survival [23]. Further data have revealed high levels of miR-21 and miR-181b in premalignant lesions (colonic adenomas) [26], indicating that changes in miRNA expression may be an early step in the process of malignant transformation. Yet, to our knowledge this is the first study to show that miRNA expression in benign tissue from the tumor margin has prognostic value in rectal cancer.
The ability of miRNA to predict rectal cancer recurrence after local excision has substantial clinical significance. Currently, no reliable prognostic indicators exist for early-stage rectal cancers. Assessment of a miRNA profile, for example at the time of a rectal lesion biopsy, potentially could identify patients at high risk for cancer recurrence following local excision. Such knowledge would help guide surgeons’ choice of therapy (local excision versus radical resection). The local recurrence rate of 10–40% after local excision [24] makes it essential to correctly identify high-risk patients so that they may undergo a more aggressive procedure. On the other hand, patients who are found to be at low risk may be able to undergo a less invasive procedure than previously planned, thereby sparing the significant morbidity associated with radical surgery and improving quality of life [3, 10, 32]. In the future, miRNA expression profiles may even be used to guide the need for adjuvant and neoadjuvant therapy [25]. Furthermore, it has been shown that oncogenic miRNAs can be inhibited in vivo to decrease cellular proliferation, making miRNAs a novel target for future therapies [19, 21].
These original findings should be interpreted while taking into account the study’s limitations. First, this study’s size was relatively limited at 17 patients. Second, slightly over half of the tumors in this study were clinically staged as T2. Although this was not considered against standard of care during our study period, the number of T2 tumors in this study likely overestimates the number that would have undergone local excision today. This change in accepted management occurred largely due to studies published near the end of our enrollment period which showed that the T2 recurrence rate was too great to advise local excision as the standard of care [7, 14, 27]. Still, this does not seriously detract from the main point of our study, being that there may be prognostic value in the molecular profile of histologically benign tissue at the surgical margin. The study’s third limitation is that overall rates of recurrence were somewhat high (25% for T1 and 22% for T2 tumors). However, these rates were within the 10–40% range reported by other studies published during our enrollment period [6, 22, 24]. Still, since the method of clinical staging was not standardized in our study, it is possible that suboptimal staging accounts for these high recurrence rates. Fourth, quantitative data measuring the distance of the analyzed benign tissue from malignant tissue was not available due to the manner in which the tissue was obtained and archived. Therefore, a conclusion could not be made regarding whether or not the miRNA effect is distance-dependent. Instead, we are only able to conclude that histologically benign tissue was found to be molecularly different than histologically malignant tissue. A final limitation is that although the results presented here do support the possibility of using the miRNA profile as a prognostic marker, it remains uncertain how to best utilize this information in clinical practice. Further studies are needed to clarify the role of miRNA as a prognostic indicator in rectal cancer.
In future studies, our lab plans to investigate the biological relevance of miRNAs found to be upregulated in the benign tissue of recurrent patients. We plan to inhibit and overexpress miRNAs in vitro in order to clarify their role in tumor growth. Demonstrating a causal role in carcinogenesis would increase the validity of using miRNAs in prognosis. Other studies have already shown success in inhibiting tumor growth (both in vitro and in vivo) through ‘antagomirs’ that inhibit specific oncogenic miRNAs [19, 29].
In conclusion, this the first study to show that the miRNA profile of histologically normal tissue can provide important prognostic information regarding risk of recurrence in patients with early-stage rectal cancer. Although much uncertainty regarding the clinical utility of miRNA still exists, it seems likely that further research in this area will be vital in reducing recurrence rates after local excision of rectal cancer.
ACKNOWLEDGEMENTS
We would like to thank the Department of Surgery NIH T35 Short Term Training Grant DK 062709-0401 for providing the funding for this project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin. Chem. 2009;55:623–631. doi: 10.1373/clinchem.2008.112805. [DOI] [PubMed] [Google Scholar]
- 3.Bentrem DJ, Okabe S, Wong WD, Guillem JG, Weiser MR, Temple LK, Ben-Porat LS, Minsky BD, Cohen AM, Paty PB. T1 adenocarcinoma of the rectum: transanal excision or radical surgery? Ann. Surg. 2005;242:472–477. doi: 10.1097/01.sla.0000183355.94322.db. discussion 477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braakhuis BJ, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A genetic explanation of Slaughter's concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63:1727–1730. [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Chakravarti A, Compton CC, Shellito PC, Wood WC, Landry J, Machuta SR, Kaufman D, Ancukiewicz M, Willett CG. Long-term follow-up of patients with rectal cancer managed by local excision with and without adjuvant irradiation. Ann. Surg. 1999;230:49–54. doi: 10.1097/00000658-199907000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang AJ, Nahas CS, Araujo SE, Nahas SC, Marques CF, Kiss DR, Cecconello I. Early rectal cancer: local excision or radical surgery? J. Surg. Educ. 2008;65:67–72. doi: 10.1016/j.jsurg.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N. Engl. J. Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 9.Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol. Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Endreseth BH, Myrvold HE, Romundstad P, Hestvik UE, Bjerkeset T, Wibe A Norwegian Rectal Cancer Group. Transanal excision vs. major surgery for T1 rectal cancer. Dis. Colon Rectum. 2005;48:1380–1388. doi: 10.1007/s10350-005-0044-6. [DOI] [PubMed] [Google Scholar]
- 11.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 12.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 13.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 14.Gimbel MI, Paty PB. A current perspective on local excision of rectal cancer. Clin. Colorectal Cancer. 2004;4:26–35. doi: 10.3816/ccc.2004.n.007. discussion 36-7. [DOI] [PubMed] [Google Scholar]
- 15.Greither T, Grochola LF, Udelnow A, Lautenschlager C, Wurl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int. J. Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 16.Ibanez de Caceres I, Cairns P. Methylated DNA sequences for early cancer detection, molecular classification and chemotherapy response prediction. Clin. Transl. Oncol. 2007;9:429–437. doi: 10.1007/s12094-007-0081-9. [DOI] [PubMed] [Google Scholar]
- 17.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 19.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 20.Liu M, Chen H. The role of microRNAs in colorectal cancer. J. Genet. Genomics. 2010;37:347–358. doi: 10.1016/S1673-8527(09)60053-9. [DOI] [PubMed] [Google Scholar]
- 21.Manne U, Shanmugam C, Bovell L, Katkoori VR, Bumpers HL. miRNAs as biomarkers for management of patients with colorectal cancer. Biomark Med. 2010;4:761–770. doi: 10.2217/bmm.10.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellgren A, Sirivongs P, Rothenberger DA, Madoff RD, Garcia-Aguilar J. Is local excision adequate therapy for early rectal cancer? Dis. Colon Rectum. 2000;43:1064–1071. doi: 10.1007/BF02236551. discussion 1071-4. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen BS, Jorgensen S, Fog JU, Sokilde R, Christensen IJ, Hansen U, Brunner N, Baker A, Moller S, Nielsen HJ. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin. Exp. Metastasis. 2011;28:27–38. doi: 10.1007/s10585-010-9355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paty PB, Nash GM, Baron P, Zakowski M, Minsky BD, Blumberg D, Nathanson DR, Guillem JG, Enker WE, Cohen AM, Wong WD. Long-term results of local excision for rectal cancer. Ann. Surg. 2002;236:522–529. doi: 10.1097/00000658-200210000-00015. discussion 529-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitz KJ, Hey S, Schinwald A, Wohlschlaeger J, Baba HA, Worm K, Schmid KW. Differential expression of microRNA 181b and microRNA 21 in hyperplastic polyps and sessile serrated adenomas of the colon. Virchows Arch. 2009;455:49–54. doi: 10.1007/s00428-009-0804-0. [DOI] [PubMed] [Google Scholar]
- 27.Sengupta S, Tjandra JJ. Local excision of rectal cancer: what is the evidence? Dis. Colon Rectum. 2001;44:1345–1361. doi: 10.1007/BF02234796. [DOI] [PubMed] [Google Scholar]
- 28.Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and Prognostic Value of MicroRNA-21 and MicroRNA-155 in Colorectal Cancer. Oncology. 2010;79:313–320. doi: 10.1159/000323283. [DOI] [PubMed] [Google Scholar]
- 29.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 30.Slattery ML, Wolff E, Hoffman MD, Pellatt DF, Milash B, Wolff RK. MicroRNAs and colon and rectal cancer: differential expression by tumor location and subtype. Genes Chromosomes Cancer. 2011;50:196–206. doi: 10.1002/gcc.20844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 32.Stamos MJ, Murrell Z. Management of early rectal T1 and T2 cancers. Clin. Cancer Res. 2007;13:6885s–6889s. doi: 10.1158/1078-0432.CCR-07-1150. [DOI] [PubMed] [Google Scholar]
- 33.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. SciUS. A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waldman SA, Terzic A. Translating MicroRNA discovery into clinical biomarkers in cancer. JAMA. 2007;297:1923–1925. doi: 10.1001/jama.297.17.1923. [DOI] [PubMed] [Google Scholar]
- 35.Wang V, Wu W. MicroRNA-based therapeutics for cancer. BioDrugs. 2009;23:15–23. doi: 10.2165/00063030-200923010-00002. [DOI] [PubMed] [Google Scholar]
- 36.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]