1. Introduction
The discovery of Anthozoa homologs of the green fluorescent protein (GFP) from jellyfish Aequorea victoria, which emit not only green but also yellow, orange, and red fluorescence, provided a powerful boost for in vivo labeling due to the colors and biochemical features never before encountered in GFP variants.1,2 GFP and several Anthozoa fluorescent proteins (FPs) have been developed into monomers suitable for protein tagging, such as: (i) permanently fluorescent conventional blue FP, yellow FPs, orange FPs, red FPs (RFPs), (ii) permanently fluorescent GFPs and RFPs with a large Stokes shift fluorescence emission (LSS-FPs), (iii) irreversibly photoactivatable/photoswitchable GFPs and RFPs,3,4 and (iv) fluorescent timers (FTs).5,6 Among various fluorescent probes, the most valuable for deep-tissue and whole-body imaging are the red-shifted FPs because of reduced autofluorescence, low light-scattering, and minimal absorbance at longer imaging wavelengths. The mechanisms of formation of the GFP-like chromophore and its transformations were studied and described very well, however only a few reviews of those for RFPs are available 2,7.
Several mechanisms of the autocatalytic and photoinduced formation of the red chromophores have been proposed.7 Because of the complexity of the red chromophore transformations, no general integrating scheme has been suggested. Despite the numerous data available, there are long lasting contradictions about the formation of the red chromophore. From the discovery of a DsRed FP, the formation of a DsRed-like chromophore was commonly suggested to occur through a green GFP-like intermediate form,8 and only several recent publications uncovered that the formation of the DsRed-like chromophore occurs via a TagBFP-like blue intermediate form, not via the GFP-like one.
More than 140 crystal structures are currently available for FPs of different classes, and some of them are for the same FP in a different state or containing different mutations. An overview of the structural data together with FP spectral and photochemical properties illustrate the relationship between the FPs’ structure and function.
Here, we focus on a description of the chromophores in RFPs, suggest the mechanisms of the red chromophores formation and its further modifications, and attempt to discover general postulates for this complex chemistry. We also provide insights into how the red chromophore chemistry and the RFP crystal structures are translated into RFPs function. Lastly, we descuss major applications of RFPs in the modern imaging techniques.
2. Chromophore diversity in red fluorescent proteins
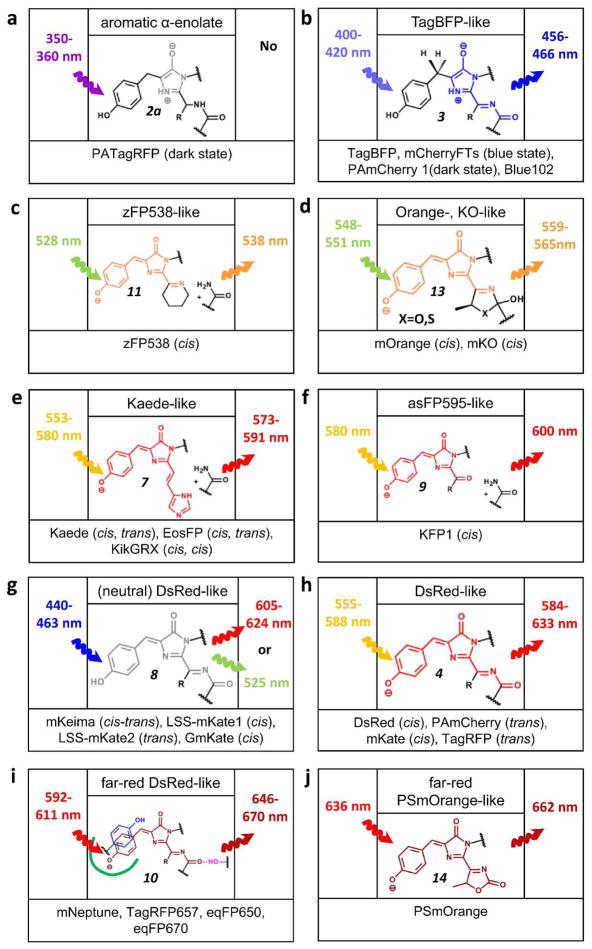
Recent advances in structural and biochemical studies of RFPs provide a better understanding of the molecular mechanisms of red chromophore autocatalytic synthesis and its further modifications, presenting a unique opportunity for researchers to manipulate spectral properties of RFPs. The post-translational chemistry of RFPs is diverse and complex, and covers mainly the autocatalytic and photoinduced formation of zFP538-,9 Orange-,10 KO-,11 Kaede-,12–14 asFP595-,15 neutral DsRed-,16–19 DsRed-,8,20–23 far-red DsRed-24–26 and far-red PSmOrange-like27 chromophores via intermediate forms such as aromatic α-enolate28 and TagBFP-like21,22,29 intermediates (Figure 1 and Table 1). We consider RFP being far-red if it has the excitation/emission maxima above 590 nm/630 nm, respectively.
Figure 1.
Chromophores in RFPs are shown in an order of their fluorescence emission color (from a through i). Excitation and emission maxima of respective chromophore for indicated FPs are shown on left and right sides, respectively. Configuration of a double bond between the Tyr-ring and imidazolinone is shown in the brackets. For Kaede-like chromophore configuration of the double bond between His-ring and imidazolinone is shown in the second position in the brackets. Numbering of the chromophore structures follows that in Figures 2 and 4. References and PDB ID numbers corresponding to the respective chromophores are listed in Table 1.
Table 1.
Structural data available for the discussed in the text blue, orange, red and far-red fluorescent proteins and their progenitors.
| Parental protein/Organism | FP name | Chemical structure of chromophore | PDB ID number of crystal structure | References |
|---|---|---|---|---|
| Kaede/Trachyphyllia geoffroyi stony coral | Kaede | Figure 1e | 2GW4 | 56,90 |
| EosFP/Lobophyllia hemprichii stony coral | EosFP | 2BTJ | 13,84 | |
| IrisFP | 2VVJ | 74 | ||
| KikG/Favia favus stony coral | KikGR | 2DDD | 14,150 | |
| asFP595/Anemonia sulcata sea anemone | KFP | Figure 1f | 1XQM, 1XMZ | 15,57,151 |
| Keima/Montipora sp. stony coral | mKeima | Figure 1g | 3IR8, 2WHU, 2WHT, 2WHS | 16,17,147 |
| KO/Fungia concinna stony coral | mKO | Figure 1d | 2ZMW, 3MGF, 2ZMU | 11,152 |
| DsRed/Discosoma sp. mushroom coral | mOrange | Figure 1d | 2H5O | 10,93 |
| DsRed | Figure 1h | 1GGX, 1G7K, 1ZGO, 1ZGP, 1ZGQ | 92,103,153 | |
| mStrawberry | 2H5P, 2H5R | 10,93 | ||
| mCherry | 2H5Q | 10,93 | ||
| PAmCherry | 3KCT | 21,60 | ||
| PSmOrange | Figure 1j | none | 27 | |
| eqFP611/Entacmaea quadricolor sea anemone | eqFP611 | Figure 1h | 1UIS | 154,155 |
| d1eqFP611 | Figure 1h | 3E5T | 76 | |
| d2RFP630 | Figure 1i | 3E5V | ||
| RFP639 | 3E5W | |||
| HcRed/Heteractis crispa sea anemone | HcRed | Figure 1i | 1YZW | 156 |
| eqFP578/Entacmaea quadricolor sea anemone | TagBFP | Figure 1b | 3M24 | 22,36 |
| LSSmKate1 | Figure 1g | 3NT9 | 18,44 | |
| LSSmKate2 | 3NT3 | 18,44 | ||
| GmKate | 3SVS, 3SVR, 3SVO | 19 | ||
| TagRFP | Figure 1h | 3M22 | 22 | |
| mKate | 3BX9, 3BXA, 3BXB | 23,117 | ||
| mNeptune | Figure 1i | 3IP2 | 24 | |
| TagRFP657 | none | 25 | ||
| eqFP650 | none | 26 | ||
| eqFP670 | none |
2.1. Common blue intermediate in the formation of red chromophores
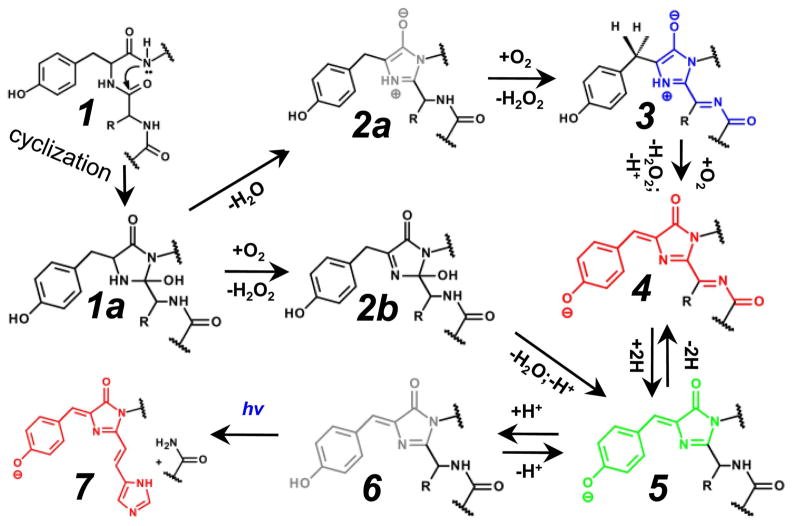
An autocatalytic mechanism of the formation of red chromophores is more complex than that of the GFP-like chromophore because one to three additional double bonds are present in chemical structure of the red chromophore as compared with those present in GFP-like green chromophore. Since only some of these additional double bonds were already present in the chromophore tripeptide, additional posttranslational steps are required for their formation. Many of the currently known RFPs share the DsRed-like chromophore (structure 4 in Figures 1 and 2)8 after the first protein where it has been found. DsRed-like chromophore has the same structure as GFP-like chromophore but with additional N-acylimine group (C=N-C=O). Several versions of the autocatalytic chemical mechanism for the DsRed-like chromophore formation have been recently suggested such as “via-GFP”, “via-TagBFP”, and “branched pathway model”.7,30 All these mechanisms suggest as the first step a cyclization of the chromophore-forming tripeptide (Figure 2, 1→1a) in a way similar to that suggested for the GFP-like chromophore, involving catalysis by its Arg96 residue that is conservative among FPs.31–34
Figure 2.
Mechanisms of autocatalytic formation of the chromophores in fluorescent proteins are shown. The color of the chemical structures corresponds to the spectral range of the chromophore fluorescence emission, where the gray color denotes the non-fluorescent state. Many of the indicated steps may also be photo-induced. Chromophores are presented in a cis-isoform. Possible trans-isoforms are not shown. The hv symbol designates UV-light.
The earliest “via-GFP” hypothesis suggested that DsRed-like chromophore formation occurs through a GFP-like green intermediate 5.8 Specifically, this hypothesis suggested that the oxidation of Cα-Cβ bond of the Tyr chromophore occurs first and the N-acylimine formation occurs afterwards. This “via-GFP” mechanism could not explain the results of the kinetic study which reported that the blue-absorbing form 3 is the true intermediate during DsRed-like chromophore formation and the GFP-like green form 5 is the dead-end product.35 It was recently confirmed that the DsRed-like chromophore is indeed formed through the autocatalytic posttranslational modification of a blue TagBFP-like chromophore 3.5,21,22,29,36 The chemical structure of this TagBFP-like intermediate (found in the dark state of PAmCherry121 first and later in TagBFP36 and in the blue form of FTs5) was determined according to the strong evidence coming from X-ray, mass-spectrometry, and mutagenesis data.21,22,29
According to the “via-TagBFP” mechanism, the N-acylimine is formed first as a result of the tripeptide cyclization followed by dehydration/oxidation steps (Figure 2, steps 1a→2a→3) or, alternatively, oxidation/dehydration steps (Figure 2, steps 1a→2b→3). Step 3→4 with the oxidation of the Cα-Cβ bond of Tyr chromophore occurs afterwards. Of the two alternative mechanisms suggested for the formation of N-acylimine, the mechanism including 2b intermediate is similar to that suggested for GFP chromophore.37 During N-acylimine formation (step 2a→3 or 1a→2b), the Glu222 (here and below amino acids numbering follows that for GFP, see Figure 3) amino acid was suggested to be a general base for the proton abstraction either from Cα atom of the Tyr chromophore or from the Cα atom of the first residue in a chromophore-forming tripeptide.21 This residue can also protonate N1 atom of the 5-member ring of chromophore 4-imidazolone. Arg96 may be stabilizing the negative charge on the O-atom of 4-imidazolone ring. The general bases for the proposed proton abstraction from the Cβ atom of Tyr66 chromophore in the final oxidation step 3→4 are suggested to be the tandems of Glu222/Lys69 or Glu222/Arg69 (in the case of mCherry and FTs21,29, respectively), and Glu222/Arg203 (in the case of PAmCherrys21). Hence, the suggested general bases for proton abstraction in RFPs are different from Arg96 which was supposed to be the base during the GFP-like chromophore formation.37 The formation of unstable peroxy adduct during the 3 to 4 oxidation step was proposed.29 The formation of a similar hydroperoxy intermediate has been earlier suggested for maturation of the GFP-like chromophore.38
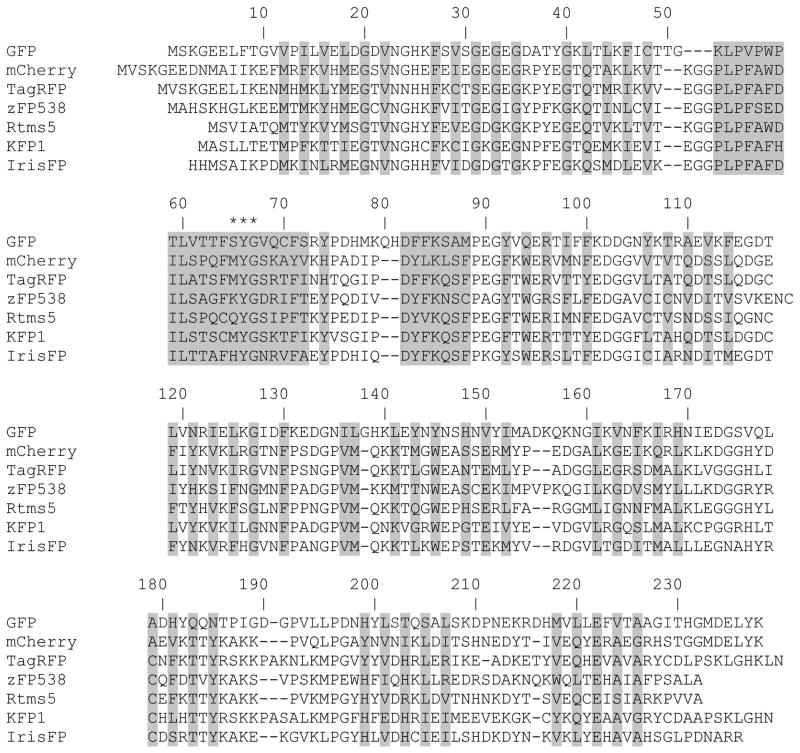
Figure 3.
Alignment of amino acid sequences for selected RFPs relative to Aequorea victoria GFP. Alignment numbering follows that of GFP. Residues buried inside of FPs’β-barrels are shaded. Asterisks indicate the residues that form the chromophore.
Recently, a “branched pathway model” for the DsRed chromophore formation based on the kinetic studies has been proposed.7,39 This model denies formation of the GFP-like green intermediate during the DsRed chromophore maturation. To explain kinetics of the hydrogen peroxide production during the protein maturation, the “branched pathway model” postulates that all oxidation steps occur before the formation of the blue intermediate 3 39. This is at a variance with the experimental observations that after the formation of the blue form 3 there is no blue-tored chromophore transition in the absence of oxygen. For example, oxygen was shown to be required for the blue-to-red maturation of FTs29 and for the photoactivation of PAmCherrys.21 The “branched pathway model” of the DsRed-like chromophore formation also introduces a new cationic chemical structure for the blue intermediate 3. However, quantum mechanical calculations revealed that the cationic structure of the blue form 3 is unlikely because it should have an excitation maximum at ~230 nm.40 Based on the same calculations, the anionic or zwitter-ionic forms of the blue intermediate 3, suggested in the “via-TagBFP” mechanism, should have excitation maxima at ~400 nm, which coincides with that observed for mTagBFP and blue-emitting derivatives of RFPs.40 Thus, the “via-TagBFP” mechanism explains all currently available data and is the preferred chemical scheme for the formation of the DsRed-like chromophores.
The GFP-like intermediate 5 is an uncommon intermediate during the formation of red chromophore, which has only been observed in the case of z2FP574 red fluorescent protein. It has been suggested that the red chromophore of z2FP574 arises from a coupled oxidation-decarboxylation of Asp, the first amino acid of the chromophore-forming DYG tripeptide.41,42 The “green” anionic GFP-like form 5 was shown to be the actual intermediate which is further oxidized to DsRed-like chromophore 4 according to the chemistry of β-keto acid decarboxylation. It has been proposed that the formation of the zFP574-like chromophore occurs via two consequent steps of oxidation and decarboxylation of the GFP-like chromophore,43 not a coupled one as suggested earlier.41 The formation of asFP595 chromophore was suggested to occur via a protonated GFP-like intermediate, however, more evidence is needed to distinguish between the GFP- and TagBFP-like intermediates. Indeed, both absorbance maxima of the observed intermediate form at 420 nm and 20 Da loss in molecular mass of resulting chromopeptide as compared to the peptide before posttranslational modification can be attributed to either GFP- or TagBFP-like intermediates with no preference.
2.2. Chemical transitions in the formed red chromophores
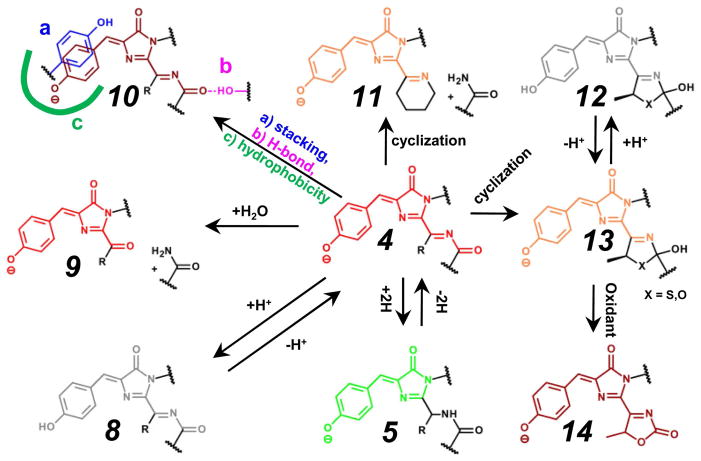
Once the red chromophore 4 is formed, it can undergo several chromophore transitions, such as protonation-deprotonation, as well as cyclization at the N-acylimine group and “greening” (Figure 4).
Figure 4.
Major autocatalytic modifications of the red chromophore in FPs and their interaction with the immediate surroundings are shown. The color of the chemical structures of chromophores corresponds to the spectral range of the chromophore fluorescence emission, where gray color denotes the non-fluorescent state. Many of the indicated chromophore transformations may also be photo-induced. Chromophores are presented in a cis-form. Possible cis-trans isomerizations are not shown. Transition 4→10a, stacking interactions with tyrosine ring are shown, however, a similar interaction with Phe, Arg, His, or Trp residues are also possible. Transition 4→10c, a green line denotes hydrophobic amino acid’s surrounding of the chromophore. Transition 13→14, an oxidant is molecular oxygen or chemical compound such as potassium ferricyanide or benzoquinone.
Protonation of the DsRed-like chromophore 4 at the hydroxyl group of the Tyr chromophore results in the formation of the neutral (protonated) form 8 absorbing around 440–460 nm, as seen for mKeima and LSSmKates18,44 RFPs with a large Stokes shift. This protonation is a result of the presence of the hydrogen bond (H-bond) network, which includes the hydroxyl group of the Tyr ring of the chromophore and strong proton donors such as Glu, Asp with lower pKa values (pKa~4.5) than that for phenolic group of chromophore (pKa~7.1–7.8).45,46 The water molecules and amino acids with proton donor-acceptor properties could be the link between Glu or Asp as a proton donor and the hydroxyl group of the chromophore Tyr as an acceptor. The 440–460 nm absorbing form 8 in an excited state can undergo an excited-state proton transfer (ESPT), thus generating the intermediate red anionic excited state 4 that first emits a red photon similar to an anionic RFP-like chromophore and then goes back to the protonated state.47 Examples of ESPT pathways include proton wires that consist of chromophore Tyr hydroxyl→ Asp167 for LSSmKate1,18 chromophore Tyr hydroxyl→ Ser165→ Asp167 for LSSmKate2,18 or chromophore Tyr hydroxyl→ Ser148 → Glu165 for mKeima.16 The basis for the ESPT relies on the fact that the pKa value of the phenolic group of the chromophore in the excited state dramatically decreases, reaching ~3,18,44 which explains why in the excited state it can protonate very strong proton donors such as Glu and Asp.48 For the ESPT to occur, the geometry of the hydrogen bond network around phenolic group of the chromophore must satisfy two conditions: the right distance between proton donor/acceptor, and the right angle between the hydrogen donor and acceptor. These parameters can affect the rate of the proton transfer, and even completely block this process.47,48 The short H-bond between chromophore and Asp148 was suggested to be responsible for the ultrafast (<170 fs) ESPT observed in S65T/H148D GFP mutant.49 A more optimal geometry of the proton transfer pathway in red fluorescent protein mKeima was suggested to be responsible for a faster rate of proton transfer of ~4 ps as compared with those for GFP variants with slower proton transfer rates of ~12 ps. The replacement of protons with deuterium ions slows down the rate of the proton transfer and results in an appearance of emission of the protonated chromophore.16,18 The more protons are involved in the proton wire, the greater the deuterium isotopic effect is. Proton relay can be blocked by Glu-Arg salt bridges, threonine side-chain rotation50 or calcium ions, as have been suggested for Ca2+ indicators.51
Additional N-acylimine group of the DsRed-like chromophore allows for the transitions different from those for the GFP-like chromophore. Further cyclization at the N-acylimine group in the DsRed-like chromophore results in formation of an mOrange-like or mKO-like 13 6,10 or zFP538-like 11 9 chromophores exhibiting orange emission. This cyclization is initiated by the first residue of the chromophore-forming tripeptide. A 5-membered oxazole ring of mOrange is formed as a result of the intramolecular attack of the OH-group of Thr65 at the carbonyl carbon of N-acylimine with no backbone cleavage.10 Deprotonation of the OH-group of Thr65 was suggested to be the prerequisite for the attack on the carbonyl carbon of residue 64.10 We propose that the formation of 5-membered ring of mKO follows the same mechanism. In zFP538, the third 6-membered ring is formed as a result of intramolecular nucleophilic attack of NH2-group of Lys65 residue on N-acylimine with the backbone cleavage before chromophore.9 Further hydrolysis of N-acylimine group of DsRed-like chromophore results in main chain break immediately preceding the chromophore tripeptide and formation of the asFP595-like chromophore 9.15
The photoconversion of the DsRed-like chromophore 4 into GFP-like chromophore 5, called “greening”, was demonstrated for mKate and HcRed using high-power irradiation at 405 nm and 561 nm.52 The mechanism of this photoconversion is not clear but could include the N-acylimine reduction or degradation with the formation of GFP-like chromophore 5. DsRed-like chromophore 4 can be photoconverted to far-red shifted red fluorescence species upon high-intensity irradiation with 532 nm light.53 This photoconversion occurs through a two-photon absorption process with cis-trans isomerization of the chromophore and decarboxylation of Glu222 via a Kolbe-like mechanism.53 The bathochromic shift was suggested to be caused by a different immediate environment of the red chromophore in its trans-state vs. its cis-state. The light-induced Kolbe-like mechanism for the Glu222 decarboxylation was earlier described for GFP.54 It consists of (i) an electron transfer from the carboxylic group of Glu222 to the neutral chromophore in the excited state; (ii) a decarboxylation with the formation of an electron-deficient carbon radical, and (iii) a radical recombination that may involve either a direct transfer of an electron and a proton from the chromophore or a transfer of a hydrogen radical from the chromophore.
2.3. Transformation of red chromophores into orange chromophores
mOrange-, zFP538-, and mKO-like orange chromophores can be attributed to the same group because they have an additional cycle as compared to the DsRed-like red chromophore. The mOrange- and mKO-like 13 and zFP538-like 11 chromophores are suggested to form via the DsRed-like chromophore 4, as described above. mOrange-, mKO- and zFP538-like chromophores should be able to undergo similar transformations observed for the GFP- and DsRed-like chromophores, such as cis-trans isomerization of Tyr ring and its phenolic group protonation-deprotonation accompanied by the ESPT process. To date, these transformations have not been observed but provide strong basis for the development of FPs with a large Stokes shift and reversibly switchable behavior in orange part of light spectrum. Because of the presence of the additional cycle, the mOrange-like chromophore was shown to be able to undergo different transformations from those observed for GFP- and DsRed-like chromophores. Illumination of mOrange-like chromophore 13 with either 458 nm or 488 nm laser generated farred species 14.52 The far-red shifted absorbance/emission maxima of these far-red species were around 610 nm/650 nm, respectively, and suggested the formation of the chromophore that is different from the DsRed-like chromophore 4. This photoconversion was shown to be a multiphoton process that required higher light power, suggesting that at least two or more steps occur during photoconversion.52
Intense irradiation of the mKO-like chromophore 13 with 436 nm light from mercury lamp can induce the 13-to-5 conversion resulting in the “greening” process.55 Illumination with 514 or 543nm laser resulted in a minor red-shift in the emission of mKO.52 In contrast to mOrange-like chromophore, the illumination of the mKO-like chromophore did not result in formation of far-red species 14,52 suggesting unique properties for the mKO-like chromophore.
3. Photo-induced transformations in red chromophores
Photochemical behavior of RFPs was discovered relatively recently. After a sample of the green Kaede protein had been exposed to direct sunlight, it had turned red.56 Kindling property of the asFP595 reversibly switchable RFP was found after its illumination with light of different wavelength.57 In a relatively short period of time many different light-inducible processes were discovered in RFPs, including the formation of chromophores with new chemical structures, alteration of the chromophore’s immediate environment and contacts via decarboxylation or conformational change of the neighboring amino acids, and alteration of the chromophore’s configuration resulting from its cis-trans isomerization.
3.1. Light adds more options to chromophore transitions
Chemistry of the chromophore phototransitions is more complex than that for autocatalytic transformations. Photoactivation light delivers additional energy (one, two, or three photons) to the chromophore, enabling chromophore modifications via a photo-chemical reaction that otherwise would not have occurred autocatalytically due to a high energy barrier. Thermal energy consumed during autocatalytic chromophore transformations can be substituted with light energy. For example, the maturation of DsRed chromophore 4 can be accelerated using illumination with UV-violet light35. The blue-to-red transition that occurs over time in mCherry-derived FTs can be also accelerated with violet light.5 Since the wavelength of the photoactivation light can be regulated, researchers can selectively deliver energy to the differently absorbing forms of chromophores and thus selectively induce different transformations. For example, illumination of PATagRFP 350 nm absorbing form 2a with ~350 nm light results in the accumulation of TagBFP-like intermediate form 3,28 while illumination with ~400 nm light results in the form’s further transformation into form 4 with the DsRed-like chromophore.
Low-intensity photoactivation light of 10−4–10−2 W/cm2 can efficiently accelerate chromophore transformations that require the energy of one photon such as cis-trans isomerization of DsRed-like chromophore, which is responsible for the reversible photoswitching of rsTagRFP.58 The light with higher intensity of 10−1–103 W/cm2 causes photoconversion of the PSmOrange27 and PATagRFP28 proteins; this photoconversion requires absorbance of two photons. These two-photon photoconversion processes consist of two consequent one-photon processes with the photon absorbance by two different forms of the chromophore. The higher intensity is possibly required due to the suboptimal wavelength of light for the forms of the chromophore having different absorbance. High intensity light of ~105–107 W/cm2 can induce modifications of the chromophore with the consumption of three photons. For example, illumination of DsRed with a two-photon excitation source at 750 nm causes its fluorescence change from red to green.59 It has been suggested that the red-to-green photoconversion was a result of three-photon absorption.59
3.2. Diversity of photo-transformations in DsRed-like chromophore
The DsRed-like red chromophore 4 formation could be induced by irradiation with violet light21, as is the case of PAmCherrys60 and PATagRFP28. The red chromophores formed as a result of the illumination of these FPs are the same as the DsRed-like chromophore 4 but the mechanisms of their formation are different. In the original dark ground state PAmCherrys have the TagBFP-like chromophore 3, which converts into the DsRed-like chromophore 4 in response to UV-illumination. PATagRFP stops its maturation at a cyclized and dehydrated chromophore 2a, while during UV-light irradiation it undergoes the transition through the TagBFP-like intermediate 3 into the DsRed-like product 4.
The “redding” of the GFP-like chromophore 5 could be photoinduced under anaerobic conditions (“anaerobic redding”)61,62 or both aerobic and anaerobic conditions in the presence of oxidants (“oxidative redding”) for a number of green FPs from the GFP-family of Hydrozoa, Anthozoa, and Crustacea classes.63 The mechanism of “anaerobic redding” is not clear but potentially represents a GFP-like chromophore 5 photoreduction with the formation of a stable oxygen-sensitive radical state of the GFP chromophore.63 The two-electron oxidation mechanism of “oxidative redding” with the absorbance of one-photon has been proposed.63 This mechanism includes two-steps: first, the GFP-like chromophore 5 in an excited state donates one electron to an oxidant molecule with the formation of a short-lived cation-radical intermediate, which then reacts with an electron acceptor to form the DsRed-like chromophore 4. This “oxidative redding” process can be exploited for the development of fluorescent probes for the measurement of redox potential in a live cell.
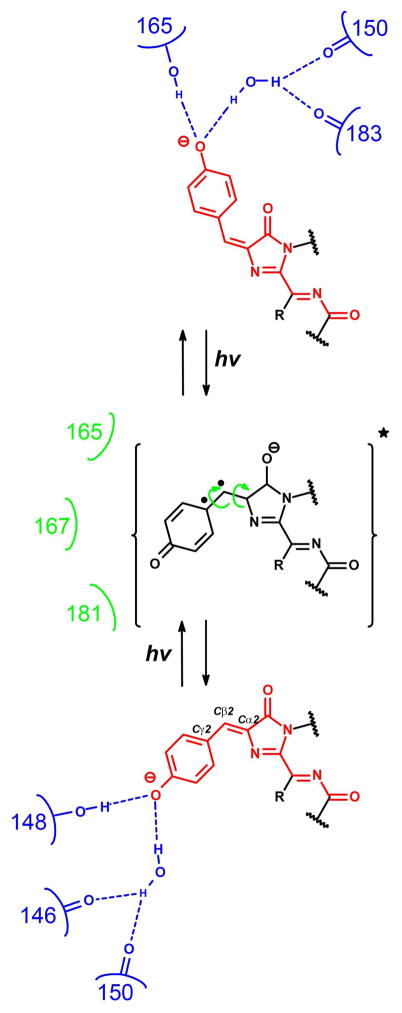
The formal rotation of a Tyr ring of the chromophore around Cα2-Cβ2 double bond is called cis- trans isomerization (Figure 5). This isomerization process was shown to be light-inducible and can also be driven via the alteration of pH.
Figure 5.
Cis-trans isomerization of the red chromophore in fluorescent proteins induced by light and regulated by surrounding amino acids. The suggested intermediate (data from ref. 78) representing chromophore in excited state is shown in brackets. Residues stabilizing chromophore in its cis- or trans-configuration via hydrogen bonds (dash lines) are shown in blue color. Hydroxyl group of the Tyr-ring of the chromophore can be protonated in cis- or trans-configuration of the chromophore. Residues hindering movement of Tyr ring by direct or indirect steric clashing are shown in green color. Electrons on the high-energy orbitals are shown as dots.
The pH-dependent cis-trans transition was observed on crystal structures of mKate,23 eqFP578, Katushka64 and Rtms5/H148S,65 and suggested for KFP.66 The protonation-deprotonation of conserved Glu215 close to chromophore was suggested to be involved in promoting of the cis-trans isomerization of Rtms5/H148S. The pH-induced cis-trans isomerization of KFP chromophore was shown to be due to the changes in the hydrogen bond network around the chromophore including side chains of Cys64 and Ser165.66
Light-inducible cis-trans isomerization of the DsRed-like chromophore structure 4 is the more commonly observed phenomenon, and is thought to be responsible for the reversible behavior of photoswitchable cyan mTFP0.7,67 green Dronpa68 and its derivatives,69–71 red asFP59557 and its KFP derivative,72 rsCherrys,73 rsTagRFP,58 and green-to-red Iris-FP74. The synthetic GFP-like cis-chromophores in the solution were shown to be more stable (on the energy of about one hydrogen bond) than their trans-counterparts, perhaps due to the steric clashing between Tyr ring of the trans-chromophore and oxygen atom of the imidazolinone 5-member ring.75 In contrast to the solution, the red chromophore inside of the β-barrel of FPs can be stabilized in both cis- and trans-configurations. The side chains of the amino acid residues at the positions 148 and 165 were shown to stabilize the chromophore in its cis- or trans-configuration, respectively, by making a direct H-bond with the hydroxyl of the chromophore in RFPs such as mKate,23 TagRFP,22 Katushka64, KFP, eqFP611,76 mStrawberry and mCherry10 (Figure 5). The water molecule, which is coordinated by amino acids in positions 146, 205 and 150, 183, can perform the same function and primarily or additionally stabilize the cis- or trans-configuration of the chromophore, respectively, in various proteins such as mKate,23 mOrange, mCherry,10 PAmCherry in ON state,21 Katushka,64 and KFP/S165V in ON state.77
According to the quantum calculations for KFP chromophore within molecular cluster modeling protein environment, the light-induced cis-trans isomerization of the red chromophore is accompanied by the absorption of a photon which initiates a movement of electrons from the π-π electron orbital of double bond to the higher-energy orbitals, with the formation of the chromophore in the excited state with a temporal Cα2-Cβ2 single bond (Figure 5).78 Similar results on quantum calculations for the GFP and KFP chromophores in vacuo have been described previously.79,80 Data are contradictory regarding the protonation state of the chromophore during its cis-trans isomerization but the fact that the chromophore in excited state has low pKa value of ~3 (ref. 18,44) supports the concept that the chromophore is deprotonated during isomerization, with the structure shown in Figure 5. Molecular dynamics simulations visualized that trans-to-cis isomerization of the chromophore occurs by means of both 90° rotation of the Tyr phenolic ring around Cβ2-Cγ2 bond and 180° rotation of Cβ2 CH-group around Cα2-Cβ2 and Cβ2-Cγ2 single bonds.81 After Tyr-ring and CH-group movements, the Cα2-Cβ2 double bond reforms and locks the Tyr phenolic ring back into its position. This isomerization occurs in a few picoseconds. The movement of the Tyr phenolic ring in excited state can be hindered by direct or indirect steric clashing with amino acid residues at positions 165, 167 and 181 in RFPs such as rsCherrys,73 KFP,82 rsTagRFP58 and IrisFP.74 The replacement of the bulky amino acids at the positions 165, 167 and 181 facilitates cis-trans transition. Hence, the cis-trans transition of the chromophore phenolic ring can be regulated both by the steric clashing with surrounding amino acids and by the hydrogen bond formation with a hydroxyl group of Tyr of the chromophore (Figure 5).
NMR study in solution of Dronpa FP suggests another mechanism of reversible photoswitching of FPs.83 Light-inducible changes in the interactions between the chromophore and β-barrel are suggested to be responsible for the switching between the ON and OFF states rather than cis-trans isomerization of the chromophore phenolic ring. The rigid attachment of the anionic chromophore to the protein matrix via hydrogen bonding favors fluorescence in the ON state. In contrast, a protonation of the chromophore hydroxyl moiety results in a loss of interactions with β-barrel and higher flexibility of the chromophore in the OFF state, which decreases its fluorescence.
3.3. Chromophore formation in Kaede-like proteins requires light
A Kaede-like chromophore 7 is characteristic for the red state of green-to-red irreversibly photoswitchable fluorescent proteins (PS-FPs) such as Kaede,12 EosFP/tdEosFP/mEos2,84,85 Dendra/Dendra2,86–88 KikGR/mKikGR,85 and mClavGR1/mClavGR2.89 The key feature of this type of FPs is that they have a His65 residue at the first position of the chromophore-forming tripeptide X65-Tyr66-Gly67. Another unique property of Kaede-like FPs is that formation of a Kaede-like red chromophore 7 requires UV or UV-violet light irradiation, and is not possible via an autocatalytic reaction.
In the original green fluorescent state, Kaede-like PS-FPs have an anionic GFP-like chromophore 5. It has been shown that the photoconversion process starts from the neutral form of GFP-like chromophore 6.56 The UV-light illumination excites the protonated GFP-like form 6 of Kaede-like PS-FP, and the His65 residue is protonated via a hydrogen network that catalyzes the formal β-elimination reaction, resulting in a peptide cleavage between the Nα and the Cα of His65 (Figure 2).90 The competitive E1- and E1cb-type β-elimination mechanisms of the red Kaede-like chromophore formation were suggested to involve deprotonation at Cβ of His65 by Glu222 followed by N-Cα bond cleavage at His65.91 According to the X-ray structure of KikGR protein, the imidazole ring of His65 residue flips around Cα-Cβ bond in KikGR protein supporting E1-mechanism of photoinduced β-elimination.14 Because His65 residue of the chromophore takes part in the formation of the red Kaede-like chromophore, possible chemical transformations of the Kaede-like chromophore are potentially limited.
Reversible cis-trans isomerization of Kaede-like chromophore involving rotation of the Tyr66 ring around Cα2 –Cβ2 bond was proven to occur in an IrisFP according to crystal structure data.74 This cis-trans isomerization was observed for both green 5 and red 7 states. Switching ON and OFF lights were different for green and red states, and coincided with the absorbance maxima of neutral and anionic forms of GFP-like chromophore in the green state and with those of neutral and anionic forms of Kaede-like chromophore in the red state. This evidence supports the notion that all types of chromophores can undergo photoinduced cis-trans isomerization process via a similar mechanism. Another feature of Kaede-like chromophore is the possibility of its cis-trans isomerization via the rotation of the His65 imidazole ring around the Cα-Cβ double bond. This isomerization has not yet been observed, however its possibility is supported by the fact that the His65 imidazole ring is present in trans- or cis-configuration in Kaede, EosFP or KikGR Kaede-like PS-FPs, respectively.14 The cis-configuration of His65 Cα-Cβ double bond is responsible for ~16 nm bathochromic shift of the emission maxima observed in KikGR protein as compared with that for Kaede protein having trans-configuration of the His-65 Cα-Cβ double bond.
4. How general are the principles of chromophore transformations?
Based on numerous data in the published literature and on availability of respective fluorescent proteins, three conclusions can be formulated (Figures 2 and 4): (i) many chemical transitions between chromophore structures can either occur autocatalytically, or photochemically, or can be blocked; (ii) many chromophore structures can either be in a fluorescent state (which absorbs and emits light) or in a chromo state (which absorbs but does not emit light), (iii) autocatalytical vs. photoinduced vs. blocked and fluorescent vs. chromo choices are mainly determined by the amino acid residues in the chromophore and its surrounding environment (the so-called molecular determinants of color35). The conclusions (i)–(iii) present an excellent basis for the development of new FPs optimized for different applications.
4.1. Different fluorescent proteins made on the basis of DsRed and its derivatives
The statements (i)–(iii) can be illustrated using RFPs with the DsRed-like chromophore, DsRed92 and its monomeric versions, collectively called mFruits (mOrange, mCherry, mStrawberry, etc.).93 Originally, the chromophore in these proteins autocatalytically develops/matures from the structure 1 via 2a and 3 to 4 (Figure 2).10 In the mCherry’s mutant mCherry/M65L/L84W/S148I, called Blue102, the chromophore stops maturation at structure 3.29 In the mCherry-derived Fluorescent Timers (FTs),5 which change its fluorescence from blue to red over time, the blue forms have chromophores with the structure similar to structure 3, and the blue-to-red autocatalytic maturation from 3 to 4 was slowed down to various degrees in FastFT, MediumFT, and SlowFT variants.5 It has been shown that the blue-to-red transition in FTs could also be induced with violet light. After maturation, chromophores of photoactivatable mCherry variants, called PAmCherrys, are in the dark OFF state absorbing at 400 nm with no detectable blue emission60 and have chemical structures similar to 3.21 After irradiation with violet light, the PAmCherry chromophores photo-chemically convert into structures similar to 4 (in a trans-isoform).21 An example of the chromo state of the chromophore structure 4 is the OFF (dark) state of the reversibly photoswitchable mCherry variants, called rsCherry and rsCherryRev.73 Introduction of strong proton donors in a close proximity to chromophore 4 (such as Glu or Asp at positions 158 or 160) in mCherry, mStrawberry, and mOrange causes a formation of a protonated (neutral) form 8, which absorbs at 434–457 nm and fluoresces at 560–610 nm, owing to the ESPT (see below).18 A water-mediated or direct hydrogen bond formation between the acylimine oxygen of the chromophore and the side chain of amino acid at the position 16 (Figure 2b, structure 10) was shown to be responsible for the formation of the far-red chromophore 10 in mPlum94,95 and mRouge96 proteins. The π-stacking interactions between chromophore and aromatic surrounding amino acids, such as Tyr203 in mRojoA96 and mGrapes24 and Tyr203 and Phe65 in E2-Crimson,97 can also cause formation of the chromophore 10. Mutations at the first amino acid in the chromophore-forming tripeptide and amino acids around the chromophore resulted in the mFruits variants having different chromophores, such as 4 and 13, with excitation/emission shifted for 47 nm/57 nm, respectively.10,93
4.2. Various fluorescent probes derived from TagRFP
Regulation of chromophore transformations via mutagenesis is not limited to the DsRed derivatives. Another subfamily of RFPs, based on TagRFP, exhibits those transformations as well. In the original TagRFP, the chromophore autocatalytically transforms from structure 1 via 2a, 3 to 4 (in a trans-isoform).98 In the TagRFP’s far-red mutants, called mKate/mKate298,99 and mNeptune,24 the chromophore 4 exhibits a π-cation stacking with Arg203 (another type of stacking, not as depicted in 10).23 In the TagRFP’s mutant, called TagBFP36, the chromophore stops maturation at structure 3.22 In the dark state, the chromophore of the photoactivatable TagRFP variant, PATagRFP, most likely has the chemical structure 2a.28 After irradiation with UV-violet light, the PATagRFP chromophore photochemically transforms via a detectable structure 3 into 4.28 In the mKate’s mutants, called LSSmKates, chromophore that absorbs around 460 nm light is a neutral (protonated) DsRed-like chromophore (structure 8).18,44 In the excited state, the ESPT occurs from a hydroxyl group of Tyr66, resulting in an anionic (deprotonated) DsRed-like chromophore (structure 4), which then emits a low-energy red photon.18 A similar ESPT phenomenon also occurs in LSS-GFPs, such as Sapphire100 and Ametrine,101 where structure 6 absorbs but structure 5 emits fluorescence. Cis-trans photoconversion, including 4 and 8 structures, is suggested to be responsible for the reversible photoswitching of the TagRFP’s variant, called rsTagRFP58. TagRFP657, another mKate mutant which has 5 residues substituted for aromatic amino acids,25 has both absorbance and emission shifted for 23–24 nm toward far-red, as compared to mKate, suggesting that its DsRed-like chromophore has the extended π-π stacking and highly hydrophobic microenvironment (possibly structure 10). The chromo states (structures 6, 8, and 12) are not necessarily caused by a cis-trans chromophore isomerization, but may result from the modifications of the chromophore’s environment in the same isoform that decrease quantum yield (QY).73 In the latter case, the protonation state of the chromophore is anionic, and has similar absorbance as the fluorescent one. Nonetheless, the cis-trans chromophore isomerization may cause: (a) transitions between the fluorescent and chromo states in reversibly switchable FP (RS-FP), such as KFP,72 Dronpa,68 and rsTagRFP;58 and/or (b) shifts in the absorbance/emission wavelengths, such as those observed in the cases of near-red TagRFP (trans) vs. far-red mKate/mKate2 (cis), Dronpa,68 and rsTagRFP.58
5. Structure-function relationship in the fluorescent proteins
Understanding of the relationship between FPs’ structure and main characteristics such as absorbance/emission spectra, photochemical behavior, brightness, photostability, and pH stability is important for the development of optimal FPs with given properties. The available crystal structures of FPs and the biochemical data provide information to suggest a correlation between FPs’ structure and some of the characteristics described above (Table 1).
5.1. Chromophore structure is the major player
The most important characteristics of FPs, such as absorbance/emission spectra and QY, are mainly pre-determined by the chemical structure of the chromophore and additionally adjusted by the surrounding environment.
A larger number of conjugated double bonds in a chromophore correlates with longer absorbance/emission wavelengths of FPs (Table 1), and is the most important determinant of excitation/emission color of FPs.102 A larger number of double bonds provides for better delocalization of the electron density that results in a lower energy of the chromophore in its excited state and a decreased difference in the energy between its excited and ground states. The lower the difference in the energy between these two states, the lower the energy of both the absorbed and emitted photons. According to quantum mechanical calculations8 and X-ray data,20,103 the red shift of DsRed-like chromophore 4 vs. GFP-like one 5 is quantitatively accounted by the additional conjugation of two N-acylimine double bonds with GFP-like chromophore 5.
An another factor that affects the excitation/emission wavelength is the polarizability of the double bond that increases in the following order: C=C < C=N < C=O. In alkaline water, the synthetic chromophores having GFP-like chromophore with additional C=C, 104 C=N, 105 or C=O 46 double bond in conjugation have absorbance maxima at 460 nm, 486 nm, and 520 nm, respectively. The most polarizable double bond C=O in the synthetic chromophore of asFP595 FP results in a more profound bathochromic shift.46 Cis- or trans-configuration of the chromophore can also slightly affect its absorbance maxima. Trans-isoform of synthetic GFP-like chromophore was shown to be about 5–10 nm red shifted as compared with cis-counterpart.75
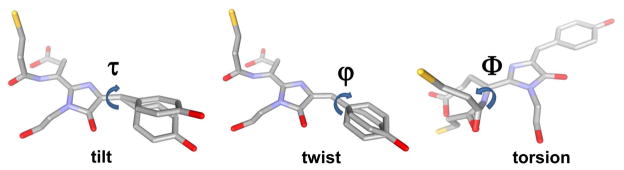
Planarity is necessary for an efficient conjugation in the system of double bounds. Distortion of coplanarity of the rings and N-acylimine group within a red chromophore occurs relative to a Cα2-Cβ2 (tilt-angle), Cβ2-Cγ2 bond (twist-angle) and peptide bond (torsion-angle) of the Tyr66 chromophore (Figure 6). A more planar ring of Tyr66 of the chromophore in terms of its lower tilt- and twist- angles values results in a higher QY of mFruits10 and GFP variants.49 According to the available X-ray data for all RFPs like DsRed,103 mCherry,10 and TagRFP,22 the C=O group of the N-acylimine group in the DsRed-like chromophore is out of chromophore plane that can hinder their effective conjugation with the chromophore and results in a reduced bathochromic shift. The orange-to-far-red photoconvertable PSmOrange protein was recently developed by our group.27 Its far-red form has the excitation/emission maxima at 633 nm/659 nm, respectively, which is the most far-red shifted excitation for an RFP as compared to that for other known RFPs. The PSmOrange chromophore 14 in the far-red state has the same number of double bonds in conjugation as the DsRed-like chromophore 4 but exhibits an additional 5-member cycle and a main-chain break before the chromophore-tripeptide.93 The bathochromic excitation/emission shift of the photoconverted PSmOrange chromophore, as compared to the DsRed-like chromophore, could be explained by a coplanarity of the C=O group of N-acylimine in PSmOrange with a chromophore plane (with an optimal torsion angle). The coplanarity of the C=O group with the chromophore has been suggested because the main-chain break removes the restriction for rotation of this group toward the chromophore plane, thus allowing it to become a completely conjugated part of the 5-member ring of the chromophore.93
Figure 6.
Characteristics of chromophore planarity in terms of tilt (τ) and twist (ϕ) and torsion (Ψ) angles. Tilt and twist angles correspond to rotation of the Tyr-ring of the chromophore around the Cα2-Cβ2 and Cβ2-Cγ2 bonds, respectively. Torsion angle is the dihedral angle defined by atoms X65:Cβ1 - X65:Cα1 - X65:N1 - X64:C, where X65 and X64 denote the residue 65 of the chromophore-forming tripeptide and the residue 64 preceding the chromophore, respectively.
RFPs with larger QYs have increased durations of the chromophore in the excited state that provides additional time for dark-state conversion to occur, thus resulting in the increased dark-state conversion photobleaching or blinking.106 Hence, the chromophores with higher QYs may be more susceptible to photochromism or blinking as compared to the chromophores having lower QYs.
5.2. Support from an immediate environment of the chromophore
The excitation/emission wavelength, QY, photostability, pH-stability and two-photon absorbance of a chromophore are additionally modified by its surrounding amino acids (Table 1). We can distinguish several mechanisms responsible for this impact on chromophore functionality, such as stacking and electrostatic interactions, protonation, hydrogen bonding, hydrophobicity and density of the amino acid packing.
The π-π stacking interactions between a Tyr ring of a chromophore and aromatic amino acids, such as Tyr, were shown to be responsible for the bathochromic shift (of ~20 nm) of both emission and excitation of yellow EYFP107, and red TagRFP657,25 E2-Crimson,97 mNeptune24 and mRojoA.96 Polarizability of a Tyr phenolic ring is probably the most important factor that contributes to the red shift. Phenolic ring of Tyr chromophore can also make cation-π stacking interactions with the guanidinium group of Arg. This type of interactions was also shown to be responsible for the bathochromic shift in mKate.23
It has been suggested that a positive charge on His203 near the phenolate moiety reduces the extent of charge delocalization between the hydroxybenzyl ring and the imidazolinone ring, thus decreasing both excitation and emission wavelengths of amFP486 cyan protein.108 Therefore, the placement of an electron-donating groups around a phenolic ring of the chromophore should lead to red shifts in absorbance and emission.108 The displacement of a positively charged Lys69 residue resulted in the disturbance of its electrostatic interactions with DsRed-like chromophore in mCherry and mStrawberry RFPs, and led to far-red shift of their excitation/emission.10 A similar displacement of Lys69 and/or positive charge deletion at position 84 resulted in a red shift of both absorption and emission maxima in a K84M mutant of DsRed.20 Quantum chemical calculations demonstrated that the position of charged or polar residues around the chromophore controls the direction and strength of the electric field, which may result in large shifts in spectral bands.109,110 A R69K mutation in TagRFP-T, which affects the position of the positive charge, was suggested to decrease irreversibly photobleaching of TagRFP-T via transient changes in the conformation or protonation state of the chromophore that results in decreased excitation of the chromophore in the dark-state.106
The direct or water-mediated hydrogen bond formation between C=O carbonyl group of N-acylimine of red chromophore and amino acid residues at positions 16 and 31 was observed in far-red shifted RFPs such as mPlum95 and mNeptune.24 The dynamic Stokes shift of emission wavelength in mPlum was suggested to result from the both the strengthening of the Glu16-chromophore hydrogen bond interaction in the excited state, which stabilizes the excited state of the chromophore, and from the specific excited-state rearrangement between the chromophore and the hydrogen bond forming residue Glu16.95,111 Conformational flexibility of water-mediated hydrogen bonding was suggested to cause bathochromic shifts in both the excitation and emission maxima of mNeptune.24
Protonation-deprotonation of Glu222 which in turn can protonate imidazolinone nitrogen of red chromophore was proposed to account for the far-red shift in the excitation/emission maxima of mCherry and mStrawberry.10 It has been suggested that in some cases, the chromophore protonation is responsible for its conversion to the transient dark state during RFP photobleaching.106 It was also proposed that a different protonation state of Glu222 is responsible for pH dependence of the fluorescence flickering in single-molecule spectroscopy of mRFP1, mCherry, and mStrawberry.112
Hydrophobicity around the chromophore was suggested to be critical for the bathochromic shift in FPs, since the destabilization of the negative charge localization at the oxygen atom of the chromophore’s phenolate group can raise the energy of the ground state relative to the excited state.96 Hydrophobic Phe residue in the first position of the DsRed-like chromophore-forming tripeptide results in the far-red shift of fluorescence spectra of E2-Crimson by ~20 nm.97 Substantially more hydrophobic environments in mCherry and mStrawberry as compared with that of the progenitor DsRed were suggested to lead to a net shift of electron density toward the imidazolinone moiety, and shifted excitation/emission of these FPs to far-red wavelengths.10
Rigidity of the chromophore is necessary for the high QY. Flexibility of the chromophore can be accompanied by the non-radiative deactivation pathways of the chromophore in the excited state, such as cis-trans isomerization of the chromophore, rotation of the chromophore Tyr-ring around Cβ2-Cγ2 bond, and thermal motion of other groups of the chromophores.78,81 Introduction of bulky amino acids around the chromophore, hydrogen bond formation with the surrounding amino acids, and stabilizing stacking interactions can dramatically decrease flexibility of the chromophore and improve the FP’s QY. Stacking interactions of the chromophore with His203 improves QY of such FPs as amFP486 cyan protein (in about 100-fold)108 and asFP595 red protein by stabilizing chromophore in coplanar conformation. As described above, on the one hand, amino acid residues at positions 146, 148, 150, 165 and 183 can form hydrogen bonds with the chromophore phenolic group and stabilize chromophore in cis- or trans-coplanar configuration improving its QY (Figure 5). On another hand, the introduction of bulky amino acids around the chromophore at positions 165, 167 and 181 should block non-activated decays from the excited state via cis-trans chromophore isomerization, also increasing QY of FPs (Figure 5).
Tight packing of amino acids around the chromophore can increase its photostability, due to better shielding of the chromophore from the environment. Excited GFPs can donate electrons to appropriate electron acceptors, such as biologically relevant FAD, FMN and NAD+, and cause photochemical reactions.63 The removal of all potentially redox-active compounds from the medium dramatically increases photostability of GFPs.113 In one study, illumination of TagRFP and mOrange in anaerobic conditions improved their photostability 8.7- and 25-fold, respectively.114 Other light-induced chemical reactions, such as decarboxylation of the neighboring to the chromophore amino acid in DsRed53 and formation of the far-red shifted and green species during illumination of orange and red FPs,27,52 can dramatically increase the FPs’ susceptibility to light. Hence, access restriction of oxidants and oxygen to the chromophore by means of tight packing around the chromophore could improve the RFPs’ photostability. Another way to block the oxygen-dependent reactions affecting photostability was implemented in mOrange2. The introduction of two mutations Q63H and F100Y in mOrange was suggested to cause a rearrangement of the side chains in the vicinity of the chromophore, and hindered the critical oxidation step leading to loss of fluorescence114 and formation of a far-red species, possibly containing the PSmOrange chromophore 14.27,52
Additionally, tight packing of amino acids around the chromophore can reduce chromophore’s mobility and undesirable cis-trans isomerization of the chromophore, and as a result may prevent its blinking in terms of the dark-state photoconversion on the 10−4 s timescale.106,114 Introduction of bulky amino acids close to the chromophore fills up a cavity around it, and was shown to contribute to decreased photobleaching of TagRFP-T, affecting its dark-state conversion rate.106,114
Among all RFPs, the RFPs with a large Stokes shift (LSS-RFPs) have extremely high pH-stability of red fluorescence reported to date.18,44 High pH-stability of LSS-RFPs can be attributed to the neutral state of the chromophore which is stabilized by the protonation of the chromophore phenolic group with carboxylic groups of surrounding amino acids at positions 148, 165 or 167.
In contrast to one-photon absorption, the efficiency of two-photon absorption of RFPs was shown to be highly sensitive to the electric field around the chromophore, which is formed by amino acid residues of its microenvironment.115 Two-photon absorption of RFPs may be increased by changing certain charged amino acids or hydrogen bonding network in the direction of the excited state chromophore dipole.
5.3. Role of the structural elements distant from the chromophore
Crystal structures of FPs have common features that include a β-barrel shape, an oligomeric structure, and a water-filled opening in the β-barrel. These features affect many functions, such as maturation speed, photostability, pH stability, thermodynamic stability, intracellular stability, toxicity, and phototoxicity (Table 1). Here, we will attempt to determine the relationship between FPs’ crystal structure and FPs’ function.
The chromophore of the FPs is located deep inside the β-barrel, which protects it from the surrounding medium. The excitation/emission spectra of the synthetic Kaede45 and asFP59546 chromophores in water are >100 nm blue-shifted as compared to those for their respective FPs. Only in basic polar (hydrophilic) aprotic DMF and/or DMSO solvents the excitation/emission spectra of synthetic chromophores are similar to their respective FPs.45,46 Hence, the chromophore’s environment properties within FPs appear to be close to basic, polar, and aprotic properties of DMF and DMSO solvents. Synthetic chromophores in solution have 102–104-fold lower QY as compared to their respective FPs.
Computational study of the GFP-like chromophore in vacuo revealed that the decay of the fluorescent state occurs via a barrierless rotation of the fluorophore phenyl moiety via a single bond Cβ2-Cγ2 (twist-angle in Figure 6) rather than through a cis-trans isomerization of Cα2-Cβ2 bond (tilt-angle in Figure 6).79 A similar deactivation pathway was computed for DsRed-like chromophore in vacuo.116 The main difference between the GFP- and DsRed-like chromophores was related with the presence of an electronegative N-acylimine substituent present in the red chromophore, which suppresses the cis-trans photo-isomerization of the Cα2-Cβ2 bond and favors rotation of the chromophore’s phenyl moiety around the Cβ2-Cγ2 bond.116 The quantum calculations for the red chromophore of reversibly-switchable KFP in protein environment suggest that the fast radiationless relaxation to the ground state also occurs due to the rotation of the phenolic fragment of the chromophore.78 Hence, the β-barrel surrounding may decrease non-radiative decays from the excited state, resulting in high QY of FPs via stabilization of the chromophore in planar conformation and prevention of the movement and conformational changes of its phenolic ring.
According to the crystal structure data and biochemical data, many of wild-type RFPs cloned from original coral species are tetramers or dimers, or have the tendency to undergo oligomerization. Tetrameric structure of FPs can affect location and function of the labeled protein, and can result in the formation of intracellular aggregates, which are toxic for the cells and explain different cytotoxicity of FPs. For example, a relatively low death rate of cells expressing dimeric Katushka and monomeric EGFP as compared with cells expressing tetrameric DsRed-express indicated that FPs may exert different toxic effects on living cells because of their tendency to aggregate.117 Many efforts have been made to disrupt the oligomeric interface in RFPs such as DsRed,118 eqFP611,119 and eqFP578.98 Tetrameric/dimeric structure of the FPs provides better shielding of the chromophore from the environment such as oxygen and other oxidants that can result in the chromophore’s degradation during light illumination. Indeed, as compared to other monomeric RFPs, the tetrameric DsRed-Express had higher photostability under photobleaching with both a confocal microscope with laser and wide-field microscope with a mercury arc lamp.117 Correlation between the tetrameric interface of the FPs and their high stability in terms of longer intracellular half-life and susceptibility to denaturation reagents and elevated temperatures has been found based on the comparison of stabilities of tetrameric DsRed and monomeric EGFP both in cultured cells and in vitro.120 With a few exceptions, longer intracellular lifespan of the proteins correlates with their higher thermal stability in bacteria121–124 and eukaryotic cells125. Presumably, proteins are selected for degradation by cellular proteolytic components that recognize the unfolded polypeptide chain.
Dronpa, TurboGFP and KillerRed contain a water-filled opening at the β-barrel between strands β7 and β1071,126,127. Cleft-like structure between β-sheets (β7 and β10) is conserved in many other FPs, such as zGFP506, zYFP538, zRFP574,42,128 and mKate.23 The amino acids at positions 145–147 and 203–205 appear to be responsible for the pronounced cleft. The opening may relay environmental conditions, like pH, to the chromophore. Quenching by water molecules entering the water-filled channel may also contribute to the diminished QY.129 The opening may facilitate oxygen conveyance to a premature chromophore and/or promote the dehydrogenation step of chromophore maturation, providing abstracted proton transport outside the β-barrel, thus speeding up the chromophore’s maturation.126 For example, a pore found in TurboGFP was suggested to be essential for fast maturation of the chromophore.96 On another hand, better accessibility of oxygen or other oxidants to the chromophore through a water-filled opening can result in poor photostability of the FP. One important difference of KillerRed FP from other FPs is that its water channel directly reaches the chromophore and provides a so-called “proton wire”.127 This channel is suggested to open solvent access to the methylene/imidazolinone moieties of the chromophore, likely to allow generation and release of harmful reactive oxygen species (ROS) out of the protein. This feature of the KillerRed pore enables inactivation of specific proteins through the chromophore assisted light inactivation (CALI) effect.130 The release of harmful ROS out of the FP’s β-barrel through a water-filled pore may explain phototoxicity of other FPs resulting in the inhibition of cell division during and after cell imaging with FPs.97,131
6. Red fluorescent proteins in advanced imaging
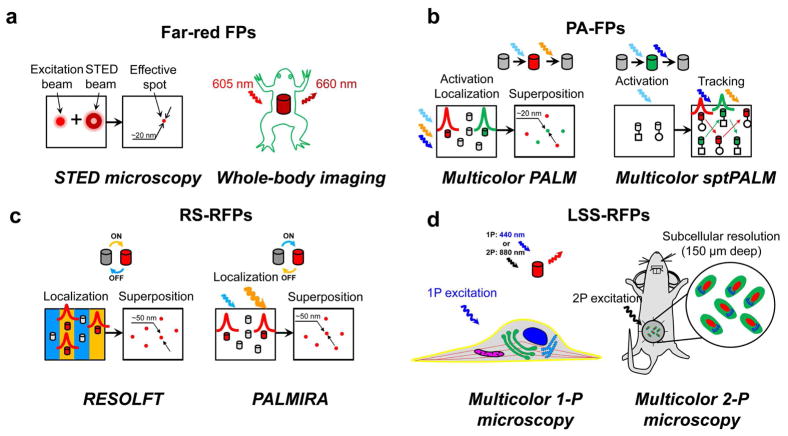
Design of RFPs with new properties stimulated the development of several modern approaches in fluorescence microscopy such as whole-body imaging, multicolor one- and two-photon microscopy with a single excitation wavelength, and super-resolution techniques including stimulated emission depletion (STED) microscopy and photoactivated localization microscopy (PALM) (Figure 7). These imaging approaches utilize FPs with significantly different properties.
Figure 7.
Applications of RFPs in modern fluorescence microscopy techniques. (a) Use of far-red shifted FPs in super-resolution STED microscopy and the whole-body imaging. On the left, doughnut-shaped depletion beam confining excitation beam to an effective spot. On the right, whole-body imaging of frog expressing Katushka far-red FP. (b) Dark-to-red and dark-to-green PA-FPs in multicolor super-resolution PALM and sptPALM microscopy. On the top, photoactivation/photobleaching cycles for PA-RFP and PA-GFP. Red and green Gaussian-shaped curves stand for center localization procedure. Red and green arrows show movement of PA-RFP and PA-GFP particles, respectively. (c) Reversibly switchable FPs (RS-FPs) in RESOLFT and PALMIRA super-resolution STED-based techniques. On the top, photoswitching cycles for KFP and rsCherryRev. Red and green Gaussian-shaped curves stand for center localization procedure. Blue and yellow thick strokes denote grooves of OFF- and ON-light, respectively. (d) Utilization of LSS-RFPs in combination with conventional cyan, green and/or yellow FPs in one- and two-photon imaging of the cell using single wavelength of excitation. On the left, plasma membrane (yellow, CFP), endoplasmic reticulum (cyan, mMiCy), Golgi (green, GFP), microtubules (red, YFP), nucleus (dark blue, dKeima570) and mitochondria (purple, mKeima) visualized by respective FPs. On the right, two-photon imaging of tumor cells in a mouse through optical window; nucleus (red, LSSmKate), cytoplasma (green, GFP) and Golgi (cyan, CFP) are labeled with respective FP. FP molecules are shown as cylinders colored according to their emission colors. Colored waved arrows denote the respective activation/switching, excitation or emission light.
6.1. Far-red shifted FPs in STED nanoscopy and whole-body imaging
STED microscopy is based on stimulated depletion of fluorescence of FPs by high power light.132 STED microscope uses a red excitation beam (Figure 7a, red circle), which is superimposed by a doughnut-shaped far-red STED beam that instantly quenches excited FPs, confining the fluorescence emission to a spot with the size of 20–70 nm, which substantially overcomes the resolution limit of >200 nm set by diffraction (Figure 7a, red dot). Conceptually, theresolution in STED microscopy is “infinite” ( i.e., down to a molecule), and is inversely proportional to the intensity of the STED quenching light.132 In practice, the STED resolution is limited by the photodamage of the sample, which usually sets the intensity limit on the depletion beam, particularly for biological samples.133,134
STED imaging of fixed cells has been demonstrated using far-red shifted FPs such as TagRFP65773 and E2-Crimson.101 Super-resolution images of cell structures were recorded with ~3-fold improved resolution over conventional diffraction limited confocal images. The main limitation for the application of RFPs in STED microscopy is their ~10-fold lower photostability as compared to synthetic dyes, which can emit up to 105–106 photons before photobleaching.134 Excitation maxima of far-red shifted FPs are most compatible with the commercial STED microscopes having 635–640 nm laser for FP excitation and 760–750 nm laser for emission depletion (Figure 7a). Far-red FPs are also beneficial for STED microscopy since the far-red shifted excitation/probing light is less phototoxic to live cells.
Whole-body and deep-tissue imaging also exploit far-red shifted FPs with excitation/emission compatible with a near infrared window (NIRW) in biological tissues. The wavelength range between 650–900 nm is called NIRW. Absorbance of oxy-hemoglobin, deoxy-hemoglobin, melanin and water are minimal in this range. Light scattering from cellular components is also reduced. To date, the most far-red shifted FPs, such as Katushka,117 mKate2, tdKatushka2,99 E2-Crimson,135 mNeptune,24 TagRFP657,25 eqFP650, and eqFP67026 share a farred shifted DsRed-like chromophore 10, and exhibit the excitation and emission maxima limited to about 610 and 670 nm, respectively. Among these far-red shifted FPs, eqFP650 has the highest fluorescence signal in whole-mouse imaging, and may be currently considered as the preferred far-red probe for in vivo experiments.26 Another far-red FP, called Katushka and expressed in a frog, allowed better imaging of the heart as compared to DsRed-Express RFP, which has shorter emission wavelength.117 Katushka also allowed noninvasive whole-body detection of Cre-mediated recombination events in tissues located deep within a transgenic mouse.136 Fluorescence of far-red FP Neptune can be detected in a living mouse even in the liver, where hemoglobin absorbance is particularly high due to the presence of heavily vascularized structures.24 However, increased Stokes shift of emission of far-red FPs results in a substantial drop in QY, possibly due to the dynamic nature of this shift. A dynamic Stokes shift of emission of mPlum was shown to be the result of a specific excited-state rearrangement between the chromophore and strongly interacting surrounding amino acids, suggesting high flexibility of the chromophore’s environment.111 This flexibility may result in low QY of mPlum. Further increase in QY and in far-red shift of excitation could be beneficial for applications such as the whole-body imaging.
6.2. Photoactivatable RFPs in multicolor PALM microscopy
PALM is a microscopy technique that relies on sequential stochastic photoactivation and localization of PA-FPs molecules separated by more than 200 nm to create super-resolution images.137 After all photoactivated molecules are photobleached, localization of centroid positions of individual PA-FPs of different colors on a diffraction-limited image (Figure 7b, dots) is accomplished with a precision well beyond the diffraction limit (up to ~20 nm). This procedure is repeated thousands of times to build a 2D super-resolution image. Because the resolution in the PALM technique is proportional to the square of the number of photons emitted per molecule, PA-FPs with higher single-molecule brightness should be optimal.138 Consequently, tandem green-to-red tdEosFP having high brightness has turned out to be one of the most popular PA-FPs for PALM.139 Development of future PA-FPs emitting large number of photons for shorter period of time (i.e., having a high photon rate) could increase the acquisition speed of PALM images.
Single particle tracking PALM (sptPALM) relies on a similar procedure, except that sptPALM tracks molecules localized in many PALM images in order to draw particle trajectories resulted from particle movement.140,141 Analysis of these trajectories allows calculating the diffusion coefficients for each particle and plotting the diffusion coefficient maps, which help to distinguish fractions of particles with different mobility. sptPALM was successfully applied to map the dynamics of single particles labeled with EosFP140 and PATagRFP28 photoactivatable proteins in live mammalian cells. PA-FPs with higher photostability are preferred for sptPALM experiments since the high photostability allows the particle traking for a longer time.
A two-color PALM and sptPALM have been demonstrated in fixed and live cells using dark-to-red photoactivatable PAmCherry and PATagRFP proteins in combination with dark-to-green PAGFP protein.1–4,28,60 Two-color PALM in fixed cells was also reported using the pairs Dronpa/EosFP and PSCFP2/EosFP.142 Using these FP pairs, two-color PALM images of cellular organelles, cytoskeletal elements, extracellular adhesion proteins, and membrane protein clusters were collected with ~25–75 nm resolution. Since green-to-red EosFP and cyan-to-green PSCFP2 photoconvertable proteins have green and cyan fluorescence in the original state, respectively, it complicates two-color PALM imaging procedure, thus, making utilization of PA-FPs with a dark original state more beneficial for two-color PALM. Engineering of future PA-FPs with new colors is required for PALM techniques.
A combination of PALM with single-photon simultaneous multiphase interferometry resulted in invention of an interferometric PALM microscopy (iPALM).143 The iPALM technique has enabled a 3D-imaging of cellular ultrastructures with 10 nm vertical and 20 nm lateral resolutions using mKikGR, tdEosFP, and mEosFP PA-FPs.143,144
6.3. Application of reversibly switchable RFPs to super-resolution techniques
Reversibly switchable FPs (RS-FPs) have become popular probes for advanced versions of STED and PALM techniques. In order to achieve sub-diffraction resolution in STED microscopy, the intensity of depletion light should be higher than the intensity of saturation light of the transition between fluorescent ON and non-fluorescent OFF states when 50% of the molecules are in the ON state and 50% of the molecules are in the OFF state.132 In conventional FPs, the transition from the first excited to the ground electron levels is reached at very high light intensities of ~100 MW/cm2, which are harmful to live cells.132,145
The STED modification with RS-FPs, called RESOLFT, utilizes the transition between ON and OFF states, which is connected to cis-trans-isomerization of the chromophore.145 The saturation intensity for this transition has a much lower value of ~0.1 W/cm2, therefore RESOLFT has the advantage over the STED approach with standard FPs, and can be applied for super-resolution imaging of live cells. RESOLFT concept was demonstrated with asFP595 red RS-FP, where the resolution of 50–100 nm was achieved.145 Currently available RS-FPs are limited by low photostability and brightness, therefore their further utilization in STED microscopy primarily depends on the improvement of these key properties.
PALM with independently running acquisition (PALMIRA) is based on a dynamic equilibrium which is formed during image acquisition when most of the molecules are in the dark OFF state, and a small fraction of the molecules are in the bright ON state.146 This equilibrium is reached by adjusting the power of ON and OFF light. The fraction of the molecules in fluorescent state is being kept at a low percent in order to avoid overlapping of diffraction limited spots in the PALMIRA image. Localization of the centers of individual photoswitchable molecules in the image and reconstruction of the final PALMIRA image from thousands of images are accomplished similarly to the standard PALM. PALMIRA imaging of live cells expressing red RS-FPs such as rsCherryRev and rsCherry achieved ~75 nm resolution, resulting in a 4-fold improvement in resolution as compared to the conventional fluorescence microscopy.73
6.4. RFPs with large Stokes shifts in multicolor fluorescence microscopy
Multicolor one- and two-photon fluorescence microscopy using single excitation wavelength relies on FPs having similar one- or two-photon excitation but different emission maxima. A single wavelength excitation of FPs provides an advantage since it enables the researchers to perform imaging of several targets simultaneously to clarify the dynamic interactions of proteins and subcellular structures. Simultaneous six-color one-photon imaging of subcellular structures in Vero cells has been demonstrated using single 458 nm laser line and LSSRFPs, dKeima570 and mKeima in a combination with cyan FPs, CFP and mMiCy, EGFP, and yellow FP, YFP.147 Dual-color two-photon fluorescent imaging of cellular organelles was performed using two alternative pairs mKeima/ECFP and mKeima/EGFP and two-photon excitation at 880 nm and 960 nm, respectively.148,149 Three-color two-photon imaging of cellular compartments was demonstrated using LSSmKates.44 Two-photon excitation at 870 nm was found to be the optimal wavelength for simultaneous excitation of LSSmKates, ECFP and EGFP.
7. Conclusions
Fluorescent proteins have become the indispensable genetically-encoded tools for various biomedical applications. The continuing progress in the development and characterization of RFPs provides the information sufficient to assemble the chromophore’s puzzles together into a general scheme of chemical transformations of the red chromophore. Majority of the Tyr-containing red chromophores are formed via the TagBFP-like blue intermediate containing the N-acylimine group. In RFPs, similar transitions between chromophore structures can either occur autocatalytically or be photo-induced or be blocked. Moreover, the chromophores that share the same structure can be in either fluorescent or dark states. Autocatalytical vs. photoinduced vs. blocked and fluorescent vs. chromo choices are mainly determined by amino acid residues in the chromophore and its immediate environment. The primary chromophore chemical structure, as well as the surrounding amino acids’ immediate environment, predetermines the function of RFPs. Several structural features of the RFPs are translated into the RFPs functions. Important developments of modern imaging approaches consist of the super-resolution microscopy, deep-tissue imaging, intravital and two-photon microscopy. These novel techniques demand RFPs with quite different properties, which are yet to be developed. The general scheme of the chromophore transformations covered in this review presents the basis to design advanced red fluorescent probes for existing and emerging fluorescence imaging approaches.
Table 2.
Relationship between the structural elements of the fluorescent proteins and their characteristics illustrated with examples.
| Structural element | Property of FP | Structural feature affecting this property | Observed effect | Examples of | Ref. | |
|---|---|---|---|---|---|---|
| Suboptimal property | Optimal property | |||||
| Chromophore | Absorbance/emission maxima | Larger number of double bonds in conjugation | Red shift in absorbance/emission | mOrange-like chromophore 13 with 7 double bonds, Ex/Em at 540–548 nm/561–565 nm | DsRed-like chromophore 4 with 8 double bonds, Ex/Em at 555–610 nm/583–670 nm | 93 |
| Higher polarizability of double bonds | zFP538-like chromophore 11 with C=N-group, Ex/Em at 528 nm/538nm | asFP595-like chromophore 9 with C=O-group, Ex/Em at 580 nm/600 nm | 9,15 | |||
| Quantum yield (QY) | Higher planarity terms of tilt-, twist-, and torsion-angles | Large QY | mCherry with non-planar DsRed- like chromophore 4 (twist-angle = 11.3) has QY=0.22 | mOrange with planar mOrange-like chromophore 13 (twist-angle = 1.5) has QY=0.69 | 10 | |
| Smaller size | Citrine and mWasabi with smaller GFP-like chromophore 5, QY=0.76- 0.80 | mKate2 and TagRFP with larger DsRed-like chromophore 4, QY=0.40–0.48 | 2 | |||
| Chromophore immediate microenvironment | Absorbance/emission maxima | Stacking interactions with chromophore | Red shift in absorbance/emission | mCherry with DsRed-like chromophore 4, Ex/Em at 588 nm/611 nm | mCherry/I203Y with DsRed-like chromophore 10, Ex/Em at 598 nm/618 nm | 96 |
| Hydrophobic interaction with chromophore | mCherry/Q163M with DsRed-like chromophore 10, Ex/Em at 590 nm/615 nm | 96 | ||||
| H-bond with N-acylimine group of chromophore | mPlum with DsRed-like chromophore 10, Ex/Em at 590 nm/649 nm | 95 | ||||
| Positive charge near Tyr ring of chromophore | Blue shift in absorbance/emission | DsRed with DsRed-like chromophore 4, Ex/Em at 558 nm/583 nm | 10 | |||
| QY | Rigidity of chromophore | Large QY | TagRFP with rigid DsRed-like chromophore 4, QY=0.48 | rsTagRFP with flexible DsRed-like chromophore 4, QY=0.11 | 58 | |
| Photostability | Tight packing of amino acids around chromophore | High photostability | TagRFP with DsRed-like chromophore 4, bleaching half-time is 37 s | TagRFP-T with DsRed-like chromophore 4, bleaching half-time is 337 s | 114 | |
| Photoconversion of chromophore | Low photostability | mOrange-like chromophore 13 undergoes orange-to-far-red photoconversion, less photostable | mKO-like chromophore 13 does not undergo photoconversion, more photostable | 11 | ||
| pH stability | Direct H-bond between enol- group of chromophore and Arg residue | High pH stability | Anionic mOrange-like chromophore 13, with direct H-bond with Ser residue, pKa=6.5 | TagBFP-like chromophore 3, with direct H-bond with Arg residue, pKa=2.7 | 10,22 | |
| H-bond between enol-group of chromophore and carboxylic group of adjacent residue | LSSmKates with neutral DsRed- like chromophore 8, with H-bond with Glu or Asp residues, pKa=2.7–3.2 | 18 | ||||
| Two-photon absorption | Higher electric field around chromophore | High two-photon absorption | mStrawberry with DsRed-like chromophore 4, electric field intensity of 1.4 debye, two-photon cross-section σ2=18 GM | tdTomato with DsRed-like chrmophore 4, electric field intensity of 3.7 debye, two-photon cross-section σ2=138 GM | 115 | |
| Structural elements that are far away from chromophore | Absorbance/emission maxima | β-barrel | Red shift in absorbance/emission | Synthetic Kaede-like chromophore 7 in water, Ex/Em=490 nm/unknown | Kaede with Kaede-like chromophore 7, Ex/Em=572/582 nm | 45,56 |
| QY | Large QY | Synthetic Kaede-like chromophore 7 in basic DMF, QY=0.005 | Kaede with Kaede-like chromophore 7, QY=0.33 | |||
| Cytotoxicity | Tetrameric structure | High cytotoxicity | Monomeric EGFP, low cytotoxiciy | Tetrameric DsRed-express, high cytotoxicity | 117 | |
| Photostability | High photostability | Monomeric mCherry, bleaching half-time is 630 s | Tetrameric DsRed-express, bleaching half-time is 3,510 s | 117 | ||
| Stability | High stability | Monomeric EGFP, low stability in terms of intracellular half-life and susceptibility to denaturation reagents | Tetrameric DsRed, high stability in terms of intracellular half-life and susceptibility to denaturation reagents | 120 | ||
| Phototoxicity | Wide water-filled opening at β-barrel | High phototoxicity | DsRed2 with narrow water-filled opening, low phototoxicity | KillerRed with wide water-filled opening, ~10,000-fold higher phototoxicity than that of DsRed2 | 127 | |
| Maturation rate | Fast maturation | TurboGFP/V205L, narrow water- filled pore, maturation half-time is 42 min | TurboGFP, wide water-filled pore, maturation half-time is 24 min | 126 | ||
Acknowledgments
This work was supported by the grants GM073913 and CA164468 from the National Institutes of Health to V.V.V.
Biographies

Fedor Subach received an M.S. degree in Chemistry at Moscow State University and pursued a Ph.D. degree in Bioorganic Chemistry at the same university in Department of Chemistry of Natural Compounds under supervision of Dr. Elizaveta Gromova. His doctoral thesis focused on studies of interaction of restriction endonuclease EcoRII with DNA. After completing of his graduate work, Dr. Subach has joined the Dr. Vladislav Verkhusha laboratory at Albert Einstein College of Medicine in New York as a postdoc. His postdoctoral work is focused on development of photoactivatable fluorescent proteins for super-resolution imaging and fluorescent timers for determining of age of intracellular events. He has received the Dennis Shields postdoctoral award. Dr. Subach is currently an instructor at Albert Einstein College of Medicine.

Vladislav Verkhusha received an M.S. degree in Biophysics from Moscow University of Physics and Technology and a Ph.D. degree in Chemical Kinetics and Catalysis from Moscow State University. Dr. Verkhusha obtained his postdoctoral training in Osaka Bioscience Institute and later he worked as a research scientist in research centers of Japan Science and Technology Corporation in Kyoto and Tokyo. In 2002, Dr. Verkhusha was appointed an assistant professor at University of Colorado, where he developed FRET imaging techniques and photoactivatable probes. From 2006, Dr. Verkhusha is an associate professor and then a professor at Albert Einstein College of Medicine in New York. Main research interests of Dr. Verkhusha include design of fluorescent proteins and molecular biosensors, development of high-throughput screening and super-resolution imaging approaches.
References
- 1.Davidson MW, Campbell RE. Nat Methods. 2009;6:713. doi: 10.1038/nmeth1009-713. [DOI] [PubMed] [Google Scholar]
- 2.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Physiol Rev. 2010;90:1103. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 3.Nienhaus GU, Wiedenmann J. Chemphyschem. 2009;10:1369. doi: 10.1002/cphc.200800839. [DOI] [PubMed] [Google Scholar]
- 4.Lippincott-Schwartz J, Patterson GH. Trends Cell Biol. 2009;19:555. doi: 10.1016/j.tcb.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subach FV, Subach OM, Gundorov IS, Morozova KS, Piatkevich KD, Cuervo AM, Verkhusha VV. Nat Chem Biol. 2009;5:118. doi: 10.1038/nchembio.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsuboi T, Kitaguchi T, Karasawa S, Fukuda M, Miyawaki A. Mol Biol Cell. 2009;21:87. doi: 10.1091/mbc.E09-08-0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wachter RM, Watkins JL, Kim H. Biochemistry. 2010;49:7417. doi: 10.1021/bi100901h. [DOI] [PubMed] [Google Scholar]
- 8.Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. Proc Natl Acad Sci USA. 2000;97:11990. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remington SJ, Wachter RM, Yarbrough DK, Branchaud B, Anderson DC, Kallio K, Lukyanov KA. Biochemistry. 2005;44:202. doi: 10.1021/bi048383r. [DOI] [PubMed] [Google Scholar]
- 10.Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Biochemistry. 2006;45:9639. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- 11.Kikuchi A, Fukumura E, Karasawa S, Mizuno H, Miyawaki A, Shiro Y. Biochemistry. 2008;47:11573. doi: 10.1021/bi800727v. [DOI] [PubMed] [Google Scholar]
- 12.Mizuno H, Mal TK, Tong KI, Ando R, Furuta T, Ikura M, Miyawaki A. Mol Cell. 2003;12:1051. doi: 10.1016/s1097-2765(03)00393-9. [DOI] [PubMed] [Google Scholar]
- 13.Nienhaus K, Nienhaus GU, Wiedenmann J, Nar H. Proc Natl Acad Sci USA. 2005;102:9156. doi: 10.1073/pnas.0501874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsutsui H, Shimizu H, Mizuno H, Nukina N, Furuta T, Miyawaki A. Chem Biol. 2009;16:1140. doi: 10.1016/j.chembiol.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Quillin ML, Anstrom DM, Shu X, O’Leary S, Kallio K, Chudakov DM, Remington SJ. Biochemistry. 2005;44:5774. doi: 10.1021/bi047644u. [DOI] [PubMed] [Google Scholar]
- 16.Henderson JN, Osborn MF, Koon N, Gepshtein R, Huppert D, Remington SJ. J Am Chem Soc. 2009;131:13212. doi: 10.1021/ja904665x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Violot S, Carpentier P, Blanchoin L, Bourgeois D. J Am Chem Soc. 2009;131:10356. doi: 10.1021/ja903695n. [DOI] [PubMed] [Google Scholar]
- 18.Piatkevich KD, Malashkevich VN, Almo SC, Verkhusha VV. J Am Chem Soc. 2010;132:10762. doi: 10.1021/ja101974k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, Byrnes LJ, Shui B, Röhrig UF, Singh A, Chudakov DM, Lukyanov S, Zipfel WR, Kotlikoff MI, Sondermann H. PLoS ONE. 2011;6:e23513. doi: 10.1371/journal.pone.0023513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. Proc Natl Acad Sci USA. 2001;98:462. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subach FV, Malashkevich VN, Zencheck WD, Xiao H, Filonov GS, Almo SC, Verkhusha VV. Proc Natl Acad Sci USA. 2009;106:21097. doi: 10.1073/pnas.0909204106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subach OM, Malashkevich VN, Zencheck WD, Morozova KS, Piatkevich KD, Almo SC, Verkhusha VV. Chem Biol. 2010;17:333. doi: 10.1016/j.chembiol.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pletnev S, Shcherbo D, Chudakov DM, Pletneva N, Merzlyak EM, Wlodawer A, Dauter Z, Pletnev V. J Biol Chem. 2008;283:28980. doi: 10.1074/jbc.M800599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin MZ, McKeown MR, Ng HL, Aguilera TA, Shaner NC, Campbell RE, Adams SR, Gross LA, Ma W, Alber T, Tsien RY. Chem Biol. 2009;16:1169. doi: 10.1016/j.chembiol.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morozova KS, Piatkevich KD, Gould TJ, Zhang J, Bewersdorf J, Verkhusha VV. Biophys J. 2010;99:L13. doi: 10.1016/j.bpj.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shcherbo D, Shemiakina, Ryabova AV, Luker KE, Schmidt BT, Souslova EA, Gorodnicheva TV, Strukova L, Shidlovskiy KM, Britanova OV, Zaraisky AG, Lukyanov KA, Loschenov VB, Luker GD, Chudakov DM. Nat Methods. 2010;7:827. doi: 10.1038/nmeth.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subach OM, Patterson GH, Ting LM, Wang Y, Condeelis JS, Verkhusha VV. Nat Methods. 2011;8:771. doi: 10.1038/nmeth.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subach FV, Patterson GH, Renz M, Lippincott-Schwartz J, Verkhusha VV. J Am Chem Soc. 2010;132:6481. doi: 10.1021/ja100906g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pletnev S, Subach FV, Dauter Z, Wlodawer A, Verkhusha VV. J Am Chem Soc. 2010;132:2243. doi: 10.1021/ja908418r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piatkevich KD, Verkhusha VV. Curr Opin Chem Biol. 2010;14:23. doi: 10.1016/j.cbpa.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barondeau DP, Putnam CD, Kassmann CJ, Tainer JA, Getzoff ED. Proc Natl Acad Sci USA. 2003;100:12111. doi: 10.1073/pnas.2133463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sniegowski JA, Lappe JW, Patel HN, Huffman HA, Wachter RM. J Biol Chem. 2005;280:26248. doi: 10.1074/jbc.M412327200. [DOI] [PubMed] [Google Scholar]
- 33.Sniegowski JA, Phail ME, Wachter RM. Biochem Biophys Res Commun. 2005;332:657. doi: 10.1016/j.bbrc.2005.04.166. [DOI] [PubMed] [Google Scholar]
- 34.Wood TI, Barondeau DP, Hitomi C, Kassmann CJ, Tainer JA, Getzoff ED. Biochemistry. 2005;44:16211. doi: 10.1021/bi051388j. [DOI] [PubMed] [Google Scholar]
- 35.Verkhusha VV, Chudakov DM, Gurskaya NG, Lukyanov S, Lukyanov KA. Chem Biol. 2004;11:845. doi: 10.1016/j.chembiol.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 36.Subach OM, Gundorov IS, Yoshimura M, Subach FV, Zhang J, Gruenwald D, Souslova EA, Chudakov DM, Verkhusha VV. Chem Biol. 2008;15:1116. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pouwels LJ, Zhang L, Chan NH, Dorrestein PC, Wachter RM. Biochemistry. 2008;47:10111. doi: 10.1021/bi8007164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wachter RM. Acc Chem Res. 2007;40:120. doi: 10.1021/ar040086r. [DOI] [PubMed] [Google Scholar]
- 39.Strack RL, Strongin DE, Mets L, Glick BS, Keenan RJ. J Am Chem Soc. 2010;132:8496. doi: 10.1021/ja1030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bravaya KB, Subach OM, Korovina N, Verkhusha VV, Krylov AI. J Am Chem Soc. 2012;134:2807. doi: 10.1021/ja2114568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pakhomov AA, Martynov VI. Biochemistry. 2007;46:11528. doi: 10.1021/bi700721x. [DOI] [PubMed] [Google Scholar]
- 42.Pletneva N, Pletnev V, Tikhonova T, Pakhomov AA, Popov V, Martynov VI, Wlodawer A, Dauter Z, Pletnev S. Acta Crystallogr D Biol Crystallogr. 2007;63:1082. doi: 10.1107/S0907444907042461. [DOI] [PubMed] [Google Scholar]
- 43.Ivashkin PE, Lukyanov KA, Lukyanov S, Yampolsky IV. J Org Chem. 2011;76:2782. doi: 10.1021/jo200150b. [DOI] [PubMed] [Google Scholar]
- 44.Piatkevich KD, Hulit J, Subach OM, Wu B, Abdulla A, Segall JE, Verkhusha VV. Proc Natl Acad Sci USA. 2010;107:5369. doi: 10.1073/pnas.0914365107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yampolsky IV, Kislukhin AA, Amatov TT, Shcherbo D, Potapov VK, Lukyanov S, Lukyanov KA. Bioorg Chem. 2008;36:96. doi: 10.1016/j.bioorg.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yampolsky IV, Remington SJ, Martynov VI, Potapov VK, Lukyanov S, Lukyanov KA. Biochemistry. 2005;44:5788. doi: 10.1021/bi0476432. [DOI] [PubMed] [Google Scholar]
- 47.Kennis JT, Larsen DS, van Stokkum IH, Vengris M, van Thor JJ, van Grondelle R. Proc Natl Acad Sci USA. 2004;101:17988. doi: 10.1073/pnas.0404262102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiehler J, Jung G, Seebacher C, Zumbusch A, Steipe B. Chembiochem. 2003;4:1164. doi: 10.1002/cbic.200300595. [DOI] [PubMed] [Google Scholar]
- 49.Shu X, Kallio K, Shi X, Abbyad P, Kanchanawong P, Childs W, Boxer SG, Remington SJ. Biochemistry. 2007;46:12005. doi: 10.1021/bi7009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agmon N. Biophys J. 2005;88:2452. doi: 10.1529/biophysj.104.055541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T, Campbell RE. Science. 2011;333:1888. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kremers GJ, Hazelwood KL, Murphy CS, Davidson MW, Piston DW. Nat Methods. 2009;6:355. doi: 10.1038/nmeth.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Habuchi S, Cotlet M, Gensch T, Bednarz T, Haber-Pohlmeier S, Rozenski J, Dirix G, Michiels J, Vanderleyden J, Heberle J, De Schryver FC, Hofkens J. J Am Chem Soc. 2005;127:8977. doi: 10.1021/ja047023o. [DOI] [PubMed] [Google Scholar]
- 54.van Thor JJ, Gensch T, Hellingwerf KJ, Johnson LN. Nat Struct Biol. 2002;9:37. doi: 10.1038/nsb739. [DOI] [PubMed] [Google Scholar]
- 55.Goedhart J, Vermeer JE, Adjobo-Hermans MJ, van Weeren L, Gadella TW., Jr PLoS One. 2007;2:e1011. doi: 10.1371/journal.pone.0001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. Proc Natl Acad Sci USA. 2002;99:12651. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lukyanov KA, Fradkov AF, Gurskaya NG, Matz MV, Labas YA, Savitsky AP, Markelov ML, Zaraisky AG, Zhao X, Fang Y, Tan W, Lukyanov SA. J Biol Chem. 2000;275:25879. doi: 10.1074/jbc.C000338200. [DOI] [PubMed] [Google Scholar]
- 58.Subach FV, Zhang L, Gadella TW, Gurskaya NG, Lukyanov KA, Verkhusha VV. Chem Biol. 2010;17:745. doi: 10.1016/j.chembiol.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marchant JS, Stutzmann GE, Leissring MA, LaFerla FM, Parker I. Nat Biotechnol. 2001;19:645. doi: 10.1038/90249. [DOI] [PubMed] [Google Scholar]
- 60.Subach FV, Patterson GH, Manley S, Gillette JM, Lippincott-Schwartz J, Verkhusha VV. Nat Methods. 2009;6:153. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elowitz MB, Surette MG, Wolf PE, Stock J, Leibler S. Curr Biol. 1997;7:809. doi: 10.1016/s0960-9822(06)00342-3. [DOI] [PubMed] [Google Scholar]
- 62.Sawin KE, Nurse P. Curr Biol. 1997;7:R606. doi: 10.1016/s0960-9822(06)00313-7. [DOI] [PubMed] [Google Scholar]
- 63.Bogdanov AM, Mishin AS, Yampolsky IV, Belousov VV, Chudakov DM, Subach FV, Verkhusha VV, Lukyanov S, Lukyanov KA. Nat Chem Biol. 2009;5:459. doi: 10.1038/nchembio.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pletneva NV, Pletnev VZ, Shemiakina II, Chudakov DM, Artemyev I, Wlodawer A, Dauter Z, Pletnev S. Protein Sci. 2011 doi: 10.1002/pro.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Battad JM, Wilmann PG, Olsen S, Byres E, Smith SC, Dove SG, Turcic KN, Devenish RJ, Rossjohn J, Prescott M. J Mol Biol. 2007;368:998. doi: 10.1016/j.jmb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 66.Rusanov AL, Mironov VA, Goryashenko AS, Grigorenko BL, Nemukhin AV, Savitsky AP. J Phys Chem B. 2011;115:9195. doi: 10.1021/jp1094245. [DOI] [PubMed] [Google Scholar]
- 67.Henderson JN, Ai HW, Campbell RE, Remington SJ. Proc Natl Acad Sci USA. 2007;104:6672. doi: 10.1073/pnas.0700059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ando R, Mizuno H, Miyawaki A. Science. 2004;306:1370. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 69.Ando R, Flors C, Mizuno H, Hofkens J, Miyawaki A. Biophys J. 2007;92:L97. doi: 10.1529/biophysj.107.105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andresen M, Stiel AC, Folling J, Wenzel D, Schonle A, Egner A, Eggeling C, Hell SW, Jakobs S. Nat Biotechnol. 2008;26:1035. doi: 10.1038/nbt.1493. [DOI] [PubMed] [Google Scholar]
- 71.Stiel AC, Trowitzsch S, Weber G, Andresen M, Eggeling C, Hell SW, Jakobs S, Wahl MC. Biochem J. 2007;402:35. doi: 10.1042/BJ20061401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chudakov DM, Belousov VV, Zaraisky AG, Novoselov VV, Staroverov DB, Zorov DB, Lukyanov S, Lukyanov KA. Nat Biotechnol. 2003;21:191. doi: 10.1038/nbt778. [DOI] [PubMed] [Google Scholar]
- 73.Stiel AC, Andresen M, Bock H, Hilbert M, Schilde J, Schonle A, Eggeling C, Egner A, Hell SW, Jakobs S. Biophys J. 2008;95:2989. doi: 10.1529/biophysj.108.130146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adam V, Lelimousin M, Boehme S, Desfonds G, Nienhaus K, Field MJ, Wiedenmann J, McSweeney S, Nienhaus GU, Bourgeois D. Proc Natl Acad Sci USA. 2008;105:18343. doi: 10.1073/pnas.0805949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Voliani V, Bizzarri R, Nifosi R, Abbruzzetti S, Grandi E, Viappiani C, Beltram F. J Phys Chem B. 2008;112:10714. doi: 10.1021/jp802419h. [DOI] [PubMed] [Google Scholar]
- 76.Nienhaus K, Nar H, Heilker R, Wiedenmann J, Nienhaus GU. J Am Chem Soc. 2008;130:12578. doi: 10.1021/ja8046443. [DOI] [PubMed] [Google Scholar]
- 77.Andresen M, Wahl MC, Stiel AC, Grater F, Schafer LV, Trowitzsch S, Weber G, Eggeling C, Grubmuller H, Hell SW, Jakobs S. Proc Natl Acad Sci USA. 2005;102:13070. doi: 10.1073/pnas.0502772102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shelaev I, Mironov V, Rusanov A, Gostev F, Bochenkova A, Sarkisov O, Nemukhin A, Savitsky A. Laser Phys Lett. 2011;8:469. [Google Scholar]
- 79.Martin ME, Negri F, Olivucci M. J Am Chem Soc. 2004;126:5452. doi: 10.1021/ja037278m. [DOI] [PubMed] [Google Scholar]
- 80.Olsen S, Smith SC. J Am Chem Soc. 2008;130:8677. doi: 10.1021/ja078193e. [DOI] [PubMed] [Google Scholar]
- 81.Schafer LV, Groenhof G, Boggio-Pasqua M, Robb MA, Grubmuller H. PLoS Comput Biol. 2008;4:e1000034. doi: 10.1371/journal.pcbi.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chudakov DM, Feofanov AV, Mudrik NN, Lukyanov S, Lukyanov KA. J Biol Chem. 2003;278:7215. doi: 10.1074/jbc.M211988200. [DOI] [PubMed] [Google Scholar]
- 83.Mizuno H, Mal TK, Walchli M, Kikuchi A, Fukano T, Ando R, Jeyakanthan J, Taka J, Shiro Y, Ikura M, Miyawaki A. Proc Natl Acad Sci USA. 2008;105:9227. doi: 10.1073/pnas.0709599105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, Rocker C, Salih A, Spindler KD, Nienhaus GU. Proc Natl Acad Sci USA. 2004;101:15905. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. Nat Methods. 2009;6:131. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Nat Biotechnol. 2006;24:461. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 87.Chudakov DM, Lukyanov S, Lukyanov KA. Nat Protoc. 2007;2:2024. doi: 10.1038/nprot.2007.291. [DOI] [PubMed] [Google Scholar]
- 88.Adam V, Nienhaus K, Bourgeois D, Nienhaus GU. Biochemistry. 2009;48:4905. doi: 10.1021/bi900383a. [DOI] [PubMed] [Google Scholar]
- 89.Hoi H, Shaner NC, Davidson MW, Cairo CW, Wang J, Campbell RE. J Mol Biol. 2010;401:776. doi: 10.1016/j.jmb.2010.06.056. [DOI] [PubMed] [Google Scholar]
- 90.Hayashi I, Mizuno H, Tong KI, Furuta T, Tanaka F, Yoshimura M, Miyawaki A, Ikura M. J Mol Biol. 2007;372:918. doi: 10.1016/j.jmb.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 91.Li X, Chung LW, Mizuno H, Miyawaki A, Morokuma K. J Phys Chem B. 2010;114:16666. doi: 10.1021/jp1101779. [DOI] [PubMed] [Google Scholar]
- 92.Baird GS, Zacharias DA, Tsien RY. Proc Natl Acad Sci USA. 2000;97:11984. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Nat Biotechnol. 2004;22:1567. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 94.Wang L, Jackson WC, Steinbach PA, Tsien RY. Proc Natl Acad Sci USA. 2004;101:16745. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shu X, Wang L, Colip L, Kallio K, Remington SJ. Protein Sci. 2009;18:460. doi: 10.1002/pro.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chica RA, Moore MM, Allen BD, Mayo SL. Proc Natl Acad Sci USA. 2010;107:20257. doi: 10.1073/pnas.1013910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Strack RL, Hein B, Bhattacharyya D, Hell SW, Keenan RJ, Glick BS. Biochemistry. 2009;48:8279. doi: 10.1021/bi900870u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, Gaintzeva A, Lukyanov KA, Lukyanov S, Gadella TW, Chudakov DM. Nat Methods. 2007;4:555. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 99.Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, Lukyanov S, Zaraisky AG, Davidson MW, Chudakov DM. Biochem J. 2009;418:567. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zapata-Hommer O, Griesbeck O. BMC Biotechnol. 2003;3:5. doi: 10.1186/1472-6750-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ai HW, Hazelwood KL, Davidson MW, Campbell RE. Nat Methods. 2008;5:401. doi: 10.1038/nmeth.1207. [DOI] [PubMed] [Google Scholar]
- 102.Remington SJ. Curr Opin Struct Biol. 2006;16:714. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 103.Tubbs JL, Tainer JA, Getzoff ED. Biochemistry. 2005;44:9833. doi: 10.1021/bi0472907. [DOI] [PubMed] [Google Scholar]
- 104.He X, Bell AF, Tonge PJ. Org Lett. 2002;4:1523. doi: 10.1021/ol0200403. [DOI] [PubMed] [Google Scholar]
- 105.Yampolsky IV, Balashova TA, Lukyanov KA. Biochemistry. 2009;48:8077. doi: 10.1021/bi900719x. [DOI] [PubMed] [Google Scholar]
- 106.Dean KM, Lubbeck JL, Binder JK, Schwall LR, Jimenez R, Palmer AE. Biophys J. 2011;101:961. doi: 10.1016/j.bpj.2011.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wachter RM, Elsliger MA, Kallio K, Hanson GT, Remington SJ. Structure. 1998;6:1267. doi: 10.1016/s0969-2126(98)00127-0. [DOI] [PubMed] [Google Scholar]
- 108.Henderson JN, Remington SJ. Proc Natl Acad Sci USA. 2005;102:12712. doi: 10.1073/pnas.0502250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Topol I, Collins J, Savitsky A, Nemukhin A. Biophys Chem. 2011;158:91. doi: 10.1016/j.bpc.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 110.Hasegawa JY, Ise T, Fujimoto KJ, Kikuchi A, Fukumura E, Miyawaki A, Shiro Y. J Phys Chem B. 2010;114:2971. doi: 10.1021/jp9099573. [DOI] [PubMed] [Google Scholar]
- 111.Abbyad P, Childs W, Shi X, Boxer SG. Proc Natl Acad Sci USA. 2007;104:20189. doi: 10.1073/pnas.0706185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hendrix J, Flors C, Dedecker P, Hofkens J, Engelborghs Y. Biophys J. 2008;94:4103. doi: 10.1529/biophysj.107.123596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bogdanov AM, Bogdanova EA, Chudakov DM, Gorodnicheva TV, Lukyanov S, Lukyanov KA. Nat Methods. 2009;6:859. doi: 10.1038/nmeth1209-859. [DOI] [PubMed] [Google Scholar]
- 114.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Nat Methods. 2008;5:545. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Drobizhev M, Makarov NS, Tillo SE, Hughes TE, Rebane A. Nat Methods. 2010;8:393. doi: 10.1038/nmeth.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Olsen S, Smith SC. J Am Chem Soc. 2007;129:2054. doi: 10.1021/ja066430s. [DOI] [PubMed] [Google Scholar]
- 117.Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, Chudakov DM. Nat Methods. 2007;4:741. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 118.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. Proc Natl Acad Sci USA. 2002;99:7877. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kredel S, Oswald F, Nienhaus K, Deuschle K, Rocker C, Wolff M, Heilker R, Nienhaus GU, Wiedenmann J. PLoS One. 2009;4:e4391. doi: 10.1371/journal.pone.0004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Verkhusha VV, Kuznetsova IM, Stepanenko OV, Zaraisky AG, Shavlovsky MM, Turoverov KK, Uversky VN. Biochemistry. 2003;42:7879. doi: 10.1021/bi034555t. [DOI] [PubMed] [Google Scholar]
- 121.Daniel RM, Cowan DA, Morgan HW, Curran MP. Biochem J. 1982;207:641. doi: 10.1042/bj2070641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kwon WS, Da Silva NA, Kellis JT., Jr Protein Eng. 1996;9:1197. doi: 10.1093/protein/9.12.1197. [DOI] [PubMed] [Google Scholar]
- 123.Inoue I, Rechsteiner M. J Biol Chem. 1994;269:29241. [PubMed] [Google Scholar]
- 124.Parsell DA, Sauer RT. J Biol Chem. 1989;264:7590. [PubMed] [Google Scholar]
- 125.Inoue I, Rechsteiner M. J Biol Chem. 1994;269:29247. [PubMed] [Google Scholar]
- 126.Evdokimov AG, Pokross ME, Egorov NS, Zaraisky AG, Yampolsky IV, Merzlyak EM, Shkoporov AN, Sander I, Lukyanov KA, Chudakov DM. EMBO Rep. 2006;7:1006. doi: 10.1038/sj.embor.7400787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pletnev S, Gurskaya NG, Pletneva NV, Lukyanov KA, Chudakov DM, Martynov VI, Popov VO, Kovalchuk MV, Wlodawer A, Dauter Z, Pletnev V. J Biol Chem. 2009;284:32028. doi: 10.1074/jbc.M109.054973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pletneva NV, Pletnev SV, Chudakov DM, Tikhonova TV, Popov VO, Martynov VI, Wlodawer A, Dauter Z, VZP Russ J Bioorg Khim. 2007;33:421. doi: 10.1134/s1068162007040048. [DOI] [PubMed] [Google Scholar]
- 129.Carpentier P, Violot S, Blanchoin L, Bourgeois D. FEBS Lett. 2009;583:2839. doi: 10.1016/j.febslet.2009.07.041. [DOI] [PubMed] [Google Scholar]
- 130.Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, Merzlyak EM, Shkrob MA, Lukyanov S, Lukyanov KA. Nat Biotechnol. 2006;24:95. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- 131.Baker M. Nat Methods. 2010;7:782. doi: 10.1038/nmeth1010-782. [DOI] [PubMed] [Google Scholar]
- 132.Hell SW. Science. 2007;316:1153. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 133.Willig KI, Kellner RR, Medda R, Hein B, Jakobs S, Hell SW. Nat Methods. 2006;3:721. doi: 10.1038/nmeth922. [DOI] [PubMed] [Google Scholar]
- 134.Leung BO, Chou KC. Appl Spectrosc. 2011;65:967. doi: 10.1366/11-06398. [DOI] [PubMed] [Google Scholar]
- 135.Strack RL, Hein B, Bhattacharyya D, Hell SW, Keenan RJ, Glick BS. Biochemistry. 2009;48:8279. doi: 10.1021/bi900870u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dieguez-Hurtado R, Martin J, Martinez-Corral I, Martinez MD, Megias D, Olmeda D, Ortega S. Genesis. 2010;49:36. doi: 10.1002/dvg.20685. [DOI] [PubMed] [Google Scholar]
- 137.Patterson G, Davidson M, Manley S, Lippincott-Schwartz J. Annu Rev Phys Chem. 2010;61:345. doi: 10.1146/annurev.physchem.012809.103444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gould TJ, Verkhusha VV, Hess ST. Nat Protoc. 2009;4:291. doi: 10.1038/nprot.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nienhaus GU, Nienhaus K, Holzle A, Ivanchenko S, Renzi F, Oswald F, Wolff M, Schmitt F, Rocker C, Vallone B, Weidemann W, Heilker R, Nar H, Wiedenmann J. Photochem Photobiol. 2006;82:351. doi: 10.1562/2005-05-19-RA-533. [DOI] [PubMed] [Google Scholar]
- 140.Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J. Nat Methods. 2008;5:155. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 141.Manley S, Gillette JM, Lippincott-Schwartz J. Methods Enzymol. 2010;475:109. doi: 10.1016/S0076-6879(10)75005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shroff H, Galbraith CG, Galbraith JA, White H, Gillette J, Olenych S, Davidson MW, Betzig E. Proc Natl Acad Sci USA. 2007;104:20308. doi: 10.1073/pnas.0710517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, Manley S, Sougrat R, Waterman CM, Kanchanawong P, Davidson MW, Fetter RD, Hess HF. Proc Natl Acad Sci USA. 2009;106:3125. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nature. 2010;468:580. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hofmann M, Eggeling C, Jakobs S, Hell SW. Proc Natl Acad Sci USA. 2005;102:17565. doi: 10.1073/pnas.0506010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Egner A, Geisler C, von Middendorff C, Bock H, Wenzel D, Medda R, Andresen M, Stiel AC, Jakobs S, Eggeling C, Schonle A, Hell SW. Biophys J. 2007;93:3285. doi: 10.1529/biophysj.107.112201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kogure T, Karasawa S, Araki T, Saito K, Kinjo M, Miyawaki A. Nat Biotechnol. 2006;24:577. doi: 10.1038/nbt1207. [DOI] [PubMed] [Google Scholar]
- 148.Kogure T, Kawano H, Abe Y, Miyawaki A. Methods. 2008;45:223. doi: 10.1016/j.ymeth.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 149.Kawano H, Kogure T, Abe Y, Mizuno H, Miyawaki A. Nat Methods. 2008;5:373. doi: 10.1038/nmeth0508-373. [DOI] [PubMed] [Google Scholar]
- 150.Tsutsui H, Karasawa S, Shimizu H, Nukina N, Miyawaki A. EMBO Rep. 2005;6:233. doi: 10.1038/sj.embor.7400361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wilmann PG, Petersen J, Devenish RJ, Prescott M, Rossjohn J. J Biol Chem. 2005;280:2401. doi: 10.1074/jbc.C400484200. [DOI] [PubMed] [Google Scholar]
- 152.Karasawa S, Araki T, Nagai T, Mizuno H, Miyawaki A. Biochem J. 2004;381:307. doi: 10.1042/BJ20040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wall MA, Socolich M, Ranganathan R. Nat Struct Biol. 2000;7:1133. doi: 10.1038/81992. [DOI] [PubMed] [Google Scholar]
- 154.Wiedenmann J, Schenk A, Rocker C, Girod A, Spindler KD, Nienhaus GU. Proc Natl Acad Sci USA. 2002;99:11646. doi: 10.1073/pnas.182157199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Petersen J, Wilmann PG, Beddoe T, Oakley AJ, Devenish RJ, Prescott M, Rossjohn J. J Biol Chem. 2003;278:44626. doi: 10.1074/jbc.M307896200. [DOI] [PubMed] [Google Scholar]
- 156.Wilmann PG, Petersen J, Pettikiriarachchi A, Buckle AM, Smith SC, Olsen S, Perugini MA, Devenish RJ, Prescott M, Rossjohn J. J Mol Biol. 2005;349:223. doi: 10.1016/j.jmb.2005.03.020. [DOI] [PubMed] [Google Scholar]