Abstract
Introduction
Craving is being considered for inclusion in the Diagnostic and Statistical Manual (DSM) DSM-5. However, little is known of its genetic underpinnings – specifically, whether genetic influences on craving are distinct from those influencing DSM-IV alcohol dependence.
Method
Analyses were conducted in a sample of unrelated adults ascertained for alcohol dependence (N=3976). Factor analysis was performed to examine how alcohol craving loaded with the existing DSM-IV alcohol dependence criteria. For genetic analyses, we first examined whether genes in the dopamine pathway, including dopamine receptor genes (DRD1, DRD2, DRD3, DRD4) and the dopamine transporter gene (SLC6A3), which have been implicated in neurobiological studies of craving, as well as alpha-synuclein (SNCA), which has been previously found to be associated with craving, were associated with alcohol craving in this sample. Second, in an effort to identify novel genetic variants associated with craving, we conducted a genomewide association study (GWAS). For variants that were implicated in the primary analysis of craving, we conducted additional comparisons - to determine if these variants were uniquely associated with alcohol craving as compared with alcohol dependence. We contrasted our results to those obtained for DSM-IV alcohol dependence, and also compared alcohol dependent individuals without craving to non-dependent individuals who also did not crave alcohol.
Results
Twenty-one percent of the full sample reported craving alcohol. Of those reporting craving, 97.3% met criteria for DSM-IV alcohol dependence with 48% endorsing all 7 dependence criteria. Factor analysis found a high factor loading (0.89) for alcohol craving. When examining genes in the dopamine pathway, single nucleotide polymorphisms (SNPs) in DRD3 and SNCA were associated with craving (p<0.05). There was evidence for association of these SNPs with DSM-IV alcohol dependence (p<0.05) but less evidence for dependence without craving (p>0.05), suggesting that the association was due in part to craving. In the GWAS, the greatest evidence of association with craving was for a SNP in the integrin alpha D (ITGAD) gene on chromosome 7 (rs2454908; p=1.8×10−6). The corresponding p-value for this SNP with DSM-IV alcohol dependence was similar (p=4.0×10−5) but was far less with dependence without craving (p=0.02), again suggesting the association was due to alcohol craving. Adjusting for dependence severity (number of endorsed criteria) attenuated p-values but did not eliminate association.
Conclusions
Craving is frequently reported by those who report multiple other alcohol dependence symptoms. We found that genes providing evidence of association with craving were also associated with alcohol dependence; however, these same SNPs were not associated with alcohol dependence in the absence of alcohol craving. These results suggest that there may be unique genetic factors affecting craving among those with alcohol dependence.
Introduction
Genetic studies of alcohol dependence are at an exciting crossroad. With the emergence of the proposed DSM-5 criteria for diagnosing substance use disorders (dsm5.org), investigators with genetically informative data have the unique opportunity to revisit important hypotheses regarding the genetic underpinnings of specific components of alcohol use disorders.
Three important changes to the diagnosis of substance, including alcohol, use disorders are proposed in DSM-5: (a) abuse and dependence may no longer be considered independent entities. Instead, a uni-dimensional score of 11 (abuse and dependence) criteria is proposed to diagnose moderate (2–3 criteria) and severe (4 or more criteria) substance use disorders; (b) “recurrent legal problems” may no longer be part of the criteria, due to its low frequency and weak correlation with the remaining abuse and dependence criteria; and (c) a new criterion, craving, has been suggested for addition to the existing criteria (O’Brien, 2011). While most investigators with data collected using DSM-IV criteria will experience limited problems with (a) and (b), the addition of a new criterion (c) can pose challenges for studies that have not collected data on craving. Particularly for large-scale genetic studies where it is often infeasible to conduct repeat data collection, the possibility exists that the addition of this new criterion will compromise genetic examination of the proposed DSM-5 defined substance use disorders. This report examines what additional genetic information craving contributes that cannot be gleaned from examining DSM-IV dependence criteria.
For DSM-5, craving is proposed to be defined as “strong desire or urge to use a specific substance”. The preliminary rationale for the addition of craving arose from three sources – its inclusion in the International Classification of Disease (ICD-10) (World Health Organization, 2007) definition of substance use disorders, neurobiological experiments supporting cue-elicited craving (Childress et al., 1999; Kalivas & O’Brien, 2008; Volkow et al., 2006; Kalivas & Volkow, 2005), and clinical observations of its significance in relapse (Monti et al., 1990; O’Brien, 2005; Flannery et al., 2001). There is little doubt that craving is an integral aspect of substance use disorders – numerous epidemiological (Mewton, Slade, McBride, Grove, & Teesson, 2010; Keyes, Krueger, Grant, & Hasin, 2011) and emergency room (Cherpitel et al., 2010) studies report high factor loadings for alcohol craving, indicating considerable correlation between self-reported craving and the underlying liability to substance use disorders as indexed by DSM-IV dependence criteria However, whether it adds unique information to the diagnosis of substance use disorder remains unclear.
In psychometric analyses, craving is considered a “difficult” item – that is, it is infrequently endorsed and even though it discriminates between individuals with and without substance use disorders, it does so primarily in individuals already at high liability to develop the disorder. In other words, those who report craving are typically already classified as substance dependent due to endorsement of multiple (often more than 3) dependence criteria; craving further discriminates these most severely affected individuals (i.e. those with multiple other symptoms) (Bohn, Krahn, & Staehler, 1995; Bucholz et al., 1996). Consequently, the addition of craving as a criterion does not produce a significant increase in those diagnosed with alcohol use disorders (Mewton et al., 2010; Agrawal, Heath, & Lynskey, 2011; Keyes et al., 2011; Cherpitel et al., 2010). This opens the door to the discussion of whether craving is best assessed using a single item or whether the architecture of craving is better suited to a multi-dimensional assessment in order to provide unique information beyond the existing definitions of dependence.
What does this mean for genetic studies of alcohol dependence? It is well established that genetic factors play a substantial role in the etiology of substance use disorders, regardless of the type of diagnostic classification used. Therefore, it is widely accepted that the proposed DSM-5 definitions of substance use disorder will also be heritable. However, the addition of this new item raises the question: are there novel genetic influences on craving? Even though phenotypic correlations between craving and other dependence criteria are high, this does not rule out the possibility that at least some genetic pathways that contribute to craving may be distinct from those influencing other aspects of substance dependence.
There are multiple routes for examining the potential role of distinct genetic influences on craving. In the literature, craving has been studied in the context of drug compulsion, and consequently as a correlate of relapse, particularly under negative emotional states (Marlatt & Gordon, 1985). Studies of cue-elicited craving report activation in brain regions that constitute dopamine pathways. For instance, Childress et al. (Childress et al., 1999), reported increases in cerebral blood flow in limbic (amygdala and anterior cingulated) regions when cocaine users were exposed to drug videos. In addition, dopamine-rich regions such as the ventral tegmental area (VTA) have been implicated in the neuroplasticity underlying the development of addiction and the emergence of craving (Kalivas & O’Brien, 2008). Accordingly, genes in the dopamine pathway may yield promising insights regarding the genetic contributions to craving.
Based on this posited neurobiological basis of cue-elicited craving, we selected a set of genes that broadly constitute the dopamine system. These included receptor-encoding genes (DRD1, DRD2, DRD3, DRD4) and the dopamine (SLC6A3, also known as DAT) transporter gene. We also selected α-synuclein (SNCA) which was previously implicated in a candidate gene association study of alcohol craving conducted on a partially overlapping sample (Foroud et al., 2007) as well as in an independent sample (Bonsch et al., 2005b). While synucleins have been primarily studied in the context of Alzheimer’s and Parkinson’s disease (Irvine, El-Agnaf, Shankar, & Walsh, 2008), α-synucleins regulate dopamine D2 synthesis (Perez et al., 2002). Bonsch and colleagues found increased α-synuclein expression in alcoholics, which was positively correlated with alcohol craving scores (Bonsch et al., 2005a; Bonsch et al., 2004). In addition to this focus on dopamine pathway genes, in an effort to identify novel genetic variants that were associated with alcohol craving, we conducted a genomewide association study (GWAS).
Methods
Sample
The sample used for this study was the Study of Addiction: Genes and Environment (SAGE: (Bierut et al., 2010)) which drew data from three independent family-based studies to create a sample of unrelated DSM-IV alcohol dependent cases and alcohol exposed controls. The parent studies were family-based and ascertained for alcohol dependence (Collaborative Study of the Genetics of Alcoholism, COGA, (Begleiter et al., 1995)), nicotine dependence (Collaborative Study of the Genetics of Nicotine Dependence, COGEND, (Bierut et al., 2007)) and cocaine dependence (Family Study of Cocaine Dependence, FSCD (Bierut, Strickland, Thompson, Afful, & Cottler, 2008). The SAGE series consisted of 1899 unrelated DSM-IV alcohol dependent cases and 1938 unrelated alcohol exposed (drank at least one drink during the lifetime) controls.
Measures
All three parent studies used modified versions of the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA, Bucholz et al., 1994) to assess substance use disorders with good validity (Hesselbrock, Easton, Bucholz, Schuckit, & Hesselbrock, 1999) and reliability (Bucholz et al., 1994). Alcohol craving was evaluated with a single item. In COGA and COGEND, the item queried “In situations where you could not drink alcohol, did you ever have such a strong desire for it that you couldn’t think of anything else?” In FSCD, the item queried “Have you ever had such a strong craving for alcohol that you couldn’t think of anything else?” DSM-IV alcohol dependence was defined by the clustering of 3 or more dependence criteria in a 12 month period. Note that, consistent with DSM-IV, craving was not utilized as a criterion to diagnose DSM-IV alcohol dependence.
Genotypic data
Genotyping was done with the Illumina Human 1M beadchip by the Center for Inherited Diseases Research (CIDR) at Johns Hopkins University. A thorough data cleaning procedure on the genotype samples was applied to ensure the highest possible data quality, including using HapMap controls, detection of gender and chromosomal anomalies, hidden relatedness, population structure, missing call rates, plate effects, duplication error detection, and Hardy-Weinberg equilibrium (excluding SNPs with p < 10−6) (Laurie et al., 2009; Bierut et al., 2010). The final number of autosomal and X-chromosome markers consisted of 948,142 SNPs (only autosomal SNPs were used). The software package EIGENSTRAT/EIGENSOFT(Price et al., 2006) was used to calculate principal components reflecting continuous variation in allele frequencies representing ancestral differences in subjects. Two principal components distinguishing African-American participants from European-American participants and Hispanic and non-Hispanic subjects, were identified (Bierut et al., 2010).
Factor Analyses
Descriptive statistics were computed in SASv9 (SAS Institute, 1999). As the items assessing craving in COGA/COGEND and FSCD were slightly different, a multi-group confirmatory factor model was used to determine (a) the factor loading (discrimination) and threshold (difficulty) for craving in the full SAGE sample and (b) to examine whether these parameters varied across the 3 contributing parent studies. Item response modeling was conducted in MPlus v5 (Muthen & Muthen, 2007).
Statistical Analysis
In the primary analysis, individuals endorsing craving (with or without DSM-IV alcohol dependence) were designated as affected and those who did not report craving (whether or not they met criteria for DSM-IV alcohol dependence) were designated as unaffected. Secondary analyses were performed to examine the association of particular SNPs with alcohol dependence without craving. For this analysis, individuals meeting criteria for DSM-IV alcohol dependence who did not endorse the proposed DSM-5 craving item were designated as affected and those who did not report craving and did not meet criteria for DSM-IV dependence were designated as unaffected. Results were also compared to those from our previous GWAS for DSM-IV alcohol dependence (Bierut et al., 2010).
PLINK (Purcell et al., 2007) was used to conduct all autosomal association analyses using logistic regression models. Genotype was coded log-additively (AA, Aa/aA, aa) and covariates representing sex, age at interview, whether participants were drawn from COGA or FSCD (COGEND serving as the reference group) and the first two principal components to account for ethnic variation in allele frequencies, were also included. As our primary analysis, we examined SNPs in SNCA, which has been previously studied in relation to alcohol craving, as well as genes in the dopamine pathway (DRD1, DRD2, DRD3, DRD4, SLC6A3). For the analysis of these genes of a priori significance, we set a liberal threshold of p < 0.05. Next, we conducted a genomewide search – as these analyses are exploratory, statistical significance in the GWAS was set at p < 5 × 10−8. For both association analyses, primary analyses were conducted to identify SNPs associated with alcohol craving. For SNPs that were implicated in the primary analysis of craving, secondary analyses were performed examining other comparisons. These included an analysis of only non-craving individuals, contrasting those with and without alcohol dependence, as well as an analysis of DSM-IV alcohol dependence and an adjustment for severity of alcohol dependence.
RESULTS
Sample characteristics
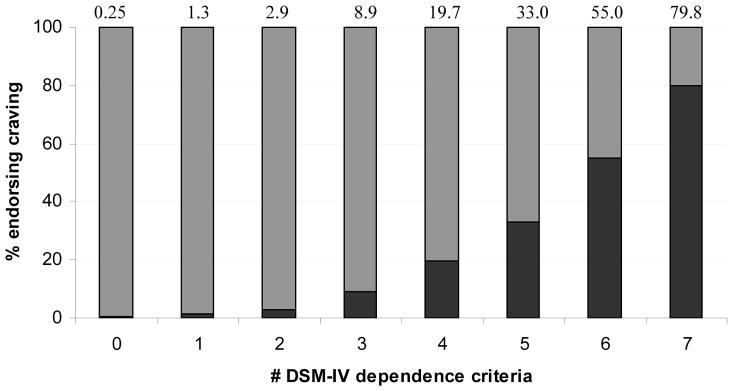
Table 1 provides the characteristics of those endorsing craving compared with those who did not endorse it. An overwhelming proportion (97.3%) of those endorsing craving met criteria for DSM-IV dependence. Nearly 48% of those who reported alcohol craving also endorsed all 7 DSM-IV dependence criteria. COGA contributed a majority of the craving cases for alcohol. Alternatively, as shown in Figure 1, endorsement of craving increased with increasing number of dependence criteria endorsed with nearly 80% of those who reported all 7 DSM-IV criteria also reporting craving
Table 1.
Sample characteristics for 3,976 individuals from the Study of Addiction: Genes and Environment.
| Craving (N=841) | No Craving (N=3135) | ||
|---|---|---|---|
|
| |||
| Alcohol Dependence (N=1080) | No Alcohol Dependence (N=2055) | ||
|
|
|||
| Sex (Male) | 59.2% | 61.8% | 31.2% |
|
| |||
| Mean Age | 39.4 [SD 9.2] | 38.6 [SD 9.3] | 39.3 [SD 9.1] |
|
| |||
| European-American | 67.5% | 62.9% | 74.0% |
|
| |||
| Hispanic | 4.4% | 3.6% | 3.1% |
|
| |||
| COGA | 57.1% | 38.8% | 26.6% |
|
| |||
| FSCD | 25.1% | 31.9% | 29.8% |
|
| |||
| COGEND | 17.8% | 29.4% | 45.2% |
|
| |||
| Meet criteria for DSM-IV dependence | 97.3% | 100% | 0% |
|
| |||
| Mean number of dependence criteria | 5.95 [SD 1.3] | 4.5 [SD 1.3] | 0.6 [SD 0.8] |
SD – Standard Deviation
FIGURE 1.
Percentage (dark shading) of those with 0 to 7 DSM-IV dependence criteria who also endorsed alcohol craving.
Factor analysis
Standardized factor loadings for all dependence criteria (0.80–0.92) and for craving (0.89) were high in the full sample (Table 2). When factor loadings and thresholds (estimates of prevalence) for craving were allowed to vary across COGA, COGEND and FSCD, factor loadings remained consistently high while thresholds varied modestly across samples. A 4 degree of freedom test resulted in a significant chi-square difference (Δχ2= 27.8) although this difference was entirely attributed to the threshold differences. Furthermore, excluding craving from the factor analysis did not perturb the factor loadings for the DSM-IV dependence criteria.
Table 2.
Factor loadings and endorsement prevalence for craving relative to DSM-IV dependence criteria in 3976 subjects from SAGE.
| Loading | Prevalence (%) | |
|---|---|---|
| Craving | .89 | 21 |
| DSM-IV criteria | ||
| Tolerance | .80 | 47 |
| Withdrawal | .91 | 25 |
| Larger/Longer | .87 | 61 |
| Quit attempts | .87 | 46 |
| Time Spent | .89 | 28 |
| Give up activities | .92 | 29 |
| Use despite problems | .88 | 46 |
The factor loadings are drawn from a confirmatory single factor model fit in MPlus.
Genes in the dopamine pathway
Table 3 shows the results for SNPs in genes in the dopamine pathway. Only SNPs with a p-value less than 0.05 for craving are shown and only for this subset of SNPs were comparisons with DSM-IV alcohol dependence and dependence without craving made. Setting a liberal threshold of p < 0.05, SNPs in DRD2, DRD3 and SNCA showed evidence for association with craving. Comparing the results for these SNPs with those for DSM-IV alcohol dependence, SNPs in DRD3 and SNCA appeared to have independent genetic effects on craving, infrequently showing a p-value < 0.05 for alcohol dependence while for DRD2, a number of SNPs associated with craving had a p-value < 0.05 for alcohol dependence. With some exception, the SNPs associated with craving did not show p-values < 0.05 for dependence without craving.
Table 3.
Genes in the dopamine pathway (only SNPs at p ≤ 0.05 for craving are shown).
| SNP | Craving (with or without alcohol dependence). | DSM-IV dependence | Dependence without craving | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Position (basepair) | O.R. | Lower C.I. | Upper C.I. | P-value | O.R. | P-value | O.R. | P-value | |
| DRD3, chromosome 3, 27 SNPs | |||||||||
| rs9828046 | 115340953 | 0.71 | 0.53 | 0.94 | 1.65E-02 | 0.78 | 2.93E-02 | 0.93 | 5.71E-01 |
| rs2630349 | 115356062 | 1.19 | 1.00 | 1.40 | 4.61E-02 | 1.01 | 9.04E-01 | 0.97 | 7.23E-01 |
| rs167770 | 115362252 | 0.82 | 0.73 | 0.93 | 2.07E-03 | 0.97 | 6.15E-01 | 1.09 | 1.79E-01 |
| rs226082 | 115363703 | 0.82 | 0.72 | 0.93 | 1.82E-03 | 0.98 | 7.65E-01 | 1.10 | 1.26E-01 |
| rs324029 | 115364313 | 0.82 | 0.73 | 0.93 | 1.93E-03 | 0.98 | 6.80E-01 | 1.09 | 1.66E-01 |
| SNCA, chromosome 4, 31 SNPs | |||||||||
| rs35101723 | 90866209 | 0.32 | 0.11 | 0.91 | 3.23E-02 | 0.41 | 1.09E-02 | 0.53 | 9.00E-02 |
| rs6834765 | 90903013 | 0.83 | 0.69 | 0.98 | 3.14E-02 | 0.96 | 5.51E-01 | 1.05 | 6.07E-01 |
| rs3796661 | 90906530 | 0.76 | 0.58 | 0.98 | 3.63E-02 | 0.83 | 8.66E-02 | 0.92 | 4.99E-01 |
| rs11944331 | 90909352 | 0.84 | 0.71 | 0.99 | 3.23E-02 | 0.98 | 7.38E-01 | 1.09 | 3.10E-01 |
| rs3775439 | 90928764 | 0.86 | 0.74 | 0.99 | 3.28E-02 | 0.93 | 2.68E-01 | 1.02 | 7.55E-01 |
| DRD2, chromosome 11, 34 SNPs | |||||||||
| rs1076560 | 112788898 | 1.20 | 1.02 | 1.41 | 2.72E-02 | 1.13 | 8.72E-02 | 1.08 | 3.71E-01 |
| rs2734838 | 112791711 | 0.88 | 0.78 | 0.99 | 3.55E-02 | 0.87 | 8.60E-03 | 0.88 | 3.92E-02 |
| rs1079727 | 112794392 | 1.21 | 1.03 | 1.42 | 2.22E-02 | 1.13 | 1.04E-01 | 1.06 | 4.74E-01 |
| rs4986918 | 112800452 | 0.55 | 0.34 | 0.89 | 1.48E-02 | 1.08 | 6.89E-01 | 1.30 | 1.81E-01 |
| rs1076563 | 112801119 | 0.87 | 0.77 | 0.98 | 2.67E-02 | 0.87 | 7.15E-03 | 0.88 | 3.68E-02 |
| rs1116313 | 112801317 | 0.87 | 0.77 | 0.98 | 2.04E-02 | 0.86 | 6.01E-03 | 0.88 | 3.46E-02 |
| rs12364051 | 112810524 | 0.88 | 0.78 | 1.00 | 4.29E-02 | 0.88 | 2.14E-02 | 0.89 | 6.90E-02 |
| rs17529477 | 112822277 | 0.84 | 0.73 | 0.96 | 1.31E-02 | 0.91 | 9.24E-02 | 0.97 | 6.21E-01 |
O.R. Odds-ratio; Lower/Upper C.I. – lower and upper 95% confidence interval on odds-ratio. See supplemental table for minor allele frequency. Position in base pair and gene based on NCBI build 36.3
GWAS
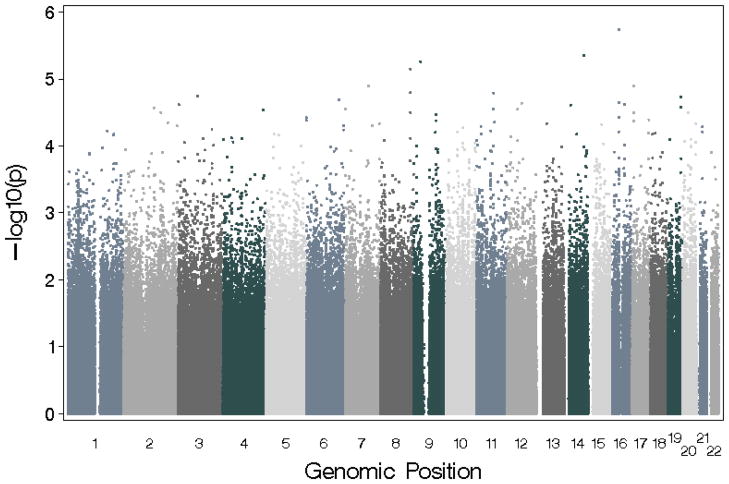
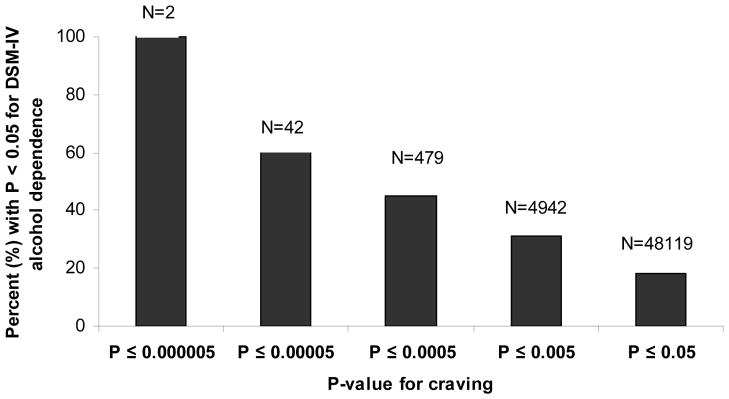
Figure 2 and Table 4 summarize the results of the autosomal genomewide analysis using the craving for alcohol phenotype. The best association for craving was found for SNP rs2454908 (p-value=1.83 × 10−6) on chromosome 16. Table 4 also compares results for craving with those for DSM-IV alcohol dependence and with dependence without craving. For rs2454908, the odds-ratio and p-value for craving and DSM-IV dependence were comparable; however, the p-value for this SNP was 0.018 when examining dependence without craving. Of the 30 most significant SNPs for craving, 20 SNPs also had p-values less than 0.05 for DSM-IV alcohol dependence with or without craving, but only rs2454908 had a p-value less than 0.05 for dependence without craving. The overlap between alcohol dependence and craving dropped steadily as the threshold for association with craving was made less stringent (Figure 3).
FIGURE 2.
Manhattan plot showing genomewide association study (GWAS) results for alcohol craving.
Table 4.
Top 30 signals from GWAS of alcohol craving with corresponding results for the top SNPs from GWAS of DSM-IV alcohol dependence and dependence without craving.
| Chromosome | SNP | Position (basepair) | Gene | Craving | DSM-IV dependence | Dependence without craving | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| O.R. | Lower C.I. | Upper C.I. | P-value | O.R. | P-value | O.R. | P-value | ||||
| 16 | rs2454908 | 31318686 | ITGAD | 0.74 | 0.65 | 0.83 | 1.83E-06 | 0.80 | 4.04E-05 | 0.86 | 1.86E-02 |
| 14 | rs11849247 | 87778256 | KCNK10 | 1.67 | 1.34 | 2.08 | 4.44E-06 | 1.31 | 1.05E-02 | 1.15 | 2.70E-01 |
| 9 | rs3849883 | 30629205 | 2.41 | 1.65 | 3.51 | 5.45E-06 | 1.51 | 3.91E-02 | 1.05 | 8.51E-01 | |
| 8 | rs2469518 | 133225649 | KCNQ3 | 0.77 | 0.68 | 0.86 | 7.06E-06 | 0.87 | 8.31E-03 | 0.96 | 4.95E-01 |
| 7 | rs6466086 | 105550662 | 1.41 | 1.21 | 1.65 | 1.26E-05 | 1.17 | 2.47E-02 | 1.04 | 6.10E-01 | |
| 17 | rs34870220 | 7025635 | 1.81 | 1.39 | 2.36 | 1.26E-05 | 1.34 | 2.20E-02 | 1.00 | 9.99E-01 | |
| 8 | rs1991718 | 133230506 | KCNQ3 | 0.77 | 0.69 | 0.87 | 1.60E-05 | 0.85 | 1.33E-03 | 0.92 | 1.71E-01 |
| 11 | rs2447947 | 75621188 | 1.29 | 1.15 | 1.44 | 1.60E-05 | 1.13 | 1.48E-02 | 1.02 | 7.51E-01 | |
| 3 | rs4561856 | 87742811 | 0.73 | 0.63 | 0.84 | 1.79E-05 | 0.91 | 1.44E-01 | 1.04 | 5.53E-01 | |
| 19 | rs198967 | 56104833 | KLK4 | 2.02 | 1.47 | 2.79 | 1.84E-05 | 1.17 | 3.30E-01 | 0.83 | 3.31E-01 |
| 6 | rs7762384 | 145081988 | UTRN | 0.59 | 0.46 | 0.75 | 2.03E-05 | 0.98 | 8.35E-01 | 1.24 | 6.15E-02 |
| 16 | rs17855121 | 30911670 | STX1B2 | 1.32 | 1.16 | 1.50 | 2.21E-05 | 1.17 | 6.79E-03 | 1.06 | 4.15E-01 |
| 12 | rs721793 | 66720281 | 1.40 | 1.20 | 1.64 | 2.28E-05 | 1.25 | 2.50E-03 | 1.07 | 4.40E-01 | |
| 16 | rs767505 | 55770151 | NIP30 | 0.70 | 0.60 | 0.83 | 2.36E-05 | 0.78 | 3.10E-04 | 0.87 | 7.39E-02 |
| 3 | rs11707897 | 2589626 | CNTN4 | 0.68 | 0.56 | 0.81 | 2.41E-05 | 0.82 | 8.00E-03 | 0.93 | 4.02E-01 |
| 14 | rs4983262 | 27190834 | 0.75 | 0.65 | 0.86 | 2.44E-05 | 0.88 | 3.02E-02 | 0.98 | 8.11E-01 | |
| 19 | rs198966 | 56103822 | KLK4 | 1.85 | 1.39 | 2.46 | 2.61E-05 | 1.17 | 2.73E-01 | 0.87 | 4.17E-01 |
| 2 | rs1901439 | 134153941 | 0.69 | 0.58 | 0.82 | 2.67E-05 | 0.87 | 5.15E-02 | 1.05 | 5.68E-01 | |
| 7 | rs17132242 | 2145988 | MAD1L1 | 1.50 | 1.24 | 1.81 | 2.80E-05 | 1.16 | 9.93E-02 | 0.96 | 7.04E-01 |
| 11 | rs10899187 | 75643975 | 1.31 | 1.15 | 1.48 | 2.81E-05 | 1.17 | 6.56E-03 | 1.06 | 3.97E-01 | |
| 12 | rs3782323 | 48769188 | SMARCD1 | 0.76 | 0.67 | 0.86 | 2.83E-05 | 0.90 | 6.65E-02 | 1.02 | 7.54E-01 |
| 4 | rs2666055 | 179581414 | 1.44 | 1.22 | 1.72 | 2.90E-05 | 1.16 | 6.63E-02 | 1.03 | 7.81E-01 | |
| 8 | rs1595411 | 133228607 | KCNQ3 | 0.78 | 0.70 | 0.88 | 3.12E-05 | 0.85 | 1.17E-03 | 0.92 | 1.37E-01 |
| 2 | rs687415 | 166011844 | TTC21B | 1.41 | 1.20 | 1.65 | 3.15E-05 | 1.19 | 2.53E-02 | 1.06 | 5.34E-01 |
| 20 | rs8114108 | 24980075 | ACSS1 | 0.59 | 0.46 | 0.76 | 3.15E-05 | 0.92 | 4.53E-01 | 1.11 | 3.82E-01 |
| 17 | rs12947697 | 7030341 | 1.76 | 1.35 | 2.30 | 3.19E-05 | 1.30 | 4.17E-02 | 0.96 | 8.12E-01 | |
| 9 | rs10115971 | 100064708 | 1.35 | 1.17 | 1.56 | 3.37E-05 | 1.18 | 1.16E-02 | 1.07 | 4.08E-01 | |
| 16 | rs17790906 | 30477160 | MGC13138 | 1.50 | 1.24 | 1.82 | 3.73E-05 | 1.24 | 1.80E-02 | 1.05 | 6.48E-01 |
| 6 | rs6899567 | 157595 | 0.69 | 0.58 | 0.82 | 3.76E-05 | 0.93 | 3.35E-01 | 1.07 | 4.53E-01 | |
| 17 | rs12603684 | 74735394 | 1.33 | 1.16 | 1.53 | 4.02E-05 | 1.16 | 1.88E-02 | 1.03 | 6.45E-01 | |
O.R. Odds-ratio; Lower/Upper C.I. – lower and upper 95% confidence interval on odds-ratio. See supplemental table for minor allele frequency. Position in base pair and gene based on NCBI build 36.3
Figure 3.
Proportion of SNPs with a p-value ≤ 0.05 for DSM-IV alcohol dependence amongst SNPs with varying p-values for alcohol craving.
Adjusting for severity of alcohol dependence
As indicated in Figure 1, craving was more common in those endorsing a greater number of alcohol dependence criteria, an index of severity. We re-ran association analyses for SNPs in Tables 3 and 4 including number of dependence criteria (0–7) as a covariate. Adjustment for symptom count, as expected, attenuated p-values, in some cases. For genes in the dopamine pathway, after correction, the 3 DRD3 SNPs with the lowest p-values (rs167770, rs226082 and rs324029) continued to show p-values less than 0.05. Three SNPs in SNCA also remained associated (rs6834765, rs11944331 and rs3775439) with p-values indicating a greater degree of significance upon adjustment for severity. For the GWAS, only 2 SNPs (rs1595411 and rs10115971) were no longer associated with craving at p ≤ 0.05 after adjusting for severity. For example, after accounting for number of alcohol dependence criteria, rs2454908 retained a p-value of 0.0002.
Discussion
Craving, reflecting strong desire or urge to use a drug, is being proposed for inclusion in DSM-5. In the present analyses, while we did not identify any genomewide association signals for craving, we found that a number of SNPs that were associated with alcohol craving at p-values less than 1 × 10−6 also showed nominal association with DSM-IV alcohol dependence, but not with dependence excluding craving. Several SNPs in dopamine-related genes did exhibit differing results when comparing craving and DSM-IV alcohol dependence albeit at far lower levels of statistical significance. While imposing a high statistical threshold yielded significant overlap across SNPs for craving and for alcohol dependence, lowering this threshold and specifically focusing on genes of a priori interest indicated potential biological specificity for craving.
We selected genes in the dopamine pathways because multiple imaging studies have implicated dopamine-rich brain regions in experiments evoking cue-elicited craving for cocaine (Volkow et al., 2006; Childress et al., 1999) and other drugs (Kalivas & O’Brien, 2008; Kalivas & Volkow, 2005). This limbic activation suggests that genes in dopamine pathways might be of salience to craving but not all aspects of alcohol dependence. Despite the significant overlap between DSM-IV alcohol dependence and craving in our sample, we saw some support for differential effects of SNPs on craving, alcohol dependence and dependence without craving. We also found support for signals in SNCA, a gene previously implicated in a partially overlapping sample (Foroud et al., 2007). However, the SNPs identified from this study were not correlated with those identified by Foroud and colleagues.
Multiple SNPs in DRD3 were associated with craving alone, which supports the role of dopamine pathway genes in craving. Dopamine receptors in the striatum are down-regulated in alcohol dependent individuals (Volkow et al., 1996), and this reduction in striatal D2/3 receptors was shown to be associated with increased craving in alcoholics (Heinz et al., 2004). These authors subsequently demonstrated that the increased alcohol craving was inversely related to dopamine synthesis and binding in the same subjects (Heinz et al., 2005). Similarly, Tupala and colleagues reported reduced D2/D3 binding in the nucleus accumbens and amygdala among late-onset type 1 alcoholics, who have been hypothesized to have a deficit in dopamine availability (Tupala et al., 2001). A more general study revealed that decreased striatal D2/3 availability in healthy controls was associated with greater impulsivity and craving for stimulants (Buckholtz et al., 2010). D3 knock-out mice also show intense withdrawal from ethanol (Narita, Soma, Tamaki, Narita, & Suzuki, 2002). In fact dopamine activation has been proposed to treat addictions in general, (Blum, Liu, Shriner, & Gold, 2011) and more specifically, as a therapeutic target to reduce craving and relapse in the “dopamine-impoverished addicted brain” (Diana, 2011). Our results, despite not achieving genomewide levels of statistical significance, are promising in that they support the results reported above.
In reviewing the present findings, it should also be recognized that craving experienced as a consequence of cue exposure may differ from self-reported “strong desire or urge” to use alcohol, which is the phenotype used in this study (Carter & Tiffany, 1999; Kozlowski, Mann, Wilkinson, & Poulos, 1989; Marlatt & Gordon, 1985). Furthermore, craving is commonly reported during abstinence and can be an aspect of withdrawal, but whether anticipatory pleasure from alcohol use, cue-induced urges, desires to drink during withdrawal and self-reported craving are overlapping, or even related, constructs has not been well-explored (Moss, 2011). There is also evidence that craving for some drugs (e.g. cocaine) is a significant contributor to relapse (O’Brien, 2005) with one study reporting that alcohol craving was associated with time to relapse after residential treatment (Oslin, Cary, Slaymaker, Colleran, & Blow, 2009). However, this effect was not noted for alcohol in one large study (Leggio, Ray, Kenna, & Swift, 2009) and the effects were found to be even weaker when considering self-reported craving (Drummond, 2001; Drummond, Litten, Lowman, & Hunt, 2000; Lowman, Hunt, Litten, & Drummond, 2000).
It should also be noted that cue-elicited craving may be accentuated in individuals experiencing negative emotional states, particularly in high risk situations. In the context of the cognitive behavioral relapse prevention model (Marlatt & Gordon, 1985), under situations of stress (Larimer, Palmer, & Marlatt, 1999; Hendershot, Witkiewitz, George, & Marlatt, 2011), the anticipatory pleasure (i.e. positive reinforcement, preexisting expectancies) associated with drug-taking is heightened, intensifying craving. In fact, stress-induced craving has been found to be a more potent predictor of relapse than drug-cue induced craving (Sinha, Garcia, Paliwal, Kreek, & Rounsaville, 2006; Sinha et al., 2011). Therefore, our association signals in dopamine pathway genes, which have previously been implicated in cue-induced craving, may have shown modest p-values for self-reported craving, in part, due to the well-documented variations between cue-induced, stress-induced and self-reported craving.
The most significant association for alcohol craving in this GWAS was an intronic SNP, rs2454908, in the integrin, alpha D (ITGAD) gene which belongs to the CD11 antigen-like family of genes. To the best of our knowledge, the only potentially functionally relevant SNP that is in moderate linkage disequilibrium with this marker is in the 3′ untranslated regulatory region of the neighboring Integrin, alpha X (ITGAX) gene. How this SNP relates to alcohol craving is unknown. A number of SNPs in the potassium voltage-gated channel, KQT-like subfamily, member 3 (KCNQ3) gene were also associated with craving. The protein encoded by this gene, along with the protein product of KCNQ2 and KCNQ5, form potassium M channels that are critical in neuronal excitability and variants in KCNQ5 were previously identified in an African-American subset of individuals with alcohol dependence (Kendler et al., 2011). Of particular relevance to our study, Koyama et al (Koyama, Brodie, & Appel, 2007) have reported that ethanol induced excitation of dopamine neurons in the Ventral Tegmental Area (VTA) can be attributed to inhibition of currents in M-channels.
An important observation in these data was that (a) nearly all those who endorsed craving met criteria for DSM-IV alcohol dependence and (b) craving was more commonly reported in those with a greater number of alcohol dependence symptoms, which is consistent with a prior latent class analysis in an extended subset of these data (Foroud et al., 1998). These findings, despite being drawn from a study ascertained for alcoholism, are consistent with the extant epidemiological literature that notes that craving, while being an excellent item when distinguishing between those at high versus lower liability to alcohol dependence, is only effective at making this distinction in individuals at high risk. In the context of psychometrics, the craving item has high discrimination (i.e. good factor loading) and high difficulty/severity (i.e. infrequently endorsed by low risk individuals). For instance, in the National Longitudinal Alcohol Epidemiologic Survey (NLAES), craving cohered well with other DSM-IV alcohol dependence criteria, had the second highest discrimination and also the highest severity (with the exception of the abuse criterion of legal problems, which is proposed for exclusion in DSM-5). This indicates that the utility of craving will be most noticeable in distinguishing individuals with multi-symptom alcohol dependence but less so in those at low to moderate risk. That craving will add additional alcohol use disorder cases to DSM-5 diagnosis remains uncertain as, in our study and other epidemiological investigations (e.g. (Agrawal et al., 2011; Keyes, Krueger, Grant, & Hasin, 2010)), those reporting craving already met the burden of diagnosis. Therefore, as currently conceptualized craving, doesn’t significantly modify the epidemiology of alcohol use disorders but adds predictive utility in high risk individuals. Despite this significant overlap between craving and severity of alcohol dependence, a number of our candidate gene and a majority of the GWAS signals retained p-values ≤ 0.05 after adjusting for severity. This would suggest further specificity of these SNPs in the etiology of craving.
Some limitations of our study should be noted. First, our sample drew from parent studies with differing ascertainment criteria and this may have contributed to heterogeneity in those endorsing craving. Our factor analyses, however, indicated that heterogeneity is attributable to the relative frequency of endorsement of craving across the parent studies, but not in its relationship with dependence. Second, as our sample was designed to be enriched for alcohol and drug dependent individuals, results from our study may not generalize to the population. Nonetheless, phenotypic similarities with epidemiological studies are apparent. Third, none of the association signals surpassed genomewide significance and hence, our comparison across craving, dependence without craving and DSM-IV dependence, which relied on p-values, may be confounded with a high rate of false positive findings. Replication will be necessary before we can be confident in these findings. Although 97% of individuals endorsing the craving item were alcohol dependent, the observation that some SNPs uniquely associated with craving while other SNPs were only associated with alcohol dependence demonstrate both the heterogeneity of the alcohol dependence diagnosis and the potential specificity of the craving item. Similar results were reported by Foroud et al (2007) in that SNPs in SNCA were associated with craving but not with alcohol dependence.
Perhaps most importantly, the definition of craving is known to have a significant impact on its psychometric relationship with the construct of dependence – we used a single item that approximates the proposed DSM-5 criteria, however more work is required to outline whether this definition of craving is optimal for diagnostic purposes and for genetic analysis. The single craving item was employed in both a family-based study of European subjects in which it was associated with SNPs in SNCA (Foroud et al, 2007) as well as in a family-based linkage study of Mission Indians, which estimated the heritability of craving using this singe item to be 65% (Ehlers & Wilhelmsen, 2005).
Although a multi-dimensional craving assessment such as the Desires for Alcohol Questionnaire (DAQ) might offer a more powerful model providing overlapping and perhaps additional independent association results, we were unable to employ this measure in the current study because the questionnaire was only administered to a small subset of one participating study. However, Kramer et al (Kramer et al., 2010) have studied the DAQ and found it to correlate well with this single craving item. Whether the 3 facets identified in that study (Strong Desires/Intentions, Negative Reinforcement and Positive Reinforcement and ability to control drinking) yield differing genetic signals should be investigated when larger samples with genetic data become available.
Overall, in our sample ascertained for alcohol dependence, we found that craving was strongly associated with severity of dependence. This may indicate its potential role in relapse. However, based on our work and that of numerous others, there is a significant need for research to further clarify how craving should be operationalized for diagnostic and research purposes. The genomic analyses presented here lead to two important conclusions – first, much larger sample sizes will be required to detect genes of small effect for craving. Despite the relatively high prevalence for craving in our sample (relative to epidemiological cohorts), we did not succeed in identifying signals of genomewide significance. Second, despite the overlap in some of the top signals across craving and alcohol dependence, there was some evidence of specificity in association when examining genes in the dopamine pathway and further, our GWAS signals for craving do not appear to be entirely attributable to the high degree of overlap between craving and severity of alcohol dependence. There needs to be a continued effort to examine candidate genes that may be particularly relevant to craving – while we explore one major family of genes in this study, multiple others in GABA-ergic, glutamatergic and opioidergic pathways have also been posited as participating in the neurobiology of craving (Kalivas & Volkow, 2005). While we could not conclude the extent to which the specificity of our association signals for the dopamine genes was due to false positive or negative findings, future studies may wish to focus on these genes with additional measures of craving.
Highlights.
A majority of those endorsing craving meet criteria for DSM-IV alcohol dependence.
Genes in the dopamine pathway show association with alcohol craving. The association between these variants and alcohol dependence is also evident but not with dependence without craving.
Genomewide association studies (GWAS) do not identify genomewide significant SNPs.
There is overlap in genetic signals for craving and for DSM-IV alcohol dependence.
Acknowledgments
Role of funding sources: Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). Other support includes DA23668 and ABMRF/Foundation for Alcohol Research (A.A.). Funding bodies played no role in data analysis or manuscript preparation.
Footnotes
Contributors: AA, LW, HJE and TF conceived analyses, revised and refined hypotheses and wrote and edited all drafts. AA analyzed data. KKB, JK, SK, MTL and MS provided expertise on phenotypic analyses and clinical significance. HJE, JIN, TF, LW and JT provided expertise on genetic analyses. All authors reviewed the manuscript prior to submission.
Conflicts of Interest: LJB is listed as inventor on the patent “Markers for Addiction” (US 20070258898): covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction and acted as a consultant for Pfizer, Inc. in 2008.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agrawal A, Heath AC, Lynskey MT. DSM-IV to DSM-5: the impact of proposed revisions on diagnosis of alcohol use disorders. Addiction. 2011;106:1935–1943. doi: 10.1111/j.1360-0443.2011.03517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begleiter H, Reich T, Hesselbrock V, Porjesz B, Li TK, Schuckit M, et al. The collaborative study on the genetics of alcoholism. Alcohol Health Res World. 1995;19:228–236. [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Strickland JR, Thompson JR, Afful SE, Cottler LB. Drug use and dependence in cocaine dependent subjects, community-based individuals, and their siblings. Drug Alcohol Depend. 2008;95:14–22. doi: 10.1016/j.drugalcdep.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum K, Liu Y, Shriner R, Gold MS. Reward circuitry dopaminergic activation regulates food and drug craving behavior. Curr Pharm Des. 2011;17:1158–1167. doi: 10.2174/138161211795656819. [DOI] [PubMed] [Google Scholar]
- Bohn MJ, Krahn DD, Staehler BA. Development and initial validation of a measure of drinking urges in abstinent alcoholics. Alcohol Clin Exp Res. 1995;19:600–606. doi: 10.1111/j.1530-0277.1995.tb01554.x. [DOI] [PubMed] [Google Scholar]
- Bonsch D, Greifenberg V, Bayerlein K, Biermann T, Reulbach U, Hillemacher T, et al. Alpha-synuclein protein levels are increased in alcoholic patients and are linked to craving. Alcohol Clin Exp Res. 2005a;29:763–765. doi: 10.1097/01.alc.0000164360.43907.24. [DOI] [PubMed] [Google Scholar]
- Bonsch D, Lederer T, Reulbach U, Hothorn T, Kornhuber J, Bleich S. Joint analysis of the NACP-REP1 marker within the alpha synuclein gene concludes association with alcohol dependence. Hum Mol Genet. 2005b;14:967–971. doi: 10.1093/hmg/ddi090. [DOI] [PubMed] [Google Scholar]
- Bonsch D, Reulbach U, Bayerlein K, Hillemacher T, Kornhuber J, Bleich S. Elevated alpha synuclein mRNA levels are associated with craving in patients with alcoholism. Biol Psychiatry. 2004;56:984–986. doi: 10.1016/j.biopsych.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret RJ, Cloninger RC, Dinwiddie SH, Hesselbrock V, Nurnberger JI, et al. A New, Semi-Structured Psychiatric Interview For Use In Genetic Linkage Studies. Journal for the Studies on Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Heath AC, Reich T, Hesselbrock VM, Kramer JR, Nurnberger JI, Jr, et al. Can we subtype alcoholism? A latent class analysis of data from relatives of alcoholics in a multicenter family study of alcoholism. Alcohol Clin Exp Res. 1996;20:1462–1471. doi: 10.1111/j.1530-0277.1996.tb01150.x. [DOI] [PubMed] [Google Scholar]
- Buckholtz JW, Treadway MT, Cowan RL, Woodward ND, Li R, Ansari MS, et al. Dopaminergic network differences in human impulsivity. Science. 2010;329:532. doi: 10.1126/science.1185778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter BL, Tiffany ST. Meta-analysis of cue-reactivity in addiction research. Addiction. 1999;94:327–340. [PubMed] [Google Scholar]
- Cherpitel CJ, Borges G, Ye Y, Bond J, Cremonte M, Moskalewicz J, et al. Performance of a craving criterion in DSM alcohol use disorders. J Stud Alcohol Drugs. 2010;71:674–684. doi: 10.15288/jsad.2010.71.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childress AR, Mozley PD, McElgin W, Fitzgerald J, Reivich M, O’Brien CP. Limbic activation during cue-induced cocaine craving. Am J Psychiatry. 1999;156:11–18. doi: 10.1176/ajp.156.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diana M. The dopamine hypothesis of drug addiction and its potential therapeutic value. Front Psychiatry. 2011;2:64. doi: 10.3389/fpsyt.2011.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DC. Theories of drug craving, ancient and modern. Addiction. 2001;96:33–46. doi: 10.1046/j.1360-0443.2001.961333.x. [DOI] [PubMed] [Google Scholar]
- Drummond DC, Litten RZ, Lowman C, Hunt WA. Craving research: future directions. Addiction. 2000;95(Suppl 2):S247–S255. doi: 10.1080/09652140050111816. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wilhelmsen KC. Genomic scan for alcohol craving in Mission Indians. Psychiatr Genet. 2005;15:71–75. doi: 10.1097/00041444-200503000-00012. [DOI] [PubMed] [Google Scholar]
- Flannery BA, Roberts AJ, Cooney N, Swift RM, Anton RF, Rohsenow DJ. The role of craving in alcohol use, dependence, and treatment. Alcohol Clin Exp Res. 2001;25:299–308. [PubMed] [Google Scholar]
- Foroud T, Bucholz KK, Edenberg HJ, Goate A, Neuman RJ, Porjesz B, et al. Linkage of an alcoholism-related severity phenotype to chromosome 16. Alcohol Clin Exp Res. 1998;22:2035–2042. [PubMed] [Google Scholar]
- Foroud T, Wetherill LF, Liang T, Dick DM, Hesselbrock V, Kramer J, et al. Association of alcohol craving with alpha-synuclein (SNCA) Alcohol Clin Exp Res. 2007;31:537–545. doi: 10.1111/j.1530-0277.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- Heinz A, Siessmeier T, Wrase J, Buchholz HG, Grunder G, Kumakura Y, et al. Correlation of alcohol craving with striatal dopamine synthesis capacity and D2/3 receptor availability: a combined [18F]DOPA and [18F]DMFP PET study in detoxified alcoholic patients. Am J Psychiatry. 2005;162:1515–1520. doi: 10.1176/appi.ajp.162.8.1515. [DOI] [PubMed] [Google Scholar]
- Heinz A, Siessmeier T, Wrase J, Hermann D, Klein S, Grusser SM, et al. Correlation between dopamine D(2) receptors in the ventral striatum and central processing of alcohol cues and craving. Am J Psychiatry. 2004;161:1783–1789. doi: 10.1176/appi.ajp.161.10.1783. [DOI] [PubMed] [Google Scholar]
- Hendershot CS, Witkiewitz K, George WH, Marlatt GA. Relapse prevention for addictive behaviors. Subst Abuse Treat Prev Policy. 2011;6:17. doi: 10.1186/1747-597X-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA--a comparison with the SCAN. Addiction. 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- Irvine GB, El-Agnaf OM, Shankar GM, Walsh DM. Protein aggregation in the brain: the molecular basis for Alzheimer’s and Parkinson’s diseases. Mol Med. 2008;14:451–464. doi: 10.2119/2007-00100.Irvine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivas PW, O’Brien C. Drug addiction as a pathology of staged neuroplasticity. Neuropsychopharmacology. 2008;33:166–180. doi: 10.1038/sj.npp.1301564. [DOI] [PubMed] [Google Scholar]
- Kalivas PW, Volkow ND. The neural basis of addiction: a pathology of motivation and choice. Am J Psychiatry. 2005;162:1403–1413. doi: 10.1176/appi.ajp.162.8.1403. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Kalsi G, Holmans PA, Sanders AR, Aggen SH, Dick DM, et al. Genomewide association analysis of symptoms of alcohol dependence in the molecular genetics of schizophrenia (MGS2) control sample. Alcohol Clin Exp Res. 2011;35:963–975. doi: 10.1111/j.1530-0277.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes KM, Krueger RF, Grant BF, Hasin DS. Alcohol craving and the dimensionality of alcohol disorders. Psychol Med. 2010:1–12. doi: 10.1017/S003329171000053X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes KM, Krueger RF, Grant BF, Hasin DS. Alcohol craving and the dimensionality of alcohol disorders. Psychol Med. 2011;41:629–640. doi: 10.1017/S003329171000053X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama S, Brodie MS, Appel SB. Ethanol inhibition of m-current and ethanol-induced direct excitation of ventral tegmental area dopamine neurons. J Neurophysiol. 2007;97:1977–1985. doi: 10.1152/jn.00270.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski LT, Mann RE, Wilkinson DA, Poulos CX. “Cravings” are ambiguous: ask about urges or desires. Addict Behav. 1989;14:443–445. doi: 10.1016/0306-4603(89)90031-2. [DOI] [PubMed] [Google Scholar]
- Kramer JR, Chan G, Hesselbrock VM, Kuperman S, Bucholz KK, Edenberg HJ, et al. A principal components analysis of the abbreviated Desires for Alcohol Questionnaire (DAQ) J Stud Alcohol Drugs. 2010;71:150–155. doi: 10.15288/jsad.2010.71.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimer ME, Palmer RS, Marlatt GA. Relapse prevention. An overview of Marlatt’s cognitive-behavioral model. Alcohol Res Health. 1999;23:151–160. [PMC free article] [PubMed] [Google Scholar]
- Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut L, Bhangale T, et al. Quality control and quality assurance in genotypic data for genome-wide association studies. 2009. Under Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggio L, Ray LA, Kenna GA, Swift RM. Blood glucose level, alcohol heavy drinking, and alcohol craving during treatment for alcohol dependence: results from the Combined Pharmacotherapies and Behavioral Interventions for Alcohol Dependence (COMBINE) Study. Alcohol Clin Exp Res. 2009;33:1539–1544. doi: 10.1111/j.1530-0277.2009.00982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowman C, Hunt WA, Litten RZ, Drummond DC. Research perspectives on alcohol craving: an overview. Addiction. 2000; 95(Suppl 2):S45–S54. doi: 10.1080/09652140050111636. [DOI] [PubMed] [Google Scholar]
- Marlatt GA, Gordon JR. Relapse Prevention. New York: Guilford Press; 1985. [Google Scholar]
- Mewton L, Slade T, McBride O, Grove R, Teesson M. An evaluation of the proposed DSM-5 alcohol use disorder criteria using Australian national data. Addiction. 2010 doi: 10.1111/j.1360-0443.2010.03340.x. [DOI] [PubMed] [Google Scholar]
- Monti PM, Abrams DB, Binkoff JA, Zwick WR, Liepman MR, Nirenberg TD, et al. Communication skills training, communication skills training with family and cognitive behavioral mood management training for alcoholics. J Stud Alcohol. 1990;51:263–270. doi: 10.15288/jsa.1990.51.263. [DOI] [PubMed] [Google Scholar]
- Moss HB. Does Research Support “Craving” as a Core Symptom of Substance Use Disorders in DSM-5? Psychiatric Times. 2011 http://www.psychiatrictimes.com/substance-abuse/content/article/10168/1775007.
- Muthen LK, Muthen BO. Mplus User’s Guide. 5. Los Angeles, CA: Muthen & Muthen; 2007. [Google Scholar]
- Narita M, Soma M, Tamaki H, Narita M, Suzuki T. Intensification of the development of ethanol dependence in mice lacking dopamine D(3) receptor. Neurosci Lett. 2002;324:129–132. doi: 10.1016/s0304-3940(02)00235-5. [DOI] [PubMed] [Google Scholar]
- O’Brien C. Addiction and dependence in DSM-V. Addiction. 2011;106:866–867. doi: 10.1111/j.1360-0443.2010.03144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien CP. Anticraving medications for relapse prevention: a possible new class of psychoactive medications. Am J Psychiatry. 2005;162:1423–1431. doi: 10.1176/appi.ajp.162.8.1423. [DOI] [PubMed] [Google Scholar]
- Oslin DW, Cary M, Slaymaker V, Colleran C, Blow FC. Daily ratings measures of alcohol craving during an inpatient stay define subtypes of alcohol addiction that predict subsequent risk for resumption of drinking. Drug Alcohol Depend. 2009;103:131–136. doi: 10.1016/j.drugalcdep.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez RG, Waymire JC, Lin E, Liu JJ, Guo F, Zigmond MJ. A role for alpha-synuclein in the regulation of dopamine biosynthesis. J Neurosci. 2002;22:3090–3099. doi: 10.1523/JNEUROSCI.22-08-03090.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute. SAS User Guide, Version 8.2. Cary, NC: SAS Institute Inc; 1999. [Google Scholar]
- Sinha R, Fox HC, Hong KI, Hansen J, Tuit K, Kreek MJ. Effects of adrenal sensitivity, stress- and cue-induced craving, and anxiety on subsequent alcohol relapse and treatment outcomes. Arch Gen Psychiatry. 2011;68:942–952. doi: 10.1001/archgenpsychiatry.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R, Garcia M, Paliwal P, Kreek MJ, Rounsaville BJ. Stress-induced cocaine craving and hypothalamic-pituitary-adrenal responses are predictive of cocaine relapse outcomes. Arch Gen Psychiatry. 2006;63:324–331. doi: 10.1001/archpsyc.63.3.324. [DOI] [PubMed] [Google Scholar]
- Tupala E, Hall H, Bergstrom K, Sarkioja T, Rasanen P, Mantere T, et al. Dopamine D(2)/D(3)-receptor and transporter densities in nucleus accumbens and amygdala of type 1 and 2 alcoholics. Mol Psychiatry. 2001;6:261–267. doi: 10.1038/sj.mp.4000859. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Fowler JS, Logan J, Hitzemann R, Ding YS, et al. Decreases in dopamine receptors but not in dopamine transporters in alcoholics. Alcohol Clin Exp Res. 1996;20:1594–1598. doi: 10.1111/j.1530-0277.1996.tb05936.x. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Telang F, Fowler JS, Logan J, Childress AR, et al. Cocaine cues and dopamine in dorsal striatum: mechanism of craving in cocaine addiction. J Neurosci. 2006;26:6583–6588. doi: 10.1523/JNEUROSCI.1544-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. International Statistical Classification of Diseases and Related Health Problems 10th Revision. 10. Geneva, Switzerland: 2007. [Google Scholar]