Abstract
Over-expression of the major myelin proteolipid protein (PLP) is detrimental to brain development and function and is the most common cause of Pelizaeus-Merzbacher disease. MicroRNAs (miRNA), small non-coding RNAs have been shown to play critical roles in oligodendrocyte lineage. In this study, we sought to investigate whether miRNAs control PLP abundance. To identify candidate miRNAs involved in this regulation, we have examined differentiation-induced changes in the expression of miRNAs in the oligodendroglial cell line, Oli-neu, and in EGFP+ oligodendrocytes ex vivo. We have identified 145 miRNAs that are expressed in oligodendrocyte cell lineage progression. Dicer1 expression decreases in differentiated oligodendrocytes and knock down of Dicer1 results in changes in miRNAs similar to those associated with differentiation. To identify miRNAs that control the PLP expression, we have selected miRNAs whose expression is lower in differentiated vs. undifferentiated Oli-neu cells and that have ≥ 1 binding site(s) in the PLP 3′UTR. The PLP 3′UTR fused to the luciferase gene reduces the activity of the reporter, suggesting that it negatively regulates message stability or translation. Such suppression is relieved by knock down of miR-20a. Over-expression of miR-20a decreases expression of the endogenous PLP in primary oligodendrocytes and of the reporter gene. Deletion or mutation of the putative binding site for miR-20a in the PLP 3′UTR abrogated such effects. Our data indicate that miRNA expression is regulated by Dicer1 levels in differentiated oligodendrocytes and that miR-20a, a component of the cluster that controls OL cell number, regulates PLP gene expression through its 3′ UTR.
Keywords: microRNA, oligodendrocytes, proteolipid protein
Introduction
Proteolipid protein (PLP) and the alternatively spliced isoform, DM20, account for approximately 50% of central nervous system myelin protein content (Macklin et al. 1987). PLP expression is activated as part of the oligodendrocyte differentiation program (Baumann and Pham Dinh 2001) and is regulated transcriptionally and post-transcriptionally (Gardinier et al. 1986; Sporkel et al. 2002; Wang et al. 2008). Alternative selection of polyadenylation sites in the PLP 3′ UTR gives rise to three PLP/DM20 mRNAs (1.6 kb, 2.4 kb and 3.2 kb), which are expressed with a distinct temporal pattern (Gardinier et al. 1986). The 3.2 kb message is expressed at post-natal day 3, followed by 2.4 kb message, while the short, 1.6 kb message is detected at the peak of myelination (Gardinier et al. 1986; Wight et al. 1993; Fuss et al. 2000). The data suggest that the expression of the 1.6 and 2.4 kb messages is under negative regulation at early stages of development. The full length PLP 3′ UTR regulates stability of a reporter gene, indicating that it contains regulatory elements involved in post-transcriptional and translational regulation of PLP (Mallon and Macklin 2002).
microRNAs (miRNA) are small non-coding RNAs that control cell fate, development and differentiation by post-transcriptional regulation of gene expression (Ambros 2004; Bartel 2004; Bartel 2009). miRNAs are negative regulators of gene expression by binding to recognition motifs, generally present in the 3′ untranslated region (UTR) of target mRNAs, leading to translational repression or degradation of the target mRNAs (Ambros 2004; Bartel 2004; Bartel 2009). Mature miRNAs are generated from a primary transcript through multiple processing steps in the nucleus and a final maturation in the cytoplasm through the activity of Dicer, a dsRNA endonuclease (He and Hannon 2004). Changes in Dicer expression represent an important regulatory mechanism of miRNA abundance and function (Yang and Lai 2011).
miRNAs play critical roles in oligodendrocyte (OL) lineage progression (Lin and Fu 2009; Shin et al. 2009; Dugas et al. 2010; Zhao et al. 2010). Conditional ablation of Dicer1 in oligodendrocyte progenitor cells (OPC) and immature OLs severely impairs the progression of OPCs to differentiated OLs by removing a miRNA-mediated inhibition of negative regulators of OL differentiation (Shin et al. 2009; Dugas et al. 2010; Zhao et al. 2010). Ablation of Dicer1 at later stages of OL differentiation resulted in demyelination and inflammation, demonstrating that miRNAs regulate myelin stability (Shin et al. 2009). Profiling of miRNAs has identified a number of miRNAs that are preferentially expressed in OLs and differentially regulated during OL lineage progression and are likely to regulate OL gene expression (Lau et al. 2008). In particular, the 17–92 miRNA cluster was conditionally knocked out and shown to regulate OL cell number (Budde et al. 2010). Myelin gene expression is intrinsic to OL differentiation; however, a direct role of miRNAs in regulating myelin protein expression in the developing mammalian CNS has not been examined.
We have investigated whether miRNAs participate in the post-transcriptional regulation of PLP through the 3′UTR. We have characterized the expression of miRNAs in the oligodendrocyte cells, Oli-neu, and in primary mouse OLs isolated from the developing brain and have examined the role of differentially expressed miRNAs in the regulation of the PLP 3′UTR. We present the analysis of miRNAs identified in the current work and compare our findings with previously published profiles. We show that reduced Dicer1 level in differentiated OLs accounts for most changes in miRNA expression and that miR-20a, a component of the cluster that controls OL cell number, regulates PLP gene expression through its 3′ UTR. This finding demonstrates functional diversity of miRNAs co-expressed in the same cluster.
Methods
Plasmids
The reporter constructs were generated by PCR cloning of the relevant portions of PLP 3′UTR (Figure 4A), inserted into the vector psiCHECK™-2 (Promega) at the restriction enzymes sites XhoI and Not I. The primer set used for the reporter construct UTRPLP-1.6 is: 5′ aggcctctcgagCTCCCATAGAAACTCCCCTTTGTC 3′ (forward primer, underlined are the restriction enzyme sites for cloning.) and 5′ gtcgacgcggccgcTCCTCCAGGAAATTCGATCAGG 3′ (reverse primer). The primer set used for the reporter construct UTRPLP-0.6 is: 5′ aggcctctcgagCTCCCATAGAAACTCCCCTTTGTC 3′ (forward primer) and 5′ gtcgacgcggccgcTGGGCAAAGCTATATTGCTCTGCT 3′ (reverse primer). The primer set used for the reporter construct UTRPLP-2.2 is: 5′ aggcctctcgagCATTGTAGATTTGTGCTGTCATTC 3′ (forward primer) and 5′ gtcgacgcggccgcGATTTTATAAAATACATTTTGTAAGATGAGT 3′ (reverse primer). The mutation (deletion/replacement, Figure 5) was introduced into the reporter construct UTRPLP-1.6 by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). All plasmid DNAs were verified by sequencing.
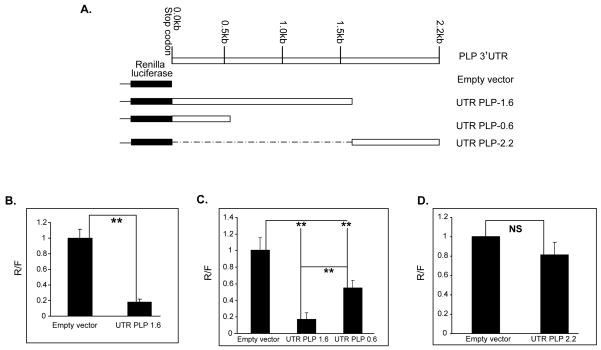
Figure 4. Mouse PLP 3′ UTR contains regulatory elements that control PLP gene expression.
A. Schematics of the luciferase-PLP 3′UTR containing constructs. The UTRPLP-1.6 and UTRPLP-0.6 were generated by cloning the 1.6 kb and 0.6 kb (GenBank Accession #NM_011123) sequences downstream of the PLP stop codon in the vector psiCHECK™-2 (Promega), respectively. The UTRPLP-2.2 contains the 0.6 kb sequences of the mouse PLP 3′UTR downstream of the 1.6 fragment, which are uniquely present in the 3.2 kb PLP mRNA (GenBank Accession #AK134554). Dashed line represents deleted portion from the 3.2 kb transcript 3′UTR. B, C and D. Bar graphs showing the luciferase activity derived from the constructs shown in A. Luciferase activity was assayed 48 hrs (B) and 72 hrs (C and D) after transfection into Oli-neu cells. The data are expressed as ratio of the Renilla luciferase to the firefly luciferase activity (R/F), and the R/F of the empty vector is set as 1. Error bars represent SD. n≥3. ** P<0.01 (Student’s t test); ns, not significant.
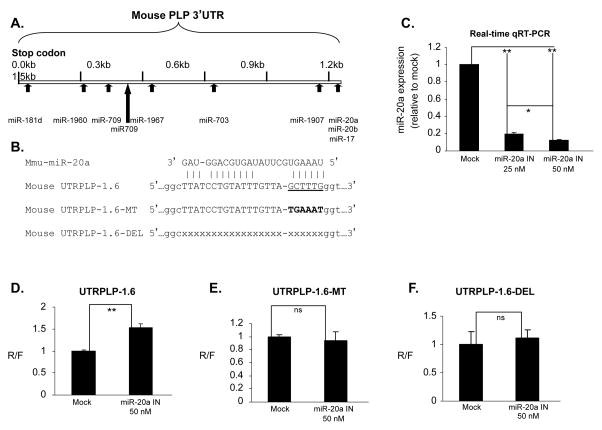
Figure 5. Knock down of miR-20a increases PLP 3′UTR-luciferase reporter expression in Oli-neu cells.
A. miRNA binding motifs in the PLP 3′UTR. Candidate miRNAs shown in Figure 2C were scanned against the PLP 3′UTR (GenBank Accession # NM_011123) using RNA22 bioinformatics tool. Ten of these miRNAs are predicted to have ≥ 1 putative binding sites in PLP 3′UTR. B. Shown are the sequences for mouse miR-20a and the binding motifs in PLP 3′UTR included in construct UTRPLP-1.6 and 2 mutants that disrupt the miR-20a binding site. C. Bar graph shows dose dependent decrease in miR-20a levels in Oli-neu cells transfected with increasing amount of miR-20a inhibitor (IN) (n=3). The decrease in miR-20a levels is expressed as fold change over that of control inhibitor (Mock) treated cells set as 1. Error bars represent SD. ** P<0.01 and * P<0.05 (Student’s t test). D, E and F. miR-20a regulates luciferase activity through a specific motif site in the PLP 3′UTR. Oli-neu cells transfected with the UTRPLP-1.6 (D), UTRPLP-16-MT (E) and UTRPLP-1.6-DEL (F) were treated with miR-20a inhibitor (IN) and control inhibitor (Mock) (Applied Biosystems). Luciferase activity from cells treated with control inhibitor is set as 1. Error bars represent SD (n=3). **P<0.01 (Student’s t test). The luciferase activity of UTRPLP-1.6-MT and UTRPLP-1.6-DEL was not decreased by the miR-20a inhibitor.
Cell culture and FACS analysis
Oli-neu cells were grown in SATO medium as described (Jung et al. 1995), and differentiated in 1mM dbcAMP for 10 days. L-cells, Hela cells and mouse primary astrocytes cells were cultured in DMEM medium supplemented with 10% FBS. Cell suspensions were prepared from the CNP-EGFP mouse brains (kind gift of Dr. V Gallo) and EGFP+ OLs were isolated by Fluorescent Activated Cell Sorting (FACS), as previously described (Yuan et al. 2002; Belachew et al. 2003; Zhu et al. 2012). Primary mouse oligodendrocyte progenitor cells (OPC) were enriched from mixed brain cultures prepared from C57BL/6 brains at post-natal day 1, as previously described (Zhu et al. 2012). After removal of microglia by differential adhesion and astrocytes by immunopanning, enriched OPC were transfected in OM-5 medium. After transfection, OPC were cultured in chemically defined medium containing 10 ng/ml biotin, 5 ug/ml insulin, 2 mM glutamine, 100 units/ml penicillin and 100 units/ml streptomycin, 1X B27, and 5% Hyclone FBS in DMEM and 30 % of B104 conditioned medium (Huang et al. 2002).
Nuclear extracts and Western blot
Nuclear extracts were prepared from Oli-neu cells grown in either growth or differentiation medium, by using the NEP kit (Piercenet), as previously described (Wang et al. 2007; Wang and Cambi 2009). For western blot analysis 25.0 μg of cytoplasmic and nuclear extracts prepared from undifferentiated and 10-day differentiated Oli-neu cells were separated on 7.5% SDS-PAGE, transferred onto PVDF membranes and probed with antibodies to mouse Dicer1 (1:1000, ABCam) and hnRNP L (1:2000, ABCam), α-Tubulin (1: 3000, Sigma), and GAPDH (1:1000, AbCam), detected by HRP-conjugated secondary antibody (GE) diluted 1:2000 and developed with enhanced chemiluminescence (GE) (Wang et al. 2007).
miRNA expression profile
miRNA arrays were performed by a commercial provider (LC Sciences, Houston, TX). Total RNAs including miRNAs were extracted from Oli-neu cells undifferentiated, differentiated for 10-days, treated with mock and Dicer1 siRNAs. Oli-neu cells were treated with mock and Dicer1 siRNAs for 3 days at a concentration of 100nM. Total RNA including miRNAs was prepared from primary oligodendrocytes FACS-purified from CNP-EGFP mouse brains at post-natal day 1 and 21. Five μg of Oli-neu RNAs and three μg of primary oligodendrocytes RNAs were submitted to LC Sciences. miRNAs were labeled by Cy3 and probed to mouse miRNA array (miRBase Version 14.0, LC Sciences).
miRNA extraction and real-time RT-PCR
Total RNAs were extracted with the miRNeasy mini kits according to the manufacturer’s instructions (Qiagen). Taqman-based miRNA qRT-PCR was carried out using the reagents from Applied Biosystems and U6 snRNA was amplified to serve as the internal normalization control. qRT-PCR was performed using the StepOne real-time PCR system (Applied Biosystems) in the University of Kentucky Spinal Cord and Brain Injury Research Center core facility, as described (Wang et al. 2008), and data was analyzed by the StepOne™ Software v2.0 (Applied Biosystems). Relative miRNA levels were determined by comparing threshold cycles for individual miRNA products normalized with U6 snRNA using the 2−ΔΔCT method (Livak and Schmittgen 2001). Endogenous PLP/DM20 mRNA expression was quantitated by real-time qRT-PCR, as previously described (Wang et al. 2008).
Transfection, miRNA inhibition and overexpression analyses, and luciferase assays
Plasmid DNAs, miRNA precursors and anti-sense inhibitors were cotransfected into Oli-neu cells using the siPORT Amine reagents (Applied Biosystems), as previously described (Wang et al. 2007). OPC were transfected in suspension with miRNA precursors for 6 hrs and total RNA was extracted 66 hrs after transfection. The expression of the target miRNAs was monitored by real-time qRT-PCR using Taqman probes and primer sets specific for the targeted miRNAs (Applied Biosystems). Endogenous Luciferase assays were performed using the Dual-Luciferase® Reporter System (Promega) on a Lumat LB 9507 Luminometer (EG&G Berthold, Germany). The Renilla luciferase activity was normalized by that of the firefly luciferase expressed from the same vector. All the luciferase assays were performed in ≥ 3 independent experiments, with ≥ 3 repeats in each experiment unless specifically described.
Results
Dicer1 regulates differentiation-induced changes of miRNA expression in oligodendrocytes
Profiling the expression of miRNAs in mouse oligodendrocytes
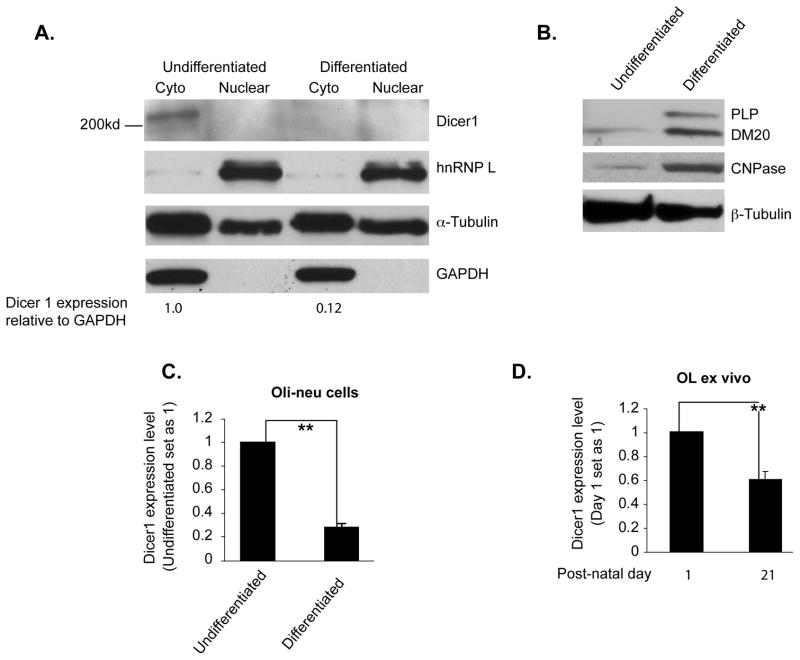
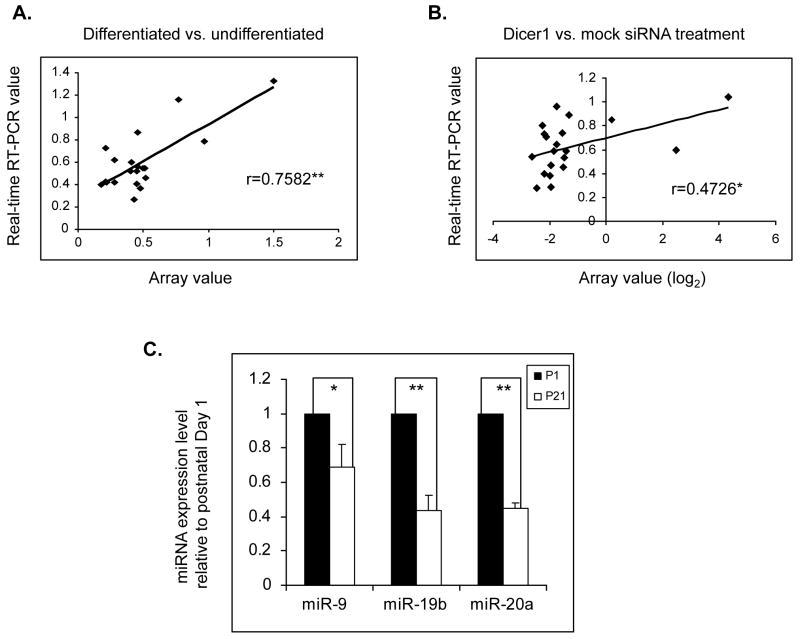
miRNAs expression levels are regulated during OL differentiation, resulting in functional changes in their targets (Lin and Fu 2009; Shin et al. 2009; Dugas et al. 2010; Zhao et al. 2010). Since Dicer1 regulates the final step of miRNA maturation, we investigated whether Dicer1 expression is down regulated in differentiated OLs and accounts for changes in miRNA levels. We have measured Dicer1 protein and message in Oli-neu cells, an oligodendrocyte cell line that replicates OL differentiation and PLP gene regulation (Jung et al. 1995; Wang et al. 2007) and in EGFP+ oligodendrocytes isolated from brains of the CNP-EGFP mouse (Yuan et al. 2002; Zhu et al. 2012). Dicer1 protein levels are >80% lower in Oli-neu cells differentiated for 10 days vs. undifferentiated Oli-neu cells (0.12 vs. 1.0, Fig. 1A). Expression of CNPase and PLP/DM20 is increased in Oli-neu cells differentiated for 10 days, indicating lineage progression (Fig. 1B). Dicer1 transcript levels were reduced > 50% in P21 EGFP+ OLs (Fig. 1D), when most EGFP+ cells are mature and express high levels of PLP/DM20 and Sirtuin 2 (Zhu et al. 2012) compared to EGFP+ oligodendrocytes isolated at post-natal (P) day 1, when most EGFP+ cells are progenitor cells (Yuan et al. 2002; Belachew et al. 2003; Zhu et al. 2012) (Fig. 1D). In keeping with these data, Dicer1 transcript level was decreased by ~80% in differentiated compared to undifferentiated Oli-neu cells (Fig. 1C) accounting for the reduction in Dicer1 protein. The data suggest that a decrease in Dicer1 expression accompanies OL differentiation and may regulate miRNA expression in differentiated OLs. To more directly demonstrate that Dicer1 regulates miRNA expression in differentiated OLs, we have determined whether knock down of Dicer1 in Oli-neu cells induces changes in miRNA expression similar to those occurring in differentiated Oli-neu cells. We have characterized the miRNA expression profiles of Oli-neu cells undifferentiated and differentiated for 10 days, and of undifferentiated Oli-neu cells treated with mock and Dicer1 siRNAs (Fig. 2). Dicer1 message was decreased more than 60% in silenced vs. mock treated Oli-neu cells (Fig. 2A). Total RNA containing miRNAs was extracted from undifferentiated, 10-day differentiated, mock and Dicer1 siRNA treated undifferentiated Oli-neu cells, labeled with Cy3, and probed to mouse miRNA array (miRBase Version 14.0, LC Sciences). For each of the 4 profiles, approximately 100 miRNAs were identified with a normalized signal of >500, which is the cut-off value for selection of miRNAs in the post-array verification (LC Sciences, Table 1S). To compare changes in miRNA profiles in the different groups, we defined as an Oli-neu cell-expressed miRNA only miRNAs, which have a normalized signal of >500 in at least one of the 4 profiles. We identified 145 miRNAs that are expressed in Oli-neu cells (Table S1). The fold changes (in log2) of miRNAs in differentiated vs. undifferentiated and Dicer1 vs. mock siRNA treated Oli-neu cells were calculated for each of the 145 miRNAs. Overall, there is a good correlation (r=0.3669, P<0.01) between the ratio of miRNA expression levels in differentiated vs. undifferentiated and Dicer1 silenced vs. mock treated Oli-neu cells (Fig. 2B).
Figure 1. Dicer1 is downregulated in differentiated oligodendrocyte cells.
A. Western blot analysis of Dicer1 expression in 10-day differentiated and undifferentiated Oli-neu cells (n=2). hnRNPL serves as control for loading accuracy of nuclear proteins, GAPDH for cytoplasmic proteins and α-tubulin for both. Dicer1 band detected in the cytoplasm was measured by densitometry and corrected by the GAPDH. The value is indicated below each lane. B. Western blot analysis of PLP/DM20 and CNPase expression in 10-day differentiated and undifferentiated Oli-neu cells (n=2). PLP/DM20 and CNPase are used as control for differentiation of Oli-neu cells and β-tubulin as loading control. C. Real-time RT-PCR analysis of Dicer1 expression in undifferentiated and 10-day differentiated Oli-neu cells. The expression of Dicer1 in differentiated Oli-neu cells is expressed as percent of that in undifferentiated cells, which is set as 1 (n=4). ** p<0.01 (Student’s t test). D. Real-time RT-PCR analysis of Dicer1 expression in EGFP+ OL. The expression of Dicer1 at P21 is expressed as percent of that at P 1 which is set as 1 (n=4). ** p<0.01 (Student’s t test).
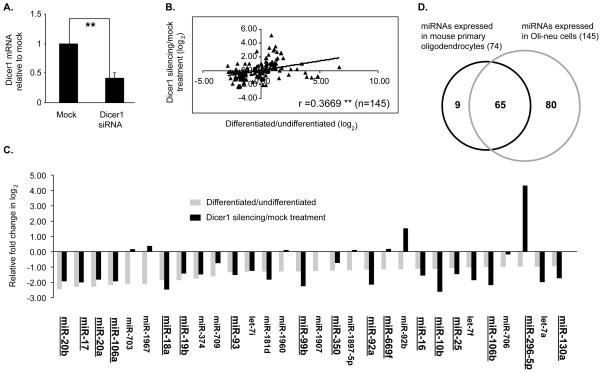
Figure 2. Expression profile of miRNAs in mouse oligodendrocytes.
A. Bar graph showing the expression levels of Dicer1 mRNA after treatment with mock and mouse Dicer1 siRNAs (n=4). **P <0.01 (Student’s t test). B. Shown is the correlation between the miRNA expression fold change in differentiated vs. undifferentiated Oli-neu cells and that in Dicer1 vs. mock siRNA treated Oli-neu cells. r = Pearson’s correlation coefficient (n=145). **p<0.01 (Student’s t test). C. Comparison of relative expression levels of miRNAs in differentiated/undifferentiated and Dicer1/mock siRNA-treated Oli-neu cells. Shown are the bottom 30 miRNAs with a ≥1 fold (in log2) decrease in expression in differentiated vs. undifferentiated (gray bars). Also shown is the fold change in Dicer1 vs. mock siRNA treated Oli-neu cells (black bars). Underlined and enlarged are miRNAs selected for further validation by qRT-PCR. D. Venn diagram showing the overlap of miRNAs expressed in mouse primary oligodendrocyte cells and in Oli-neu cells.
We show a comparison of the bottom 30 miRNAs with >1 fold (log2) decrease in differentiated vs. undifferentiated Oli-neu cells with those caused by siRNA-mediated Dicer1 knockdown (Figure 2C). Twenty two of the thirty miRNAs are similarly regulated in the two groups. Eight miRNAs are increased by Dicer1 knock down compared to a decrease in differentiated Oli-neu cells. The decrease in miRNAs in Dicer1-silenced Oli-neu cells largely mirrors the changes in differentiated OLs, suggesting that Dicer1 plays an important role in regulating miRNA expression in differentiated OLs.
To demonstrate that the differentially expressed miRNAs in Oli-neu cells are similarly regulated in differentiated OLs in vivo, we performed miRNA profiles in EGFP+ OPCs isolated from P1 and mature OLs, isolated from P21 brains. We have identified 74 miRNAs with a normalized signal above background noise in at least one of these 2 profiles for EGFP+ OLs (Table S2). The data show that 88% (65 out of 74) of the miRNAs identified in primary OLs are present in the 145 miRNAs expressed in Oli-neu cells (Fig. 2D), thus, demonstrating that Oli-neu cell-expressed miRNAs replicate the in vivo miRNA expression profile.
Real-time RT-PCR validation
We have validated with qRT-PCR 17 miRNAs (Figure 2C, underlined and enlarged miRNAs), that decreased >1 fold (log2) in differentiated vs. undifferentiated Oli-neu cells and for which Taqman probes and PCR primer sets are available from the manufacturer (Applied Biosystems) (Fig. 2C). miR-29a and miR-9 were included in the qRT-PCR assays because they were previously shown to regulate PMP22 gene expression in Schwann cells and OLs (Lau et al. 2008; Verrier et al. 2009). miR-1 was also included as a negative control since its expression level is similar to background noise. The fold change detected by qRT-PCR correlates well with the changes detected by miRNA arrays in both differentiated vs. undifferentiated and Dicer1 vs. mock silenced Oli-neu cells (Fig. 3A and B, raw data are shown in Tables S3–4). Additionally, we sought to verify whether these miRNAs are also downregulated in primary OLs. We selected miR-19b, miR-20a and miR-9 for verification. miR-19b and miR-20a are members of the miR-17–92 cluster, which regulates OL number in mouse developing brains (Budde et al. 2010). Expression levels of miR-19b, miR-20a and miR-9 are reduced in P21 OLs vs. P1 OPCs (Fig. 3C). We conclude that miRNAs are similarly regulated in Oli-neu cells and differentiated OLs in vivo, thus validating the use of Oli-neu cells for studies on the PLP 3′ UTR.
Figure 3. Validation of individual miRNA expression by real-time qRT-PCR.
A. Shown is the correlation between the fold changes in expression of the 20 selected miRNAs in differentiated vs. undifferentiated Oli-neu cells assessed by real-time qRT-PCR and microarray analysis, respectively. The 20 selected miRNAs are miR-1, miR-9 and miR-29a, along with the 17 miRNAs shown Figure 2C (underlined and enlarged). r = Pearson’s correlation coefficient (n=20). **P<0.01 (Student’s t test). B. Shown is the correlation between the fold changes in expression of the 20 selected miRNAs in A, in Dicer1 vs. mock siRNAs treated Oli-neu cells assessed by real-time qRT-PCR and microarray analysis, respectively. r = Pearson’s correlation coefficient (n=20). *p<0.05 (Student’s t test). C. Real-time qRT-PCR analysis of miRNA expression in primary oligodendrocytes. Expression levels of the selected miRNAs were quantitated in primary oligodendrocytes isolated from CNP-GFP brains at postnatal day 1 and 21. The levels of each miRNA at P21 is expressed as a percent of that at P 1 which is set as 1 (n=4). *P<0.05 and ** P<0.01.
Comparison with published miRNA arrays in OLs
A number of studies have reported miRNA profiles in rodent OLs (Lau et al. 2008; Shin et al. 2009; Budde et al. 2010; Dugas et al. 2010; Lehotzky et al. 2010; Zhao et al. 2010). We compared Oli-neu cell miRNA profile with the published miRNA array data in the rat primary oligodendrocytes (Lau et al. 2008), which was the first published rodent miRNA expression profile in which each of the 98 rat primary OL expressed miRNAs was verified by real-time qRT-PCR (Lau et al. 2008). The rat miRBase version 9.1 was used for the rat studies, while we have used the mouse miRBase version 14.0 (LC Sciences). We first identified the miRNAs that are in common between rat miRBase version 9.1 and mouse miRBase version 14.0. Of the 145 Oli-neu expressed-miRNAs, 81 are novel miRNAs that were not present in the rat miRBase version 9.1; the remainder 64 miRNAs are present in both. Of the 98 rat primary OL-expressed miRNAs (Lau et al. 2008), 3 miRNAs are specific to rat and not present in mouse OLs while 95 miRNAs are common to both databases (Figure S1A). Interestingly, 44 miRNAs are expressed in both rat and mouse OLs. They account for ~46.3% (44/95) and ~68.8% (44/64) of the total miRNAs that are common to both rat and mouse databases (Fig. S1A). The correlation in fold change of the 44 shared miRNAs in differentiated vs. undifferentiated Oli-neu cells compared to A2B5−GalC+ vs. A2B5+GalC− rat primary OLs (Lau et al. 2008), was significant (r(44)=0.4697, P<0.01, Fig. S1B). A similar correlation was found between the Oli-neu miRNA profile and that of in vitro cultured rat oligodendrocyte cell line CG4 cells (Lehotzky et al. 2010) (data not shown). Importantly, the Oli-neu cell miRNA profile contains all the members of the miR 17–92 cluster (Table 1S), which regulates OL cell number (Budde et al. 2010). Furthermore, ~45% of the miRNAs discovered in our arrays are intronic miRNAs (data not shown), and this percentage is similar to that found in rat array (Lau et al. 2008). Taken together, the mouse oligodendrocyte-expressed miRNAs in our studies overlap well those published for rat OLs. In addition, greater than 50% of the miRNAs in our arrays are novel miRNAs that were not present in the early miRBase version 9.1.
Expression of PLP gene is post-transcriptionally regulated
Next, we have investigated whether miRNAs regulate PLP gene expression by base pairing with motifs in the PLP 3′UTR. The mouse PLP mRNA transcript gives rise to three mRNA isoforms by alternative selection of polyadenylation signals within the PLP 3′UTR: 1.6 kb, 2.4 kb and 3.2 kb comprising a 3′UTR of 0.6 kb, 1.6 kb and 2.2 kb, respectively (Gardinier et al. 1986). Since previous studies showed that different PLP mRNAs exhibit essentially similar stability in oligodendrocytes (Mallon and Macklin 2002), we focused our analysis on the mouse 1.6 kb 3′ UTR. We cloned these sequences (GenBank Accession # NM_011123) downstream from the Renilla luciferase gene in the vector psiCHECK™-2 (Promega), (UTRPLP-1.6) (Figure 4A). Oli-neu cells were transfected with UTRPLP-1.6 and the control empty vector, and Renilla luciferase activity was measured after 48 hrs and normalized by the activity of the firefly luciferase expressed from the same vector. The luciferase activity for UTRPLP-1.6, expressed as percent of that of the empty vector, was more than 80% reduced (Figure 4B), indicating that the PLP 3′UTR contains regulatory elements that repress luciferase expression. Similar reduction in luciferase activity was detected in Oli-neu cell extracts harvested 24, 72 and 96 hrs after transfection (Fig. S2). Interestingly, ~2-fold increase in the luciferase activity of UTRPLP-1.6 was observed in differentiated vs. undifferentiated Oli-neu cells (Fig. S2C). These results indicate that growth conditions, such as cell density and length of culture in mitogens, do not affect PLP 3′UTR-mediated suppression. In contrast, oligodendrocyte differentiation appears to decrease the negative regulation possibly through changes in miRNAs or other RNA binding proteins.
The luciferase activity of UTRPLP-1.6 was also reduced in mouse L cells (0.12 vs. 1.0), primary mouse astrocytes (0.42 vs. 1.0), and Hela cells (0.26 vs. 1.0) (Fig. S3), suggesting that the inhibitory effect of the 3′UTR is not cell type specific. We ascertained that miR-20a is expressed in L and HeLa cells and show that it is expressed similarly to Oli-neu cells in L cells and at higher levels in HeLa cells (Fig. S4). miR-20a was shown to be expressed in astrocytes (Budde et al. 2010). The magnitude of the inhibitory effect varies in the different cell lines compared to Oli-neu (Fig. 4B, Fig. S3). This difference is of unclear significance.
To more precisely define the PLP 3′UTR sequences that are required for suppression, we cloned the 0.6 kb of PLP 3′UTR downstream of the PLP stop codon (UTRPLP-0.6), which correspond to the 1.6 kb mRNA in the psiCHECK™-2 (Fig. 4A). Luciferase activity of the UTRPLP-0.6 was decreased by ~50% compared to the control vector (Fig. 4C), but was three-fold higher than the luciferase activity of UTRPLP 1.6 (0.55 vs. 0.17, Fig. 4C). The data suggest that inhibitory sequences are spatially distributed in the 1.6 kb 3′UTR. By contrast, sequences downstream of the 1.6 kb UTR, which are exclusively present in the 3.2 kb PLP mRNA (GenBank Accession #AK134554), cloned in the psiCHECK2 vector (Fig. 4A, upper panel, UTRPLP-2.2), did not affect the luciferase activity compared to the control vector (Fig. 4D). Taken together, these data indicate that the 1.6 kb PLP 3′UTR harbors sequences that negatively regulate the expression of luciferase in a non cell type specific fashion, while the sequences downstream of the 1.6 kb appear to have little or no effect.
miRNAs regulate the PLP 3′UTR
To determine whether miRNAs mediate the inhibitory effect of UTRPLP-1.6, we have examined miRNAs identified by our microarray analysis in OLs. Of the miRNAs whose expression is >1-fold (log2) lower in differentiated vs. undifferentiated Oli-neu cells (Fig. 2C), we selected miRNAs that have 1 or more binding site(s) in the UTRPLP-1.6 as predicted by the bioinformatics tool RNA22 (Miranda et al. 2006) (Fig. 5A). As shown in Figure 5A, there are binding sites for 10 miRNAs in the PLP 3′UTR, 3 of which are located in the first 0.6 kb downstream of the stop codon and 7 are downstream of the 0.6 kb sequences. We tested miR-17, miR-20a, miR-20b, miR-1907 and miR-1967 for their efficacy in mediating the PLP 3′UTR repression of luciferase activity since precursor mimics or anti-sense inhibitors (Applied Biosystems) were not commercially available for the other 5 miRNAs.
We co-transfected Oli-neu cells with UTRPLP-1.6 and precursor mimics or anti-sense inhibitors that target individual miRNAs (Applied Biosystems). No significant change in luciferase activity was observed by over and/or under-expression of miR-17, miR-20b, miR-1907 and miR-1967 in Oli-neu cells (data not shown). By contrast, knockdown and overexpression of miR-20a caused significantly changed luciferase activity. Seventy two hours after co-transfection of UTRPLP-1.6 and miR-20a antisense inhibitors into Oli-neu cells, miR-20a expression was quantified by real-time qRT-PCR in total RNA, and luciferase activity was assayed in protein lysates. The endogenous miR-20a was reduced >80% by the anti-sense inhibitors, and reduction was concentration-dependent (Fig. 5C). Knockdown of miR-20a increased the luciferase activity derived from UTRPLP-1.6 by 50% compared to mock treated cells (Fig. 5D). To demonstrate that the effect of knock down of miR-20a is sequence specific, we either deleted the miR-20a binding motif (UTRPLP-1.6-DEL) or mutated the 6 nucleotides that form duplex with the seed sequence of miR-20a (UTRPLP-1.6-MT, Figure 5B). Knock down of miR-20a did not affect the luciferase activity from UTRPLP-1.6-DEL and –MT (Fig. 6E and F), suggesting that the miR-20a binding motif is required for the miR-20a mediated suppression.
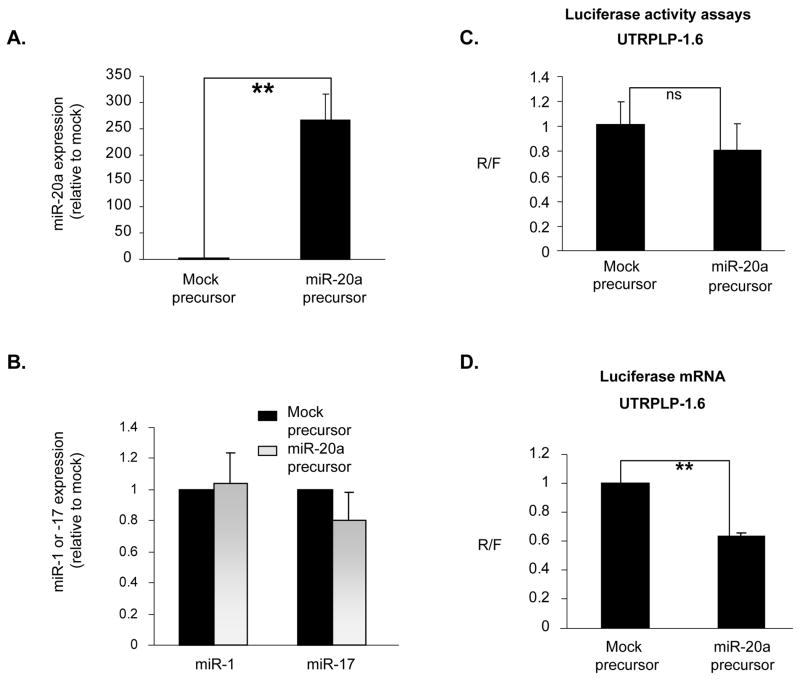
Figure 6. Over-expression of miR-20a reduces PLP 3′UTR-luciferase reporter expression in Oli-neu cells.
A. The bar graph shows that miR-20a expression is elevated ~250 fold after treatment with the exogenous miR-20a precursor mimics. Error bars represent SD (n=4). **, P<0.01. B. Bar graph shows that miR-1 and -17 expression is not affected by the miR-20a precursor mimics treatment compared to mock. Error bars represent SD (n=4). C. Bar graph shows luciferase activity in Oli-neu cells treated with mock and miR-20a precursor mimics. Error bars represent SD (n=3). ns, not significant. D. Bar graph shows real-time qRT-PCR quantitation of the luciferase mRNA level in Oli-neu cells treated with mock and miR-20a precursor mimics. Error bars represent SD (n=4). **P<0.01 (Student’s t test).
To determine whether over-expression of miR-20a decreased the luciferase activity of the UTRPLP-1.6, we co-transfected either miR-20a precursor mimics or a mock precursor mimic (Applied Biosystems) with the UTRPLP-1.6 into Oli-neu cells. The expression of miR-20a was increased more than 250-fold in miR-20a precursors vs. mock (precursor mimics) treated Oli-neu cells (Fig. 6A). As control of specificity we show that miR-1 and miR-17, which belongs to the same cluster as miR-20a, are not affected (Fig. 6B). Over-expression of miR-20a caused only a modest (20%) decrease in the luciferase activity compared to mock treated cells (Fig. 6C). The reduction is not significant (Fig. 6C). However, the luciferase mRNA measured by real-time qRT-PCR and normalized to the firefly luciferase mRNA was decreased by 40% in Oli-neu cells over-expressing miR-20a compared to mock treated cells (Fig. 6D, p<0.05). These data suggest that the luciferase assay may not be sufficiently sensitive to detect changes induced by exogenous miR-20a precursors, especially in consideration of the high levels of endogenous expression of miR-20a (Table S1).
Together, the data show that miR-20a regulates the PLP 3′ UTR mediated suppression of luciferase expression through specific binding of its recognition motif.
miR-20a controls endogenous PLP gene expression in oligodendrocytes
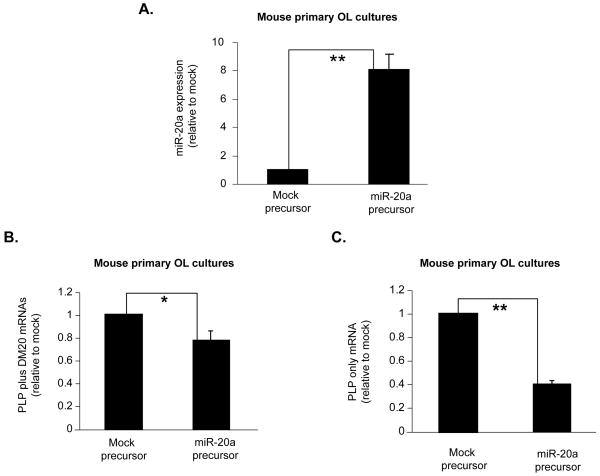
To firmly establish that miR-20a plays an important role in regulating levels of PLP/DM20 message, we over-expressed either miR-20a precursor mimic or a mock precursor mimic (Applied Biosystems) in primary mouse oligodendrocytes and measured the endogenous PLP and DM20 mRNAs by real time qRT-PCR. Concomitant to an eight-fold increase in miR20a levels in oligodendrocytes transfected with miR-20a mimic (Fig. 7A), there was a 25% reduction in total PLP and DM20 messages and 60% reduction in PLP message (Fig. 7B and C) compared to mock treated oligodendrocytes. We conclude that miR-20a regulates PLP gene expression in oligodendrocytes.
Figure 7. Endogenous PLP mRNA expression is decreased by miR-20a in mouse primary oligodendrocytes.
A. The bar graph shows that miR-20a expression is increased in primary OL culture after treatment with exogenous miR-20a precursor mimics. Error bars represent SD (n=4). ** P<0.01. B. Bar graph shows real-time qRT-PCR quantitation of the endogenous PLP plus DM20 mRNAs in primary OL treated with mock and miR-20a precursor mimics. Error bars represent SD (n=4). * P<0.05. C. Bar graph shows real-time qRT-PCR quantitation of the endogenous PLP specific mRNA in primary OL treated with mock and miR-20a precursor mimics. Error bars represent SD (n=4). **P<0.01 (Student’s t test).
Discussion
In this study, we report that Dicer1 is down-regulated in differentiated OLs, concomitantly with the expression of myelin proteins. Differentiation-induced downregulation of Dicer1 is responsible in large part for changes in miRNA expression profiles in differentiated vs. undifferentiated OLs. Furthermore, we show that miR-20a participates in the regulation of the major myelin protein, PLP, by exerting a negative effect through the 3′UTR. This is the first report that miRNAs regulate myelin gene expression in OLs.
Three PLP mRNA isoforms are generated by alternative utilization of polyadenylation signals within the PLP 3′UTR. The 1.6 kb contains the upstream 0.6 kb of the 3′UTR, the 2.4 kb contains 1.6 kb of the 3′ UTR and 3.2 kb contains 2.2kb of the 3′ UTR (Mallon and Macklin 2002). We show that both 0.6 and 1.6 kb of the PLP 3′UTR inhibit the reporter luciferase activity in a non cell specific fashion. In contrast, previous studies (Mallon and Macklin 2002) had shown that the PLP 3′UTR enhances the stability of the luciferase reporter gene mRNA and reduces the endogenous PLP mRNA expression, suggesting a possible competition for trans-acting acting factors (Mallon and Macklin 2002). In Oli-neu cells, over-expression of either 1.6 or 0.6 kb 3′UTR did not affect the expression of the endogenous PLP (data not shown). The apparent discrepancy between results of the current and the previous study may reflect the use of different cell lines and experimental systems in the two studies. Stably transfected C6 cell lines and transgenic mouse lines expressing the EGFP fused to the PLP 3′ UTR were used in the published studies, while we have transiently transfected Oli-neu cells with the dual-luciferase reporter assay system. The internal normalization control derived from the same vector minimizes experimental variations caused by transfection efficiency, cell viability and cell lysis efficiency.
We think that the negative effect of the PLP 3′UTR, characterized in this paper, and the positive effect, shown by Mallon et al., are both active in vivo. By virtue of using different experimental systems, we might have been able to separately identify and characterize each effect. We would like to propose that the negative effect is a general mechanism that suppresses PLP expression in non-glial cells and in oligodendrocyte progenitors. Release of this negative regulation, driven by reduced levels of miR-20a, would allow the cell-specific factors expressed in OL to positively regulate the PLP message in OL but not in non-glial cells. Our data show that the negative effect of PLP 3′UTR is lower, albeit of only two-fold, in differentiated vs. undifferentiated Oli-neu cells (Fig. S2), consistent with a differentiation-mediated regulation. The increase in luciferase does not match the dramatic increase in PLP message in the developing brain (Gardinier et al. 1986; Wight et al. 1993; Fuss et al. 2000). In brain, the increase in PLP/DM20 message results from multiple mechanisms: transcriptional activation, message stability regulated by the 3′UTR through miR-20a (shown in this manuscript) and transacting factors, likely to be cell-specific and possibly developmentally regulated (as suggested by Macklin’s paper). In the experimental paradigm of a luciferase assay in Oli-neu cells, all these levels of regulation cannot be faithfully recapitulated. Furthermore, transcription and 3′ RNA processing are tightly coupled processes (Darzacq et al. 2007; Perales and Bentley 2009) and are regulated by chromatin organization (Spies et al. 2009). This level of regulation is not present with a chimeric construct in transient transfections.
Interestingly, we have found that inhibition is mediated by the 0.6 and 1.6 UTR. Our data, therefore, could indicate that there may be positively acting regulatory sequences in the longer mRNA. The latter finding is of particular interest in light of the presence of the 3.2 kb mRNA early in development and the relative stability of this message throughout development (Gardinier et al. 1986). In contrast, the shorter mRNAs are expressed at late time point in post-natal brain development, consistent with the hypothesis that inhibitory sequences are developmentally regulated by changes in regulatory factors and miRNAs. Our data would support this possibility.
miRNAs have emerged as major post-transcriptional regulators of gene expression, largely by interacting with motifs in the 3′UTR of target RNAs. Here, we show that miR-20a regulates the reporter luciferase activity through a binding motif in the 1.6 kb UTR. miR-20a is part of the 17–92 cluster, which contains miR-17, -18a, -19b, -20a, -20b and -92a (Olive et al. 2009; van Haaften and Agami 2010). miR-17 and -20a are highly enriched in OLs, as shown in this and a previous study (Lau et al. 2008; Budde et al. 2010). Conditional knock out of the miR-17–92 cluster causes a reduction in OL cell number, which is rescued by miR-17 and -19b (Budde et al. 2010). Although miR-20a has been shown to be highly expressed in OLs, a function for this miRNA was not investigated. Thus, this is a first report of a function of miR-20a in OLs. The binding site for miR-20a in PLP 3′UTR overlaps with that for miR-17, miR-20b and miR-106a (Fig. 5). The recognition motifs of these four miRNAs share the seed sequence (nt 2 to 8), but differ in either two or three nucleotides outside of the seed region. This raises the possibility that the anti-sense inhibitors that target miR-20a may inhibit the expression of miR-17, miR-20b and miR-106a and deletion or mutation of the putative binding site for miR-20a may also disrupt the interaction of these miRNAs with PLP 3′UTR. This appears to be unlikely since the overexpression of miR-17 and miR-20b had no effect on the luciferase activity (data not shown), whereas the overexpression of miR-20a alone (Fig. 6C, D) was sufficient to decrease luciferase activity. Our data demonstrate for the first time a role of miRNAs in the regulation of PLP expression and show that such regulation is mediated by RNA degradation rather than translational inhibition,
Although not examined in this study, it is likely that additional mechanisms and factors contribute to the inhibitory effect mediated by the PLP 3′ UTR. There are putative binding motifs for RNA binding proteins, namely polypyrimidine tract binding protein, heterogeneous nuclear ribonucleoproteins H and F, and AU-rich elements binding factors, which are well known to regulate polyA site selection and RNA 3′ end processing (Millevoi et al. 2009; Millevoi and Vagner 2010). Importantly, the expression of these RNA binding factors changes in developing OLs in vitro (Wang et al. 2007, and unpublished data) and in vivo (unpublished data, Zhu, Papa and Cambi), supporting the notion that they may also regulate message stability.
While previous studies have shown that miRNA expression in oligodendrocytes is developmentally regulated, the mechanism by which miRNA expression is regulated was not investigated (Lin and Fu 2009; Shin et al. 2009; Dugas et al. 2010; Zhao et al. 2010). We show that Dicer1 expression is decreased in differentiated OLs in vivo and in vitro. By contrast previous studies have shown that Dicer1 is up-regulated in differentiated OLs in vitro (Dugas et al. 2010; Zhao et al. 2010). There are notable differences in the cell systems used in these studies. We have used FACS sorted OLs ex vivo which directly reflect the in vivo environment without exposure to growth factors. It is difficult to predict whether the OLs cultured in vitro for 4 days and analyzed in previous studies represent a population similar to the OLs isolated by flow cytometry at day P21 and examined in this study. Interestingly, Dicer1 is also down-regulated in differentiated Schwann cells (SC) compared to SC progenitors (Verrier et al. 2009), suggesting that downregulation of Dicer1 may be a common mechanism by which myelin producing cells of the CNS and PNS regulate miRNAs expression.
Although changes in miRNAs induced by Dicer1 knock down mirror those induced by differentiation, a number of miRNAs are up-regulated by Dicer knock down rather than down- regulated (Fig. 2). This partial overlap may indicate that Dicer1 independent pathways regulate miRNAs biogenesis in differentiated OLs (Cifuentes et al. 2010). Furthermore, knockdown of Dicer1 may derepress the expression of genes that harbor these miRNAs clusters, resulting in their increased expression. A case in point are miR-703 and -1967 (Fig. 2), which are within the introns of protein coding genes, miR-703 located in intron 2 of 1700007G11Rik, and miR-1967 in intron 1 of AC114005.5-001 (ENSMUST00000127664) (unpublished observation). The expression pattern and function of these protein coding genes in OLs have not been explored. Although additional mechanisms are likely to regulate miRNAs in differentiated OLs, our studies support the conclusion that reduced expression of Dicer1 represents an important regulatory mechanism.
Finally, while comparison with previous array results shows that there is overlap between ours and previous studies, greater than 50% of miRNAs identified in our study are novel miRNAs that were released in mouse miRbase version 14.0. Interestingly, many of these novel miRNAs are up-regulated in differentiated oligodendrocyte cells (data not shown). The functional relevance of these miRNAs remains to be investigated. In summary, we show that miRNAs are potential regulators of the PLP gene expression. Since more than 60% of Pelizaeus-Merzbacher disease (PMD) cases are due to duplication of the PLP gene causing over-expression of the PLP/DM20 proteins (Mimault et al. 1999; Hobson et al. 2002; Inoue 2005; Garbern 2007; Karim et al. 2007; Boespflug-Tanguy et al. 2008), miR-20a characterized in this study and other miRNAs to be discovered may represent therapeutic targets for interventions (Weinberg and Wood 2009; Zeng 2009) aimed at reducing expression of PLP gene in PMD.
Supplementary Material
A. Venn diagram showing the overlap of miRNAs expressed in rat primary oligodendrocyte cells and in Oli-neu cells. B. Shown is the correlation between the miRNA expression fold change in differentiated vs. undifferentiated Oli-neu cells and that in A2B5−GalC+ vs. A2B5+GalC− rat primary Ols. r = Pearson’s correlation coefficient (n=44). **P<0.01 (Student’s t test).
A. The bar graph shows the luciferase activity derived from the UTRPLP-1.6 in Oli-neu cells harvested 24 hrs after transfection and cultured in SATO medium. B. The bar graph shows the luciferase activity derived from the UTRPLP-1.6 in Oli-neu cells harvested 72 hrs after transfection and cultured in SATO medium. C. The bar graph shows the luciferase activity derived from UTRPLP-1.6 in Oli-neu cells cultured in SATO medium without (undifferentiated) or with 20nM dbcAMP (differentiated), and harvested 4-day after transfection. The data are expressed as ratio of the Renilla luciferase to the firefly luciferase activity (R/F), and the R/F of the empty vector is set as 1. Error bars represent SD. n≥3. *P<0.05 and **P<0.01 (Student’s t test).
Both the UTRPLP-1.6 and the empty vector were transfected into the indicated cell lines. The luciferase activity was assayed seventy two hours after transfection. The data are expressed as ratio of the Renilla luciferase to the firefly luciferase activity (R/F), and the R/F of the empty vector is set as 1. A. The bar graph shows the luciferase activity of the UTRPLP-1.6 transfected into HeLa cells compared to the empty vector (n=3). B. The bar graph shows the luciferase activity of the UTRPLP-1.6 transfected into mouse L cells compared to the empty vector (n=2). C. The bar graph shows the luciferase activity of the UTRPLP-1.6 transfected into primary astrocytes compared to the empty vector (n=3). Error bars represent SD. **P<0.01 (Student’s t test).
A. Representative RT-PCR analysis of mature miR-20a levels in HeLa, L and Oli-neu cells. U6 snRNA was amplified as loading control. Negative control is a PCR reaction without RT template added. B. The bar graph shows the mature miR-20a level±SD (n=3). The data are expressed as fold over the Oli-neu cells set at 1. *P<0.05 (Student’s t test). ns, not significant.
Acknowledgments
Funding: This work was supported by a grant of the University of Kentucky Research Foundation to EW and FC, European Leukodystrophy Association (2008-013I1) to FC and EW, and NIH/NINDS (RO1NS053905) to FC.
The authors thank Dr. V. Gallo for the CNP-GFP mouse. We also thank the members of the Cambi laboratory for preparing mouse EGFP+ oligodendrocyte cells and Ms. Jennifer Strange for the excellent technical assistance with FACS isolation of oligodendrocytes.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev. 2001;81(2):871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- Belachew S, Chittajallu R, Aguirre AA, Yuan X, Kirby M, Anderson S, Gallo V. Postnatal NG2 proteoglycan-expressing progenitor cells are intrinsically multipotent and generate functional neurons. J Cell Biol. 2003;161(1):169–186. doi: 10.1083/jcb.200210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boespflug-Tanguy O, Labauge P, Fogli A, Vaurs-Barriere C. Genes involved in leukodystrophies: a glance at glial functions. Curr Neurol Neurosci Rep. 2008;8(3):217–229. doi: 10.1007/s11910-008-0034-x. [DOI] [PubMed] [Google Scholar]
- Budde H, Schmitt S, Fitzner D, Opitz L, Salinas-Riester G, Simons M. Control of oligodendroglial cell number by the miR-17–92 cluster. Development. 2010;137(13):2127–2132. doi: 10.1242/dev.050633. [DOI] [PubMed] [Google Scholar]
- Campagnoni AT, Macklin WB. Cellular and molecular aspects of myelin protein gene expression. Mol Neurobiol. 1988;2(1):41–89. doi: 10.1007/BF02935632. [DOI] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, Wolfe SA, Giraldez AJ. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328(5986):1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT, Barres BA. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65(5):597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss B, Mallon B, Phan T, Ohlemeyer C, Kirchhoff F, Nishiyama A, Macklin WB. Purification and analysis of in vivo-differentiated oligodendrocytes expressing the green fluorescent protein. Dev Biol. 2000;218(2):259–274. doi: 10.1006/dbio.1999.9574. [DOI] [PubMed] [Google Scholar]
- Garbern JY. Pelizaeus-Merzbacher disease: Genetic and cellular pathogenesis. Cell Mol Life Sci. 2007;64(1):50–65. doi: 10.1007/s00018-006-6182-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardinier MV, Macklin WB, Diniak AJ, Deininger PL. Characterization of myelin proteolipid mRNAs in normal and jimpy mice. Mol Cell Biol. 1986;6(11):3755–3762. doi: 10.1128/mcb.6.11.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hobson GM, Huang Z, Sperle K, Stabley DL, Marks HG, Cambi F. A PLP splicing abnormality is associated with an unusual presentation of PMD. Ann Neurol. 2002;52(4):477–488. doi: 10.1002/ana.10320. [DOI] [PubMed] [Google Scholar]
- Huang Z, Tang XM, Cambi F. Down-regulation of the retinoblastoma protein (rb) is associated with rat oligodendrocyte differentiation. Mol Cell Neurosci. 2002;19:250–262. doi: 10.1006/mcne.2001.1077. [DOI] [PubMed] [Google Scholar]
- Inoue K. PLP1-related inherited dysmyelinating disorders: Pelizaeus-Merzbacher disease and spastic paraplegia type 2. Neurogenetics. 2005;6(1):1–16. doi: 10.1007/s10048-004-0207-y. [DOI] [PubMed] [Google Scholar]
- Jung M, Kramer E, Grzenkowski M, Tang K, Blakemore W, Aguzzi A, Khazaie K, Chlichlia K, von Blankenfeld G, Kettenmann H, et al. Lines of murine oligodendroglial precursor cells immortalized by an activated neu tyrosine kinase show distinct degrees of interaction with axons in vitro and in vivo. Eur J Neurosci. 1995;7(6):1245–1265. doi: 10.1111/j.1460-9568.1995.tb01115.x. [DOI] [PubMed] [Google Scholar]
- Karim SA, Barrie JA, McCulloch MC, Montague P, Edgar JM, Kirkham D, Anderson TJ, Nave KA, Griffiths IR, McLaughlin M. PLP overexpression perturbs myelin protein composition and myelination in a mouse model of Pelizaeus-Merzbacher disease. Glia. 2007;55(4):341–351. doi: 10.1002/glia.20465. [DOI] [PubMed] [Google Scholar]
- Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28(45):11720–11730. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehotzky A, Lau P, Tokesi N, Muja N, Hudson LD, Ovadi J. Tubulin polymerization-promoting protein (TPPP/p25) is critical for oligodendrocyte differentiation. Glia. 2010;58(2):157–168. doi: 10.1002/glia.20909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ST, Fu YH. miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis Model Mech. 2009;2(3–4):178–188. doi: 10.1242/dmm.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Macklin WB, Campagnoni CW, Deininger PL, Gardinier MV. Structure and expression of the mouse myelin proteolipid protein gene. J Neurosci Res. 1987;18(3):383–394. doi: 10.1002/jnr.490180302. [DOI] [PubMed] [Google Scholar]
- Mallon BS, Macklin WB. Overexpression of the 3′-untranslated region of myelin proteolipid protein mRNA leads to reduced expression of endogenous proteolipid mRNA. Neurochem Res. 2002;27(11):1349–1360. doi: 10.1023/a:1021623700009. [DOI] [PubMed] [Google Scholar]
- Millevoi S, Decorsiere A, Loulergue C, Iacovoni J, Bernat S, Antoniou M, Vagner S. A physical and functional link between splicing factors promotes pre-mRNA 3′ end processing. Nucleic Acids Res. 2009;37(14):4672–4683. doi: 10.1093/nar/gkp470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38(9):2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimault C, Giraud G, Courtois V, Cailloux F, Boire JY, Dastugue B, Boespflug-Tanguy O. Proteolipoprotein gene analysis in 82 patients with sporadic Pelizaeus-Merzbacher Disease: duplications, the major cause of the disease, originate more frequently in male germ cells, but point mutations do not. The Clinical European Network on Brain Dysmyelinating Disease. Am J Hum Genet. 1999;65(2):360–369. doi: 10.1086/302483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Nave KA, Lai C, Bloom FE, Milner RJ. Splice site selection in the proteolipid protein (PLP) gene transcript and primary structure of the DM-20 protein of central nervous system myelin. Proc Natl Acad Sci U S A. 1987;84(16):5665–5669. doi: 10.1073/pnas.84.16.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17–92. Genes Dev. 2009;23(24):2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Shin JY, McManus MT, Ptacek LJ, Fu YH. Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann Neurol. 2009;66(6):843–857. doi: 10.1002/ana.21927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporkel O, Uschkureit T, Bussow H, Stoffel W. Oligodendrocytes expressing exclusively the DM20 isoform of the proteolipid protein gene: myelination and development. Glia. 2002;37(1):19–30. doi: 10.1002/glia.10014. [DOI] [PubMed] [Google Scholar]
- van Haaften G, Agami R. Tumorigenicity of the miR-17–92 cluster distilled. Genes Dev. 2010;24(1):1–4. doi: 10.1101/gad.1887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier JD, Lau P, Hudson L, Murashov AK, Renne R, Notterpek L. Peripheral myelin protein 22 is regulated post-transcriptionally by miRNA-29a. Glia. 2009;57(12):1265–1279. doi: 10.1002/glia.20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Cambi F. Heterogeneous nuclear ribonucleoproteins H and F regulate the proteolipid protein/DM20 ratio by recruiting U1 small nuclear ribonucleoprotein through a complex array of G runs. J Biol Chem. 2009;284(17):11194–11204. doi: 10.1074/jbc.M809373200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Dimova N, Cambi F. PLP/DM20 ratio is regulated by hnRNPH and F and a novel G-rich enhancer in oligodendrocytes. Nucleic Acids Res. 2007;35(12):4164–4178. doi: 10.1093/nar/gkm387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Dimova N, Sperle K, Huang Z, Lock L, McCulloch MC, Edgar JM, Hobson GM, Cambi F. Deletion of a splicing enhancer disrupts PLP1/DM20 ratio and myelin stability. Exp Neurol. 2008;214(2):322–330. doi: 10.1016/j.expneurol.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Weinberg MS, Wood MJ. Short non-coding RNA biology and neurodegenerative disorders: novel disease targets and therapeutics. Hum Mol Genet. 2009;18(R1):R27–39. doi: 10.1093/hmg/ddp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wight PA, Duchala CS, Readhead C, Macklin WB. A myelin proteolipid protein-LacZ fusion protein is developmentally regulated and targeted to the myelin membrane in transgenic mice. J Cell Biol. 1993;123(2):443–454. doi: 10.1083/jcb.123.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43(6):892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Chittajallu R, Belachew S, Anderson S, McBain CJ, Gallo V. Expression of the green fluorescent protein in the oligodendrocyte lineage: a transgenic mouse for developmental and physiological studies. J Neurosci Res. 2002;70(4):529–545. doi: 10.1002/jnr.10368. [DOI] [PubMed] [Google Scholar]
- Zeng Y. Regulation of the mammalian nervous system by microRNAs. Mol Pharmacol. 2009;75(2):259–264. doi: 10.1124/mol.108.052118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Mi QS, Xin M, Wang F, Appel B, Lu QR. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65(5):612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Zhao L, Wang E, Dimova N, Liu G, Feng Y, Cambi F. The QKI-PLP pathway controls SIRT2 abundance in CNS myelin. Glia. 2012;60(1):69–82. doi: 10.1002/glia.21248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Venn diagram showing the overlap of miRNAs expressed in rat primary oligodendrocyte cells and in Oli-neu cells. B. Shown is the correlation between the miRNA expression fold change in differentiated vs. undifferentiated Oli-neu cells and that in A2B5−GalC+ vs. A2B5+GalC− rat primary Ols. r = Pearson’s correlation coefficient (n=44). **P<0.01 (Student’s t test).
A. The bar graph shows the luciferase activity derived from the UTRPLP-1.6 in Oli-neu cells harvested 24 hrs after transfection and cultured in SATO medium. B. The bar graph shows the luciferase activity derived from the UTRPLP-1.6 in Oli-neu cells harvested 72 hrs after transfection and cultured in SATO medium. C. The bar graph shows the luciferase activity derived from UTRPLP-1.6 in Oli-neu cells cultured in SATO medium without (undifferentiated) or with 20nM dbcAMP (differentiated), and harvested 4-day after transfection. The data are expressed as ratio of the Renilla luciferase to the firefly luciferase activity (R/F), and the R/F of the empty vector is set as 1. Error bars represent SD. n≥3. *P<0.05 and **P<0.01 (Student’s t test).
Both the UTRPLP-1.6 and the empty vector were transfected into the indicated cell lines. The luciferase activity was assayed seventy two hours after transfection. The data are expressed as ratio of the Renilla luciferase to the firefly luciferase activity (R/F), and the R/F of the empty vector is set as 1. A. The bar graph shows the luciferase activity of the UTRPLP-1.6 transfected into HeLa cells compared to the empty vector (n=3). B. The bar graph shows the luciferase activity of the UTRPLP-1.6 transfected into mouse L cells compared to the empty vector (n=2). C. The bar graph shows the luciferase activity of the UTRPLP-1.6 transfected into primary astrocytes compared to the empty vector (n=3). Error bars represent SD. **P<0.01 (Student’s t test).
A. Representative RT-PCR analysis of mature miR-20a levels in HeLa, L and Oli-neu cells. U6 snRNA was amplified as loading control. Negative control is a PCR reaction without RT template added. B. The bar graph shows the mature miR-20a level±SD (n=3). The data are expressed as fold over the Oli-neu cells set at 1. *P<0.05 (Student’s t test). ns, not significant.