Abstract
We report photo-catalytic H2 production by hydrogenase (H2ase)-quantum dot (QD) hybrid assemblies. Quenching of the CdTe exciton emission is observed, consistent with electron transfer from quantum dot to H2ase. GC analysis shows light driven H2 production in the presence of a sacrificial electron donor with an efficiency of 4%, which is likely a lower limit to these hybrid systems. FTIR was employed for direct observation of active site reduction in unprecedented detail for photo-driven H2ase catalysis with sensitivity towards both H2ase and sacrificial electron donor. Photosensitization with Ru(bpy)32+ shows distinct FTIR photo- reduction properties generating all states along the steady-state catalytic cycle with minimal H2 production indicating slow, sequential one electron reduction steps. Comparing H2ase activity and FTIR results of both systems shows that QDs bind more efficiently for electron transfer and the final enzyme state is different for the two sensitizers. The possible origins of these differences and their implications for the enzymatic mechanism are discussed.
H2ases catalyze the two reactions that are fundamental for a viable hydrogen-based economy: the reduction of protons in water to hydrogen and the oxidation of hydrogen to protons One class of H2ases, denoted [NiFe] based on the metal content of the active site, is tolerant of, or reversibly inhibited by O2, and consequentially has been heavily studied for biotechnology applications.1–5 Recent research has explored using light to drive the chemistry of H2ase enzymes with a variety of photosensitizers, including photosystem I, ruthenium sensitized TiO2 and QDs.6–12 Light induced H2 generation and ET has been characterized, but no one to date has used this approach to rapidly initiate turnover for mechanistic studies. Triggering enzyme turnover with light may provide exquisite control of the complex catalytic cycle (SI-1), which opens the possibility of directly observing short lived intermediates by infrared spectroscopy through the CO and CN− ligands bound to iron in the active site.
Herein we present a hybrid photo-catalyst that couples H2ase from Thiocapsa roseopersicina (Tr) and mercaptopropionic acid (MPA) capped CdTe QDs to efficiently drive hydrogen production using visible light. This [NiFe] H2ase was selected for its overall chemical stability towards various buffers, pH and ionic strength as well as its exceptional thermal stability, high tolerance towards O2 and reversible reactivation.4 FTIR spectroscopy provides direct evidence of active site reduction as well as sacrificial electron donor (SED) consumption and GC analysis confirms highly efficient enzyme turnover. Comparison with Ru(bpy)32+ sensitized H2ase reveals striking differences that we attribute in part to the efficiency of photo-reduction which may have important implications for the catalytic mechanism.
Understanding the interaction between the QD and enzyme surface is of critical importance to design ET active binding.13 Electrostatic binding was used as the simplest approach to attaching the QD photosensitizer to the enzyme. Since the crystal structure of the Tr H2ase has not been determined, homology modeling (SI-2) was performed to assess possible binding sites of the CdTe QD. 14,15 The model shows a positively charged region around the small subunit near the distal and medial FeS clusters. We hypothesize the latter as the most likely binding site for the negatively charged CdTe QD. Binding at this site should orient the nanoparticle optimally on the enzyme surface for interfacial ET to the distal or medial FeS cluster.13
QD photoluminescence quantum efficiency (PLQE) has been shown previously to be sensitive to molecules and proteins adsorbed to the QD in nanoparticle assemblies.6,16,17 We use this property of QDs to investigate the nature of H2ase-QD binding interaction and non-radiative contributions to excitonic quenching from H2ase adsorbed on the QD surface as shown in Figure 1.
Figure 1.
Photoluminescence titration spectra of 500 nM CdTe QDs with H2ase (red = 0 μM H2ase, purple = 4 μM H2ase) in 100 mM phosphate buffer pH = 7.5. Inset shows integrated photoluminescence intensity as a function of H2ase concentration.
Titration of QDs with H2ase shows quenching of the PLQE. We attribute the decrease in PLQE to a non-radiative ET quenching mechanism that directly reduces the distal FeS cluster. The observed behavior is likely due to higher interfacial ET efficiency (decreases PLQE) over the proposed surface passivation effect (increases PLQE).16,17 We postulate that surface passivation is indeed occurring, but that the high ET efficiency obscures the predicted increase in PLQE by surface passivation. The PL quenching does not show saturation over the accessible range of H2ase concentrations, preventing detailed analysis of the binding constant and free energy, but salt screening effects, vide infra, corroborate electrostatic binding. The titration indicates a binding constant < 106 M−1 (SI) or that the binding is not one to one.
Encouraged by evidence for ET in the PL titration, FTIR experiments were performed in an attempt to observe photo-reduction at the active site. Light titrations, monitored by FTIR difference spectroscopy, were used to follow the active site reduction through frequency shifts in the CO and CN− bands observed after enzyme reduction (Figure 2).
Figure 2.
(a) Light titrations probed by FTIR spectroscopy of 500 μM H2ase, 1 mM QDs and 50 mM ascorbate in 100 mM phosphate buffer pH 7.4. Red = 0 s illumination, purple = 12.5 seconds of illumination. (b) Comparison of peak to peak absorbance difference from ascorbate to dehydroascorbate (blue circles) and Nir-B to Nia-S (red squares). Linear fits from 0 – 10 s of illumination give slopes of 3.3 × 10−8 and 8.5 × 10−8 for H2ase and ascorbate respectively.
Aerobic reduction by QD excitation yields bleached bands at 2091, 2079 and 1944 cm−1. These bands are nearly identical to the CN− and CO frequencies associated with the oxidized Nir-B state of [NiFe] from D. gigas.18 Induced absorbances at 2084 cm−1, 2075 cm−1 and 1930 cm−1 also match well with the one-electron reduced Nia-S state, indicating light initiated formation of the catalytically active state.18 These difference spectra represent the first infrared characterization of [NiFe] H2ase from Thiocapsa roseopersicina and verify its similarity to other well-studied [NiFe] H2ases.18–20 Aerobic reduction typically results in rapid re-oxidation to the Nir-B state in electron rich conditions. 4,21,22 Due to the long time scale (minutes) of the steady state FTIR difference measurements in Figure 2(a), no signal would be observed if this were the case, since the initial and final states would be the same. We thus conclude, either photo-reduction results in rapid and complete reductive O2 consumption in the experimental cell, as has been proposed by Zadvornyy et al., or that re-oxidation is kinetically hindered.11 The formation of further reduced states such as Nia-C and Nia-SR was not observed even for a short illumination times (100 ms), as expected for multi-electron reduction followed by rapid H2 evolution (sub-millisecond) that re-oxidizes the enzyme to Nia-S faster than the timescale of the difference FTIR method.
The consumption of the sacrificial electron donor, ascorbate, and subsequent formation of dehydroascorbate is observable in the mid-IR by following the carbonyl modes. The rate of ascorbate consumption observed (Figure 2(b)) is approximately three times the rate of H2ase reduction. We conclude that the apparent single electron reduction process observed in the steady state FTIR spectrum is actually the end product of a more complex cycle involving full reduction of Nir-B to Nia-SR followed by rapid H2 evolution to form Nia-S. This interpretation is corroborated by anaerobic steady-state light titrations where the enzyme is activated under H2 (SI-6). In these experiments Nia-C is observed to bleach as a function of illumination time with concomitant reforming of the Nia-S state and some Nia-SR along with SED consumption. The net result is the same as the aerobic case: multi-electron reduction results in H2 evolution to re-form the catalytically active oxidized state.
To determine photo-catalytic H2 production efficiency, gas chromatography was used to quantify H2 production (Figure 3). Rapid H2 production is observed with a peak of 81 nmoles produced in 40 s of illumination. Based on the H2 production after absorbance of 2.07 × 1018 photons, 4% of absorbed photons are converted to proton reducing equivalents with an enzyme TON of 92 (explicit calculations of efficiency and TON are laid out in the SI). The efficiency is drastically reduced by electrostatic screening in high ionic strength solutions, as shown in comparison to the same system in artificial seawater (Figure 3). This observation indicates engineering better electrostatic interactions will likely increase the overall efficiency. Relative to similar work in the literature, our system has the disadvantage of an enzyme naturally biased towards H2 oxidation, but with the significant advantages of O2 tolerance and a better basis for mechanistic studies.23 The system also shows some photo-decomposition after long illumination times as observed by Brown et al. for similar systems, likely due to oxidation of surface ligands of the thiolate capped QDs.23
Figure 3.
GC assay of H2 production versus photons absorbed. Red circles represent 1 μM H2ase, 0.5 μM QD, 50 mM ascorbate in 100 mM phosphate buffer pH = 7.4. Blue triangles represent 1 μM H2ase, 0.5 μM QD, 50 mM ascorbate in 50 mM TRIS buffered seawater pH = 7.4.
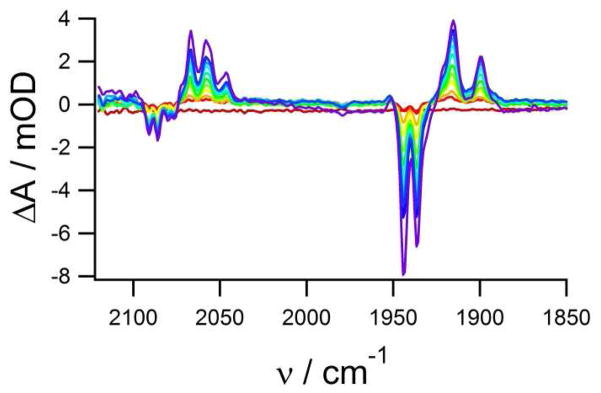
The inability to observe intermediates Nia-C and Nia-SR in the QD-H2ase light initiated difference measurements means that under these conditions turnover is very efficient, consequently there is no buildup of partially reduced intermediates. Since the instantaneous fluence and duration of the laser pulse (10 ns) are large enough to produce multiple excitation and exciton generation/dissociation events (not limited by the rate of oxidation of the SED), we postulate that there are multiple electron transfer events into the protein, resulting in rapid enzyme reduction and turnover. To test this hypothesis, we compared the light driven turnover of H2ase using Ru(bpy)32+, an intrinsic single electron photo-reductant. Light titrations of Ru(bpy)32+ sensitized H2ase shown in figure 4, provide evidence for light-induced production of every known redox intermediate of the enzyme (each CO peak corresponds to a separate state). The amplitudes of the FTIR difference features increase linearly with illumination time over the entire light titration, indicating that the photoreduction rate is constant throughout the titration.
Figure 4.
Laser-induced FTIR light titration of Ru(bpy)32+ sensitized H2ase. 27 mM Ru(bpy)32+, 500 μM H2ase and 100 mM ascorbate in 100 mM phosphate buffer pD = 7.4. Red = dark spectrum, purple = difference FTIR after 12 s of laser illumination.
Two CO bleaches are observed, one corresponding to Nir-B, and one that is 6 cm−1 blue-shifted from the previously observed Nia-S state. This shift is likely due to the spectral crowding of positive features around bleaching bands, which shifts the apparent peak position away from the adjacent positive feature. Bleaching of the CN− bands assigned to Nir-B and Nia-S is also observed, indicating that the bleach at 1937 cm−1 is in fact Nia-S. Induced absorbances are observed at 1915 cm−1 (Nir-S), 1899 cm−1 (Ni-L*) and 1951 cm−1 (Nia-C), with a shoulder growing in at 1921 cm−1 that is associated with the fully reduced state Nia-SR.24,25 A small amount of SED consumption is observed confirming its involvement in the re-reduction of the Ru(bpy)33+, but the amount of ascorbate oxidation is too small to quantify by FTIR spectroscopy. For the reaction conditions employed, the bimolecular reaction of Ru(bpy)33+ with ascorbate is much higher than that of reduction by H2O/OH−(108 s−1 versus 10−3 s−1 for pseudo first order rate constants respectively).26,27 Regeneration of Ru(bpy)32+ may occur on a fast timescale, but due to the lower binding affinity or lower ET efficiency of Ru(bpy)32+-H2ase complexes relative to H2ase-QDs, the likelihood of multiple reduction events from a single Ru(bpy)32+ is very low.
GC analysis of H2 produced using photosensitization with Ru(bpy)32+ (SI-5) shows markedly lower solar to hydrogen conversion efficiency of 0.02%. This is likely due to non-specific binding and possibly preferential electrostatic binding to the large subunit not electronically connected to the active site. No H2 production is observed without ascorbic acid, H2ase or Ru(bpy)32+, evidence that each component is obligatory for H2ase turnover.
The key differences between the QD and Ru(bpy)32+ photo-driven enzyme reduction can be summarized as follows: First, QD binding is electrostatic, is screened at high salt concentration and quenches QD PL. Ru(bpy)32+ is non-specifically bound or bound in ET inactive sites on the enzyme surface, based on homology modeling and PL titrations (data not shown). Secondly, H2 production with QDH2ase hybrids is reasonably efficient, whereas Ru(bpy)32+ is 100x less efficient per photon absorbed. And finally, light induced difference FTIR measurements shows very different populations of intermediate states. Photo-reduction with QDs results in formation of the Nia-S state only whereas Ru(bpy)32+ generates all known steady state intermediates. These are fundamentally different end points in photo-reduction, and the Ru(bpy)32+ spectra don’t evolve to the QD-H2ase spectra under long illumination times. The accumulation of a distribution of intermediates is correlated with inefficient enzyme turnover.
The origins of the light titration differences are not completely understood, but certainly have to do with the fundamental differences between Ru(bpy)32+ and QDs. One possible explanation is that the observed differences are purely a consequence of the mode of photosensitizer binding. Homology modeling suggests positively charged Ru(bpy)32+ may bind non-preferentially for ET. This non-preferential binding could make observation of intermediates much more likely since the fundamental reduction events are slower. The mode of binding may also influence the flux of electrons entering the enzyme through the FeS chain versus a more direct route, which in turn could influence the turnover, for example by modulating the efficiency of coupled proton transfers. Finally, QDs may be capable of delivering multiple electrons from multiple photons without requiring hole filling.28,29 In contrast, Ru(bpy)32+ can only deliver a single electron and then must be regenerated by the SED (no faster than bimolecular diffusion). Thus QDs may produce multiple reducing equivalents on a timescale that is fast relative to the TOF of the enzyme. This observation raises the possibility that efficient turnover requires fast multi-electron reduction, and that the partially reduced steady state intermediates are a consequence of slow single electron reduction and are not productive. Further experiments will be required to determine the source of the observed differences.
In summary we have presented direct spectroscopic and chromatographic evidence of efficient QD photo-driven enzyme reduction and H2 production using an O2 tolerant [NiFe] H2ase. We have also demonstrated the power of these QD-H2ase assemblies for studying very fast and complex redox chemistry of enzymes using light triggers that could open up new doors for sub-turnover temporal spectroscopic resolution. The strikingly different photo-reduction behaviors observed between QD and Ru(bpy)32+ sensitized H2ases are likely due to multi-photon, multi-electron pathways in QD assemblies that are not possible in the case of Ru(bpy)32+. We intend to further elucidate the mechanism of light-driven H2ase turnover using time resolved IR and transient absorbance experiments capable of directly probing ET rates and catalytic intermediates.
Supplementary Material
Acknowledgments
Funding Sources
This project was funded by U. S. Army Research Laboratory and the U. S. Army Research Office under grant number 54635CH as well as the NIH under grant number GM068036.
The authors would like to thank Julius O. Campeciño for protein purification work.
ABBREVIATIONS
- FTIR
Fourier transform infrared
- H2ase
hydrogenase
- FeS
iron sulfur cluster
- QD
quantum dot
- PL
photoluminescence
- PLQE
photoluminescence quantum efficiency
- TON
turnover number
- SED
sacrificial electron donor
- ET
electron transfer
- MPA
mercaptopropionic acid
- [NiFe]
nickel iron active site
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors claim no conflicts of interest
Supporting Information. Experimental details on homology modeling, QD synthesis, H2ase expression and purification, GC calibration, Ru(bpy)32+ photo-catalytic H2 production, anaerobic FTIR light titrations and explicit efficiency calculations can be found in the SI. This material is available free of charge via the Internet at http://pubs.acs.org.
Supporting Information Placeholder
References
- 1.Fisher HF, Krasna AI, Rittenberg D. J Biol Chem. 1954;209:569. [PubMed] [Google Scholar]
- 2.Ludwig M, Cracknell JA, Vincent KA, Armstrong FA, Lenz O. J Biol Chem. 2009;284:465. doi: 10.1074/jbc.M803676200. [DOI] [PubMed] [Google Scholar]
- 3.Pandelia ME, Nitschke W, Infossi P, Giudici-Orticoni MT, Bill E, Lubitz W. P Natl Acad Sci USA. 2011;108:6097. doi: 10.1073/pnas.1100610108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gogotov IN, Zorin NA, Serebriakova LT, Kondratieva EN. Biochim Biophys Acta. 1978;523:335. doi: 10.1016/0005-2744(78)90036-0. [DOI] [PubMed] [Google Scholar]
- 5.Shomura Y, Yoon KS, Nishihara H, Higuchi Y. Nature. 2011;479:253. doi: 10.1038/nature10504. [DOI] [PubMed] [Google Scholar]
- 6.Brown KA, Dayal S, Ai X, Rumbles G, King PW. J Am Chem Soc. 2010;132:9672. doi: 10.1021/ja101031r. [DOI] [PubMed] [Google Scholar]
- 7.Reisner E, Powell DJ, Cavazza C, Fontecilla-Camps JC, Armstrong FA. J Am Chem Soc. 2009;131:18457. doi: 10.1021/ja907923r. [DOI] [PubMed] [Google Scholar]
- 8.Krassen H, Schwarze A, Friedrich B, Ataka K, Lenz O, Heberle J. Acs Nano. 2009;3:4055. doi: 10.1021/nn900748j. [DOI] [PubMed] [Google Scholar]
- 9.Nedoluzhko AI, Shumilin IA, Nikandrov VV. J Phys Chem-Us. 1996;100:17544. [Google Scholar]
- 10.Cuendet P, Rao KK, Gratzel M, Hall DO. Biochimie. 1986;68:217. doi: 10.1016/s0300-9084(86)81086-0. [DOI] [PubMed] [Google Scholar]
- 11.Zadvornyy OA, Lucon JE, Gerlach R, Zorin NA, Douglas T, Elgren TE, Peters JW. J Inorg Biochem. 2012;106:151. doi: 10.1016/j.jinorgbio.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Lubner CE, Grimme R, Bryant DA, Golbeck JH. Biochemistry-Us. 2010;49:404. doi: 10.1021/bi901704v. [DOI] [PubMed] [Google Scholar]
- 13.Madden C, Vaughn MD, Diez-Perez I, Brown KA, King PW, Gust D, Moore AL, Moore TA. J Am Chem Soc. 2012;134:1577. doi: 10.1021/ja207461t. [DOI] [PubMed] [Google Scholar]
- 14.Ogata H, Kellers P, Lubitz W. J Mol Biol. 2010;402:428–444. doi: 10.1016/j.jmb.2010.07.041. [DOI] [PubMed] [Google Scholar]
- 15.Sali A, Blundell TL. J Mol Biol. 1993;234:779. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 16.Mattoussi H, Mauro JM, Goldman ER, Anderson GP, Sundar VC, Mikulec FV, Bawendi MG. J Am Chem Soc. 2000;122:12142. [Google Scholar]
- 17.Ipe BI, Niemeyer CM. Angew Chem Int Edit. 2006;45:504. doi: 10.1002/anie.200503084. [DOI] [PubMed] [Google Scholar]
- 18.deLacey AL, Hatchikian EC, Volbeda A, Frey M, FontecillaCamps JC, Fernandez VM. J Am Chem Soc. 1997;119:7181. [Google Scholar]
- 19.Bleijlevens B, van Broekhuizen FA, De Lacey AL, Roseboom W, Fernandez VM, Albracht SPJ. J Biol Inorg Chem. 2004;9:743. doi: 10.1007/s00775-004-0570-z. [DOI] [PubMed] [Google Scholar]
- 20.Bagley KA, Duin EC, Roseboom W, Albracht SPJ, Woodruff WH. Biochemistry-Us. 1995;34:5527. doi: 10.1021/bi00016a026. [DOI] [PubMed] [Google Scholar]
- 21.Jones AK, Lamle SE, Pershad HR, Vincent KA, Albracht SPJ, Armstrong FA. J Am Chem Soc. 2003;125:8505. doi: 10.1021/ja035296y. [DOI] [PubMed] [Google Scholar]
- 22.Armstrong FA, Albracht PJ. Philos T Roy Soc A. 2005;363:937. doi: 10.1098/rsta.2004.1528. [DOI] [PubMed] [Google Scholar]
- 23.Brown KA, Wilker MB, Boehm M, Dukovic G, King PW. J Am Chem Soc. 2012;134:5627. doi: 10.1021/ja2116348. [DOI] [PubMed] [Google Scholar]
- 24.Whitehead JP, Gurbiel RJ, Bagyinka C, Hoffman BM, Maroney MJ. J Am Chem Soc. 1993;115:5629. [Google Scholar]
- 25.Kellers P, Pandelia ME, Currell LJ, Gorner H, Lubitz W. Phys Chem Chem Phys. 2009;11:8680. doi: 10.1039/b913635e. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh PK, Brunschwig BS, Chou M, Creutz C, Sutin N. J Am Chem Soc. 1984;106:4772. [Google Scholar]
- 27.Macartney DH, Sutin N. Inorg Chim a-Article. 1983;74:221. [Google Scholar]
- 28.Vanhouten J, Watts RJ. J Am Chem Soc. 1976;98:4853. [Google Scholar]
- 29.Zhu H, Song N, Rodriguez-Cordoba W, Lian T. J Am Chem Soc. 2012;134:4250. doi: 10.1021/ja210312s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.