Abstract
Data processing in proteomics can be a challenging endeavor, requiring extensive knowledge of many different software packages, all with different algorithms, data format requirements, and user interfaces. In this article we describe the integration of a number of existing programs and tools in Taverna Workbench, a scientific workflow manager currently being developed in the bioinformatics community. We demonstrate how a workflow manager provides a single, visually clear and intuitive interface to complex data analysis tasks in proteomics, from raw mass spectrometry data to protein identifications and beyond.
Scientific workflow managers have become popular in bioinformatics as they are well-suited for assembling different specialized software modules or scripts into an overall data flow, typically a directed acyclic graph, taking the data through consecutive steps of analysis. Additionally, workflow managers such as Kepler (1, 2) and Taverna Workbench (3, 4) provide visualization and a graphical user interface for designing and executing analytical process flows.
Data analysis in mass spectrometry and proteomics is inherently a multistep process, typically involving several of the following steps: m/z calibration, chromatographic time alignment, intensity normalization, compound identification by database search or de novo interpretation, quantitation, addressing the protein inference problem, merging and/or comparing data from different samples or time points, multivariate statistics, and mapping of data to Gene Ontology annotations or biological networks.
Many free (both as in “beer” and as in “speech”) modules and software for analyzing peptide and protein mass spectrometry data already exist. For instance, more than 150 different algorithms and software packages are categorized and listed on http://en.wikipedia.org/wiki/Mass_spectrometry_software and http://www.ms-utils.org. Matthiesen has reviewed algorithms and software for analysis of mass spectrometry data and computational proteomics (5), as well as freely available software tools (6). In this environment of many existing, more or less specialized tools, scientific workflow managers are directly applicable and useful. Here we will describe a number of ways in which existing and novel proteomics analysis scenarios can be implemented in a scientific workflow manager.
Theoretical Background
Workflows
A workflow describes a collection of processing units connected to each other through data and control connections, together constituting a process. In a workflow, inputs are processed in consecutive steps by processing units, whereby data flows from one processing unit to another until all processing units have finished their task and the output is finalized.
Processing units in a workflow are responsible for generating, modifying and/or consuming data. They represent conventional software, such as algorithms or binary executables, as well as software for the retrieval of data from specialized hardware, e.g. a mass spectrometer. Data that is to be consumed or modified is received through input ports, and generated or modified data is returned through output ports.
Processing units are bound together by data and/or control connections. Data connections are responsible for transporting data from the output port of one processing unit to the input port of another. Despite their seemingly passive task, they are active entities responsible for adapting or wrapping data from the output port data format to the format defined by the input ports. Control connections are connections that police data and processing flows. Using logical control mechanisms, these connections are able to steer the data flow within a workflow, much like a train switch is able to steer the direction of a train.
The rationale behind using workflows and flexible data formats for stringing together independent modules is akin to core aspects of the Unix philosophy expressed by McIlroy (7) in 1978 as (1) “Make each program do one thing well. To do a new job, build afresh rather than complicate old programs by adding new features.” and (2) “Expect the output of every program to become the input to another, as yet unknown, program. Don't clutter output with extraneous information. Avoid stringently columnar or binary input formats. Don't insist on interactive input.”
In the bioinformatics domain, workflows and workflow management systems, which are computer systems responsible for the design, execution, and monitoring of a workflow, have become increasingly popular and are already widely used in many research groups. Most scientific data analyses are conducted by gathering data followed by a set of data processing steps, such as filtering, transformation, statistical analysis and data mining. Workflows enable the user to automate those steps in a simple, comprehensive, and visual manner.
Additionally, workflows allow for easy recreation, reuse, verification and augmentation of experiments or analyses by others. Most workflow managers allow workflows and user-defined inputs to be stored and loaded, thus each execution of the workflow on the same data produces the same results, provided no randomization or modification in processing units is introduced. Furthermore, workflows promote reuse, because workflow management systems allow embedding of workflows. This saves the researcher from having to recreate a workflow when it only needs to be extended with additional pre- or postprocessing steps. Embedding of workflows allows for abstraction as well, (graphically) hiding the details of embedded workflows within the larger workflow. This increases the understandability of a workflow.
Finally, some scientific workflow managers support methods and protocols for easy and transparent parallel and remote computing, which may speed up workflow execution significantly. By using parallel computing, multiple processing units can do their work without having to share or compete for processor resources, removing bottlenecks in the analysis. Supporting remote computing allows processes that require significant computation time, access to large databases, or configuration and installation of large software packages to be run on a remote location, lessening the burden on the local work station. This enables the utilization of computer clusters or other specialized hardware, such as a Field Programmable Gate Array (8), without requiring the workflow user to have in-depth knowledge of operating such hardware.
Over time, a considerable number of workflow management systems have been developed for bioinformatics (9), some of which were recently discussed and compared by Curcin and Ghanem (10). We have chosen the Taverna Workbench workflow management system on the basis of its wide range of protocol support for both local and remote computing, its ease of use, and the large shared content provided by its online community.
Alternatives to Workflows
Workflows are but one of many means by which analyses can be performed. Depending on factors such as computer experience, required performance, professional environment and functional requirements, other automated analyses methods, i.e., scripting languages or dedicated packages may also be used. We will briefly discuss and compare these alternatives with workflows, keeping in mind that most workflow managers support one or more scripting languages.
Scripting languages are high-level programming languages that allow the user to program control over applications and other functionalities such as in- and output, user interface, Internet and networking, and operating system. Although scripting languages resemble conventional programming languages to a great deal, the main difference between them is that programming languages are compiled before execution, whereas scripting languages are interpreted during execution. This usually makes scripts a little slower in execution, but gives them greater runtime flexibility. Examples of well-known scripting languages include AWK (11), JavaScript, Perl (12), PHP (13), and Python (14).
Dedicated packages are software that assist or guide the user in a small number of domain-specific tasks. The software is created to assist users in common tasks within a restricted knowledge domain, and usually refers to concepts within that knowledge domain using specific ontologies or jargon. In proteomics, a well-known specialized software package is the TPP1 (15), which assists the user in the identification, validation, and quantification of proteins, as well as interpretation of results.
When performing analyses or in silico experiments, the researcher has the choice between using workflows, using scripts and when available, specialized software. Intrinsically, this choice is often guided by expertise and personal preferences. External factors often play a role as well; corporations and research facilities usually set a standard for experimentation, whereby tools for experimentation and analysis are often predefined.
When choosing between specialized software, scripting, or workflows for solving a particular data analysis problem, there are a number of attributes that are important to keep in mind, such as user-friendliness, learnability, performance, and modifiability. Table I shows a comparison between the three types of automated analysis with respect to these attributes.
Table I. Attribute scores for workflows and workflow alternatives. + indicates poor, ++ indicates average and +++ indicates good performance in a certain area.
| Attributes | Dedicated software | Scripting | Workflows |
|---|---|---|---|
| User-friendliness | ++/+++ | + | +++ |
| Learnability | +++ | + | ++ |
| Performance | ++ | +++ | ++ |
| Modifiability | + | +++ | +++ |
The user-friendliness of an analysis method is apart from its complexity also partially dependent on its GUI. Because scripting languages usually do not offer much more than a command-line interface, the user-friendliness of scripting languages is generally poor. User-friendliness of specialized software ranges between average and good; they usually come with a GUI, but can be very complex, because of the potentially great number of options available, Most workflow management systems have a strong emphasis on a good and simple GUI, thus user-friendliness is generally good.
Considering learnability, scripting is the most complex of all three options. Scripting languages are very technical and mathematical in nature, and mastering scripting languages requires a considerable time investment before they are well-understood. Workflows and workflow managers are easier to learn, partially because of the better user interface and the fairly limited amount of basic structures offered. The learnability of specialized systems is generally the best of the three, because they have reasonably good user interfaces and only automate a restricted amount of tasks, which are often already familiar to the users.
With respect to the execution performance, scripts are often the best choice because of the relatively little overhead. Most specialized programs and workflows have some graphical feedback or result presentation functionality to promote user-friendliness at the expense of minor performance losses.
Finally, given a script, saved workflow, or stored experiment, modifiability indicates how well this experiment can be modified. When the experiment is carried out on another data set of a similar type, this is usually not a problem. However, extending an experiment is usually not possible in dedicated software if the extension is not supported. In contrast, scripts and workflows can easily be extended for new functionalities by respectively adding a new command in the script or inserting a new processing unit in the workflow.
Given the lack of poor points combined with good user-friendliness and modifiability, we perceived workflows as the good way to perform and explore new concepts for complex data analysis in proteomics.
Remote and Distributed Computing
Remote computing is the accessing of one or more computers over a network in order to perform tasks without having physical access to those computers. Remote computing can be achieved quite easily because of the emergence of Web services, software systems designed to support interoperable machine-to-machine interaction over a network (16). With Web services, different forms of remote computing, e.g. remote procedure calls or service invocations can be done in a standardized way by using interface standards and data transport standards. The popularity of Web services is partially caused by their simplicity, but also due to the wide vendor support, both in service availability and in development tools, allowing nonexperienced users and developers to use and develop remote services.
Currently there are two styles of Web services that are widely supported: nonRESTful services and RESTful services. The nonRESTful services are Web services that depend on the Web service Description Language (17) (WSDL) standard to describe their interfaces, and the Simple Object Access Protocol (18) (SOAP) standard for communication between client and server. RESTful services do not rely on these standards, but are based on representational state transfer (19) (REST) whereby a document that represents a resource to be modified or used is passed between client and server.
Related to remote computing is the concept distributed computing, which is the term for processing a task or program by using multiple computers that communicate with each other over a network. Distributed computing is often employed to speed up computationally intensive tasks by splitting those tasks up in multiple, independent subtasks, and remotely compute those in parallel on different computers.
A popular form of distributed computing is grid computing. In essence, a computer grid is a collection of computers that are combined and connected to each other to work together on one or a few processing tasks. Each computer in the grid is assigned to work on a part of the program workload until all processing is done. Over time, a number of open source software packages have been written for the creation of grids (20, 21). Within the field of proteomics, several efforts have been made in grid-based proteomics in data sharing (22) and mass-parallelism in protein identification (23).
Recently, grid computing technology and Web service technology have been combined into a new way of computing called cloud computing. With cloud computing, software packages themselves no longer just incorporate services, but are services themselves that can be retrieved over the Internet from clouds, which are networks of servers managed by a third party. Cloud computing combines the mass parallelism of grid computing with the standardized remote access methods of Web services, allowing clients to deploy and access that software over the Internet without having to install software locally or invest in servers or grids. Currently there are a number of big corporations that offer public cloud services, including Amazon Elastic Compute Cloud (EC2) (24) and IBM's Smart Cloud (25).
Taverna Workbench
The Taverna Workbench, hereafter referred to as Taverna, is an open-source and domain independent workflow management system created by the myGrid team. Taverna has a myriad of distinctive features that enable easy design as well as fast execution of workflows, and incorporates support for many remote and parallel computing protocols. We will discuss the main features below; for a more extensive list of features supported by Taverna we refer to the online introduction (26).
The Taverna interface is easy to understand and use. Workflows can be created in a fast and efficient way through point-and-click or drag-and-drop, and the user can choose the level of detail in which a workflow is displayed. Apart from a graphical representation of the workflow, a separate window displays detailed information on all the workflow elements, i.e. processing units, connections, input ports, and output ports. Taverna incorporates a substantial amount of predefined processes, which perform basic tasks such as reading or writing files, executing programs, list handling, and string handling. In case a more complex local processing unit is needed, Taverna offers support to modify or create new services through scripts written in BeanShell, a lightweight scripting language for the Java programming language (27). With basic knowledge of Java and BeanShell functional libraries, the user can easily implement a variety of tasks. However, using workflows in Taverna does not require any knowledge of programming or scripting languages.
For remote processing, Taverna provides access to a set of remote services, such as Biomart (28), Biomoby (29), Soaplab (30), and Web services hosted by organizations such as EBI and NCBI. As of Taverna version 2.3.0 these services, as well as a variety of other life science Web services, are all grouped in the Service Catalog, a module that lists all the services available in the BioCatalogue project (31). In addition, Taverna supports the local or remote execution of RShell code (32, 33), providing access to a variety of statistical functions, both for general purpose and for bioinformatics, such as those included in the Bioconductor package (34). The incorporation of R also benefits performance, as there is a framework for parallel execution of algorithms in R (35). Beyond the freedom to design and execute one's own workflows, Taverna is integrated with an online community called myExperiment (36), where users can share workflows and other resources.
To illustrate the use of scientific workflow managers in proteomics, we have implemented a number of existing computational workflows for analysis of mass spectrometry and proteomics data in Taverna, such as a subset of TPP functionalities, and extended these workflows with additional analyses steps. To enable existing software modules such as msalign (37) or recal (38) to function in a Taverna workflow, minor modifications had to be made to some of these modules, including the standardization of input and output data formats. For simplicity and compatibility, we have chosen to work with open formats such as mzXML (39) for mass spectrometry data and pepXML/protXML (15) for peptide/protein identification and quantitation.
Example Workflows and Results
In this section we will discuss four workflows for analysis of mass spectrometry and proteomics data, whereby each succeeding workflow extends the previous one. The first workflow automates the process of peptide identification from liquid chromatography-tandem MS (LC-MS/MS) data in mzXML. The second workflow aligns and combines an LC-MS/MS data set with an accurate mass LC-MS data set. The third workflow recalibrates the LC-MS data set using peptides identified by LC-MS/MS for the best possible mass measurement accuracy. Finally, the fourth workflow converts raw FTICR and ion trap (IT) data to the mzXML standard, and executes Workflow 3 for a user-defined number of times to optimize recalibration. Note that all the workflows in the examples perform all processing locally. For a brief description of methods for incorporating remote processing of individual workflow services, see the Appendix.
For the reader to use the workflows described below, we have prepared several tutorials to guide the reader in the installation and use of Taverna, TPP and the discussed workflows. Installation of all required applications as well as the workflow package is described in Tutorial S1. Tutorial S2 provides a basic overview of Taverna, and demonstrates how users can load and execute workflows. For those readers that are interested in creating their own workflows in Taverna, we refer to Tutorial S3.
Workflow 1: From LC-MS/MS Data to Peptide Identifications
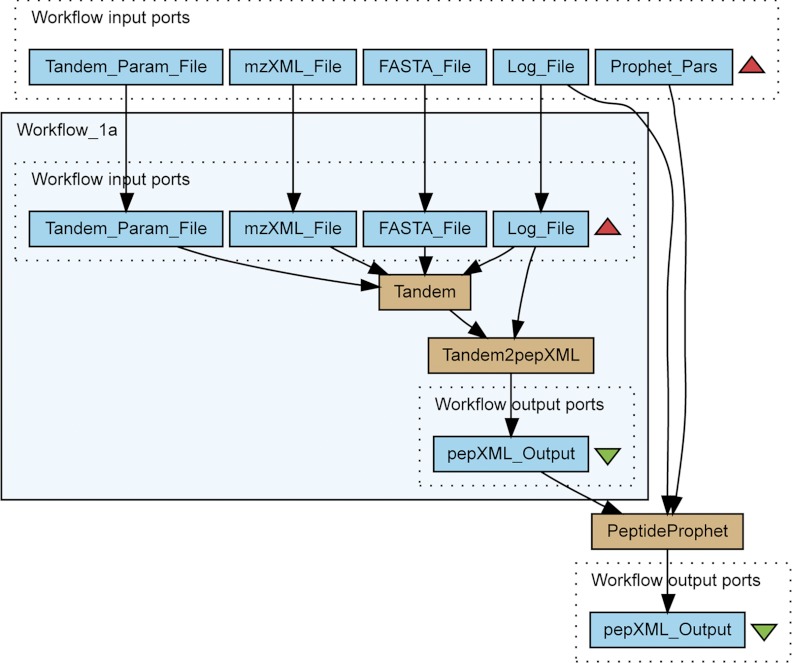
In the first workflow we automate one of the most frequently performed tasks in proteomics: the identification of peptides and proteins from LC-MS/MS data. There are a number of different programs that can perform this kind of identification, such as SEQUEST (40), Mascot (41), X!Tandem (42), OMSSA (43), MassMatrix (44), Crux (45), and MS-GFDB (46), to name only a few. Because X!Tandem is open source and already comes preinstalled in the TPP, we chose to use this search engine in the workflows described here. The resulting Workflow 1 is shown in Fig. 1.
Fig. 1.
Protein Identification and format conversion from LC-MS/MS data. The workflow contains three processing elements. The first element is the Tandem component that calls the X!Tandem executable. When completed, the resulting data is forwarded to the second processing unit, Tandem2XML, which executes the Tandem2XML tool from the TPP package. The resulting pepXML file is then transferred to the PeptideProphet component. Finally, a statistically validated pepXML file is returned to the user.
This workflow will seem familiar to TPP users, since it mimics the first steps of a typical protein data analysis in TPP. The main difference is that in Taverna workflows, user-defined inputs for all steps of the workflow are provided before execution, and each subsequent step is performed automatically.
Workflow 1 contains five input ports, namely Tandem_Param_File, mzXML_File, FASTA_File, Log_File and Prophet_Pars. The Tandem_Param_File port contains the location of the file configuration parameters for X!Tandem, which is by default in BioML (47) format. Similarly, the mzXML_File port contains the location of the mzXML file used for analysis; the FASTA_File port contains the file location of the protein database in FASTA format, and the Log_File port contains the location of the log file that keeps a record of all workflow operations. The Prophet_Pars port contains a string with command-line parameters for the PeptideProphet service. The workflow has only one output port, pepXML_Output, which contains the location of a pepXML file with the statistically validated peptide and protein identifications.
To demonstrate the extensibility of Taverna workflows we divided the functionality of Workflow 1 into two parts; Workflow_1a, which is the embedded workflow, is responsible for protein identification with X!Tandem in the Tandem service and the transformation of its results to a standard pepXML format via Tandem2pepXML. The embedded workflow functionality is extended by statistical validation of identification results in PeptideProphet (48). PeptideProphet takes as input the peptide-spectrum assignments produced by the embedded workflow, converts the raw database search scores into a discriminant score and models the two score distributions of true and false positive matches. These models, specifically the ratio of the true to the sum of the true and false distributions at a given discriminant score, are then used to estimate the probability for a given peptide-spectrum match being correct. As output, PeptideProphet returns a pepXML file containing the peptide-spectrum matches with estimated confidences.
Now we have a basic workflow for searching an LC-MS/MS data set in mzXML against a FASTA sequence file using X!Tandem, translating the results to pepXML and estimating confidences in each peptide-spectrum assignment using PeptideProphet. We can use this as a module in larger and more complex analysis workflows.
Workflow 2: Aligning Ion Trap Tandem Mass Spectrometry and Accurate Mass Data
We have previously described how LC-MS and LC-MS/MS data sets can be combined through chromatographic alignment (37, 49). There are at least two reasons for aligning low resolution ion trap MS/MS data with accurate, high resolution MS data: to replace the peptide precursor mass values in the ion trap MS/MS data with more accurate ones from the MS-only data (37, 49) and using the high-resolution MS profiles for more precise quantitation (50, 51).
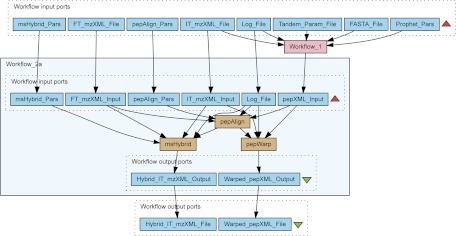
In order to align LC-MS/MS data with the accurate MS data from e.g. a TOF or an FTICR mass spectrometer, we modified and updated the msalign program. This version, renamed to pepAlign, linearly aligns MS data with peptide MS/MS data. New features include alignment using scan numbers or retention time, a faster genetic algorithm and the option to generate a gnuplot (52) data file in order to graphically depict and inspect the alignment. Once the alignment is complete, the resulting PLF is forwarded to two services, pepWarp and msHybrid, which are executed in parallel. msHybrid replaces the precursor m/z for the MS/MS scans with the more accurate values from accurate MS data. Simultaneously, pepWarp uses the PLF to modify the retention times of peptides in the pepXML file according to the chromatographic alignment, linking the two data sets and enabling the use of high-resolution MS data for quantitation (50, 51). Workflow 2 in Fig. 2 shows the process described above.
Fig. 2.
Aligning ion trap tandem mass spectrometry and accurate mass data. Workflow 2 aligns ion trap tandem mass spectrometry and accurate mass data. The alignment proceeds through the pepAlign processing unit, which produces a piecewise linear function representing the alignment, and passes it on to both pepWarp and msHybrid. The msHybrid processing unit is responsible for creating a new hybrid ion trap/FTICR mzXML file based on this alignment, whereas the pepWarp component applies the alignment to warp retention times in the pepXML file resulting from Workflow 1.
Workflow 3: Internal Calibration of FTICR Data Using Identified Peptides
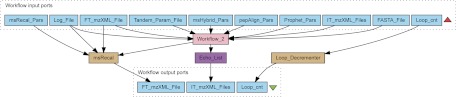
The third workflow, depicted in Fig. 3, introduces the msRecal algorithm for automatic internal calibration (38, 49) of high-resolution FTICR data. Provided a suitable recalibration function, this could in principle be any accurate and high-resolution data, for instance from a TOF or Orbitrap™ instrument.
Fig. 3.
Internal calibration of FTICR data using identified peptides. Workflow_3 uses the msRecal component for automatic internal calibration of the high-resolution MS data, here from an FTICR. A set of identified peptides from one or more aligned LC-MS/MS data sets are used for automatic internal recalibration (38) of the FT_mzXML_File data set.
The msRecal service uses the output of Workflow 2, a set of identified peptides with their retention times aligned to the FT_mzXML_File, to individually recalibrate each mass spectrum in the FT_mzXML_File using the least-squares minimization and outlier removal method previously described (38). As an output it produces an mzXML file with recalibrated m/z axes for each spectrum where possible. The Loop_cnt port enables the workflow to be iterated by a user-specified number of times.
Workflow 4: From Raw Data Sets to Iterated Recalibration
Most mass spectrometer vendors use proprietary data formats. Some manufacturers may provide a library for reading the raw data, but these typically require a commercial software license. Although some proprietary formats may be easy to reverse-engineer, this would not provide a forwards-compatible solution. Some vendors provide utilities that convert their proprietary data format to open formats such as mzXML or mzML. There is also third-party software using the vendor libraries to convert raw data into mzXML, such as ReAdW (for Thermo Fisher/Xcalibur data), trapper (Agilent/MassHunter), massWolf (Waters/MassLynx) and mzWiff (Applied Biosystems/Analyst) (53). The resulting data from all recent Bruker Daltonics electrospray instruments can be converted to mzXML or mzML by compassXport (54).
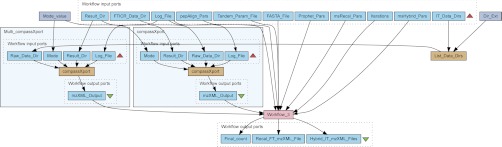
In Workflow 4, illustrated in Fig. 4, there are two instances of the compassXport service, once for the FTICR raw data, and once for the directory containing one or multiple raw ion trap LC-MS/MS data sets. To prevent the user from having to specify all the individual ion trap data sets, we inserted the List_Data_Dirs processing unit which, given a directory, finds all the raw data set subdirectories. Furthermore, the input Result_Dir contains the user-specified storage location for files generated during the workflow execution. The embedded workflow Workflow_3 was configured to stop execution when the user-specified counter Iterations reaches zero, at which point execution of Workflow 4 ends as well. By using loops in a workflow, one can easily make iterative refinements to an analysis.
Fig. 4.
Internal calibration of FTICR data using peptides identified by ion trap. The fourth and last workflow contains two instances of the compassXport service that handle the conversion of raw data to the mzXML standard. The service executes the compassXport program, after which the output is forwarded to the embedded Workflow_3. Workflow_3 is then iterated for a user-defined amount of times, using the output values of a previous iteration as inputs for the next. When the number of iterations has been reached, the output is returned to the user.
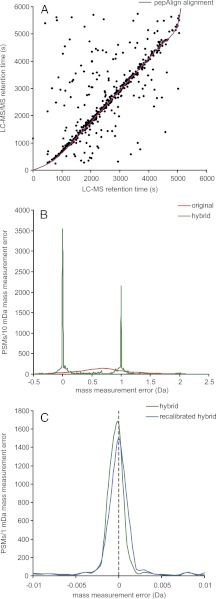
The results of applying Workflow 4 to some real ion trap and FTICR data sets is shown in Fig. 5.
Fig. 5.
Examples of workflow outputs. A pepAlign alignment of a single ion trap LC-MS/MS data set of an E. coli whole cell lysate with a high-resolution FTICR data set of a similar sample (A). After alignment, the accurate precursor mass information from the FTICR data set can be combined with the ion trap MS/MS data into a hybrid data set, resulting in drastically smaller mass measurement errors (B), here from 410 ± 330 ppm in 14,200 peptide-spectrum matches from LC-MS/MS data from 24 SDS-PAGE fractions of the same E. coli sample with PeptideProphet p > 0.95 (1.2% estimated FDR) to −0.24 ± 0.46 ppm (if considering only the central error distribution within ±1.3 ppm) of 14,247 matches with p > 0.95. Without a procedure to relocate the monoisotopic peak, there is often a +1 Da error. However, many search engines, including Mascot and X!Tandem, can allow for this when matching measured and theoretical peptide precursor masses. As would be expected, there is still a small systematic (average) error from the externally calibrated FTICR data. This systematic error can be removed by internally calibrating the FTICR spectra using peptides identified in the ion trap, reducing the relative mass measurement error to −0.03 ± 0.55 ppm, for 14,011 matches with p > 0.95 still keeping spectra that could not be recalibrated because of a lack of acceptable internal calibrants (C). Workflow 4 can be iterated an arbitrary number of times, although the improvements after the second hybrid data set with recalibrated FTICR data will be negligible in most cases. All data was searched against the UniProt E. coli K12 (MG1655) FASTA sequence database (20080926) containing 4,347 protein sequences, allowing for ±0.5 Da and parent isotope error, 0.4 Da fragment monoisotopic mass error, two missed cleavages and assuming carbamidomethylation of cysteines and proteolytic cleavage C-terminally of arginine and lysine but not N-terminally of proline.
DISCUSSION
The successful, straightforward implementation of existing and novel proteomics data processing workflows in Taverna demonstrates the applicability and soundness of this approach to proteomics bioinformatics. Although involving a short learning curve, workflow managers enable most biologists to process their data in a systematic and comprehensible manner. By hiding the details pertaining to data and input/output formats, the user can focus on the overall information flow and purpose of the analysis. Finally, and on a more philosophical note: the divide-and-conquer strategy of deconstructing a complicated analysis task into small and easily managed components which are then assembled into task-specific workflows may be said to take a page from McIlroy's first principle. The ambition that each algorithm should be implemented with the expectation to work with input from another program and provide output to yet another software using a standardized but extensible XML-based data formats even more strikingly follows the second principle: expecting the output of every program to become the input to another, avoiding stringently columnar or binary input formats (e.g. by using XML formats) while not insisting on interactive input.
Although the workflows described here all use X!Tandem, this search engine could easily be interchanged for any other that accepts data in mzXML or mzML, and outputs peptide spectrum assignments in pepXML or mzIdentML. All workflows and modules described in this article and the data used to test them are available on www.ms-utils.org/Taverna.
Acknowledgments
We thank Dr. Alex Henneman, Dr. Yassene Mohammed, Mr. Rob Marissen, Ms. Ekaterina Mostovenko, and Mr. Bjorn Victor for fruitful discussions on workflow design, Taverna, remote computing, and helpful comments on the manuscript.
APPENDIX
In this article we have mentioned several standards for remote and distributed computing, and stated how Taverna incorporates facilities for using these in workflows. Although a thorough treatise of these technologies or a tutorial on the implementation of remote, distributed versions of workflow services is beyond the scope of this article, this appendix provides a few examples on how we have implemented remote and distributed services for Taverna in our labs. These methods are well-tested and are already used in the analysis of large data sets.
Web services can be implemented in different ways. If a program is a command-line executable such as the tools used in the workflows presented in this article, the easiest way to implement a Web service is to wrap the executable in a Web service layer: On the server side, a Web service is implemented that makes a system call to run the executable with the parameters supplied via the service interface. For example, X!Tandem can be implemented as a Web service using the Axis2/C platform (55) to call the X!Tandem executable in its installation directory. An automatic WSDL generator is then used to generate a WSDL file, which exposes its interface. Finally, the service is started on the Apache web server (56), after which the Web service is then ready to be added to Taverna by supplying its network address. Note that the interface of the Web service would differ slightly from the local X!Tandem service, because we can no longer specify locations of files as parameters, but need to specify the contents of those files, upload them to the server, or supply the location where they are remotely accessible.
Web service technology is a convenient way to use remote computing. However, remote computing only increases workflow performance if the time saved by remote processing is greater than the time spent on uploading files and downloading the results, and any overhead from splitting data files and merging results. This is certainly an issue for the X!Tandem application, where the sizes of the mzXML files are often in the 100 MiB–1 GiB range. Of course, X!Tandem processing time is not solely a function of data size, but also the search parameters; if searching against a large sequence database with relatively unspecific enzyme rules and a wide mass measurement error tolerance, even a 100 MiB mzXML file can take several hours to process on a single CPU. Using parallelism to speed up the X!Tandem search can thus save a considerate amount of overall processing time, as was shown earlier by Bjornson et. al (23). When a cluster of machines is available, the parallel X!!Tandem version can be wrapped in a Web service, as discussed previously, enabling the access of a remote, transparently distributed version of Tandem in Taverna.
Footnotes
 This article contains supplemental Tutorials S1 to S3.
This article contains supplemental Tutorials S1 to S3.
AUTHOR CONTRIBUTIONS: MP and AMD initiated the work and previously adopted or developed the modules and pipelines. JSB wrote new I/O libraries for reading mzXML and pepXML, modified the existing modules and implemented the workflows in Taverna. MP and JSB prepared the manuscript and wrote accompanying tutorials, AMD critically revised it and all authors read the final manuscript.
1 The abbreviations used are:
- TPP
- Trans-Proteomic Pipeline
- GUI
- Graphical user interface
- IT
- Ion trap
- PLF
- Piecewise linear function.
REFERENCES
- 1. Altintas I., Berkley C., Jaeger E., Jones M., Ludascher B., Mock S. (2004) Kepler: an extensible system for design and execution of scientific workflows. Scientific and Statistical Database Management. 16th International Conference on Scientific and Statistical Database Management, pp. 423–424 [Google Scholar]
- 2. Barseghian D., Altintas I., Jones M. B., Crawl D., Potter N., Gallagher J., Cornillon P., Schildhauer M., Borer E. T., Seabloom E. W., Hosseini P. R. (2010) Workflows and extensions to the Kepler scientific workflow system to support environmental sensor data access and analysis. Ecol. Inform. 5, 42–50 [Google Scholar]
- 3. Oinn T., Addis M., Ferris J., Marvin D., Senger M., Greenwood M., Carver T., Glover K., Pocock M. R., Wipat A., Li P. (2004) Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics 20, 3045–3054 [DOI] [PubMed] [Google Scholar]
- 4. Hull D., Wolstencroft K., Stevens R., Goble C., Pocock M. R., Li P., Oinn T. (2006) Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 34, W729–W732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matthiesen R. (2007) Methods, algorithms and tools in computational proteomics: a practical point of view. Proteomics 7, 2815–2832 [DOI] [PubMed] [Google Scholar]
- 6. Matthiesen R. (2007) Useful mass spectrometry programs freely available on the Internet. Methods Mol. Biol. 367, 303–305 [DOI] [PubMed] [Google Scholar]
- 7. McIlroy M. D., Pinson E. N., Tague B. A. (1978) Unix Time-Sharing System Forward. Bell Syst. Tech. J. 57, 1902 [Google Scholar]
- 8. Bogdan I. A., Rivers J., Beynon R. J., Coca D. (2008) High-performance hardware implementation of a parallel database search engine for real-time peptide mass fingerprinting. Bioinformatics 24, 1498–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bioinformatics workflow management systems. Available at http://en.wikipedia.org/wiki/Bioinformatics_workflow_management_systems; accessed Sep. 21, 2011
- 10. Curcin V., Ghanem M., Guo Y. (2010) The design and implementation of a workflow analysis tool. Philos. Transact. A Math. Phys. Eng. Sci. 368, 4193–4208 [DOI] [PubMed] [Google Scholar]
- 11. Aho A. V., Kernighan B. W., Weinberger P. J. (1987) The AWK programming language, Addison-Wesley Longman Publishing Co., Inc, Boston, MA [Google Scholar]
- 12.The Perl Programming Language. Available at http://www.perl.org; accessed Sep. 21, 2011
- 13.PHP: Hypertext Preprocessor. Available at http://www.php.net; accessed Sep. 21, 2011
- 14.Python Programming Language – Official Website. Available at http://www.python.org/; accessed Sep. 21, 2011
- 15. Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1, 2005 0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(2004) Web Services Glossary. In: Haas H., Brown A., eds., W3C [Google Scholar]
- 17. W3C (2007) Web Services Description Language (WSDL) Version 2.0 Part 1: Core Language. In: Chinnici R., Moreau J.-J., Ryman A., Weerawarana S., eds., W3C [Google Scholar]
- 18. Gudgin M., Hadley M., Mendelsohn N., Moreau J. J., Nielsen H. F., Karmarkar A., Lafon Y. SOAP Version 1.2 Part 1: Messaging Framework (Second Edition), W3C Recommendation 27, April 2007. Available at http://www.w3.org/TR/2007/REC-soap12-part1-20070427/; accessed Sep. 21, 2011
- 19. Fielding R. T., Taylor R. N. (2000) Principled design of the modern Web architecture. Proceedings of the 22nd international conference on Software engineering, ACM, Limerick, Ireland [Google Scholar]
- 20. Anderson D. P. (2004) BOINC: A System for Public-Resource Computing and Storage. Proceedings of the 5th IEEE/ACM International Workshop on Grid Computing, IEEE Computer Society [Google Scholar]
- 21. Foster I., Kesselman C. (1996) Globus: A metacomputing infrastructure toolkit. Int. J. Supercomput. Appl. 11, 115–128 [Google Scholar]
- 22. Veltri P., Cannataro M., Tradigo G. (2007) Sharing mass spectrometry data in a grid-based distributed proteomics laboratory. Inf. Process. Manage. 43, 577–591 [Google Scholar]
- 23. Bjornson R. D., Carriero N. J., Colangelo C., Shifman M., Cheung K.-H., Miller P. L., Williams K. (2007) X!!Tandem, an Improved Method for Running X!Tandem in Parallel on Collections of Commodity Computers. J. Proteome Res. 7, 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amazon Elastic Compute Cloud (Amazon EC2). Available at http://aws.amazon.com/ec2/; accessed Dec. 1, 2011
- 25.IBM Smart Cloud. Available at http://www.ibm.com/cloud-computing/us/en/; accessed Dec. 1, 2011
- 26.Taverna features. Available at http://www.taverna.org.uk/introduction/taverna-features/; accessed Sep. 21, 2011
- 27. Cock P. J., Antao T., Chang J. T., Chapman B. A., Cox C. J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., de Hoon M. J. (2009) Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smedley D., Haider S., Ballester B., Holland R., London D., Thorisson G., Kasprzyk A. (2009) BioMart–biological queries made easy. BMC Genomics 10, 22 doi:10.1186/1471-2164-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilkinson M. D., Senger M., Kawas E., Bruskiewich R., Gouzy J., Noirot C., Bardou P., Ng A., Haase D., Saiz E. D., Wang D., Gibbons F., Gordon P. M. K., Sensen C. W., Carrasco J. M. R., Fernandez J. M., Shen L., Links M., Ng M., Opushneva N., Neerincx P. B. T., Leunissen J. A. M., Ernst R., Twigger S., Usadel B., Good B., Wong Y., Stein L., Crosby W., Karlsson J., Royo R., Parraga I., Ramirez S., Gelpi J. L., Trelles O., Pisano D. G., Jimenez N., Kerhornou A., Rosset R., Zamacola L., Tarraga J., Huerta-Cepas J., Carazo J. M., Dopazo J., Guigo R., Navarro A., Orozco M., Valencia A., Claros M. G., Perez A. J., Aldana J., Rojano M. M., Cruz R. F. S., Navas I., Schiltz G., Farmer A., Gessler D., Schoof H., Groscurth A., Consortium B. (2008) Interoperability with Moby 1.0 - Its better than sharing your toothbrush. Brief Bioinform. 9, 220–231 [DOI] [PubMed] [Google Scholar]
- 30. Senger M., Rice P., Oinn T. (2003) Soaplab - a unified Sesame door to analysis tools. In: Cox S., ed. Proceedings, UK e-Science, All Hands Meeting 2003, pp. 509–513 [Google Scholar]
- 31. Goble C. A., Bhagat J., Tanoh F., Nzuobontane E., Laurent T., Orlowski J., Roos M., Wolstencroft K., Aleksejevs S., Stevens R., Pettifer S., Lopez R. (2010) BioCatalogue: a universal catalogue of web services for the life sciences. Nucleic Acids Res. 38, W689–W694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li P., Castrillo J. I., Velarde G., Wassink I., Soiland-Reyes S., Owen S., Withers D., Oinn T., Pocock M. R., Goble C. A., Oliver S. G., Kell D. B. (2008) Performing statistical analyses on quantitative data in Taverna workflows: An example using R and maxdBrowse to identify differentially-expressed genes from microarray data. BMC Bioinformatics 9 doi:10.1186/1471-2105-9-334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wassink I., Rauwerda H., Neerincx P. B., van der Vet P. E., Breit T. M., Leunissen J. A., Nijholt A. (2009) Using R in Taverna: RShell v1.2. BMC Res. Notes 2, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A. J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J. Y., Zhang J. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schmidberger M., Morgan M., Eddelbuettel D., Yu H., Tierney L., Mansmann U. (2009) State of the Art in Parallel Computing with R. J. Stat. Softw. 31, 1–27 [Google Scholar]
- 36. De Roure D., Goble C., Stevens R. (2009) The design and realisation of the Virtual Research Environment for social sharing of workflows. Future Generation Computer Systems 25, 561–567 [Google Scholar]
- 37. Palmblad M., Mills D. J., Bindschedler L. V., Cramer R. (2007) Chromatographic alignment of LC-MS and LC-MS/MS datasets by genetic algorithm feature extraction. J. Am. Soc. Mass Spectrom. 18, 1835–1843 [DOI] [PubMed] [Google Scholar]
- 38. Palmblad M., Bindschedler L. V., Gibson T. M., Cramer R. (2006) Automatic internal calibration in liquid chromatography/Fourier transform ion cyclotron resonance mass spectrometry of protein digests. Rapid Commun. Mass Spectrom. 20, 3076–3080 [DOI] [PubMed] [Google Scholar]
- 39. Pedrioli P. G., Eng J. K., Hubley R., Vogelzang M., Deutsch E. W., Raught B., Pratt B., Nilsson E., Angeletti R. H., Apweiler R., Cheung K., Costello C. E., Hermjakob H., Huang S., Julian R. K., Kapp E., McComb M. E., Oliver S. G., Omenn G., Paton N. W., Simpson R., Smith R., Taylor C. F., Zhu W., Aebersold R. (2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 40. Eng J. K., McCormack A. L., Yates J. R. (1994) An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass Spectr. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 41. Perkins D. N., Pappin D. J. C., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectromery data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 42. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 43. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 44. Xu H., Freitas M. A. (2009) MassMatrix: A database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics 9, 1548–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Park C. Y., Klammer A. A., Kall L., MacCoss M. J., Noble W. S. (2008) Rapid and accurate peptide identification from tandem mass spectra. J. Proteome Res. 7, 3022–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim S., Mischerikow N., Bandeira N., Navarro J. D., Wich L., Mohammed S., Heck A. J., Pevzner P. A. (2010) The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol. Cell. Proteomics 9, 2840–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fenyo D. (1999) The biopolymer markup language. Bioinformatics 15, 339–340 [DOI] [PubMed] [Google Scholar]
- 48. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 49. Palmblad M., van der Burgt Y. E. M., Dalebout H., Derks R. J. E., Schoenmaker B., Deelder A. M. (2009) Improving mass measurement accuracy in mass spectrometry based proteomics by combining open source tools for chromatographic alignment and internal calibration. J. Proteomics 72, 722–724 [DOI] [PubMed] [Google Scholar]
- 50. Palmblad M., Mills D. J., Bindschedler L. V. (2008) Heat-shock response in Arabidopsis thaliana explored by multiplexed quantitative proteomics using differential metabolic labeling. J. Proteome Res. 7, 780–785 [DOI] [PubMed] [Google Scholar]
- 51. Palmblad M., van der Burgt Y. E. M., Mostovenko E., Dalebout H., Deelder A. M. (2010) A novel mass spectrometry cluster for high-throughput quantitative proteomics. J. Am. Soc. Mass Spectr. 21, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 52.gnuplot homepage. Available at http://www.gnuplot.info/; accessed Sep. 21, 2011
- 53. Stajich J. E. (2007) An introduction to BioPerl. Methods Mo. Biol. 406, 535–548 [DOI] [PubMed] [Google Scholar]
- 54.Bruker compassXport 3.0.3. Available at http://www.bdal.com/service-support/software-support-downloads.html; accessed Sep. 21, 2011
- 55.Apache Axis2/C - The Web Services Engine. Available at http://axis.apache.org/axis2/c/core/; accessed Dec. 1, 2011
- 56.The Apache HTTP Server Project. Available at http://httpd.apache.org/; accessed Dec. 1, 2011