Abstract
Breast cancer 1, early onset (BRCA1) hereditary breast cancer, a type of cancer with defects in the homology-directed DNA repair pathway, would benefit from the identification of proteins for diagnosis, which might also be of potential use as screening, prognostic, or predictive markers. Sporadic breast cancers with defects in the BRCA1 pathway might also be diagnosed. We employed proteomics based on one-dimensional gel electrophoresis in combination with nano-LC-MS/MS and spectral counting to compare the protein profiles of mammary tumor tissues of genetic mouse models either deficient or proficient in BRCA1. We identified a total of 3,545 proteins, of which 801 were significantly differentially regulated between the BRCA1-deficient and -proficient breast tumors. Pathway and protein complex analysis identified DNA repair and related functions as the major processes associated with the up-regulated proteins in the BRCA1-deficient tumors. In addition, by selecting highly connected nodes, we identified a BRCA1 deficiency signature of 45 proteins that enriches for homology-directed DNA repair deficiency in human gene expression breast cancer data sets. This signature also exhibits prognostic power across multiple data sets, with optimal performance in a data set enriched in tumors deficient in homology-directed DNA repair. In conclusion, by comparing mouse proteomes from BRCA1-proficient and -deficient mammary tumors, we were able to identify several markers associated with BRCA1 deficiency and a prognostic signature for human breast cancer deficient in homology-directed DNA repair.
Breast cancer associated with BRCA11 mutations accounts for 1–2% of breast cancer cases in the Western world. BRCA1 hereditary breast cancer falls into the molecular subtype of basal-like breast cancer that has a poor prognosis (1). Sporadic basal-like breast tumors represent ∼10–15% of all breast carcinomas and comprise many tumors that share key features of BRCA1-associated tumors (2). A major function of BRCA1 is its role in homology-directed double-strand break repair, a DNA repair mechanism that uses the sister chromatid as a template and therefore repairs double-strand breaks in an error-free manner. Deficiencies in homology-directed DNA repair cause high levels of genomic instability that increase the risk of tumorigenesis (3, 4). Nevertheless, BRCA1 pathway dysfunction may provide an opportunity for therapeutic intervention: preclinical models suggest an increased sensitivity to ionizing radiation and DNA (repair)-targeting agents (3). In particular, the use of poly[ADP-ribose] polymerase (PARP) inhibitors holds great promise for clinical application. First results from clinical trials support this therapeutic approach for breast cancer (5). A major clinical challenge remains the identification of patients that are likely to benefit from DNA (repair)-targeting therapy. Global analyses of molecular alterations in sporadic or hereditary breast cancer have mainly used genome and transcriptome profiling methods. These studies yielded a molecular classification of breast cancer (6). In addition, genomics and transcriptomics studies yielded a number of gene signatures that were prognostic for survival, time to distant metastasis and response to treatment (1, 6–15). Two prognostic signatures, Oncotype DX® (11) and MammaPrint® (7, 16), have currently been registered for clinical use. Recently, Vollebergh et al. (13) have found in a retrospective study that a comparative genomic hybridization BRCA1-like classifier predicts the response to intensive platinum-based chemotherapy in patients with high risk breast cancer. The classifier also identifies patients with BRCA1 loss conferred by causes other than mutations. However, the underlying gene products, which would allow for a better understanding of tumor biology and a more practical diagnostic test, remain unknown. To identify patients with BRCA1-like breast cancer, the analysis of tumor proteins may also be useful in selecting patients that might benefit from tailored therapies. Mass spectrometry-based proteomics technologies have matured to the extent that they can now identify and quantify thousands of proteins. Applying these approaches to cancer tissues provides a complementary insight in breast cancer biology and may identify novel diagnostic and prognostic protein profiles and candidate biomarkers. Protein-based biomarkers may be of particular advantage in comparison with transcript-based and genomic markers, because they can be measured in routine assays, e.g. by antibody-based methods such as immunohistochemistry and ELISA, of which the latter allow for noninvasive testing. In addition, targeted multiplex mass spectrometry is emerging as a novel quantitative strategy for measuring protein signatures in tumor tissues or blood. Proteomic studies of breast cancer cells and tissues have already shown the potential for candidate biomarkers discovery (17–22). In a promising pilot study using SELDI-TOF-MS, serum peptide profiles could distinguish women with BRCA1 mutations who developed breast cancer from those who did not (carrier), normal volunteers, and women with sporadic breast cancer with good sensitivity and specificity (23). To date, no studies employing in depth nano-LC-MS/MS-based proteomics have focused on BRCA1/2-deficient tumor tissues.
In this study, we employed state of the art mass spectrometry-based proteomics to identify proteins associated with BRCA1-deficient breast tumors. To this end, we made use of inbred mouse models that display a minimal amount of genetic variability. As a model for human breast cancers deficient in BRCA1, we analyzed mouse mammary tumors harboring conditional tissue-specific mutations in BRCA1 and p53 (24). The majority of these tumors are highly similar to their human counterpart with respect to histological and molecular characteristics and show a high level of genomic instability. For comparison, we analyzed two BRCA1-proficient reference tumor models that are genomically stable (25). We report a BRCA1 deficiency signature based on 45 proteins with DNA repair(-associated) functions that can enrich for homology-directed DNA repair-deficient tumors and identify breast cancer patients with a poor prognosis in various publicly available breast cancer gene expression data sets.
EXPERIMENTAL PROCEDURES
Materials
All of the chemicals, unless otherwise specified, were obtained from Sigma-Aldrich. HPLC solvents, LC-MS grade water, acetonitrile, and formic acid were obtained from Biosolve (Biosolve B.V., Valkenswaard, The Netherlands). Porcine sequence grade modified trypsin was obtained from Promega (Promega Benelux B.V., Leiden, The Netherlands).
Mouse Strains and Tumors
Generation of conditional mutants and K14cre transgenic mice has been described previously (24, 25). All of the animal experiments were approved by the Animal Ethics Committee of the Netherlands Cancer Institute. When grown to a size of ∼500 mm3, the tumors were dissected, snap frozen, and stored at −80 °C until use.
Tissue Homogenization and Fractionation Using Gel Electrophoresis
For homogenization, we cut a piece of ∼20 mg in a bath of liquid nitrogen in smaller parts. The proteins in the mammary tumors tissue samples were solubilized in 800 μl of 1× reducing SDS sample buffer (containing 62.5 mm Tris-HCl, 2% w/v SDS, 10% v/v glycerol, and 0.0025% bromphenol blue, 100 mm DTT, pH 6.8) using a Pellet Pestles microgrinder system (Kontes glassware, Vineland, NJ). Subsequently the proteins were denatured by heating at 100 °C for 10 min. Any insoluble debris was removed by centrifuging for 15 min at maximum speed (16.1 relative centrifugal force) in a benchtop centrifuge.
The proteins were fractionated using one-dimensional SDS-PAGE. 25 μl of each homogenized sample (containing about 50 μg of protein) was loaded on a well of a precast NuPAGE 4–12% w/v Bis-Tris 1.5-mm minigel (Invitrogen). The stacking gel contained 4% (w/v) acrylamide/Bis-Tris. Electrophoresis was carried out at 200 V in NuPAGE MES SDS running buffer (50 mm Tris base, 50 mm MES, 0.1% w/v SDS, 1 mm EDTA, pH 7.3) until the dye front reached the end of the gel. Following electrophoresis, the gels were fixed with a solution of 50% ethanol and 3% phosphoric acid. Staining was carried out in a solution of 34% methanol, 3% phosphoric acid, 15% ammonium sulfate, and 0.1% Coomassie Blue G-250 (Bio-Rad) with subsequent destaining in MilliQ water.
In-gel Digestion and Nano-LC-FT-MS
In-gel Digestion
The gel lanes were cut in 10 bands, and each band was processed for in-gel digestion according to the method of Shevchenko et al. (26). Briefly, the bands were washed and dehydrated three times in 50 mm ammonium bicarbonate, pH 7.9, 50 mm ammonium bicarbonate, and 50% ACN. Subsequently, cysteine bonds were reduced with 10 mm DTT for 1 h at 56 °C and alkylated with 50 mm iodoacetamide for 45 min at room temperature in the dark. After two subsequent wash/dehydration cycles, the bands were dried for 10 min in a vacuum centrifuge and incubated overnight with 0.06 μg/μl trypsin at 25 °C. The peptides were extracted once in 1% formic acid and subsequently twice in 50% ACN in 5% formic acid. The volume was reduced to 50 μl in a vacuum centrifuge prior to LC-MS analysis.
Nano-LC-MS/MS
Peptides were separated by an Ultimate 3000 nano-LC system (Dionex LC-Packings, Amsterdam, The Netherlands) equipped with a 20-cm × 75-μm inner diameter fused silica column custom packed with 3-μm 100 Å ReproSil Pur C18 aqua (Dr. Maisch GMBH, Ammerbuch-Entringen, Germany) as described before (27). After injection, the peptides were trapped at 30 μl/min on a 0.5-cm × 300-μm inner diameter Pepmap C18 cartridge (Dionex LC-Packings, Amsterdam, The Netherlands) at 2% buffer B (buffer A, 0.05% formic acid in MQ; buffer B, 80% ACN and 0.05% formic acid in MQ) and separated at 300 nl/min in a 10–40% buffer B gradient in 60 min. Eluting peptides were ionized at 1.7 kV in a Nanomate Triversa chip-based nanospray source using a Triversa LC coupler (Advion, Ithaca, NJ). Intact peptide mass spectra and fragmentation spectra were acquired on a LTQ-FT hybrid mass spectrometer (Thermo Fisher, Bremen, Germany). Intact masses were measured at resolution 50,000 in the ICR cell using a target value of 1 × 106 charges. In parallel, following an FT prescan, the top five peptide signals (charge states 2+ and higher) were submitted to MS/MS in the linear ion trap (3-atomic mass unit isolation width, 30-ms activation, 35% normalized activation energy, Q value of 0.25, and a threshold of 5,000 counts). Dynamic exclusion was applied with a repeat count of 1 and an exclusion time of 30 s.
LC-SRM Analyses
Independent BRCA1-deficient and -proficient mouse breast tumors (n = 5 in each group) were analyzed in triplicate on an Ultimate 3000 RSCL Nanosystem (Dionex) that was hyphenated to an QTRAP® 5500 instrument (AB SCIEX, Foster City, CA) operated in positive SRM mode and equipped with a nano-electrospray source with applied voltage of 2.404 kV and a capillary heater temperature of 225 °C. The nanoflow LC system and QTRAP® 5500 system were both controlled using Analyst 1.5.1 Software. The combined information from each SRM Information Dependent Acquisition (IDA) experiment was used to perform Mascot searches against the International Protein Index (IPI) mouse database v3.65 and MultiQuantTM software version 2.1 (AB SCIEX).
The scheduled SRM mode comprised the following parameters: SRM detection window of 420 s, target scan time of 3.0 s, curtain gas of 15, ion source gas 1 of 15, declustering potential of 80, and entrance potential of 10. Q1 resolution was set to unit, and Q3 resolution was set to unit. Pause between mass ranges was set to 1 ms. Collision cell exit potentials were set to 36 for all transitions. Peak integration was performed using MultiQuantTM software version 2.1 (AB SCIEX) software and manually reviewed.
Chromatographic separation of peptides was performed by a 68-min gradient at 300 nl/min. Solvent A (0.05% formic acid water) and solvent B (0.05% formic acid, 80% acetonitrile) were mixed at 2% B from 0 to 3 min, 15% B at 4 min, 36% B at 49 min, 99% B from 50 to 54 min, and 2% B at 55 to 68 min. The nano-LC columns were made in house and consisted of 20-cm × 75-μm inner diameter fused silica custom packed with 3-μm 100 Å ReproSil Pur C18 aqua (Dr. Maisch GMBH, Ammerbuch-Entringen, Germany) as described before (27). After injection, the peptides were trapped at 6 μl/min at 2% buffer B.
SRM Assay Development
An SRM assay for the target proteins (NCAPD2, SIN3A, BAZ1B, TOP2A, TOP2B, and PARP1) was developed using the MRMPilotTM software version 2.1 from AB SCIEX. The software requires an amino acid sequence of the protein of interest Information Dependent Acquisition (IDA), a starter method containing the LC conditions, and an empty SRM-IDA experiment. The software performs an in silico digest of the protein and creates a set of peptides that would result after full tryptic digestion. For each of these peptides, it will generate an SRM transition for the calculated m/z of the precursor ion and an appropriate fragment ion. Assay development subsequently entails verification of the peptides and CE optimization of the transitions, both in multiplexed LC-SRM analyses. During verification, the highest responding peptides/transitions at a theoretically calculated optimum CE energy are determined, as well as the identity of the peptide via SRM triggered MS/MS. During CE optimization, the transitions selected after verification are optimized during the chromatographic elution of the peptide.
Verification
A mixture of samples previously analyzed using FT-MS and indicating an abundance of the target candidates was analyzed in 10 unscheduled SRM analyses to find the highest responding tryptic peptides from the target proteins, as well as their elution time during the chromatographic run. For each peptide, 10 theoretically predicted transitions were assessed for detection response and identity. Identity was confirmed using MIDAS (MRM Initiated Detection and Sequencing) with a threshold of 500 counts for an SRM transition response to trigger two MS/MS spectra of the peptide to be acquired at rolling collision energy. Each of the 10 verification analyses was set up to detect 289 of all theoretically predicted transitions and their theoretically predicted optimum collision energy for all theoretically predicted peptides that can result after tryptic digestion of the candidate proteins. The total scan time for each cycle of the instrument during verification was 3.757 s, resulting in a dwell time of 10 ms for each transition in the unscheduled verification analyses.
CE Optimization
All of the data of unscheduled analyses were uploaded to the MRMPilot, which was set to select the five best detected transitions for each peptide and assign a chromatographic retention time to each peptide. Subsequently collision energy for each transition was optimized in 13 LC-SRM analyses, with each analysis set-up to detect 104 scheduled transitions that resulted from verification, at nine different collision energies, centered at 3-V intervals around the theoretically predicted optimum with a dwell time of 25 ms. All of the data of CE optimization cycles were uploaded to the MRMPilot, and for each peptide three transitions at the experimentally found optimum and the experimentally found retention time were included in the final assay. The final assay contained 129 scheduled transitions, three for each peptide, with one to five peptides for each of the seven candidate proteins.
Data Analysis
Protein Identification
MS/MS spectra were searched against the mouse IPI (International Protein Index) database (56,555 entries) using Sequest (version 27, revision 12), which is part of the BioWorks 3.3 data analysis package (Thermo Fisher, San Jose, CA). MS/MS spectra were searched with a maximum allowed deviation of 10 ppm for the precursor mass and 1 atomic mass unit for fragment masses. Methionine oxidation and cysteine carboxamidomethylation were allowed modifications, two missed cleavages were allowed, and the minimum number of tryptic termini was 1. After database searching, the DTA and OUT files were imported into Scaffold 1.07 (Proteome software, Portland, OR). Scaffold was used to organize the gel band data and to validate peptide identifications using the Peptide Prophet algorithm (28, 29). Only identifications with a probability of >95% were retained. Subsequently, the Protein Prophet algorithm was applied, and protein identifications with a probability of >99% with two peptides or more were retained in at least one sample. The false discovery rate for the detected proteins using this workflow is on average around 0.5%, and was not calculated again. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. For each protein identified, the total number of MS/MS spectra detected for each protein identified (spectral counts) was exported to Excel 2003 (Microsoft, Redmond, WA).
Spectral Count Normalization and Statistics
Normalization was performed as described previously (30, 31). The spectral counts of each protein were divided by the total spectral counts of all proteins within a sample. This number was multiplied with a constant equal to the average of total spectral counts of all samples to obtain a normalized spectral count value in the same range as the non-normalized spectral counts. The beta-binomial test (30) was applied to find proteins that show significant differences in spectral count numbers between the tumor group and the reference group. Proteins with a p value less than 0.05 were designated as being significant. Hierarchical clustering was carried out than 0.05 were designated as being significantusing R. For analysis of reproducibility, we calculated the average coefficient xof variation (CV) of the normalized spectral counts from overlapping proteins for three technical replicates.
SRM Data Analysis
Technical replicates were removed until CV of all triplicate analyses was <20%. Subsequently, in each remaining analysis, the ratio of the Area Under the Curve (AUC) of Transition 1/Transition 2, Transition 2/Transition 3, and Transition 1/Transition 3 was calculated. The two transitions resulting in the lowest CV percentage over all analyses were selected for further calculations; the sum of the Area Under the Curve of these two transitions was determined in each sample, and a fold change for each peptide between the groups was determined by the ratio of the summed Area Under the Curve in each group. The average of the fold changes of peptides belonging to one protein was determined for each protein. When the CV percentage of the average of the fold changes of the peptides of one protein was >10%, the transitions of these peptides were visually inspected and excluded when co-eluting false positive responses were observed that had not been detected by Multiquant smoothing and peak splitting algorithms or in-house developed R-script processing. The calculated levels for each approved peptide were normalized on the level of Tuba1b in each sample.
Pathway Analysis
The list of identified proteins was uploaded into the Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems, Redwood City, CA) as a tab-delimited text file containing International Protein Index (IPI) accession numbers, p values, and fold changes calculated with a correction factor (adding 0.5 to the spectral counts of all proteins before normalization). The proteins were uploaded and mapped to corresponding “gene objects” in the Ingenuity Pathways knowledge base. Functional analysis was performed to identify the high level biological functions that were most significantly associated to the differentially regulated proteins in the data set. Significantly regulated proteins within the high level functions are displayed graphically as nodes (proteins/gene objects) and edges (the biological relationships between the nodes). All of the edges are supported by at least one reference from the literature, textbook, or canonical information stored in the Ingenuity knowledge base. Ingenuity Pathways Analysis computes one or more p values for each specific function within a high level function according to the fit of the user's set of significant proteins. The significance of functional enrichment is computed by a Fisher's exact test. Finally, the Path Designer feature was used to create graphically rich network images. In addition, we used the COFECO tool for the mapping of significantly differentially regulated proteins to protein complexes (32). The obtained complexes were further visualized using STRING (33) and Cytoscape, respectively.
Human Gene Expression Data Sets
Identifier Mapping of Mouse Protein Symbols to Human Gene Symbols of Public Gene Expression Data Sets
To explore the diagnostic and prognostic value of the protein expression data from the mouse models, we made use of publicly available human gene expression data sets. To map the up-regulated mouse BRCA1 deficiency proteins to public data sets of human arrays, we first matched mouse gene symbols to human gene symbols using the BioMart website (http://www.biomart.org). We used layout documentation files for the various microarray platforms from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo), MIAMExpress (http://www.ebi.ac.uk/miamexpress/), or Rosetta Inpharmatics (http://bioinformatics.nki.nl/data.php) to retrieve the matching gene symbols on each platform. The following human breast cancer data sets were used: (i) van de Vijver data set (12). A validation study of a prognostic gene expression signature (MammaPrint®), which included 295 young patients with early stage breast cancer, of which 151 were lymph node negative, 226 were estrogen receptor-positive, and 110 had received adjuvant chemotherapy. We were also able to retrieve p53 mutational status for 204 tumors in this data set (data not shown). (ii) van't Veer data set (1). In this discovery study for a prognostic signature (MammaPrint®), the authors also analyzed 18 BRCA1 and 2 BRCA2 samples on the same platform used for the van de Vijver data set (12). (iii) E-UCON-1 data set (10) (subsequently referred to as the Naderi data set). This data set was used for discovery of a prognosis profile in a set of women with early stage breast cancer representative of breast cancer demographics. Of the 132 breast cancer tissues, we used a subset of 120 patients for survival analysis that had the same orientation in dye labeling concerning the reference and tumor samples and that also had associated survival data. (iv) GSE2034 data set (34) (subsequently referred to as Wang data set). This was a discovery and validation analysis of a gene signature for the prediction of breast cancer patient outcomes. It consists of 286 lymph node-negative breast cancer patients who never received adjuvant chemotherapy and of which 209 were estrogen receptor-positive. We logged the normalized intensity values and performed zero mean and unit variance normalization. (v) GSE22133 data set (8) (subsequently referred to as the Jönsson data set). This discovery data set consists of 359 breast tumors including 186 familial, of which 22 were BRCA1-mutated and 32 were BRCA2-mutated. (vi) GSE19177 data set (14) (subsequently referred to as the Waddell data set). This data set contains familial tumors only. Nineteen had a BRCA1 mutation, 30 had a BRCA2 mutation, whereas 25 did not have an identifiable mutation. One tumor was excluded from analysis because it had unknown mutational status. For all of the data sets, we used the normalized log ratios in the analyses, unless specified otherwise above.
Centroid Classification and Survival Analysis
We used a nearest centroid classifier to test the diagnostic and prognostic power of the mapped protein/gene signature on the public human gene expression data sets in combination with leave-one-out cross-validation. First, the signature protein/genes in the validation sets were identified. We used a centroid classification scheme to assess BRCA1 and homology-directed DNA repair deficiency, whereby centroids were built by taking the average expression value for each signature gene in the diagnostic groups, excluding the leave-out sample. The leave-out samples were then classified into different diagnostic groups using the nearest correlation criterion. For classification with a centroid on external data sets, genes were collapsed by taking the median across all probes. This centroid classification scheme was also used for classifications in the Kaplan-Meier survival analysis. In all data sets, patients who survived 5 years or more constituted the good prognosis group (centroid), whereas patients who survived less than 5 years were used for the poor prognosis group (centroid) (10, 12, 34). The average expression value for each signature gene in the good and poor prognosis centroid was computed without the leave-out sample. The leave-out samples were then classified into good or poor prognostic groups using the nearest correlation criterion. To see whether a gene list performed better than random, both in the diagnostic and in the survival analysis, we also ran analysis with 1,000 random gene lists of the same size using the same scheme. We only included probes on the arrays that were annotated with a gene symbol. The same scheme was applied for the prognostics mRNA-based signatures used as a comparison.
RESULTS
Protein Regulations in BRCA1-deficient versus Proficient Mouse Mammary Tumors
For comparative protein profiling, we employed a label-free workflow based on protein fractionation by gel electrophoresis coupled to nano-LC-MS/MS of in-gel digested proteins and spectral counting. Before embarking on a differential analysis, we assessed the reproducibility of our discovery workflow by analyzing three aliquots of a pooled mammary tumor lysate by gel LC-MS/MS (see supplemental Fig. 1A for gel images). In this analysis, 2,220 of 2,473 proteins (90%, see supplemental Fig. 1B for Venn diagram) were identified in all three replicates with an average CV of 24% of the normalized spectral counts, indicating a very good reproducibility for the entire discovery workflow.
To identify proteins associated with BRCA1-deficient mammary tumors, we compared the protein expression profiles in five BRCA1-deficient mammary tumors (p53−/−; BRCA1−/−, carcinoma histology) with five BRCA1-proficient tumors (two p53−/− and three p53−/−; CDH1−/− tumors, all carcinosarcomas). Whereas the carcinomas have an epithelial phenotype, the carcinosarcomas have a mesenchymal phenotype characterized by spindle cell morphology. The protein band patterns obtained after gel electrophoresis of the 10 tumor lysates and Coomassie staining were similar in terms of overall pattern and intensity (see supplemental Fig. 2A). A total of 3,545 proteins were identified across all 10 samples (see supplemental Table 1 for all identifications and spectral count data). The number of proteins identified in the BRCA1-deficient tumor samples was 3,409, with 1,894 proteins identified in all five mammary tumors (see supplemental Fig. 2B), indicating acceptable reproducibility of protein identification and quantification across different biological samples. Similar values were obtained for the five BRCA1-proficient tumors (see supplemental Fig. 2C).
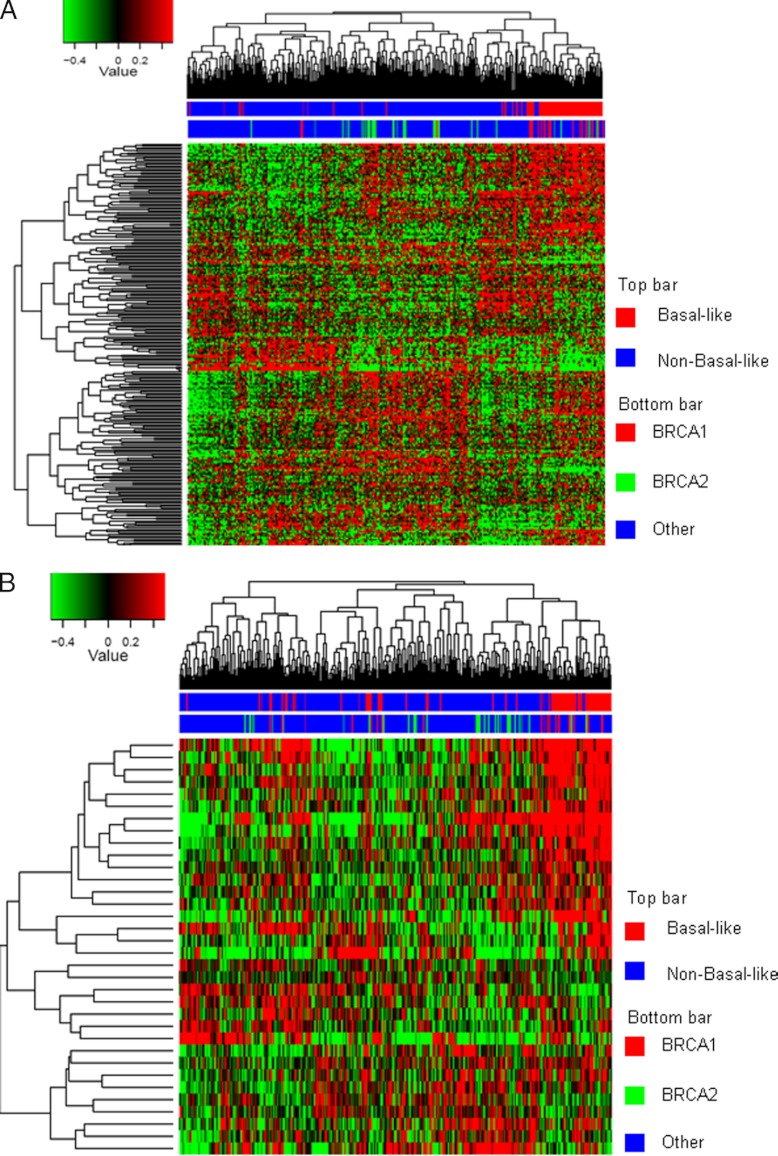
To obtain a global overview of the data set, we performed unsupervised hierarchical clustering using the normalized spectral count data from all 3,545 identified proteins. The BRCA1-deficient and proficient tumors clustered in separate groups (Fig. 1, left panel). The two different BRCA1-proficient tumor types (p53−/− and p53−/−; CDH1−/−) did not form two separate groups but were intermingled, indicating that BRCA1 status and/or histology type were the predominant factors separating the samples. Overlap analysis showed that 338 proteins were uniquely identified in the BRCA1-deficient samples, and 136 were uniquely identified in the BRCA1-proficient tumors. Statistical testing (30) revealed 801 proteins with significantly altered abundance in the BRCA1-deficient and -proficient groups (p < 0.05) of which 417 were up-regulated in the BRCA1-deficient tumors, whereas 384 were down-regulated. As expected, supervised hierarchical clustering using the 801 differential proteins (Fig. 1B) clearly showed two different groups that clustered according to BRCA1/cell type status. See supplemental Table 2 for all differential proteins. For integration with transcriptomics, we employed the data set of Liu et al. (24) containing gene expression data for the same BRCA1-proficient and -deficient mouse models as used in this study, with the exception that most of the tumors in the discovery set were mammary carcinomas. Of the 801 differential proteins, we were able to retrieve mRNA expression data for 565 proteins, of which 429 (76%) had the same direction of differential expression with 201 of these mRNAs (36%) being significantly differentially expressed (see supplemental Table 2).
Fig. 1.
Heat map and cluster analysis using protein expression data from breast tumors of genetic mouse models. A, BRCA1-deficient and -proficient tumors are clustered in separate groups using unsupervised clustering. B, supervised clustering clearly separates the BRCA1-deficient tumors from the proficient ones and shows a distinct heat map pattern for up- and down-regulated proteins.
In summary, a large proportion (23%) of the mammary tumor tissue proteome is regulated in BRCA1-deficient tumors as compared with proficient tumors. Because a large fraction of proteins showed co-regulation with a transcriptomics data set that only used BRCA1-deficient carcinomas versus BRCA1-proficient carcinomas, we conclude that the differential proteins are related mainly to BRCA1 status and only partially to cell type.
Identification of Known Markers of Human BRCA1-deficient Breast Cancer
Because BRCA1-deficient breast tumors often belong to the highly proliferative basal-like subtype, we examined the abundance of protein markers known in basal-like breast cancer in our data set. In addition, we looked for known markers of human BRCA1 deficiency. Two basal cytokeratin markers (Krt14 and Krt6b) were significantly up-regulated in the BRCA1-deficient mouse tumors (Table I). The third cytokeratin (Krt5) was up-regulated (p value = 0.066) with a fold-change of 3.2. ALDH1, a cancer stem cell marker, was exclusively detected in BRCA1-deficient mouse tumors, in accordance with previous findings (35). PCNA and KI67, two well known proliferation markers (36), were also significantly up-regulated in the BRCA1-deficient mouse tumors. These confirmatory findings underscore the value of these genetic mouse tumor models and the validity of our proteomics approach to identifying proteins associated with BRCA1-related or basal-like breast cancers in patients.
Table I. Known BRCA1/basal-like and proliferation markers associated with BRCA1 deficiency.
| Gene name | Basal, proliferation, and stem cell markers | Fold change | p value | Marker type |
|---|---|---|---|---|
| Aldh1a1 | Retinal dehydrogenase 1 | a | 0.000471 | Stem cell |
| Krt14 | Keratin, type I cytoskeletal 14 | 6.5 | 0.023115 | Basal |
| Krt5 | Keratin, type II cytoskeletal 5 | 3.7 | 0.065672 | Basal |
| Krt6b | Keratin, type II cytoskeletal 6B | a | 0.016174 | Basal |
| Pcna | Proliferating cell nuclear antigen | 1.8 | 0.001747 | Proliferation |
| Mki67 | Ki-67 protein | 79.8 | 1.18E-05 | Proliferation |
a Unique detection in BRCA1-deficient tumors.
DNA Repair Pathways and Protein Complexes Are Associated with Proteins Up-regulated in BRCA1-deficient Mammary Tumors
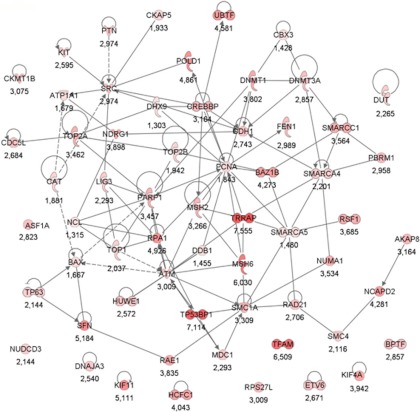
To associate biological functions with the differentially expressed BRCA1 deficiency proteins of the mouse mammary tumors, we used the software tool Ingenuity Pathway Analysis. The molecular and cellular functions associated with the up-regulated proteins in BRCA1-deficient mammary tumors are listed in Table II, with the number one function identified as DNA replication, recombination, and repair (61 proteins). See Fig. 2 for a visualization of the protein network using Ingenuity. The network contains a number of highly connected nodes (i.e., proteins), among which several are well established drug targets (i.e., TOP1, TOP2A, PARP1, and SRC). The top molecular and cellular function associated with the down-regulated proteins was cellular movement (Table II). The 61 DNA repair proteins up-regulated in BRCA1-deficient mammary tumors were involved in subfunctions like excision repair, chromatin remodeling and modification, double-strand DNA repair, and DNA damage response, among others. Moreover, canonical pathways associated with the up-regulated proteins in BRCA1-deficient tumors were involved in DNA repair, including ATM signaling, p53 signaling, and role of BRCA1 in DNA damage response (data not shown).
Table II. Molecular and cellular functions associated with proteins regulated in BRCA1-deficient breast tumors.
IPA was used to associate functions to the up- and down-regulated proteins of the BRCA1-deficient mouse tumors. IPA analysis of the up-regulated proteins identified DNA replication, recombination, and repair as the most significant up-regulated molecular and cellular function. Pathway analysis of the down-regulated proteins identified cellular movement as the most significant up-regulated molecular and cellular function.
| Name | p value | No. of proteins |
|---|---|---|
| Up-regulated proteins | ||
| DNA replication, recombination, and repair | 1.07E-10–1.08E-02 | 61 |
| Cell cycle | 1.19E-09–1.08E-02 | 73 |
| Gene expression | 8.25E-07–1.08E-02 | 73 |
| Cellular growth and proliferation | 1.90E-06–1.08E-02 | 115 |
| Cellular development | 3.77E-06–1.05E-02 | 39 |
| Down-regulated proteins | ||
| Cellular movement | 4.49E-21–1.63E-03 | 104 |
| Cell morphology | 3.92E-20–1.63E-03 | 85 |
| Cell-to-cell signaling and interaction | 6.47E-15–1.63E-03 | 82 |
| Cellular development | 2.26E-13–1.70E-03 | 88 |
| Cellular function and maintenance | 2.43E-12–1.63E-03 | 69 |
Fig. 2.
Protein network of significantly up-regulated proteins in BRCA1-deficient tumors that are associated with the top molecular and cellular function DNA replication, recombination, and repair. The nodes represent proteins, and the lines between them represent interactions. The dashed lines represent indirect interactions. Color intensity indicates fold change, which is also stated below the nodes.
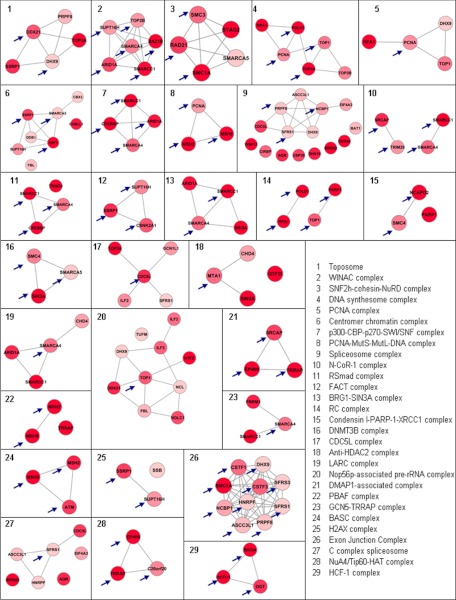
To identify protein complexes underlying the differential proteins and to further dissect the DNA repair pathways, we employed the COFECO tool (32). The up-regulated proteins were linked to 53 significant protein complexes (corrected p value <0.05, see supplemental Table 3A), of which 44 have a DNA repair(-associated) function (see supplemental Table 3A, highlighted rows). After removing the redundant protein complexes where all members were present in one of the other significant complexes, 29 DNA repair(-associated) complexes were obtained (see supplemental Table 3B). The DNA repair complexes were involved in chromosome condensation, chromosome cohesion, chromosome remodeling, RNA processing, histone methylation, histone acetylation, and the topoisomerase complex, among others. We identified also five complexes that we could not readily link to a physiological process involved in DNA repair. These non-nuclear complexes were involved in integrin cell surface interactions with laminins and collagens. Although these complexes have been implicated in evading apoptosis after DNA damage (37), we did not consider these non-nuclear complexes for further analysis. The down-regulated proteins in BRCA1-deficient tumors were not associated with any DNA repair protein complex in a COFECO analysis but instead revealed complexes involved in integrin signaling, cytoskeleton regulation, and extracellular matrix signaling among others (see supplemental Table 3C).
We focused in subsequent analyses on the proteins up-regulated in BRCA1-deficienct tumors with a link to DNA repair because we hypothesized that the up-regulation of DNA repair proteins and pathways is linked to BRCA1 status and reflects a compensatory response to the loss of BRCA1 DNA repair function. The 29 nonredundant DNA repair protein complexes associated with the up-regulated proteins in the BRCA1-deficient tumors are visualized in Fig. 3 using Cytoscape. It is apparent that many protein complexes have multiple up-regulated members. Examples of DNA repair(-associated) complexes include the BRCA1-associated complex (BASC) involved in double-stranded DNA repair (38) and the condensin I-PARP1-XRCC1 complex with established functions in single-strand DNA repair (39). In addition, five of seven members of the toposome complex including the drug targets TOP1 and TOP2A were significantly up-regulated (40). Moreover, many chromatin remodeling complexes, with a wide involvement in different types of DNA repair processes (41), were highly prevalent in our data set. Examples included the WINAC complex, the PBAF complex, the SWI/SNF complex, the GCN5-TRRAP histone acetyl-transferase complex, and the DNMT3B histone methylation complex (see supplemental Table 3). Together, the analyses pinpoint a major up-regulation of a broad range of DNA repair/chromatin remodeling pathways and protein complexes in BRCA1-deficient mammary tumors.
Fig. 3.
Nonredundant up-regulated DNA repair protein complexes identified by COFECO and visualized using STRING in Cytoscape. The nodes represent proteins, and the lines indicate their association as identified in the STRING database. Color intensity is representative of the degree of up-regulation in BRCA1-deficient proteins. Arrows indicate the most connected nodes.
Identification of a BRCA1 Deficiency DNA Repair Signature
To identify a protein signature with biological relevance for BRCA1-deficient breast tumors, we reasoned that this signature should represent the range of up-regulated DNA repair processes in these tumors and therefore contain selected up-regulated members of each of the 29 nonredundant protein complexes described above. To this end, we selected the most connected up-regulated node in each of the 29 DNA repair protein complexes (Fig. 3). This strategy may yield multiple proteins per protein complex, because some nodes show the same level of connectivity. Using this strategy, a BRCA1 deficiency signature of 45 proteins was obtained (Table III). The signature includes PARP1, involved in single-strand base repair; TRRAP, a large adaptor protein involved in histone acetylation; TOP2A, a topoisomerase; SMC1A and SMC4, involved in chromatid cohesion and condensation; BAZ1B and ATM, involved in phosphorylation of H2AX upon DNA damage; and MSH2 and MSH6, involved in mismatch repair.
Table III. List of 45 proteins in BRCA1 deficiency signature.
| Protein description | Mouse IPI identifier | Human gene symbol | Fold change | p value | Regulation comparison mRNA and proteina |
|---|---|---|---|---|---|
| Transformation/transcription domain-associated protein | IPI00330902 | TRRAP | 8.4 | <0.001 | c |
| Bromodomain adjacent to zinc finger domain protein 1B | IPI00130597 | BAZ1B | 4.0 | <0.001 | d |
| Structural maintenance of chromosomes protein 3 | IPI00132122 | SMC3 | 2.4 | <0.001 | c |
| Isoform 2 of condensin complex subunit 1 | IPI00172226 | NCAPD2 | 4.5 | <0.001 | d |
| Replication protein A 70-kDa DNA-binding subunit | IPI00124520 | RPA1 | 6.3 | <0.001 | c |
| Isoform 1 of paired amphipathic helix protein Sin3a | IPI00117932 | SIN3A | 20.0 | <0.001 | d |
| Pold1 DNA polymerase | IPI00313515 | POLD1 | 8.5 | <0.001 | c |
| Activating signal cointegrator 1 complex subunit 3-like 1 | IPI00420329 | SNRNP200 | 1.6 | 0.001 | NAe |
| Structural maintenance of chromosomes protein 1A | IPI00123870 | SMC1A | 3.2 | 0.001 | c |
| DNA topoisomerase 2α | IPI00122223 | TOP2A | 4.0 | 0.001 | NA |
| SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 | IPI00125662 | SMARCC1 | 3.0 | 0.001 | f |
| DNA topoisomerase 2β | IPI00135443 | TOP2B | 2.1 | 0.001 | d |
| Host cell factor C1 | IPI00828490 | HCFC1 | 3.5 | 0.002 | c |
| Proliferating cell nuclear antigen | IPI00113870 | PCNA | 1.8 | 0.002 | d |
| Rsf1 hepatitis B virus × associated protein | IPI00122845 | RSF1 | b | 0.002 | c |
| Casein kinase II α subunit | IPI00408176 | CSNK2A1 | 2.4 | 0.002 | NA |
| Cell division cycle 5-related protein | IPI00284444 | CDC5L | 3.1 | 0.002 | NA |
| DNA topoisomerase 1 | IPI00109764 | TOP1 | 2.2 | 0.003 | c |
| Isoform 1 of UDP-N-acetylglucosamine-peptide N-acetylglucosaminyltransferase 110-kDa subunit | IPI00420870 | OGT | 3.8 | 0.004 | d |
| Isoform 2 of E1A-binding protein p400 | IPI00229659 | EP400 | 9.7 | 0.004 | d |
| MutS homolog 6 | IPI00310173 | MSH6 | 7.1 | 0.006 | d |
| Isoform 1 of transcription intermediary factor 1β | IPI00312128 | TRIM28 | 1.9 | 0.008 | c |
| Isoform 1 of splicing factor, arginine/serine-rich 1 | IPI00420807 | SFRS1 | 1.4 | 0.008 | NA |
| Snf2-related CBP activator protein | IPI00620743 | SRCAP | b | 0.008 | NA |
| Poly(ADP-ribose) polymerase) 1 | IPI00139168 | PARP1 | 3.1 | 0.008 | c |
| SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 4 | IPI00460668 | SMARCA4 | 2.3 | 0.009 | c |
| CREB-binding protein | IPI00463549 | CREBBP | 5.5 | 0.010 | d |
| Serine-protein kinase ATM | IPI00124810 | ATM | 6.0 | 0.010 | c |
| Double-strand-break repair protein Rad21 homolog | IPI00329840 | RAD21 | 3.1 | 0.014 | d |
| Pre-mRNA-processing splicing factor 8 | IPI00121596 | PRPF8 | 1.5 | 0.014 | c |
| MRG (MORF4-related gene)-binding protein | IPI00720110 | C20ORF20 | b | 0.019 | d |
| Cleavage stimulation factor 50-kDa subunit | IPI00116747 | CSTF1 | b | 0.020 | c |
| Metastasis-associated 1 | IPI00776055 | MTA1 | b | 0.020 | c |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 | IPI00652987 | DDX21 | 2.3 | 0.020 | c |
| Isoform 1 of heterogeneous nuclear ribonucleoprotein F | IPI00226073 | HNRNPF | 1.4 | 0.022 | NA |
| Nuclear cap-binding protein subunit 1 | IPI00458056 | NCBP1 | 1.9 | 0.028 | NA |
| SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 | IPI00396739 | SMARCA5 | 1.4 | 0.030 | c |
| DNA mismatch repair protein Msh2 | IPI00118158 | MSH2 | 3.0 | 0.031 | d |
| Isoform long of splicing factor, arginine/serine-rich 3 | IPI00129323 | SFRS3 | 1.4 | 0.035 | d |
| DEAH (Asp-Glu-Ala-His) box polypeptide 9 | IPI00339468 | DHX9 | 1.3 | 0.037 | c |
| FACT complex subunit SPT16 | IPI00120344 | SUPT16H | 2.1 | 0.045 | d |
| Cleavage stimulation factor 77-kDa subunit | IPI00116929 | CSTF3 | 6.4 | 0.046 | d |
| Isoform 2 of FACT complex subunit SSRP1 | IPI00407571 | SSRP1 | 3.1 | 0.047 | d |
| Structural maintenance of chromosome protein 4 | IPI00229397 | SMC4 | 2.4 | 0.049 | d |
| AT rich interactive domain 1A | IPI00648459 | ARID1A | 3.8 | 0.049 | d |
a For mRNA regulation.
b Unique detection in BRCA1-deficient tumors.
c Significantly up-regulated mRNA.
d No significant regulation mRNA.
e NA indicates no mRNA probe on microarray.
f Significant opposite regulation mRNA.
Up-regulated Proteins Mapped to Human Transcripts Identify Human BRCA1/2-deficient Tumors
To investigate the power of the 45 protein BRCA1 deficiency signature in separating BRCA1-deficient and -proficient breast cancers in humans in comparison with all up-regulated proteins, we performed in silico analysis using publicly available gene expression data sets (Table IV). We also evaluated the specificity for BRCA2, a gene involved in the same pathway as BRCA1, to examine the ability to identify deficiency in homology-directed DNA repair in general (4). This is important because of the recent availability of drugs targeting this particular deficiency (43).
Table IV. Description of relevant information of human breast cancer gene expression data sets used for in silico validation.
| No. of patients | Patient characteristics | Clinical end point | Source | |
|---|---|---|---|---|
| van de Vijver et al. | 315 | 295 sporadic patients | Survival | http://www.rii.com/publications/ |
| van 't Veer et al. | 20 | 18 BRCA1 and 2 BRCA2 mutation carriers | NA | http://www.rii.com/publications/ |
| Wang et al. | 286 | 286 sporadic patients | Time to distant metastasis | GEO accession GSE2034: http://www.ncbi.nlm.nih.gov/projects/geo/ |
| Naderi et al. | 134 | 134 sporadic patients (120 with survival data) | Survival | Array express accession E-UCON-1: http://www.ebi.ac.uk/arrayexpress/ |
| Jönsson et al. | 359 | 22 BRCA1, 32 BRCA2, 173 sporadic and 132 non-BRCA1/2 familial patients | Survival | GEO accession GSE22133: http://www.ncbi.nlm.nih.gov/projects/geo/ |
| Waddell et al. | 75 | 19 BRCA1, 30 BRCA2, 1 unknown, and 25 non-BRCA1/2 familial patients | NA | GEO accession GSE22133: http://www.ncbi.nlm.nih.gov/projects/geo/ |
We first focused on the Jönsson data set containing 22 BRCA1-mutated and 32 BRCA2-mutated tumors and other familial and sporadic tumors (Table IV), because this whole genome gene expression data set contained the largest number of BRCA1/2-mutated tumors. Hierarchical clustering using all up-regulated proteins showed that the majority of BRCA1-mutated tumors were clustered within one branch of the dendrogram, which coincides, as expected, with the basal-like tumors (Fig. 4A). The BRCA2 samples were also clustered largely together within the middle branch of the dendrogram. Fig. 4B depicts a clustering using the BRCA1 deficiency signature. Here, a large proportion of the BRCA1 and BRCA2 falls within one branch of the dendrogram, making up approximately one-third of the tumors. Thus, the cluster analysis indicates that the 45-protein BRCA1 deficiency signature shows specificity, not only for BRCA1-mutated tumors but also for BRCA2-mutated tumors.
Fig. 4.
Hierarchical clustering of proteins mapped to gene transcripts for the Jönsson data set. A, cluster analysis using the 417 up-regulated mapped proteins. The majority of the BRCA1 patients cluster together within the basal cluster in one branch of the dendrogram. The adjacent cluster contains the majority of the BRCA2 patients. B, cluster analysis using the 45-protein BRCA1 deficiency signature. The BRCA1 and BRCA2 samples are now adjacent to each other in one cluster containing approximately one-third of all patients.
The nearest centroid classification method was employed to characterize more precisely the sensitivity and specificity of the mouse BRCA1 deficiency signature for BRCA1- and BRCA2-mutated tumors, as well as for the list of all up-regulated proteins. Table V reports the classification results on the Jönsson data set with leave-one-out cross-validation. The sensitivities for BRCA1-mutated tumors were 77 and 82% for the 417 up-regulated proteins and the BRCA1 deficiency signature, respectively. Classification for the combination of BRCA1- and BRCA2-mutated tumors yielded a similar performance: 83% sensitivity for all up-regulated proteins and 81% for the BRCA1 deficiency signature. We also assessed the performance of 1,000 sets of genes randomly sampled from the whole genome and, in a more stringent approach, from the list of DNA replication, recombination and repair, genes as defined by IPA (Table V). Although both all up-regulated proteins and the BRCA1 deficiency signature achieved similar sensitivities in classifying BRCA1/2 mutated tumors, this was only significantly better compared with random gene lists, when using all up-regulated proteins. Nevertheless, the BRCA1 deficiency signature compared favorably with both random gene lists sampled from all genes and from random DNA repair lists, showing confidence that the classification accuracies were not obtained by chance.
Table V. Performance of all 417 up-regulated proteins and the 45-protein BRCA1 deficiency signature in human gene expression data sets.
The performance for classifying BRCA1/2 patients using all 417 up-regulated proteins and the BRCA1 deficiency signature in the Jönsson et al. data set was evaluated. The performance of the expression centroids created for the 417 up-regulated protein and BRCA1 deficiency signature in the Jönsson et al. data set was applied to the combined van de Vijver et al. and van 't Veer et al. data sets and the Waddell et al. data set.
| True/predicted | BRCA1 | BRCA2 | Familial | Sporadic | Total | Sensitivity | Specificity | All genesa | DNA repair backgrounda |
|---|---|---|---|---|---|---|---|---|---|
| Jönsson et al. data set | |||||||||
| All 417 up-regulated proteins | |||||||||
| BRCA1 | 17 | 0 | 1 | 4 | 22 | 77.3% | 84.7% | 0.016 | 0.017 |
| BRCA2 | 4 | 24 | 3 | 1 | 32 | 75.0% | 85.9% | 0.286 | 0.315 |
| BRCA1/2 | 45 | 45 | 4 | 5 | 54 | 83.3% | 69.5% | 0.040 | 0.012 |
| Familial | 15 | 26 | 63 | 28 | 132 | 47.7% | 78.4% | ||
| Sporadic | 32 | 20 | 45 | 76 | 173 | 43.9% | 82.3% | ||
| BRCA1 deficiency signature | |||||||||
| BRCA1 | 18 | 0 | 1 | 3 | 22 | 81.8% | 81.3% | 0.329 | 0.243 |
| BRCA2 | 5 | 16 | 8 | 3 | 32 | 50.0% | 86.5% | 0.184 | 0.217 |
| BRCA1/2 | 39 | 39 | 9 | 6 | 54 | 72.2% | 66.6% | 0.245 | 0.178 |
| Familial | 16 | 25 | 65 | 26 | 132 | 49.2% | 72.7% | ||
| Sporadic | 42 | 19 | 53 | 59 | 173 | 34.1% | 85.6% | ||
| Combined van de Vijver et al. and van 't Veer et al. data sets | |||||||||
| All 417 up-regulated proteins | |||||||||
| BRCA1 | 17 | 0 | 1 | 18 | 94.4% | 75.8% | |||
| BRCA2 | 0 | 2 | 0 | 2 | 100.0% | 57.2% | |||
| BRCA1/2 | 19 | 19 | 1 | 20 | 95.0% | 30.2% | |||
| Sporadic | 72 | 134 | 89 | 295 | 30.2% | 95.0% | |||
| BRCA1 deficiency signature | |||||||||
| BRCA1 | 17 | 0 | 1 | 18 | 94.4% | 80.3% | |||
| BRCA2 | 0 | 2 | 0 | 2 | 100.0% | 71.1% | |||
| BRCA1/2 | 19 | 19 | 1 | 20 | 95.0% | 32.2% | |||
| Sporadic | 73 | 127 | 95 | 295 | 32.2% | 95.0% | |||
| Waddell et al. data set | |||||||||
| All 417 up-regulated proteins | |||||||||
| BRCA1 | 15 | 1 | 3 | 19 | 78.9% | 78.6% | |||
| BRCA2 | 7 | 12 | 11 | 30 | 40.0% | 91.7% | |||
| BRCA1/2 | 35 | 35 | 14 | 49 | 71.4% | 56.0% | |||
| Familial | 8 | 3 | 14 | 25 | 56.0% | 71.4% | |||
| BRCA1 deficiency signature | |||||||||
| BRCA1 | 13 | 3 | 3 | 19 | 68.4% | 77.5% | |||
| BRCA2 | 8 | 8 | 14 | 30 | 26.7% | 86.3% | |||
| BRCA1/2 | 32 | 32 | 17 | 49 | 65.3% | 52.0% | |||
| Familial | 8 | 4 | 13 | 25 | 52.0% | 65.3% |
a p value of permutation analysis using random gene lists (the fraction of a 1,000 random gene lists of the same length performing better then the up-regulated proteins or BRCA1 deficiency signature). In the case of “All genes,” sampling was done from all genes present in the human genomes that had a official gene symbol. Genes from a DNA repair background were sampled from a list generated by IPA.
In addition to leave-one-out cross-validation, we performed completely independent validation using the two other data sets containing samples with BRCA1/2 mutation status (the combined van de Vijver and van 't Veer cohorts and the Waddell cohort; Table IV). The centroids constructed from the Jönsson et al. data set can classify BRCA1/2 samples in the van de Vijver/van 't Veer cohorts with a very high sensitivity of 95% for both the up-regulated proteins and the BRCA1 deficiency signature (Table V). A large portion of sporadic samples were assigned to the BRCA1/2 class. Because the sporadic samples were not tested for BRCA1/2 dysfunction or inactivation of other components of the homologous recombination pathway, part of these mismatching predictions reported here might reflect a true deficiency in the BRCA1/2 pathway, i.e., a BRCAness phenotype (44). For the Waddell cohort, we obtained sensitivity of 79 and 68% for BRCA1 patients by the up-regulated proteins and BRCA1 deficiency signature, respectively (Table V), which is comparable with the result of 74% sensitivity reported by the authors of the data set. Our result is significant, given that the test data is completely independent from the training data, whereas internal validation was used in Waddell et al. (14).
These data show that the 417 up-regulated proteins in BRCA1-deficient mouse tumors, as well as the BRCA1 deficiency signature of 45 proteins, can classify human BRCA1-deficient breast tumors when mapped to human transcriptomics data sets. Importantly, the classification results for the mapped mouse BRCA1 deficiency protein signature were better than the results that we obtained with the published mouse transcriptome data (24) from which we also constructed a signature using the same network-based in silico approach (data not shown). For example, sensitivities of the protein signature for selecting BRCA1-deficient tumors were 81.8, 94.4, and 68.4% in the Jonsson data set and the combined Vijver and van 't Veer and Waddell data set, whereas these values were 63.6, 50.0, and 57.9% for the transcriptome signature (data not shown). The set of all up-regulated proteins achieved the best performance for diagnosing BRCA1 mutations in comparison with random (DNA repair) genes. BRCA2-deficient tumors were also classified, implying enrichment for homology-directed repair-deficient tumors in general. Moreover, the 45 protein signature and all up-regulated proteins also classify a number of familial tumors without BRCA1/2 mutation and sporadic patients as BRCA1/2-like, suggesting that these tumors might be deficient in homology-directed DNA repair.
BRCA1 Deficiency Signature Proteins Show Prognostic Power when Mapped to Human Breast Cancer Gene Expression Data Sets
To investigate whether the BRCA1 deficiency proteins and signature have prognostic power, we used the mapped mRNAs of the up-regulated proteins in the four public breast cancer gene expression data sets that have associated clinical end point data (van de Vijver, Wang, Naderi and Jönsson (8, 10, 12, 34); Table IV) to perform a Kaplan-Meier survival analysis. The Jönsson data set was the only cohort that has an enrichment of familial (BRCA1/2-related) patients. For comparison, we also performed a Kaplan-Meier analysis using two commercially available prognostic gene signatures (MammaPrint® and Oncotype DX®). In a third comparison, we used the Naderi signature (discovered in the Naderi cohort), which has been shown to also have prognostic power in both the van de Vijver and Wang cohorts (10).
The mapped list of all 417 up-regulated proteins in BRCA1-deficient tumors yielded highly significant p values for survival analysis across all data sets, but these were only significantly better than random gene lists in the van de Vijver data set. When sampling from a DNA repair gene background, no significant p values for the permutation analysis were obtained (Table VI). It is of note here that the external (commercial) gene expression-based signatures in some instances showed a similar level of underperformance when compared with random DNA repair gene lists in the sporadic data sets and were performing nonsignificantly in all permutation settings in the Jönsson cohort. Not surprisingly, the two mRNA signatures identified within their discovery cohort, MammaPrint® in the van de Vijver cohort (1, 12) and Naderi signature in the Naderi cohort (10), outperformed all other signatures within their cohort.
Table VI. Overview of Kaplan-Meier survival analysis in four breast cancer gene expression data sets that have associated clinical end point data.
The table contain data on the van de Vijver, Wang, Naderi, and Waddell cohorts. The top two rows represent the performance of all up-regulated BRCA1 deficiency proteins and the 45 protein BRCA1 deficiency signature mapped to matching gene symbols. The bottom three rows contain the performance of gene expression-derived signatures, as a means of comparison with the protein-based signatures.
| van de Vijver cohorta |
Wang cohorta |
Naderi cohorta |
Jönsson cohorta |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p valuea | # of mapped genes or proteinsb | All genesb | DNA repair backgroundb | p valuea | # of mapped genes or proteinsb | All genesb | DNA repair backgroundb | p valuea | # of mapped genes or proteinsb | All genesb | DNA repair backgroundb | p valuea | # of mapped genes or proteinsb | All genesb | DNA repair backgroundb | |
| Mouse BRCA1 deficiency proteins | ||||||||||||||||
| All 417 up-regulated proteins | 6.12E-09 | 326/417 | <0.001 | >0.5 | 0.00013 | 309/417 | 0.247 | >0.5 | 0.0229 | 262/417 | 0.133 | 0.309 | 9.80E-06 | 227/417 | 0.248 | 0.277 |
| BRCA1 deficiency signature | 1.49E-09 | 43/45 | <0.001 | 0.207 | 0.00143 | 43/45 | 0.023 | 0.440 | 0.0044 | 32/45 | 0.004 | 0.044 | 2.16E-08 | 33/45 | 0.010 | 0.008 |
| Gene expression-based signatures | ||||||||||||||||
| Naderi signature | 1.01E-08 | 50/70 | <0.001 | 0.355 | 6.30E-04 | 48/70 | 0.008 | >0.5 | 0.0003 | 60/70 | <0.001 | 0.005 | 1.44E-06 | 33/70 | 0.090 | 0.088 |
| Oncotype (commercial signature) | 2.22E-10 | 16/16 | <0.001 | 0.029 | 2.07E-05 | 14/16 | <0.001 | 0.024 | 0.0004 | 13/15 | <0.001 | 0.005 | 0.00064 | 13/16 | 0.435 | 0.359 |
| Mammaprint (commercial signature) | 1.60E-14 | 60/61 | <0.001 | <0.001 | 0.00094 | 47/61 | 0.013 | >0.5 | 0.0086 | 41/61 | 0.004 | 0.056 | 1.16E-05 | 35/61 | 0.250 | 0.233 |
a p value from Kaplan-Meier survival analysis.
b Number of proteins or genes from a signature or protein list that could be mapped to one or multiple probes on the microarray (mapped genes or proteins/all genes or protein in the list).
c p value of permutation analysis using random (DNA repair) gene lists (the fraction of a 1,000 random gene lists of the same length performing better then the gene/protein list used in survival analysis). In the case of “All genes,” sampling was done from all genes present in the human genomes that had a official gene symbol. For this reason, genes from a DNA repair background were sampled from a list generated by IPA.
The mapped BRCA1 deficiency signature has highly significant prognostic value. The Kaplan-Meier plots of the BRCA1 deficiency signature in the four breast cancer data sets is shown in Fig. 5. Performance was comparable with the gene expression-based signatures in the three sporadic cohorts (the van de Vijver, Wang, and Caldas data sets). Importantly, in the data set with an over-representation of familial (BRCA1/2) tumors (the Jönsson cohort), the mapped mouse BRCA1 deficiency signature outperformed all human gene expression-based signatures, and performance was still significant when compared with random (DNA repair) gene lists. In summary, these data demonstrate that the mouse BRCA1 deficiency protein signature, when mapped to human gene expression data has prognostic value and outperforms (commercial) gene expression-based signatures in a cohort enriched for breast cancer with defects in the homology-directed DNA repair pathway.
Fig. 5.
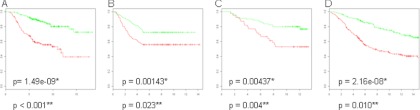
Survival analysis displaying Kaplan-Meier curves on a diverse set of public human gene expression breast cancer data using the BRCA1 deficiency signature. Green curves represent patients with poor prognosis, and red curves are patients with good prognosis. Overall survival was used as clinical end point, unless specified otherwise. A, van de Vijver data set of 295 patients. B, Wang cohort with 286 tumors using metastasis-free survival as clinical end point. C, Naderi data set with 120 early stage tumors. D, Jönsson data set containing 359 tumors, which includes 186 familial tumors, of which 54 were confirmed BRCA1/2 carriers. *, p value from Kaplan-Meier survival analysis. **, p value from permutation analysis (the fraction of a 1,000 random gene lists of the same length performing better then the BRCA1 deficiency signature.
Poor Outcome Human Breast Tumors Identified by BRCA1 Deficiency Signature Show Enrichment in p53 Mutations
p53 mutations have the capacity to disrupt the signaling between accumulated DNA damage and the induction of apoptosis. Moreover, the loss of functional p53 is often associated with BRCA1-related hereditary breast cancer in humans (45, 46). For this reason, we investigated whether the poor prognosis patients identified in the survival analysis showed a significant enrichment for p53 mutations. For the van de Vijver cohort, we were able to retrieve p53 mutational status for 204 of the 295 tumors (data not shown). Enrichment of p53 mutation in the poor prognosis patients was assessed using Fisher's exact test. Both the total list of 417 up-regulated proteins and the BRCA1 deficiency signature showed a highly significant enrichment for p53 mutations in poor prognosis patients (both p values were <10−10; Table VII). These data highlight the finding that the BRCA1 deficiency proteins and signature associate with p53 mutation as well as with survival.
Table VII. Overview and statistical analysis of enrichment for p53 mutations in poor prognosis patients versus good prognosis patients using Fisher's exact test in the van de Vijver et al. data set.
The table displays the distribution of p53 mutation status and good/bad prognosis patients within the van de Vijver data set: all 417 up-regulated BRCA-deficient proteins and the BRCA1 deficiency signature. Note that 204 of the 295 patients from the van de Vijver et al. cohort had associated p53 mutational status.
| Poor prognosis | Good prognosis | Sum | |
|---|---|---|---|
| All 417 up-regulated proteins: p value < 2.20E-16a | |||
| p53 mutation | 55 (79%) | 15 (21%) | 70 |
| Wild-type p53 | 25 (19%) | 109 (81%) | 134 |
| Sum | 80 | 124 | 204 |
| BRCA1 deficiency signature (45 proteins): p value = 2.55E-12a | |||
| p53 mutation | 49 (70%) | 21 (30%) | 70 |
| Wild-type p53 | 26 (19%) | 108 (81%) | 134 |
| Sum | 75 | 129 | 204 |
a p value from Fisher's exact test.
Protein Quantitation by Targeted Mass Spectrometry
We have selected several proteins for follow-up at the protein level: four genes/proteins that showed discordant regulations, significantly up-regulated protein levels, and down-regulated mRNA expression levels (NCAPD2, SIN3A, BAZ1B, and TOP2B) in the BRCA1-deficient breast tumors of the mouse model. We also included one gene for which no probe was available on the microarray (TOP2A) and one protein for which protein and mRNA regulation was concordant (PARP1) in the mouse model. Of these gene products, SIN3A and TOP2B had also down-regulated mRNAs in the human data set of Jönsson, whereas PARP1 was not regulated, TOP2A was up-regulated, and for NCAPD2 and BAZ1B no probes were available. First, we confirmed the protein regulations as revealed by the spectral count data in the discovery samples using an independent measure of label-free protein quantitation, i.e., the area under the curve of the extracted ion chromatograms. Second, we performed targeted mass spectrometry by SRM-MS in 10 independent mouse breast tumors, all carcinomas. The regulation of SIN3A, NCAPD2, TOP2A, TOP2B, and PARP1 was confirmed by SRM-MS in independent tumors, with all peptides being significantly up-regulated in BRCA1 deficient breast tumors, whereas only BAZ1B was not significantly up-regulated (supplemental Fig. 3). Hierarchical clustering using all peptides from the discordant proteins clearly separated this pilot validation set according to BRCA1 status (supplemental Fig. 4). In conclusion, the SRM validation of protein expression levels for which the RNA levels were discordant underscores the fact that RNA expression levels cannot always be simply translated to protein expression levels, as well as the importance of analysis of the end products of genes by proteomics.
DISCUSSION
In the present study, we aimed to identify proteins that are associated with the loss of expression of BRCA1, which is involved in homology-directed DNA repair. These proteins could potentially find use as screening, prognostic, or predictive biomarkers. To this end, we analyzed protein profiles in BRCA1-proficient and -deficient mouse breast tumors using a high resolution tandem mass spectrometry-based proteomics approach. We identified 3,545 proteins, of which 801 were significantly differentially regulated. A BRCA1 deficiency 45-protein signature was defined through the use of pathway and protein complex analysis, with good performance in human gene expression data sets enriched for BRCA1 deficiency. To our knowledge, this is the first comprehensive in depth proteomics analysis in genetic breast cancer. An overview of the discovery and data mining strategies is given in Fig. 6.
Fig. 6.
Overview of proteomics discovery, data mining, identification of a BRCA1 deficiency signature, and in silico validation in human transcriptomics data sets.
Up-regulated Proteins in BRCA1-deficient Mouse Breast Tumors Contain Basal-like Markers, Multiple Drug Targets, and DNA Repair(-associated) Proteins
As expected, we found significant up-regulation of basal-like markers that are known to occur in breast cancer of the basal-like subtype. This is in line with the fact that human BRCA1-mutated tumors belong predominantly to the basal-like breast cancer subtype. Therefore these confirmatory findings underscore the human relevance of the BRCA1-deficient mouse tumor models. BRCA1 has recently, through its function as a transcription factor, been linked to the basal transcription machinery, whereby functional BRCA1 represses transcription of basal keratins (47). In addition, we identified a number of up-regulated proliferative markers, a feature that is more prevalent in human basal-like breast cancer.
Pathway and protein complex analysis identified DNA repair and associated processes as the most important biological function associated with the up-regulated proteins of the BRCA1-deficient tumors. This is in line with previous reports that loss of functional homology-directed DNA repair through knock-out of BRCA1 might be partially compensated for by other DNA repair mechanisms (4, 48). Importantly, we found a number of therapeutic targets to be up-regulated in the BRCA1-deficient tumors, including PARP1, TOP1, TOP2A, and TOP2B. PARP1 has been shown to be a bona fide drug target for human BRCA1-mutated tumors (43). Up-regulation of the PARP1 protein may be a marker for the loss of functional homology-directed DNA repair in general and might therefore be a predictive marker for the efficacy of PARP1 inhibition. In line with this, the tumors of the BRCA1-deficient mice used in this study responded well to the PARP inhibitor olaparib, whereas the BRCA1-proficient mouse tumor models did not (49). Moreover, we also found up-regulation of the topoisomerases TOP1, TOP2A, and TOP2B drug targets for topotecan (TOP1 inhibitor) and doxorubicin (TOP2A and TOP2B inhibitor). These drugs inhibit the religation step of topoisomerases and therefore also induce indirectly DNA breaks. Higher levels of these proteins might have predictive value, because the BRCA1-deficient mouse tumors used in our experiments have been shown to be sensitive to topotecan (50) and doxorubicin (51). We detected a number of other potential drug targets. HDAC1 and HDAC2, two proteins involved in chromatin remodeling by histone deacetylation, were also up-regulated, although this was not significant (p = 0.07 and p = 0.14, respectively). At least 11 kinases were significantly up-regulated, of which established drug targets included KIT and SRC (see supplemental Table 2, IPA drug targets). Novel kinase candidate drug targets included MAPK14, CDK9, and CSFR1. Multiple proteins up-regulated in the BRCA1-deficient mice tumors act upstream of BRCA1 function in the homology-directed DNA repair pathway (ATM, BAZ1B, and TP53BP1), which may indicate an accumulation of these proteins in response to BRCA1 loss. In a previous study, Liu et al. (24) used gene expression analysis in the same mouse models as used in this study. Using gene set enrichment analysis, they reported a number of processes that were induced after BRCA1 loss, including recombinatorial repair, mitotic recombination, telomere maintenance, and transcriptional regulation (e.g. chromatin remodeling).
Mouse BRCA1 Deficiency Protein Signature with Diagnostic and Prognostic Value in Human Gene Expression Data Sets
We used an in silico validation approach to show that mouse proteins up-regulated in BRCA1-deficient tumors, including a BRCA1 deficiency signature, could classify human BRCA1 and BRCA2 tumors in cohorts that contained both sporadic and hereditary breast cancers. Using the BRCA1 deficiency signature, high sensitivities were achieved for classifying homology-directed DNA repair-deficient tumors in data sets known to be enriched for these patients (14). The BRCA1 deficiency signature also classified a considerable number of sporadic and familial tumors as BRCA1/2-like. This result may be explained by the possibility that a number of sporadic and familial tumors lacking mutations in BRCA genes might still harbor undetected deficiencies in homology-directed DNA repair and might therefore benefit from DNA-damaging agents. There is growing evidence that the majority of sporadic basal-like breast cancers have BRCA1 dysfunctionality rather than a mutation in BRCA1 itself (9, 13).
Approximately 25% of BRCA1 tumors were not picked up by our classifier. This might be explained by the fact that our mouse BRCA1 classifier is not able to capture the full heterogeneity of all BRCA1-mutated breast carcinomas. In addition, a number of BRCA1-mutated breast carcinomas might escape detection because of restoration of homology-directed double-strand break repair via loss of TP53BP1 (52, 53) or equivalent factors.
The BRCA1 deficiency DNA repair signature showed prognostic power across a wide variety of breast cancer data sets. Moreover, our mouse protein signature outperformed two commercially available prognostic RNA-based signatures (MammaPrint® and Oncotype DX®) in a data set enriched for homology repair-deficient tumors. Finally, in breast cancer, proteins with prognostic power may have predictive value as well. Examples are the hormone receptor ESR1 and the receptor tyrosine kinase ERBB2, the expression of which response to targeted therapy as well as prognosis (54).
Furthermore, patients with sporadic breast cancer identified as poor outcome by our BRCA1 deficiency signature were highly enriched for p53 mutations. Although both mouse models used to develop the BRCA1 deficiency signature were p53-deficient, this result is explained by the clinical observation that BRCA1-deficient breast cancers frequently comprise p53 mutations, and both BRCA1 and p53 alterations are enriched in triple-negative breast cancer (45, 46). The up-regulation of drug targets involved in DNA repair (PARP1, TOP1, TOP2A, and HDAC1) may indicate the predictive potential of our BRCA1 deficiency DNA repair candidates. We were not able to verify the predictive potential of our BRCA1 deficiency protein signature in large cohorts of treated breast cancers, because the therapeutic agents (PARP1 and TOP inhibition, cisplatin treatment, and histone deacetylase (HDAC) inhibition) are still in clinical trial phase, so no large scale publicly available gene expression data sets exist to date.
Several breast cancer proteomic studies have been reported to date. Biological materials used ranged from mouse tissue (21, 55) to human breast cancer cell lines and tissues (18, 20, 21, 56–58). A few studies yielded a number of markers with potential for treatment prediction. Umar et al. (19) have recently identified a protein profile in microdissected breast tumor cells putatively predictive for the efficacy of tamoxifen. Moreover, the differential up-regulation and activity of a number of kinases across a panel of breast cancer cell lines correlated with differential responsiveness to small molecule inhibitors in these cancer cell lines (59).
Concluding Remarks
Our study demonstrates that in depth high resolution proteomics of tumors tissue from different mouse models is a successful strategy to discover candidate protein biomarkers with screening, prognostic, and possibly also predictive potential for human BRCA1 and homology-directed double-strand break repair-deficient breast tumors. The proteins up-regulated in mammary tumors from mouse models with a deficiency in BRCA1 are enriched in DNA repair(-associated) proteins, which points toward a potential rescue mechanism for the loss of homology-directed DNA repair. In addition, a pathway in conjunction with protein complex analysis has proven to be a promising strategy to construct a signature that has diagnostic and prognostic potential across multiple human breast cancer gene expression data sets. This signature shows specificity for BRCA1 and homology-directed DNA repair deficiency and has high prognostic potential in breast cancer data sets enriched with homology repair deficient tumors. Several up-regulated DNA repair proteins within this signature have been shown to be drug targets in homology-directed DNA repair-deficient tumors, suggesting that they may have predictive power for tailored therapies. Because multiple drug targets are up-regulated, these tumors might also benefit from combination therapy.
Finally, we point out that the BRCA1 deficiency transcriptome signature that we obtained by mapping mouse BRCA1 deficiency-associated breast tumor proteins is novel and could not be obtained by using the published mouse transcriptome data (24) as a starting point. To date, there is only one BRCAness gene expression signature reported for ovarian cancer (42). However, this signature was developed using a publicly available ovarium cancer transcriptomics data set and with a pilot study for predictiveness based on only 10 BRCA mutated/reverted samples originating from six patients, and this signature was not externally evaluated in multiple large (BRCA1/2-deficient) breast cancer data sets. Together, these results underscore the novelty of our BRCAness transcriptome signature that we obtained by mapping mouse BRCA1 deficiency associated breast tumor proteins.
Future studies should address the value of our BRCA1 deficiency signature both at the transcriptome and proteome level for patient selection for treatment in breast cancer and other tumors types with potential homology repair deficiencies. With the advent of targeted mass spectrometry methods like SRM-MS, the signature proteins may be analyzed in pretreatment biopsies in one multiplex analysis, without the need for antibodies. Targeted multiplex analysis in aspirate fluid and plasma may highlight their potential use for noninvasive testing.
Acknowledgments
We acknowledge Anne-Lise Borresen, Anita Langerod, and Hugo Horlings for generation of and access to p53 mutation data from the van de Vijver cohort and Davide Chiasserini for generating the protein complex images using Cytoscape. We also acknowledge Piet Borst for careful reading and adjusting this manuscript.
Footnotes
* This work was supported by funds from the CenE/Van Lanschot (to M. W.) and the VUmc-Cancer Center Amsterdam (to C. R. J., T. V. P., and the proteomics infrastructure), Dutch Cancer Society project grants (to S. R. and J. J.), a Netherlands Organization for Scientific Research (NWO) Vidi grant (to S. R.), and a NWO Cancer Systems Biology Center grant to (J. J.).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- BRCA1
- Breast cancer 1, early onset
- BRCA2
- Breast cancer 2, early onset
- PARP1
- poly[ADP-ribose] polymerase 1
- MS/MS
- tandem mass spectrometry
- SRM
- selected reaction monitoring
- CE
- collision energy
- STRING
- Search Tool for the Retrieval of Interacting Genes/Proteins
- COFECO
- composite function annotation enriched by protein complex data
- p53
- tumor protein p53
- IPA
- Ingenuity Pathways Analysis
- CV
- coefficient of variance.
REFERENCES
- 1. van 't Veer L. J., Dai H., van de Vijver M. J., He Y. D., Hart A. A., Mao M., Peterse H. L., van der Kooy K., Marton M. J., Witteveen A. T., Schreiber G. J., Kerkhoven R. M., Roberts C., Linsley P. S., Bernards R., Friend S. H. (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 [DOI] [PubMed] [Google Scholar]
- 2. Turner N. C., Reis-Filho J. S. (2006) Basal-like breast cancer and the BRCA1 phenotype. Oncogene 25, 5846–5853 [DOI] [PubMed] [Google Scholar]
- 3. Jaspers J. E., Rottenberg S., Jonkers J. (2009) Therapeutic options for triple-negative breast cancers with defective homologous recombination. Biochim. Biophys. Acta 1796, 266–280 [DOI] [PubMed] [Google Scholar]
- 4. Gudmundsdottir K., Ashworth A. (2006) The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene 25, 5864–5874 [DOI] [PubMed] [Google Scholar]
- 5. Audeh M. W., Carmichael J., Penson R. T., Friedlander M., Powell B., Bell-McGuinn K. M., Scott C., Weitzel J. N., Oaknin A., Loman N., Lu K., Schmutzler R. K., Matulonis U., Wickens M., Tutt A. (2010) Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: A proof-of-concept trial. Lancet 376, 245–251 [DOI] [PubMed] [Google Scholar]
- 6. Sørlie T., Perou C. M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M. B., van de Rijn M., Jeffrey S. S., Thorsen T., Quist H., Matese J. C., Brown P. O., Botstein D., Eystein Lønning P., Børresen-Dale A. L. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U.S.A. 98, 10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Glas A. M., Floore A., Delahaye L. J., Witteveen A. T., Pover R. C., Bakx N., Lahti-Domenici J. S., Bruinsma T. J., Warmoes M. O., Bernards R., Wessels L. F., Van't Veer L. J. (2006) Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genomics 7, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jönsson G., Staaf J., Vallon-Christersson J., Ringnér M., Holm K., Hegardt C., Gunnarsson H., Fagerholm R., Strand C., Agnarsson B. A., Kilpivaara O., Luts L., Heikkilä P., Aittomäki K., Blomqvist C., Loman N., Malmström P., Olsson H., Johannsson O. T., Arason A., Nevanlinna H., Barkardottir R. B., Borg A. (2010) Genomic subtypes of breast cancer identified by array-comparative genomic hybridization display distinct molecular and clinical characteristics. Breast Cancer Res. 12, R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Joosse S. A., Brandwijk K. I., Mulder L., Wesseling J., Hannemann J., Nederlof P. M. (2011) Genomic signature of BRCA1 deficiency in sporadic basal-like breast tumors. Genes Chromosomes Cancer 50, 71–81 [DOI] [PubMed] [Google Scholar]
- 10. Naderi A., Teschendorff A. E., Barbosa-Morais N. L., Pinder S. E., Green A. R., Powe D. G., Robertson J. F., Aparicio S., Ellis I. O., Brenton J. D., Caldas C. (2007) A gene-expression signature to predict survival in breast cancer across independent data sets. Oncogene 26, 1507–1516 [DOI] [PubMed] [Google Scholar]
- 11. Paik S., Shak S., Tang G., Kim C., Baker J., Cronin M., Baehner F. L., Walker M. G., Watson D., Park T., Hiller W., Fisher E. R., Wickerham D. L., Bryant J., Wolmark N. (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351, 2817–2826 [DOI] [PubMed] [Google Scholar]
- 12. van de Vijver M. J., He Y. D., van't Veer L. J., Dai H., Hart A. A., Voskuil D. W., Schreiber G. J., Peterse J. L., Roberts C., Marton M. J., Parrish M., Atsma D., Witteveen A., Glas A., Delahaye L., van der Velde T., Bartelink H., Rodenhuis S., Rutgers E. T., Friend S. H., Bernards R. (2002) A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999–2009 [DOI] [PubMed] [Google Scholar]
- 13. Vollebergh M. A., Lips E. H., Nederlof P. M., Wessels L. F., Schmidt M. K., van Beers E. H., Cornelissen S., Holtkamp M., Froklage F. E., de Vries E. G., Schrama J. G., Wesseling J., van d V, van T. H., de B. M., Hauptmann M., Rodenhuis S., Linn S. C. (2012) An aCGH classifier derived from BRCA1-mutated breast cancer and benefit of high-dose platinum-based chemotherapy in HER2-negative breast cancer patients. Ann. Oncol., 22, 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Waddell N., Arnold J., Cocciardi S., da Silva L., Marsh A., Riley J., Johnstone C. N., Orloff M., Assie G., Eng C., Reid L., Keith P., Yan M., Fox S., Devilee P., Godwin A. K., Hogervorst F. B., Couch F., Grimmond S., Flanagan J. M., Khanna K., Simpson P. T., Lakhani S. R., Chenevix-Trench G. (2010) Subtypes of familial breast tumours revealed by expression and copy number profiling. Breast Cancer Res. Treat. 123, 661–677 [DOI] [PubMed] [Google Scholar]
- 15. Straver M. E., Glas A. M., Hannemann J., Wesseling J., van de Vijver M. J., Rutgers E. J., Vrancken Peeters M. J., van Tinteren H., Van't Veer L. J., Rodenhuis S. (2010) The 70-gene signature as a response predictor for neoadjuvant chemotherapy in breast cancer. Breast Cancer Res. Treat. 119, 551–558 [DOI] [PubMed] [Google Scholar]
- 16. Cardoso F., Piccart-Gebhart M., Van't Veer L., Rutgers E. (2007) The MINDACT trial: The first prospective clinical validation of a genomic tool. Mol. Oncol. 1, 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pavlou M. P., Kulasingam V., Sauter E. R., Kliethermes B., Diamandis E. P. (2010) Nipple aspirate fluid proteome of healthy females and patients with breast cancer. Clin. Chem. 56, 848–855 [DOI] [PubMed] [Google Scholar]
- 18. Xu X., Qiao M., Zhang Y., Jiang Y., Wei P., Yao J., Gu B., Wang Y., Lu J., Wang Z., Tang Z., Sun Y., Wu W., Shi Q. (2010) Quantitative proteomics study of breast cancer cell lines isolated from a single patient: Discovery of TIMM17A as a marker for breast cancer. Proteomics 10, 1374–1390 [DOI] [PubMed] [Google Scholar]
- 19. Umar A., Kang H., Timmermans A. M., Look M. P., Meijer-van Gelder M. E., den Bakker M. A., Jaitly N., Martens J. W., Luider T. M., Foekens J. A., Pasa-Tolić L. (2009) Identification of a putative protein profile associated with tamoxifen therapy resistance in breast cancer. Mol. Cell. Proteomics 8, 1278–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lai T. C., Chou H. C., Chen Y. W., Lee T. R., Chan H. T., Shen H. H., Lee W. T., Lin S. T., Lu Y. C., Wu C. L., Chan H. L. (2010) Secretomic and proteomic analysis of potential breast cancer markers by two-dimensional differential gel electrophoresis. J. Proteome Res. 9, 1302–1322 [DOI] [PubMed] [Google Scholar]
- 21. Celis J. E., Gromov P., Cabezón T., Moreira J. M., Ambartsumian N., Sandelin K., Rank F., Gromova I. (2004) Proteomic characterization of the interstitial fluid perfusing the breast tumor microenvironment: A novel resource for biomarker and therapeutic target discovery. Mol. Cell. Proteomics 3, 327–344 [DOI] [PubMed] [Google Scholar]
- 22. Geiger T., Cox J., Ostasiewicz P., Wisniewski J. R., Mann M. (2010) Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods 7, 383–385 [DOI] [PubMed] [Google Scholar]
- 23. Becker S., Cazares L. H., Watson P., Lynch H., Semmes O. J., Drake R. R., Laronga C. (2004) Surfaced-enhanced laser desorption/ionization time-of-flight (SELDI-TOF) differentiation of serum protein profiles of BRCA-1 and sporadic breast cancer. Ann. Surg. Oncol. 11, 907–914 [DOI] [PubMed] [Google Scholar]
- 24. Liu X., Holstege H., van der Gulden H., Treur-Mulder M., Zevenhoven J., Velds A., Kerkhoven R. M., van Vliet M. H., Wessels L. F., Peterse J. L., Berns A., Jonkers J. (2007) Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc. Natl. Acad. Sci. U.S.A. 104, 12111–12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Derksen P. W., Liu X., Saridin F., van der Gulden H., Zevenhoven J., Evers B., van Beijnum J. R., Griffioen A. W., Vink J., Krimpenfort P., Peterse J. L., Cardiff R. D., Berns A., Jonkers J. (2006) Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell 10, 437–449 [DOI] [PubMed] [Google Scholar]
- 26. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 27. Piersma S. R., Fiedler U., Span S., Lingnau A., Pham T. V., Hoffmann S., Kubbutat M. H., Jiménez C. R. (2010) Workflow comparison for label-free, quantitative secretome proteomics for cancer biomarker discovery: Method evaluation, differential analysis, and verification in serum. J. Proteome Res. 9, 1913–1922 [DOI] [PubMed] [Google Scholar]
- 28. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 29. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 30. Pham T. V., Piersma S. R., Warmoes M., Jimenez C. R. (2010) On the beta-binomial model for analysis of spectral count data in label-free tandem mass spectrometry-based proteomics. Bioinformatics 26, 363–369 [DOI] [PubMed] [Google Scholar]
- 31. Albrethsen J., Knol J. C., Piersma S. R., Pham T. V., de Wit M., Mongera S., Carvalho B., Verheul H. M., Fijneman R. J., Meijer G. A., Jimenez C. R. (2010) Subnuclear proteomics in colorectal cancer: Identification of proteins enriched in the nuclear matrix fraction and regulation in adenoma to carcinoma progression. Mol. Cell. Proteomics 9, 988–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun C. H., Kim M. S., Han Y., Yi G. S. (2009) COFECO: Composite function annotation enriched by protein complex data. Nucleic Acids Res. 37, W350-W355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jensen L. J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., Bork P., von Mering C. (2009) STRING 8: A global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Y., Klijn J. G., Zhang Y., Sieuwerts A. M., Look M. P., Yang F., Talantov D., Timmermans M., Meijer-van Gelder M. E., Yu J., Jatkoe T., Berns E. M., Atkins D., Foekens J. A. (2005) Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365, 671–679 [DOI] [PubMed] [Google Scholar]
- 35. Wright M. H., Calcagno A. M., Salcido C. D., Carlson M. D., Ambudkar S. V., Varticovski L. (2008) Brca1 breast tumors contain distinct CD44+/. Breast Cancer Res. 10, R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stuart-Harris R., Caldas C., Pinder S. E., Pharoah P. (2008) Proliferation markers and survival in early breast cancer: A systematic review and meta-analysis of 85 studies in 32,825 patients. Breast 17, 323–334 [DOI] [PubMed] [Google Scholar]
- 37. Cordes N. (2006) Integrin-mediated cell-matrix interactions for prosurvival and antiapoptotic signaling after genotoxic injury. Cancer Lett. 242, 11–19 [DOI] [PubMed] [Google Scholar]
- 38. Wang Y., Cortez D., Yazdi P., Neff N., Elledge S. J., Qin J. (2000) BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14, 927–939 [PMC free article] [PubMed] [Google Scholar]
- 39. Heale J. T., Ball A. R., Jr., Schmiesing J. A., Kim J. S., Kong X., Zhou S., Hudson D. F., Earnshaw W. C., Yokomori K. (2006) Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol. Cell 21, 837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee C. G., Hague L. K., Li H., Donnelly R. (2004) Identification of toposome, a novel multisubunit complex containing topoisomerase IIα. Cell Cycle 3, 638–647 [PubMed] [Google Scholar]
- 41. Osley M. A., Tsukuda T., Nickoloff J. A. (2007) ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat. Res. 618, 65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Konstantinopoulos P. A., Spentzos D., Karlan B. Y., Taniguchi T., Fountzilas E., Francoeur N., Levine D. A., Cannistra S. A. (2010) Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J. Clin. Oncol. 28, 3555–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fong P. C., Boss D. S., Yap T. A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O'Connor M. J., Ashworth A., Carmichael J., Kaye S. B., Schellens J. H., de Bono J. S. (2009) Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 361, 123–134 [DOI] [PubMed] [Google Scholar]
- 44. Turner N., Tutt A., Ashworth A. (2004) Hallmarks of “BRCAness” in sporadic cancers. Nat. Rev. Cancer 4, 814–819 [DOI] [PubMed] [Google Scholar]
- 45. Manié E., Vincent-Salomon A., Lehmann-Che J., Pierron G., Turpin E., Warcoin M., Gruel N., Lebigot I., Sastre-Garau X., Lidereau R., Remenieras A., Feunteun J., Delattre O., de Thé H., Stoppa-Lyonnet D., Stern M. H. (2009) High frequency of TP53 mutation in BRCA1 and sporadic basal-like carcinomas but not in BRCA1 luminal breast tumors. Cancer Res. 69, 663–671 [DOI] [PubMed] [Google Scholar]
- 46. Holstege H., Joosse S. A., van Oostrom C. T., Nederlof P. M., de Vries A., Jonkers J. (2009) High incidence of protein-truncating TP53 mutations in BRCA1-related breast cancer. Cancer Res. 69, 3625–3633 [DOI] [PubMed] [Google Scholar]
- 47. Gorski J. J., James C. R., Quinn J. E., Stewart G. E., Staunton K. C., Buckley N. E., McDyer F. A., Kennedy R. D., Wilson R. H., Mullan P. B., Harkin D. P. (2010) BRCA1 transcriptionally regulates genes associated with the basal-like phenotype in breast cancer. Breast Cancer Res. Treat. 122, 721–731 [DOI] [PubMed] [Google Scholar]
- 48. Helleday T., Petermann E., Lundin C., Hodgson B., Sharma R. A. (2008) DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer 8, 193–204 [DOI] [PubMed] [Google Scholar]
- 49. Rottenberg S., Jaspers J. E., Kersbergen A., van der Burg E., Nygren A. O., Zander S. A., Derksen P. W., de Bruin M., Zevenhoven J., Lau A., Boulter R., Cranston A., O'Connor M. J., Martin N. M., Borst P., Jonkers J. (2008) High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl. Acad. Sci. U.S.A. 105, 17079–17084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zander S. A., Kersbergen A., van der Burg E., de Water N., van Tellingen O., Gunnarsdottir S., Jaspers J. E., Pajic M., Nygren A. O., Jonkers J., Borst P., Rottenberg S. (2010) Sensitivity and acquired resistance of BRCA1: p53-deficient mouse mammary tumors to the topoisomerase I inhibitor topotecan. Cancer Res. 70, 1700–1710 [DOI] [PubMed] [Google Scholar]
- 51. Rottenberg S., Nygren A. O., Pajic M., van Leeuwen F. W., van, der, Heijden I., van de Wetering K., Liu X., de Visser K. E., Gilhuijs K. G., van Tellingen O., Schouten J. P., Jonkers J., Borst P. (2007) Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc. Natl. Acad. Sci. U.S.A. 104, 12117–12122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bouwman P., Aly A., Escandell J. M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q., Haffty B. G., Tommiska J., Blomqvist C., Drapkin R., Adams D. J., Nevanlinna H., Bartek J., Tarsounas M., Ganesan S., Jonkers J. (2010) 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17, 688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bunting S. F., Callén E., Wong N., Chen H. T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., Xu X., Deng C. X., Finkel T., Nussenzweig M., Stark J. M., Nussenzweig A. (2010) 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cianfrocca M., Goldstein L. J. (2004) Prognostic and predictive factors in early-stage breast cancer. Oncologist 9, 606–616 [DOI] [PubMed] [Google Scholar]
- 55. Sun B., Zhang S., Zhang D., Li Y., Zhao X., Luo Y., Guo Y. (2008) Identification of metastasis-related proteins and their clinical relevance to triple-negative human breast cancer. Clin. Cancer Res. 14, 7050–7059 [DOI] [PubMed] [Google Scholar]
- 56. Cha S., Imielinski M. B., Rejtar T., Richardson E. A., Thakur D., Sgroi D. C., Karger B. L. (2010) In situ proteomic analysis of human breast cancer epithelial cells using laser capture microdissection: Annotation by protein set enrichment analysis and gene ontology. Mol. Cell. Proteomics 9, 2529–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mbeunkui F., Metge B. J., Shevde L. A., Pannell L. K. (2007) Identification of differentially secreted biomarkers using LC-MS/MS in isogenic cell lines representing a progression of breast cancer. J. Proteome Res. 6, 2993–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ou K., Yu K., Kesuma D., Hooi M., Huang N., Chen W., Lee S. Y., Goh X. P., Tan L. K., Liu J., Soon S. Y., Bin Abdul Rashid S., Putti T. C., Jikuya H., Ichikawa T., Nishimura O., Salto-Tellez M., Tan P. (2008) Novel breast cancer biomarkers identified by integrative proteomic and gene expression mapping. J. Proteome Res. 7, 1518–1528 [DOI] [PubMed] [Google Scholar]
- 59. Hochgräfe F., Zhang L., O'Toole S. A., Browne B. C., Pinese M., Porta Cubas A., Lehrbach G. M., Croucher D. R., Rickwood D., Boulghourjian A., Shearer R., Nair R., Swarbrick A., Faratian D., Mullen P., Harrison D. J., Biankin A. V., Sutherland R. L., Raftery M. J., Daly R. J. (2010) Tyrosine phosphorylation profiling reveals the signaling network characteristics of Basal breast cancer cells. Cancer Res. 70, 9391–9401 [DOI] [PubMed] [Google Scholar]