Abstract
Fundamental open questions in signal transduction remain concerning the sequence and distribution of molecular signaling events among individual cells. In this work, we have characterized the intercellular variability of transforming growth factor β-induced Smad interactions, providing essential information about TGF-β signaling and its dependence on the density of cell populations and the cell cycle phase. By employing the recently developed in situ proximity ligation assay, we investigated the dynamics of interactions and modifications of Smad proteins and their partners under native and physiological conditions. We analyzed the kinetics of assembly of Smad complexes and the influence of cellular environment and relation to mitosis. We report rapid kinetics of formation of Smad complexes, including native Smad2-Smad3-Smad4 trimeric complexes, in a manner influenced by the rate of proteasomal degradation of these proteins, and we found a striking cell to cell variation of signaling complexes. The single-cell analysis of TGF-β signaling in genetically unmodified cells revealed previously unknown aspects of regulation of this pathway, and it provided a basis for analysis of these signaling events to diagnose pathological perturbations in patient samples and to evaluate their susceptibility to drug treatment.
Transforming growth factor-β (TGF-β)1 controls a diverse array of cellular processes, including cell proliferation, differentiation, apoptosis, and determination of developmental fate during embryogenesis (1, 2). TGF-β binding to the serine/threonine kinase type II receptor (TβRII) promotes the formation of a complex with the type I receptor (TβRI), whereafter the latter is phosphorylated and activated. Important substrates for the TβRI are the receptor-regulated Smads (R-Smads), Smad2 and Smad3 (3), which after C-terminal phosphorylation accumulate in the nucleus, where they form heteromeric complexes with transcriptional factors, co-repressors, and co-activators to up- or down-regulate transcription of target genes (1, 2, 4). Among the crucial limiting regulators of the TGF-β pathway are E3 ubiquitin ligases that influence the duration of Smad signaling by promoting ubiquitin-mediated proteasomal degradation of receptors and Smads. E3 ligases also promote signaling by degrading repressors of the pathway. Common mediator Smad and R-Smads form complexes with SnoN (Ski-related novel protein N) and Ski (Sloan-Kettering avian retrovirus transforming protein), transcription repressors that inhibit formation of transcriptionally active heteromeric Smad complexes or recruit co-repressor complexes to the chromatin of target genes (5–7). SnoN ubiquitination and proteasomal degradation is a required step in activation of TGF-β signaling. Thus, in response to TGF-β, SnoN in complex with activated Smad2/3 recruits E3 ligases, which mediate its ubiquitin-dependent degradation (8, 9). Earlier studies of Smad interactors have mostly relied on engineered systems of transfected overexpressing cells, with measurements made across populations of cells. Because of the limitations of such methods, important questions remain about mechanisms and kinetics of endogenous cell signaling, about the localization of complexes within different cells and compartments of the cell, and about the quantitative nature of these processes.
In this paper, we describe spatial and temporal aspects of the formation of Smad complexes in situ, under physiological conditions, using the in situ proximity ligation assay (PLA) (10). The ability to resolve and enumerate individual protein-protein interaction events has enabled us to present quantitative data of Smad complex formation and localization within compartments of single cells. Our data support and extend earlier findings about TGF-β signaling and demonstrate the potential of the in situ PLA method to reveal new mechanisms of regulation of cell signaling in genetically unmodified cells and in human tissue samples at cellular and subcellular resolution.
EXPERIMENTAL PROCEDURES
Cell Culture
Mouse embryonic fibroblasts, a human immortalized nontransformed keratinocyte epithelial cell line (HaCaT), and a mouse mammary gland cell line (NMuMG) were grown in high glucose Dulbecco's modified Eagle's medium (Sigma). Human hepatocellular liver carcinoma (HepG2) and human breast carcinoma (MDA-MB-468) cell lines were cultured in RPMI (Sigma). Media were supplemented with 10% FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin (all from Sigma). For in situ PLA experiments, the cells were seeded at a density of 10,000 cells/well onto SuperFrost Ultra Plus slides (Menzel Glaser) 48 h before treatment. For purposes of blocking basal TGF-β signaling in unstimulated cells, the low molecular weight inhibitor GW6604 (11) was added to the cells at a concentration of 5 μm 2 h prior to stimulation. After washing in PBS (137 mmol/liter NaCl, 10 mmol/liter phosphate, 2.7 mmol/liter KCl, pH 7.4), the stimulated cells were incubated in the presence or absence of TGF-β1 (10 ng/ml; BioSource/Invitrogen) in Dulbecco's modified Eagle's medium for 45 min. After stimulation, the cells were fixed in 3% paraformaldehyde (Sigma-Aldrich) for 15 min at room temperature and permeabilized for 10 min with 0.5% Triton X-100 (Sigma-Aldrich) in PBS.
Tissue Preparation
Tissue microarrays with 4-μm cores were prepared from normal colon tissue fixed in PBS-buffered 10% formalin and embedded in paraffin according to standard pathology laboratory protocols. The tissue microarrays were then baked for 30 min at 60 °C, de-paraffinized in xylene (Histolab), and incubated at 125 °C, under high pressure conditions in a pressure cooker (Biocare Medical) in 600 ml of antigen retrieval solution (Dako) for 1 min.
Preparation of Proximity Probes for Detection of Triple Complexes
A thiol-modified rolling circle amplification (RCA) primer oligonucleotide SH-AAAAAAAAAATATTGACAGAACTAGACACTT or the nonpriming oligonucleotides SH-AAAAAAAAAAATGGCCGACTCACGAATTAGA[UUU] and SH-AAAAAAAAAAGACGCTAATAGTTAAGACGCTT[UUU] ([UUU] is 2′O-methyl-RNA) (Eurogentec) were used for conjugation to antibodies against Smad2, Smad3 (made at the Ludwig Institute for Cancer Research, Uppsala), and Smad4 (Santa Cruz, sc-7966, B-8). Before conjugation, the antibodies were purified using ImmunoPure immobilized protein G (Pierce) and dialyzed overnight against PBS using a Slide-A-Lyzer dialysis cassette (Pierce). Antibodies were modified by mixing with a 30-fold molar excess of sulfosuccinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate (Pierce), freshly prepared in DMSO for 2 h at room temperature. In parallel, 300 pmol of each oligonucleotide was reduced with 10 mm dithiothreitol (Sigma) in 55 mm phosphate buffer, 150 mm NaCl and 20 mm EDTA (pH 7.2) for 1 h at 37 °C. The reactions were followed by purification and buffer exchange to 5 mm EDTA in PBS using MicroSpin G-50 columns (Amersham Biosciences). The modified antibodies and oligonucleotides were then combined in Slide-A-Lyzer MINI dialysis units (Pierce) and dialyzed against PBS overnight at 4 °C for conjugation to construct PLA probes.
In Situ PLA Protocol
Paraformaldehyde fixed cells and tissue sections were incubated with blocking solution (Olink Bioscience) for 2 h at room temperature followed by overnight incubation with primary antibodies (rabbit Smad2, 1641-1 Epitomics; rabbit Smad3, 1735-1 Epitomics; rabbit SnoN sc-9141 (H-317), Santa Cruz Biotechnology; rabbit Ski sc-9140 (H-329), Santa Cruz Biotechnology; mouse Smad2/3, 610842 BD Bioscience; goat pSmad2/3 sc-11769, Santa Cruz Biotechnology) or PLA probes (for triple complex detection) diluted 1:100 in antibody diluent (Olink Bioscience). The incubation was followed by washes in three changes for 5 min in Tris-buffered saline with Tween (TBS-T; 0.05 m Tris base, 150 mm NaCl, pH 8.4, with 0.05% Tween 20) and, for cells incubated with primary antibodies, with 2 h of incubation with anti-mouse and anti-rabbit PLA probes (Olink Bioscience) diluted 1:10 in antibody diluent (Olink Bioscience). The slides were then washed three times for 5 min each with TBS-T.
The cells and the tissue were next incubated with DuolinkII ligation solution (Olink Bioscience) at 37 °C for 30 min, followed by washes in two changes for 2 min each in TBS-T. For detection of triple complexes, three connector oligonucleotides (P-CTATTAGCGTCCAGTGAATGCGAGTCCGTCTAAGAGAGTA, P-CTATTAGCGTCAGCGATCTGCGAGACCGTATAAGAGAGTAGTACAGCAGCCGTCAAGAGTGTCTA, and P-GTTCTGTCATATTTAAGCGTCTTAA) mixed at a concentration of 62.5 nm in T4 DNA ligase buffer, 0.25 m NaCl, 0.05% Tween 20, 0.1 mm ATP, 0.37 mg/ml BSA, and 56 units/ml T4 DNA ligase in water were applied. For amplification of the ligation products and detection of complexes, DuolinkII amplification solution (Olink Bioscience) was applied on slides for 90 min at 37 °C, followed by two washes in TBS-T. To identify the localization of in situ PLA signals in nuclei and cytoplasm, the cells were counterstained with Hoechst and Alexa Fluor 488 phalloidin (Invitrogen), respectively.
Immunofluorescence
Following the in situ PLA, the cells and the tissues were washed in TBS-T and incubated with rabbit anti-histone H3 phospho-S10 (Millipore, 16-222) or anti-Ki67 (Abcam, ab15580), FITC-conjugated primary antibodies at 37 °C for 30 min, subsequently washed three times in TBS-T and mounted with SlowFade mounting medium (Invitrogen).
Image Acquisition and Analysis
The images were acquired using a Zeiss AxioPlan2 microscope with a 20×/0.8 Plan-Apo objective and an Axiocam MRm camera. The DuolinkImageTool software was used for image analysis. Fluorescent signals from RCA products were defined and counted per cell with distinction of the nucleus and the cytoplasm. For each experiment, signals per nuclei and cytoplasm from 150–200 cells were quantified.
Immunoblotting
HaCaT cells were pretreated for 2 h with GW6604 inhibitor (5 μm) diluted in DMSO and stimulated with TGF-β1 (10 ng/ml). After stimulation, the cells were lysed in 50 mm Hepes-KOH, pH 7.8, with 5 mm EDTA, 500 mm NaCl, and 1% Nonidet P-40 (all from Sigma-Aldrich). Immunoblotting experiments were performed according to standard procedures with 10% milk powder in TBS for 1 h at room temperature as blocking reagent and antibody diluent. The primary antibodies used were rabbit anti-pSmad3 (1:500; Epitomics, 1880-1), rabbit anti-Smad3 (1:1000; Epitomics, 1735-1), and mouse anti-tubulin (1:500; Sigma, T4026). The secondary anti-rabbit IgG horseradish peroxidase-labeled (NA-93) and anti-mouse IgG horseradish peroxidase-labeled (NA-931) were purchased from Amersham Biosciences and used at a 1:30,000 dilution in TBS-T.
RESULTS
Enumeration of Endogenous Complexes between Smad2, Smad3, and Smad4 during TGF-β Signaling
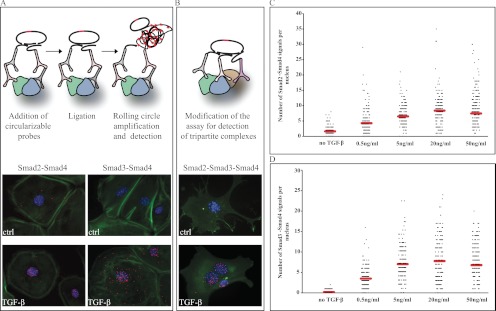
It is generally established that TGF-β signaling proceeds when R-Smads, upon phosphorylation by the receptor, form complexes with Smad4 and accumulate in the nucleus to regulate transcription of target genes. To investigate complex formation between endogenous Smad proteins in individual cells during TGF-β signaling, we used the in situ PLA method (Fig. 1A). We first examined Smad2-Smad4 and Smad3-Smad4 complex formation in mouse embryonic fibroblast cells. Increased numbers of both types of complexes, reflected by brightly fluorescent RCA products, were observed in cells with a peak at 45 min after TGF-β stimulation (Fig. 1A). The vast majority of complexes were observed in the nuclei; however, cytoplasmic complexes were also seen. No clear-cut qualitative or quantitative differences were detected between Smad2-Smad4 and Smad3-Smad4 complexes, leaving the possibility open that at least a fraction of native complexes are in fact Smad2-Smad3-Smad4 trimers, as proposed by in vitro crystallographic studies (12).
Fig. 1.
Detection of Smad2, Smad3, and Smad4 complexes in cultured cells. Mouse embryonic fibroblasts were stimulated with TGF-β (10 ng/ml) for 45 min; thereafter, complexes between Smad2-Smad4 and Smad3-Smad4 proteins (A) or between Smad2, Smad3, and Smad4 proteins (B) were determined by the in situ PLA method. Forty-eight hours after seeding the cells, signaling through the TβRI was blocked by the kinase inhibitor GW6604 in HaCaT cells (no TGF-β), followed by treatment with different concentrations of TGF-β for 45 min, fixation, and incubation with antibodies against Smad2 and Smad4 (C) or Smad3 and Smad4 (D). Formation of complexes was visualized using hybridization probes labeled with Alexa 555 (orange). The cytoplasm was stained with Alexa 488-labeled phalloidin (green), and the nuclei were stained with Hoechst (blue). The RCA signals were quantified using the DuolinkImageTool software. Mean RCA values are indicated by red lines (C and D). ctrl, control.
Next we investigated whether tri-partite complexes between Smad2, Smad3, and Smad4 proteins occur in mouse embryonic fibroblast cells, using a modified in situ PLA protocol where detection depends on binding by a set of three oligonucleotide-modified antibodies, each directed against one of the proteins in the complex. The assay revealed triple complexes between Smad2, Smad3, and Smad4 (Fig. 1B). Once again, these complexes were observed primarily in the nuclei of stimulated cells, with few specific signals in the cytoplasm. The method does not allow us to conclude whether all the Smad complexes in a given cell represent trimers or whether cells accumulate mixed populations of Smad oligomers of diverse stoichiometric compositions.
We next studied the TGF-β-induced formation of complexes between Smad3 and Smad4 in a panel of cell lines to establish the generality of this approach for measuring cellular responses to TGF-β. As expected, in Smad4-positive HepG2, HaCaT, and NMuMG cells, we consistently observed significant accumulation of Smad3-Smad4 complexes in nuclei of stimulated cells, compared with untreated cells (supplemental Fig. S1) (13). The use of several cell lines established the generality of detection of Smad complexes in cells with active TGF-β signaling.
Low Doses of TGF-β Induce Smad Complex Formation
To investigate whether formation of Smad complexes depended on the concentration of TGF-β added to the culture medium, HaCaT cells were treated for 45 min with TGF-β at concentrations ranging from 0.5 to 50 ng/ml, and we used in situ PLA to investigate assembly of complexes involving Smad2-Smad4 and Smad3-Smad4 (Fig. 1, C and D). We detected a significant increase of RCA signals in nuclei of stimulated cells using as little as 0.5 ng/ml of the ligand. The highest accumulation of complexes between Smad2 and Smad4 was observed at a concentration of 20 ng/ml (Fig. 1C). Similarly, the dose-dependent assembly of Smad3-Smad4 complexes reached a maximum at a concentration of 20 ng/ml (Fig. 1D). We therefore conclude that TGF-β causes formation of Smad complexes already at very low concentrations, increasing in a dose-dependent manner.
Formation of Smad Complexes Shows Rapid Kinetics
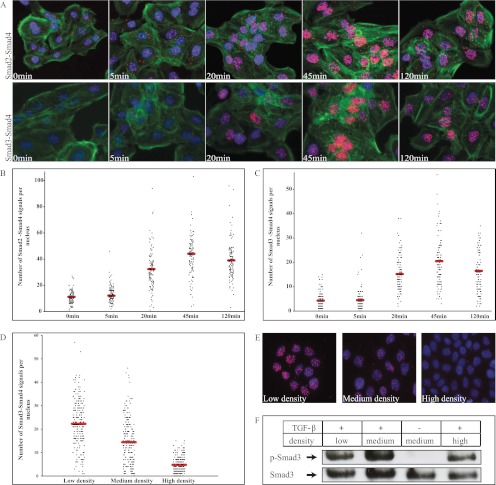
We next analyzed the kinetics of assembly of Smad2-Smad4 and Smad3-Smad4 complexes in HaCaT cells upon TGF-β treatment (Fig. 2, A–C). We observed significant accumulation of Smad2-Smad4 and Smad3-Smad4 complexes in nuclei of stimulated cells after 20 min (Fig. 2A). This increase was sustained up to 45 min after the addition of TGF-β, when the number of complexes attained a maximal value before decreasing again, but with broad intercellular variability of numbers of detected complexes, possibly reflecting the asynchronous nature of a cell population in terms of cell cycle stage or the relative position of individual cells in the population in terms of numbers of neighboring cells and developed cell-cell contacts. Almost identical kinetics of assembly was observed for Smad2-Smad4 and Smad3-Smad4 complexes (Fig. 2, B and C). The nuclear complexes observed at 0 min of TGF-β stimulation may represent constitutively active Smad complexes in the cell.
Fig. 2.
Time course of TGF-β-induced Smad complexes. HaCaT cells were stimulated with TGF-β for 5, 20, 45, and 120 min. The number of nuclear Smad2-Smad4 complexes increased significantly during the first 20 min of incubation with 10 ng/ml TGF-β (A and B). A minor increase of complexes of Smad3-Smad4 was observed already after 5 min of stimulation with TGF-β (A and C). To investigate the effect of cell density on the number of Smad interactions, HaCaT cells were seeded at three densities (50 cells/104 μm2, 30 cells/104 μm2, and 15 cells/104 μm2), and formation of the Smad3-Smad4 complexes was analyzed using in situ PLA (D and E). The cells seeded at the same densities were used for immunoblot analysis of C-terminal phosphorylation (p-Smad3) and total levels of Smad3 (F). The red signals obtained with hybridization probes labeled with Alexa 555 (orange) represent interaction between Smad2 and Smad4 and between Smad3 and Smad4. For cytoplasmic and nuclear visualization, Alexa 488-labeled phalloidin (green) and Hoechst (blue), respectively, were used (A).
Interactions between Smads Depend on the Density of Cells
To explore a possible effect of cell density on Smad complex formation, we recorded complexes of Smad3 and Smad4 in cells seeded at different densities and cultured for 24 h before stimulation with TGF-β. We investigated cells grown at high density (50 cells/104 μm2), medium density (30 cells/104 μm2), and low density (15 cells/104 μm2) and observed large variation in numbers of Smad3-Smad4 complexes (Fig. 2, D and E). In cells grown at the lowest density, the number of Smad3-Smad4 complexes was higher compared with cells seeded more densely, and in very compact cultures, Smad3-Smad4 complexes were barely detectable. We compared the in situ PLA data with the phosphorylation status of Smad3, as analyzed by immunoblotting. In contrast to the results of PLA analysis, we observed relatively similar levels of Smad3 phosphorylation among cells seeded at different densities (Fig. 2F). This suggests that cell density regulates R-Smad-Smad4 complex formation downstream of R-Smad C-terminal phosphorylation. It also raises the possibility that the well established measurement of phospho-Smad2/3 levels as an indicator of TGF-β signaling activity in cells and tissues may not always correspond to productive signaling, thus highlighting the importance of complementary technologies as used herein.
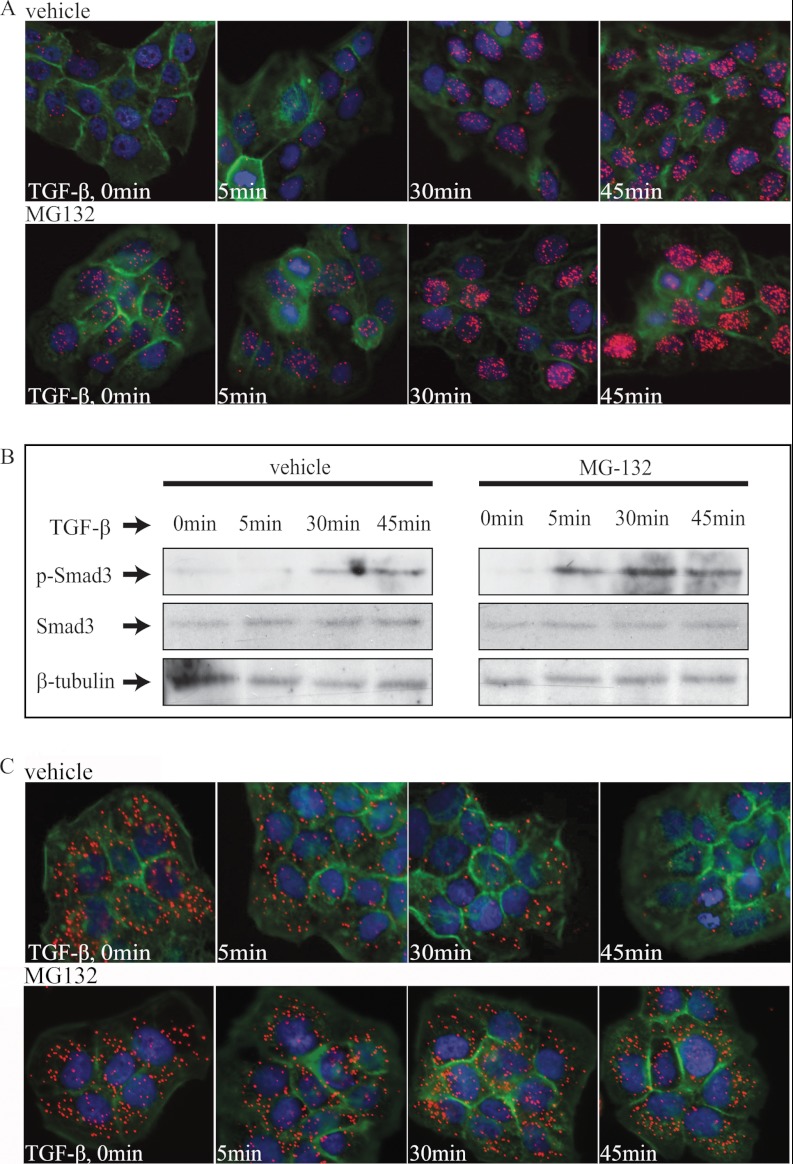
Proteasomal Degradation of Smad Complexes at Early Time Points of TGF-β Signaling
Smad complexes are subject to ubiquitin-mediated proteasomal degradation. For this reason, we explored the effect on Smad protein interactions by inhibiting degradation using the proteasomal inhibitor MG132. Interestingly, in the presence of MG132, the quantity of Smad3-Smad4 complexes was increased during the first minutes of TGF-β stimulation (Fig. 3A and supplemental Fig. S2A). Moreover, cells treated with the TβRI inhibitor GW6604 but not stimulated with TGF-β also exhibited increased levels of Smad complexes (Fig. 3A and supplemental Fig. S2A). These findings suggest that even in the absence of TGF-β stimulation, Smad3-Smad4 complexes are continuously formed at low levels and degraded immediately by proteasomes and that blocking of this degradation leads to their accumulation. Our quantitative data on Smad interactions in situ were compared again with the C-terminal phosphorylation status of R-Smads, as analyzed by immunoblotting (Fig. 3B and supplemental Fig. S2B). Phosphorylation of Smad3 was detected after 5 min of TGF-β treatment in the presence of MG-132 and peaked at 45 min. In contrast, in cells that did not receive MG-132, no phosphorylated Smad3 was observed at early time points, consistent with the data for individual cells shown in Fig. 3A. It is also worth noting that in situ PLA revealed Smad3-Smad4 complexes at very early time points of signaling (5–15 min); however, under the most common physiological conditions, the Smad complexes accumulate with the kinetics described above (Fig. 2). We then investigated possible mechanisms that may reduce the lifespan of individual R-Smad proteins under normal conditions, using in situ PLA to monitor interactions of R-Smads and SnoN or Ski in HaCaT cells. SnoN and Ski are known to bind to R-Smads and to then undergo proteasomal degradation (14–16). High numbers of cytoplasmic phospho-Smad2/3-SnoN and phospho-Smad2/3-Ski complexes were observed in cells where TβRI signaling was blocked by GW6604 (Fig. 3C and supplemental Fig. S3D). Stimulation with TGF-β caused a time-dependent decrease in phospho-Smad2/3-Ski complexes (Fig. 3C and supplemental Fig. S2C). In the presence of MG132, the quantity of phospho-Smad2/3-Ski complexes did not change with addition of TGF-β and remained constant at all time points, consistent with a role of the proteasome in regulating Ski levels. As anticipated from the literature, MG132 efficiently abolished TGF-β-dependent degradation of Ski complexes that positively correlates with increased formation of Smad complexes. This suggests that the efficient turnover of rapidly forming Smad complexes can be regulated by the activity of the Smad co-factors SnoN and Ski.
Fig. 3.
Effect of proteasomal inhibition on formation of Smad3-Smad4 complexes. A, HaCaT cells were pretreated for 2 h with GW6604 inhibitor (5 μm) to block basal TGF-β signaling and for 1 h with MG132 (25 μm) to inhibit proteasomal degradation of proteins prior to stimulation. MG132 was also present during stimulation with TGF-β for 5, 30, and 45 min. The Smad3-Smad4 complex formation was visualized using hybridization probes labeled with Alexa 555 (orange). The cytoplasm was stained with FITC Alexa 488-labeled phalloidin (green), and the nuclei were stained with Hoechst (blue). Analysis by immunoblotting revealed an enhanced C-terminal phosphorylation of Smad3 (P-Smad3) upon stimulation with TGF-β in the presence of MG132 in comparison with control (vehicle). B, the level of total Smad3 and α-tubulin remained constant at all time points. HaCaT cells were pretreated for 2 h with GW6604 inhibitor (5 μm) and for 1 h with MG132 (25 μm) and then stimulated with TGF-β at a concentration of 10 ng/ml. C, C-terminally phosphorylated Smad2/3 and Ski complexes were visualized by in situ PLA.
Smads Form Complexes with SnoN and Ski in a Cell Cycle-dependent Manner
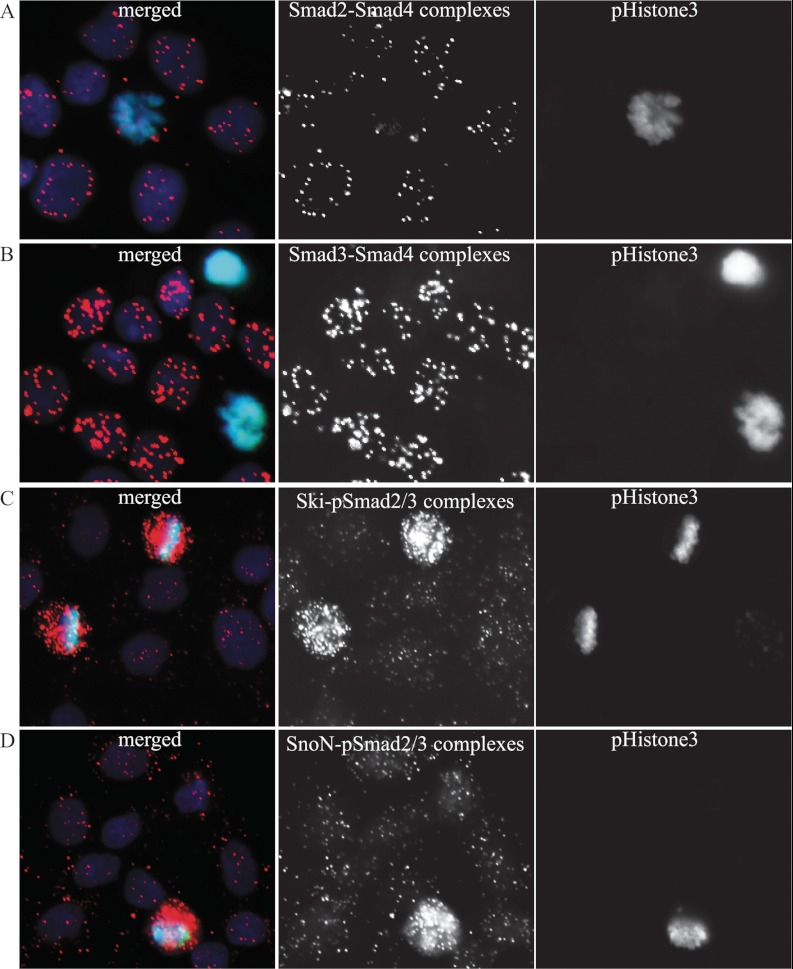
To explain the observed differences in numbers of complexes among individual cells in a population, we explored the possibility that the assembly of protein complexes may vary during the cell cycle. As a marker for mitosis, we used an antibody against a phosphorylated form of histone H3 to distinguish mitotic cells from cells in interphase. We also performed analysis of complex formation in cells sorted by the DNA content and by this separated into different cell cycle subpopulations. The analysis revealed that mitotic cells lacked signals representing complexes between Smad proteins, whereas cells in interphase responded to TGF-β treatment by forming Smad protein complexes (Fig. 4, A and B, and supplemental Fig. S4).
Fig. 4.
R-Smad-Smad4 complexes are excluded while R-Smad-Ski/SnoN complexes accumulate in mitotic cells. HaCaT cells were seeded 40 h before treatment with the TGF-β type I receptor inhibitor GW6604. Smad2-Smad4 (A), Smad3-Smad4 (B), C-terminally phosphorylated Smad2/3-Ski (C), and C-terminally phosphorylated Smad2/3-SnoN (D) complexes were visualized by in situ PLA, followed by phospho-histone 3 immunofluorescence to mark mitotic cells. Mitotic cells showed no evidence of Smad complexes in their nuclei or cytoplasm. The cells in interphase showed robust nuclear Smad complexes. Conversely, mitotic cells showed strong accumulation of nuclear pSmad2/3-Ski/SnoN complexes, whereas cells in interphase lacked the latter complexes. The nuclei were stained with Hoechst (blue), and p-histone3 was detected with Alexa 488-labeled anti-p-histone3 antibody (green). The complex formation was visualized using hybridization probes labeled with Alexa 555 (orange).
We explored the possibility that the investigated Smad complexes might be sequestered by SnoN and Ski during mitosis, because these proteins are known to be potent transcriptional repressors of TGF-β signaling (17). Interestingly, in situ PLA experiments with antibodies against SnoN, Ski, and R-Smads revealed elevated numbers of these complexes in mitotic cells (Fig. 4, C and D, and supplemental Fig. S4) and a low level of complexes in cells in interphase. Additionally, we found that in HaCaT cells, complexes of SnoN or Ski and R-Smads were predominantly cytoplasmic (supplemental Fig. S3D), and their levels depended on cell density and on the proximity of individual cells to the edge of the monolayer (supplemental Fig. S3). We thus conclude that accumulation of Smad complexes is optimal during interphase, and the SnoN/Ski co-factors may play critical roles in limiting the availability of such complexes when cells divide (supplemental Fig. S4).
Smad Complex Formation Varies among Healthy and Cancer Patient Tissue Samples
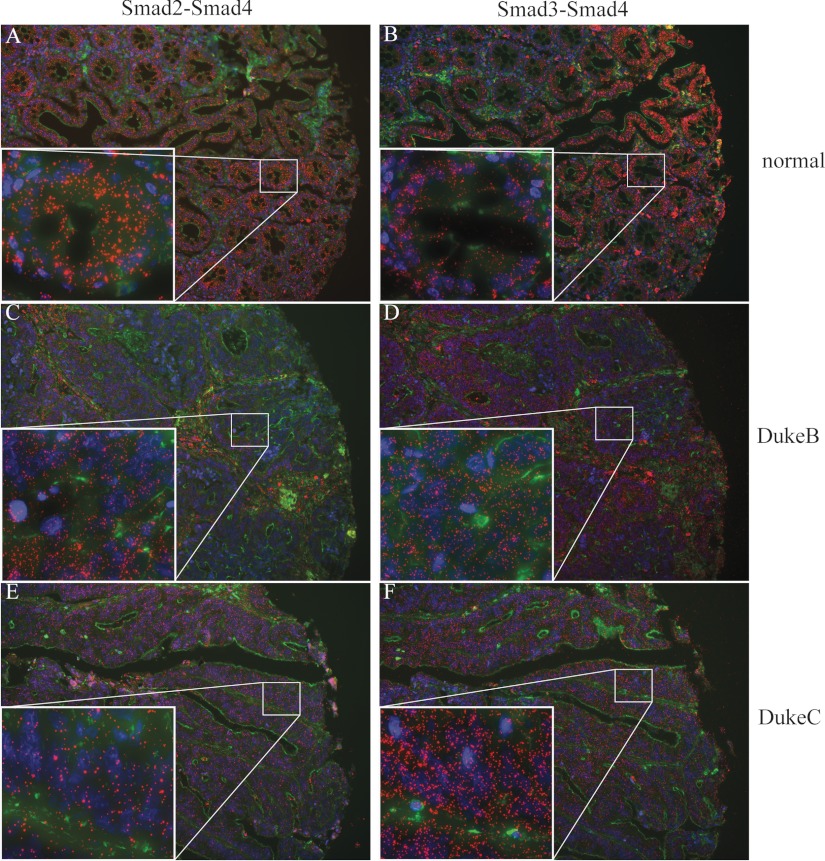
We investigated the feasibility of using in situ PLA to analyze the presence of interacting Smad proteins for diagnostic purposes in the intestinal epithelium. Colonic epithelium was chosen based on the strong evidence of the role of TGF-β signaling during normal tissue homeostasis and colon cancer development (1, 2). The data demonstrated Smad2-Smad4 and Smad3-Smad4 complexes predominantly localized in the cytoplasm of cells in formalin-fixed normal colon mucosa (Fig 5, A and B). In contrast, in colorectal tumors diagnosed as Duke B and C, stages the signals were observed both in nuclei and cytoplasm (Fig 5, C–F). The distribution of signals through the normal tissue sections was variable and localized to specific areas, whereas in samples from cancer patients the complexes were more evenly distributed, which is consistent with the generally observed more active TGF-β signaling in tumors or inflammatory lesions (1, 2). This is an interesting result that implies that the analyzed normal colonic tissue sections were captured at a stage where TGF-β signaling was rather weak or had initiated but possibly had not yet progressed at the phase where the Smad complexes accumulated also in the nucleus. Alternatively, the normal colonic mucosa samples that we have obtained from different individuals may indicate the presence of a sequestering mechanism that maintains the Smad complexes in the cytoplasm under normal homeostatic conditions. Both alternatives are now investigated more thoroughly.
Fig. 5.
Detection of Smad2, Smad3, and Smad4 complexes in human tissue samples. The presence of Smad2-Smad4 (A, C, and E) and Smad3-Smad4 (B, D, and F) complexes were analyzed in normal mucosal epithelial cells of colon tissue obtained from healthy individuals (A and B), from the adenoma adenomatous part of colorectal cancer (C and D), and in invasive adenocarcinoma (E and F). The cytoplasm was stained with FITC Alexa 488-labeled phalloidin (green), and the nuclei were stained with Hoechst (blue).
DISCUSSION
Analyses of cellular responses are generally done at the population level, ignoring sources of heterogeneity and thereby complicating interpretation of how cells coordinate responses to changes in the external and internal environments. Here we have used the in situ PLA technique to characterize responses by SMAD proteins to stimulation via TGF-β at cellular and subcellular resolution in genetically unmodified cells. We illustrate that the analyses yield new cell biological insights and provide a basis for medically relevant studies of alterations of cellular responses in tumors and in response to targeted therapies.
It is well established that TGF-β stimulates assembly of heteromeric complexes between Smad2 and Smad4, or Smad3 and Smad4 (18). Heteromeric Smad complexes are usually analyzed in cells transfected with gene constructs, and they have not been measured in single cells. Here we demonstrate a means to provide new insights in the signaling pathway by analyzing individual endogenous complexes activated by TGF-β stimulation at cellular and subcellular resolution.
Using in situ PLA, we demonstrate accumulation of heteromeric complexes of R-Smads and Smad4 in cell nuclei in response to TGF-β stimulation. Moreover, we show for the first time in situ the formation and nuclear accumulation of endogenous heterotrimeric complexes of Smad2, Smad3, and Smad4, possibly representing functional trimers. This is in agreement with previous in vitro crystallographic evidence suggesting that the major Smad complex is a trimer (12). The interfaces for trimer formation are highly preserved among the R-Smads, and it appears structurally possible for Smad4 to simultaneously engage two R-Smad molecules in a heterotrimeric complex. The formation of diverse classes of Smad complexes composed of different molecules may be a potential source of variability of nuclear actions by Smad proteins. The range of promoter recognition may be extended by Smad content-dependent binding of trimers to promoters that include different numbers of Smad boxes. The assembly of heterotrimeric Smad complexes can significantly increase the possible range of signaling complexes and thus contribute to the variety of biological activities induced by TGF-β. Although trimeric complexes were observed in the nuclei, we are not able to exclude that dimers may also be present.
The quantitative in situ analyses revealed subsets of cells with different responsiveness to TGF-β treatment, as reflected by distinct numbers of protein complexes. We observed not only that the numbers of complexes differed depending on the local cellular environment and the density of the cells but also as a function of the phase of the cell cycle (Fig. 4 and supplemental Figs. S3 and S4). The ability of TGF-β to induce assembly of Smad protein complexes was strikingly reduced in confluent cell cultures, whereas cells at the borders of monolayers or growing alone in small groups of cells exhibited greater numbers of interactions (supplemental Fig. S3). Interestingly, we did not observe any decrease in the phosphorylation status of the Smad3 C terminus in dense cells. Smad3 has been shown to be phosphorylated not only by TGF-β receptors at its C terminus, but also by cyclin-dependent kinases in the linker region, which may block the formation of complexes with other Smads and the attendant transcriptional activity (19, 20). Therefore, Smad3 can be structurally unable to bind Smad4 because of linker modifications, although phosphorylated at its C terminus. It has also been shown that additional inhibitory proteins, such as TAZ/YAP proteins that become activated by the Hippo pathway are recruited to the phosphorylated linker of Smad proteins in a cell density-dependent manner, resulting in sequestration of phosphorylated Smads and overall suppression of TGF-β signaling (21). Accordingly, the TGF-β signaling pathway can be fine-tuned by a number of mechanisms, such as the dynamics of Smad phosphorylation in response to diverse stimuli, the presence of interacting partners, and the cell context (22).
We used phosphorylated histone 3 as a mitotic marker together with PLA analysis of Smad protein interactions on FACS sorted cells, revealing that the number of complexes of Smad4 and R-Smads were reduced in cells undergoing mitotic division. This suggests a cell cycle-dependent regulation of TGF-β signaling (Fig. 4 and supplemental Fig. S4). It is interesting to find that in keratinocytes like HaCaT where TGF-β induces robust cell cycle arrest at the G1 phase (1, 2), the formation of Smad complexes appears very active at this cell cycle phase. This suggests that maximal TGF-β signaling activity correlates with the physiological effect this activity elicits in the responding epithelial cells. Our findings are also consistent with recent reports from frog embryos where Smad proteins were shown to segregate asymmetrically during mitosis and be degraded by proteasomes located in centrosomes (23).
One mechanism that could explain the lowered signaling via Smad proteins during mitosis is cytoplasmic sequestration of Smads by SnoN and Ski, which therefore could regulate the pace of signaling and responsiveness to TGF-β (24). Earlier studies have suggested that both SnoN and Ski proteins are regulated in a cell cycle-dependent manner (25, 26). It is also known that Smad3 phosphorylation at the linker or MH1 domain region may inhibit transcriptional activity of Smad3 and cause ubiquitination and proteasomal degradation mediated by Ski and SnoN. Studies on phosphoproteomes during the human cell cycle have shown that most protein phosphorylation sites in major pathways are extensively up-regulated and almost fully phosphorylated in mitotic cells (22). Therefore these proteins may be more susceptible to ubiquitination. We found that the number of complexes between SnoN, Ski, and R-Smads increases drastically in a TGF-β-independent manner in cells expressing high levels of mitotic markers. This suggests that SnoN and Ski may sequester Smad proteins and regulate TGF-β-induced transcription negatively during mitosis. Furthermore, the decrease in numbers of Ski and R-Smad complexes upon stimulation with TGF-β, which can be inhibited by MG132 treatment and the persistent cytoplasmic localization of these complexes, implies that the ability of Ski to sequester R-Smads depends on TGF-β. Taken together, our observations suggest that SnoN and Ski interfere with Smad complex formation in the absence of TGF-β stimulation, thus maintaining a low background of signaling.
We also report for the first time the detection of Smad complexes in normal and diseased human tissues (Fig. 5). The status of TGF-β signaling in the normal or cancer colonic mucosa remains poorly explored. In addition, all previous studies of TGF-β signaling in the colon relate either to stages of tumor progression or other pathological states, such as inflammatory bowel disease (27). We are in the process of analyzing TGF-β signaling using Smad-specific in situ PLA in colon and other normal tissues during mouse development, and we also characterize the distribution of Smad protein complexes in tissues from patients with colon cancer.2 The identity of the cell types that show strong Smad complexes is an intriguing question that we aim at resolving. The evidence provided here on the differences between levels of C-terminally phosphorylated R-Smads and R-Smad/Smad4 complexes (Figs. 2 and 3) suggests that in situ PLA may have advantages for analysis of active TGF-β signaling in human tissues, thus providing a valuable biomarker for this important signaling pathway.
Our results demonstrate that the ability to study TGF-β-induced Smad interactions at the level of individual cells can provide crucial information about cell signaling and its dependence on cell cycle phase and density of cell populations (28–30). We illustrate the opportunity to investigate important cell signaling events in a variety of cell types and in formalin-fixed tissue samples. Thus, analysis of Smad proteins via in situ PLA may provide new biomarkers for developmental and disease-related studies of critical signaling pathways.
Footnotes
* This work was supported by European Union Seventh Framework Programme Grants 222635 (AffinityProteome) and 241481 (Affinomics). This work was also supported by the EU-Strep project ENLIGHT (ENhanced LIGase based Histochemical Techniques), the Knut and Alice Wallenberg Foundation, and grants from the Swedish Research Council (to K. P. and U. L.), from the European Science Foundation (to A. Z.), and from the Atlantic Philanthropies/Ludwig Institute for Cancer Research Clinical Discovery Program.
 This article contains supplemental material.
This article contains supplemental material.
2 A. Zieba, M. Mignardi, O. Sieber, U. Landegren, O. Söderberg, manuscript in preparation.
1 The abbreviations used are:
- TGF-β
- transforming growth factor-β
- PLA
- proximity ligation assay
- RCA
- rolling circle amplification
- R-Smad
- receptor regulated Smad
- Smad
- small mothers against decapentaplegic
- TβRI
- TGF-β receptor type I
- TβRII
- TGF-β receptor type II.
REFERENCES
- 1. Massagué J., Blain S. W., Lo R. S. (2000) TGFβ signaling in growth control, cancer, and heritable disorders. Cell 103, 295–309 [DOI] [PubMed] [Google Scholar]
- 2. Moustakas A., Heldin C. H. (2009) The regulation of TGFβ signal transduction. Development 136, 3699–3714 [DOI] [PubMed] [Google Scholar]
- 3. Massagué J., Wotton D. (2000) Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 19, 1745–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abdollah S., Macías-Silva M., Tsukazaki T., Hayashi H., Attisano L., Wrana J. L. (1997) TβRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J. Biol. Chem. 272, 27678–27685 [DOI] [PubMed] [Google Scholar]
- 5. Akiyoshi S., Inoue H., Hanai J., Kusanagi K., Nemoto N., Miyazono K., Kawabata M. (1999) c-Ski acts as a transcriptional co-repressor in transforming growth factor-β signaling through interaction with smads. J. Biol. Chem. 274, 35269–35277 [DOI] [PubMed] [Google Scholar]
- 6. Sun Y., Liu X., Ng-Eaton E., Lodish H. F., Weinberg R. A. (1999) SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor β signaling. Proc. Natl. Acad. Sci. U.S.A. 96, 12442–12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu W., Angelis K., Danielpour D., Haddad M. M., Bischof O., Campisi J., Stavnezer E., Medrano E. E. (2000) Ski acts as a co-repressor with Smad2 and Smad3 to regulate the response to type β transforming growth factor. Proc. Natl. Acad. Sci. U.S.A. 97, 5924–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bonni S., Wang H. R., Causing C. G., Kavsak P., Stroschein S. L., Luo K., Wrana J. L. (2001) TGF-β induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol. 3, 587–595 [DOI] [PubMed] [Google Scholar]
- 9. Stroschein S. L., Bonni S., Wrana J. L., Luo K. (2001) Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN. Genes Dev. 15, 2822–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 [DOI] [PubMed] [Google Scholar]
- 11. Gellibert F., de Gouville A. C., Woolven J., Mathews N., Nguyen V. L., Bertho-Ruault C., Patikis A., Grygielko E. T., Laping N. J., Huet S. (2006) Discovery of 4-{4-[3-(pyridin-2-yl)-1H-pyrazol-4-yl]pyridin-2-yl}-N-(tetrahydro-2H-pyran-4-yl)benzamide (GW788388): A potent, selective, and orally active transforming growth factor-β type I receptor inhibitor. J. Med. Chem. 49, 2210–2221 [DOI] [PubMed] [Google Scholar]
- 12. Chacko B. M., Qin B. Y., Tiwari A., Shi G., Lam S., Hayward L. J., De Caestecker M., Lin K. (2004) Structural basis of heteromeric smad protein assembly in TGF-β signaling. Mol. Cell 15, 813–823 [DOI] [PubMed] [Google Scholar]
- 13. Pierreux C. E., Nicolás F. J., Hill C. S. (2000) Transforming growth factor β-independent shuttling of Smad4 between the cytoplasm and nucleus. Mol. Cell. Biol. 20, 9041–9054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Le Scolan E., Zhu Q., Wang L., Bandyopadhyay A., Javelaud D., Mauviel A., Sun L., Luo K. (2008) Transforming growth factor-β suppresses the ability of Ski to inhibit tumor metastasis by inducing its degradation. Cancer Res. 68, 3277–3285 [DOI] [PubMed] [Google Scholar]
- 15. Levy L., Howell M., Das D., Harkin S., Episkopou V., Hill C. S. (2007) Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol. Cell. Biol. 27, 6068–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miyazono K., Koinuma D. (2011) Arkadia: Beyond the TGF-β pathway. J Biochem. 149, 1–3 [DOI] [PubMed] [Google Scholar]
- 17. Luo K. (2004) Ski and SnoN: Negative regulators of TGF-β signaling. Curr. Opin. Genet. Dev. 14, 65–70 [DOI] [PubMed] [Google Scholar]
- 18. Nakao A., Imamura T., Souchelnytskyi S., Kawabata M., Ishisaki A., Oeda E., Tamaki K., Hanai J., Heldin C. H., Miyazono K., ten Dijke P. (1997) TGF-β receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 16, 5353–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matsuura I., Denissova N. G., Wang G., He D., Long J., Liu F. (2004) Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature 430, 226–231 [DOI] [PubMed] [Google Scholar]
- 20. Liu F., Matsuura I. (2005) Inhibition of Smad antiproliferative function by CDK phosphorylation. Cell Cycle 4, 63–66 [DOI] [PubMed] [Google Scholar]
- 21. Varelas X., Samavarchi-Tehrani P., Narimatsu M., Weiss A., Cockburn K., Larsen B. G., Rossant J., Wrana J. L. (2010) The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-β-SMAD pathway. Dev Cell 19, 831–844 [DOI] [PubMed] [Google Scholar]
- 22. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 3, ra3. [DOI] [PubMed] [Google Scholar]
- 23. Fuentealba L. C., Eivers E., Geissert D., Taelman V., De Robertis E. M. (2008) Asymmetric mitosis: Unequal segregation of proteins destined for degradation. Proc. Natl. Acad. Sci. U.S.A. 105, 7732–7737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krakowski A. R., Laboureau J., Mauviel A., Bissell M. J., Luo K. (2005) Cytoplasmic SnoN in normal tissues and nonmalignant cells antagonizes TGF-β signaling by sequestration of the Smad proteins. Proc. Natl. Acad. Sci. U.S.A. 102, 12437–12442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Macdonald M., Wan Y., Wang W., Roberts E., Cheung T. H., Erickson R., Knuesel M. T., Liu X. (2004) Control of cell cycle-dependent degradation of c-Ski proto-oncoprotein by Cdc34. Oncogene 23, 5643–5653 [DOI] [PubMed] [Google Scholar]
- 26. Wan Y., Liu X., Kirschner M. W. (2001) The anaphase-promoting complex mediates TGF-β signaling by targeting SnoN for destruction. Mol Cell 8, 1027–1039 [DOI] [PubMed] [Google Scholar]
- 27. Sancho E., Batlle E., Clevers H. (2004) Signaling pathways in intestinal development and cancer. Annu. Rev. Cell Dev. Biol. 20, 695–723 [DOI] [PubMed] [Google Scholar]
- 28. Lahav G., Rosenfeld N., Sigal A., Geva-Zatorsky N., Levine A. J., Elowitz M. B., Alon U. (2004) Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 36, 147–150 [DOI] [PubMed] [Google Scholar]
- 29. Covert M. W., Leung T. H., Gaston J. E., Baltimore D. (2005) Achieving stability of lipopolysaccharide-induced NF-κB activation. Science 309, 1854–1857 [DOI] [PubMed] [Google Scholar]
- 30. Tay S., Hughey J. J., Lee T. K., Lipniacki T., Quake S. R., Covert M. W. (2010) Single-cell NF-κB dynamics reveal digital activation and analogue information processing. Nature 466, 267–271 [DOI] [PMC free article] [PubMed] [Google Scholar]