Abstract
We report the release of mzIdentML, an exchange standard for peptide and protein identification data, designed by the Proteomics Standards Initiative. The format was developed by the Proteomics Standards Initiative in collaboration with instrument and software vendors, and the developers of the major open-source projects in proteomics. Software implementations have been developed to enable conversion from most popular proprietary and open-source formats, and mzIdentML will soon be supported by the major public repositories. These developments enable proteomics scientists to start working with the standard for exchanging and publishing data sets in support of publications and they provide a stable platform for bioinformatics groups and commercial software vendors to work with a single file format for identification data.
Protein identification in proteomics is usually performed by MS in a single stage, Peptide Mass Fingerprinting (PMF),1 or in two stages (tandem MS, MS/MS or MS2), followed by computational analysis for which a variety of software packages are available. For PMF, the data (an MS peak list) consists of the mass/charge versus intensity values for peptide ions. There are a number of software packages available for identifying proteins from PMF data by searching the peak list against a theoretical digest of a protein sequence database such as: MS-Fit (part of ProteinProspector http://prospector.ucsf.edu/), ProFound (1) and Mascot (2). Tandem MS data typically comprises mass/charge versus intensity values for fragmentation products of an individual peptide, for which there are broadly four types of computational pipelines used for interpretation: (1) a sequence database search in which mass/charge values for peptide fragments are queried against an in silico digest of a protein sequence database—Mascot (2), Sequest (3), OMSSA (4), X!Tandem (5), Phenyx (6) (2) de novo sequencing in which the software attempts to identify the complete or partial peptide sequence directly from the spectrum—PEAKS (7), Lutefisk (8), PepNovo (9), Mascot Distiller; (3) tag searching whereby software identifies short sequences of amino acids de novo (for example three amino acids in length) that are used to pre-filter a protein sequence database to reduce the database search space—PeptideSearch (10), InsPecT (11), MS-SEQ in ProteinProspector, Mascot (2), Paragon (12); (4) searches against libraries of experimental spectra that have been pre-assigned to a peptide sequence—SpectraST (13), X!Hunter (14), Bibliospec (15). The release of genome sequences for most species studied, and hence well curated protein sequence databases, means that most proteomic pipelines now use method 1, although there are many applications in which other methods still have considerable utility.
There have been developments in statistical techniques for determining whether an individual peptide-spectrum match (PSM), or a protein inferred from a set of PSMs has been correctly identified, as well as techniques for assigning significance values across a global set of identifications in shotgun experiments, such as decoy database searches (16). However, in most proteomic laboratories there remains considerable heterogeneity in the metrics used by experimentalists to determine which peptide/protein identifications are likely to be correct and there is little consensus on the best statistical approach to use. As such, different groups apply different algorithms for determining whether or not a peptide/protein is present. Differences in any part of the analysis workflow may result in different identification lists being produced and thus substantial metadata must be reported to allow critical analysis of the results.
Attempts have been made to improve the consistency and quality of proteomics data reported through minimum reporting guidelines (17, 18). Several journals recommend that authors wishing to publish must be compliant with reporting guidelines and deposit their data in a public repository. A number of public proteomics databases exist, with PeptideAtlas (19), PRIDE (20), and GPMDB (21) being the most prominent. However, search engines produce different file formats and each represents data and metadata using different terminology and levels of detail. The bioinformatics expertise required to deal with these issues may not be available to all laboratories, making it difficult for researchers to adhere to minimum reporting guidelines. Consequently, in contrast to the situation in other high-throughput omics technologies, comparatively few MS proteomics data sets are currently available in the public domain (22). Additionally, bioinformatics groups and commercial software vendors continue to support only a subset of the proprietary and open-source formats for identification data, resulting in considerable wasted effort writing bespoke file format converters and keeping existing converters compatible with rapidly changing proprietary formats.
The Proteomics Standards Initiative (PSI) of the Human Proteome Organization (HUPO) was created to facilitate community-driven standardization in proteomics data reporting, and has created several reporting requirements documents under the Minimum Information About a Proteomics Experiment (MIAPE) umbrella (18) and data format standards, including mzML for capturing mass spectra (23) and PSI-MI for molecular interactions (24). The PSI, in collaboration with instrument and software vendors, and the developers of the major open-source projects in proteomics, recognized that there was a growing need for a standard format for MS-based proteomics results, which led to the development of mzIdentML. A recent set of recommendations for mass spectrometry data quality metrics discussed the strong need to associate appropriate meta data with actual data to enable quality estimates to be made on a published dataset (25–27). The mzIdentML standard is, similar to mzML, coping with this requirement and is able to support meta data associated with the identification of peptides and proteins.
EXPERIMENTAL PROCEDURES
Early model drafts of mzIdentML were developed by examining existing formats produced by different software packages and other open formats, such as pepXML/protXML from the Institute for Systems Biology (28) and PEDRo, developed at the University of Manchester (29). The model was developed over several years in a process open to all interested parties and transparent at each stage, consisting of mailing list discussions, a code repository (http://code.google.com/p/psi-pi/), regular conference calls, and development workshops at each PSI meeting (30–33). The mzIdentML specifications were first submitted to the PSI document process in late 2008 and completed in August 2009 from which version 1.0 was released. The process ensures that specifications undergo a formal process, consisting of a public comment phase and anonymous review, similar to a journal article (34). The first software implementations identified some minor issues, particularly related to large file sizes containing some redundancy for large shotgun experiments. Here we report on version 1.1 of the standard, which has been created to reduce redundancy and was released in August 2011 from a second round of the PSI's document process. We expect that version 1.1 will be the stable release, similar to the PSI's mzML format (23). As such, the format has had input from a wide range of stakeholders and represents the consensus view of the academic and industrial research community and software vendors. The schema was tested during the process by creating example files converted from the main search engine formats, and by ensuring that the MIAPE specifications could be fulfilled. mzIdentML uses several components derived from the FuGE schema (35), which has been adapted in this context to facilitate integration with other PSI standards.
The controlled vocabulary was first developed by collecting terms from vendors of different software packages. Terms were added to a hierarchy according to logical groupings, which also facilitate the development of mappings between the schema and the CV. In common with other PSI CVs, the CV is in OBO format (http://www.geneontology.org/GO.format.shtml). New terms can be added to the CV by raising a request on the PSI website or the PSI mailing list.
RESULTS
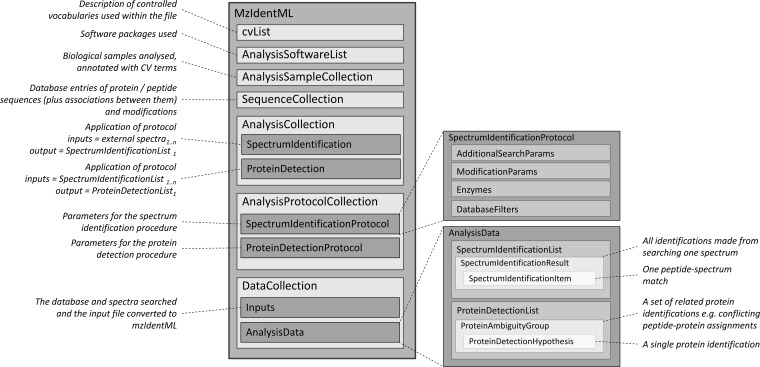
The mzIdentML format stores peptide and protein identifications based on mass spectrometry (Fig. 1) and captures metadata about methods, parameters, and quality metrics. Data are represented through a collection of protein sequences, peptide sequences (with modifications), and structures for capturing the scores associated with ranked peptide matches for each spectrum searched.
Fig. 1.
The overall structure of a typical mzIdentML file. Each file must contain one or more instances of SpectrumIdentificationList (the set of peptide identifications made by a search) and must contain zero or one ProteinDetectionList (the set of proteins identities inferred from peptide identifications).
Peptide Identifications
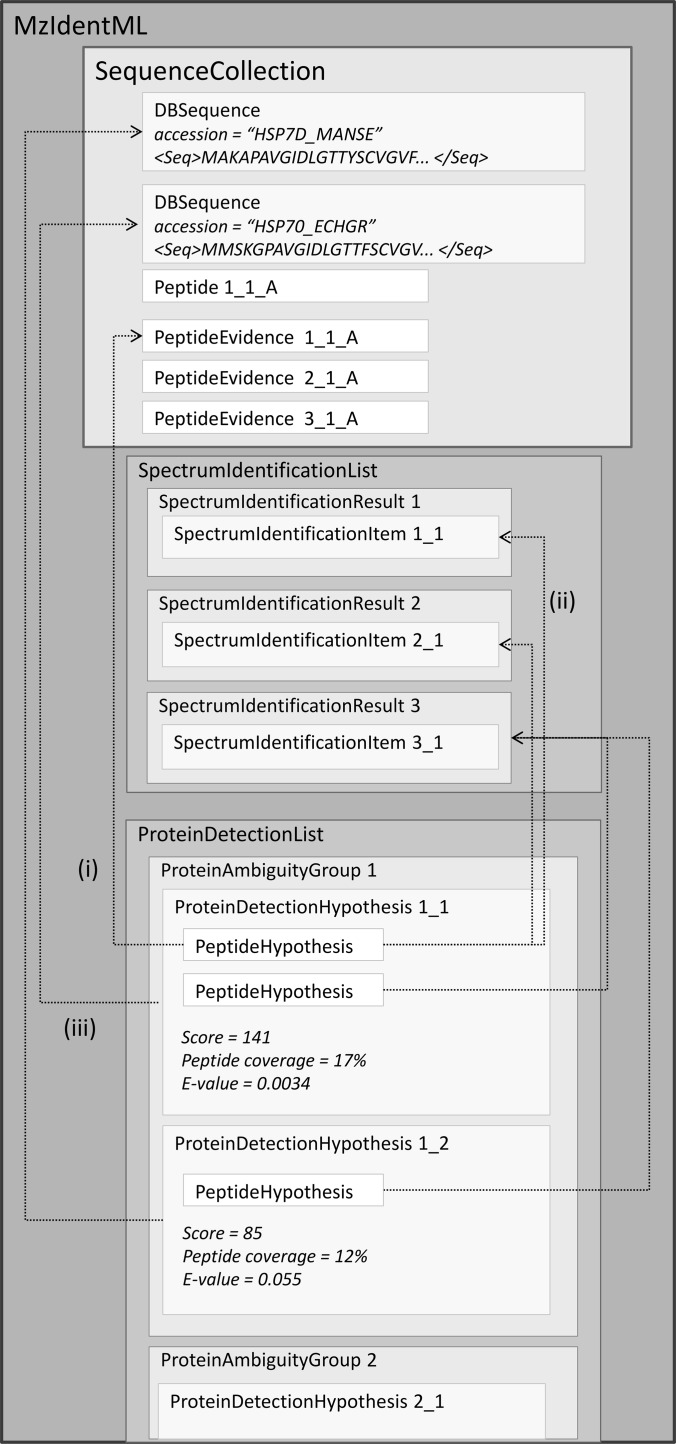
A typical peptide-spectrum match (PSM) is recorded in mzIdentML as shown in Fig. 2. A ranked set of peptides matched to the same spectrum is collected under <SpectrumIdentificationResult> with each single PSM recorded as an instance of <SpectrumIdentificationItem>. <SpectrumIdentificationItem> references the <Peptide> element, which captures a single unique representation of the peptide sequence and any modifications (see below) that have been found, to reduce file size if the same peptide has been identified multiple times. Attributes are provided on <SpectrumIdentificationItem> for the rank, peptide charge state, experimental and calculated mass/charge values. The peptide sequence could have arisen from several different protein sequences (<DBSequence>), so a many-to-many mapping (<PeptideEvidence>) is provided representing all the protein sequences in which the peptide sequences can be found. <PeptideEvidence> has attributes for the start/end positions of the peptide within the protein sequence and the flanking residues. mzIdentML makes no attempt to import the spectra that were searched because several file formats, such as the PSI's mzML format (36), already exist for this purpose. Each <SpectrumIdentificationResult> references the spectrum from which identifications have been made in an external format. As part of the documentation, guidelines are provided for unambiguously referencing a single spectrum within an mzML file or within other data formats that may be inputs to a search engine (mgf, dta, mzXML, mzData, pkl, etc.). For many use cases, it is expected that mzIdentML should be transferred in tandem with the peak list file that was searched.
Fig. 2.
Peptide identification from MS/MS represented in mzIdentML: (i) DBSequence stores database entries, such as complete protein sequences and accessions for their retrieval from external databases; (ii) Peptide holds individual peptide sequences and modifications that have been identified; (iii) PeptideEvidence instances provide the mappings between a peptide sequence and all the protein sequences from which it could have arisen; (iv) The association between SpectrumIdentificationItem and PeptideEvidence is the core result of a single PSM; and (v) SpectrumIdentificationResult captures all ranked identifications (SpectrumIdentificationItem) made from one spectrum and is mapped back to the source spectrum in an external format, such as mzML. Note, the representation of some attributes and elements has been shortened to simplify the figure, for example scores and metrics are represented in mzIdentML using CV terms to incorporate flexibility and extensibility into the schema.
Peptide and protein identifications are generally associated with some measure related to the probability of a correct identification, and it is common to use a threshold on these metrics. Where the threshold is applied can dramatically alter conclusions, and thus it is important to record it. The threshold used is specified by controlled vocabulary terms within the <SpectrumIdentificationProtocol>. <SpectrumIdentificationItem> has a Boolean attribute, passThreshold, to allow the reporting of identifications that fall below the significance threshold, which are often not considered part of the result set. The inclusion of identifications below the threshold used by the original authors allows subsequent re-analysis by others, allowing them the benefit of the full initial results. This allows a broader range of alternate analysis options, including those that might make different assumptions.
In order to assess the quality of a peptide identification made from tandem MS, it can be important to know which products of peptide fragmentation have been identified. mzIdentML uses controlled vocabulary terms to specify the types of ions that have been found (e.g. a-, b-, c-, x-, y-, z-ions and neutral losses of these) and captures data about the ions (such as mass/charge and intensity values) in a compressed array structure within <SpectrumIdentificationItem>. Because the input spectrum has been referenced in an external format, it is straightforward to write a spectrum viewer showing which product ions have been identified by the search engine, or an application to perform further statistical processing of individual PSMs.
Peptide Modifications
Modifications that have been identified on peptides are encoded in the <Modification> element (child of <Peptide>) using a combination of a controlled vocabulary term sourced from Unimod (37) or PSI-MOD (38) (for the name/molecular structure of the modification), the mass delta searched and the location of the modification within the peptide sequence. This representation should ensure that databases or tools importing files can provide consistent analysis, comparison and querying capabilities. If the modification is unknown, the export software can explicitly encode this information, using an “unknown modification” term and the mass delta. If multiple CV terms are provided within a single <Modification> element, it is understood that the modification is ambiguous but has been identified as one of those listed. Additional scores associated with modification sites should be encoded within the <SpectrumIdentificationItem> that references <Peptide> because such information is specific to a given PSM.
Protein Identifications and Protein Ambiguity Groups
In “shotgun” approaches, where proteins are digested in peptides prior to separation, the linkage from peptide identifications to protein identifications is lost. It is common for a peptide sequence to be present in more than one protein so software applications must infer the most likely protein identity from a set of peptides. mzIdentML has been designed to accommodate the ambiguity of protein inference (Fig. 3). <ProteinDetectionHypothesis> represents one possible protein identification corresponding to a <DBSequence> with one accession (with associated scores or probability values), given a set of peptide identifications, reported as references to the set of <SpectrumIdentificationItem> elements on which it was based. <ProteinAmbiguityGroup> sits above in the hierarchy, acting as a logical grouping of related hypotheses, for example where the same set of peptide sequences provides supporting evidence for more than one protein identification. This structure allows ambiguity to be communicated, preventing the data producer from having to take a final decision on which proteins are present or absent in the sample. The inclusion of p values for protein identifications, for example output by ProteinProphet (39), would allow data consumers to process the results in different ways depending on the context.
Fig. 3.
Protein identifications represented in mzIdentML. If the same set of peptide sequences provides supporting evidence for more than one protein, the proteins appear within a ProteinAmbiguityGroup. (i) Each ProteinDetectionHypothesis contains references back to the instances of PeptideEvidence on which it is based, onward references to Peptide not shown. (ii) The ProteinDetectionHypothesis element has associations to all SpectrumIdentificationItem elements that have been used for protein inference. (iii) Each ProteinDetectionHypothesis references the protein sequence (DBSequence) that has been identified.
An mzIdentML file contains at most one <ProteinDetectionList>, determined as the final result of an analysis procedure, with no intermediate results reported. In some workflows, a set of protein identifications undergo secondary statistical processing or manual validation over the initial search engine output. Such workflows are encoded in mzIdentML as one overall process that produces the final set of proteins. The design decision was taken to reduce the chance of ambiguity in how different implementers express a data set and to make it simpler for data consumers to process the results. As with peptide identifications, it is possible to report protein identifications that fall below a given threshold or those that have been determined by manual inspection to be incorrect.
Representing Specific Use Cases
Example mzIdentML documents have been made available to illustrate the wide range of proteomic analyses that are supported (http://code.google.com/p/psi-pi/source/browse/trunk/examples/1_1examples): PMF (supplemental file: mascot_pmf_example.mzid), “standard” tandem MS analysis from different search engines (supplemental files: 55merge_tandem.mzid, 55merge_omssa.mzid, 55merge_mascot_full.mzid, Sequest_example_ver1.1.mzid, Phenyx-example.mzid, Mascot_MSMS_example.mzid); and a spectral library search from SpectraST (supplemental file: spectraST.mzid). No attempt has been made to standardize the score parameters output by different search engines, instead differences between the scores and other parameters reported are documented through the use of controlled vocabulary terms. A common analysis approach is to employ multiple search engines (40–43), which can be accommodated in mzIdentML by encoding a <ProteinDetection> process that references several instances of <SpectrumIdentificationList> (one per search engine) as input, to produce a single <ProteinDetectionList> as output (Supplemental file: MPC_example_Multiple_search_engines.mzid).
The search of nucleic acid sequences requires translation of nucleic acid sequence into the corresponding amino acids. In mzIdentML, the different rules governing the translation are documented using CV terms. Example encodings of NCBI translation tables (http://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi) within <DatabaseTranslation> are provided (supplemental file: Mascot_NA_example.mzid) in which every instance of <PeptideEvidence> contains a reference to the translation table used and the reading frame.
A common experimental approach in quantitative proteomics is the use of stable isotope labeling, which typically results in heavy and light versions of amino acids. The mass of each amino acid can be reported within the <MassTable> element (supplemental file: Mascot_N15_example.mzid). In an experiment using stable isotope labeling, two tables are reported for the amino acid masses with the light or heavy isotope incorporated. Every <SpectrumIdentificationItem> element provides a reference to the appropriate mass table to demonstrate how the molecular weight has been calculated for the PSM.
Finally, the use of decoy database searching is a popular method by which the false discovery rate may be estimated (16, 44). The <PeptideEvidence> element has a Boolean attribute, isDecoy, which allows consumers of the file to calculate the false discovery rate for different score thresholds (supplemental files: 55merge_omssa.mzid and MPC_example_Multiple_search_engines.mzid).
Regarding de novo peptide sequencing results it is possible to enumerate and record all possible matches found by a de novo technique. However, this can produce very large files and we invite proposals in this area for suitable encoding of alternative results in a more compressed structure. In case of sequence-tagged searches the final results from a run can be stored in mzIdentML, but the details of tag generation and filtering cannot, except through the additional annotation of <SpectrumIdentificationItem> elements with new CV terms.
In the case of spectral library searches the recommended encoding is similar to sequence database search results (spectraST.mzid), the main difference being that rather than protein sequences represented in the <DBSequence> element, the peptide sequence for each library entry is stored here instead. Additional information about the peptide-spectrum match, such as observed modifications and consensus scores, can be stored as CV terms within each <DBSequence> entry.
Controlled Vocabulary
The group has developed controlled vocabulary terms as part of the wider PSI-Mass Spectrometry CV, which is used by mzML and other PSI formats, to ensure unambiguous reporting of search engine methods, parameters and scores, available from the project homepage (http://www.psidev.info/controlled-vocabularies). Each entry contains a definition and a specification of whether the term should be paired with a value, and if so, what the data type and unit should be. The CV therefore provides a flexible mechanism for constraining the values allowed in instance documents without hard-coding enumerations within the schema.
As an example, a <SpectrumIdentificationItem> with a Mascot ion score of 62.7 would be encoded with the following CV term:
<cvParam accession = “MS:1001171” name = “Mascot:score” cvRef = “PSI-MS” value = “62.7”>
The accession references a PSI-MS CV term that provides a formal definition of the Mascot score, specifies that a value must be given (as a double-precision floating-point data type) and that no units should be provided.
An example term that requires a unit is the fragment ion search tolerance (e.g. ± 0.5 Da) within the <SpectrumIdentificationProtocol>, encoded as two CV terms:
<cvParam accession = “MS:1001412” name = “search tolerance plus value” value = “0.5” cvRef = “PSI-MS” unit Accession = “UO:0000221” unitName = “dalton” unitCvRef = “UO”>
<cvParam accession = “MS:1001413” name = “search tolerance minus value” value = “0.5” cvRef = “PSI-MS” unit Accession = “UO:0000221” unitName = “dalton” unitCvRef = “UO”>
In this instance, the MS:1001412 CV entry specifies that a mass unit must be provided from the Unit Ontology (available from the OBO foundry, http://www.obofoundry.org/).
In addition to the CV, a mapping format has been developed by the PSI which provides formal rules for associating XML Schema elements with particular CV terms (45). The mapping file is checked by validation software to ensure that not only are correct elements provided within mzIdentML (the file is valid XML) but also that valid and sensible data values have been provided at the correct positions within the element hierarchy. The association of an XML Schema with a CV is a general problem that complicates the process of interpreting exchange formats that use CV terms, because terms are frequently used inconsistently or incorrectly. The solution developed by PSI should ensure that consistent, machine-comprehensible files are produced and provides a re-usable solution for other format developers with similar challenges, for example the PSI Molecular Interactions work group (46).
Relationship to MIAPE and MCP Guidelines
The PSI has created the MIAPE guidelines (18) that comprise a parent document and a series of technology specific modules (47–50). Each module is a minimal checklist of information that should be reported about an experiment when it is published in a journal or a data set is submitted to a repository. MIAPE is intended to ensure that the quality checking goals of journals, funding agencies and repository operators can be met. The module corresponding to mzIdentML is MIAPE-Mass Spectrometry Informatics (MSI) (47). An mzIdentML instance document can be a technically valid document without being MIAPE-compliant. supplemental Table S1 provides the mapping relationship between the items required in MIAPE-MSI (version 1.1) and what is captured by mzIdentML, including examples drawn from referenced instance documents. MIAPE-MSI compliance can be fully reached using mzIdentML except for the quantification aspects. The semantic validation software (http://psidev.info/validator) will be adapted to check whether MIAPE compliance has been reached by particular files.
Molecular and Cellular Proteomics (MCP), the Journal of Proteome Research, Proteomics, and other journals oblige or suggest authors to adhere to specific guidelines detailing information that should be submitted with manuscripts. Much of this information is currently submitted as tables of protein identifications and annotated spectra. An mzIdentML document can encapsulate all of the data required for these journals apart from the quantification requirements. supplemental Table S2 describes the conformance to these so-called “Paris guidelines” (April 2007 release, (17, 51)).
Quantification Data in mzQuantML
Numerous experimental methods have been developed for quantitative proteomics by incorporating stable isotopic labels or isobaric tags, or by label-free methods (52), and as such, the PSI Proteomics Informatics workgroup is also developing a complementary format for quantification data, called mzQuantML. The purpose of mzQuantML is to communicate data about peptide and protein abundance, such as ratios of quantitative differences across different samples or absolute measures of protein abundance. The format also contains structures for describing how data has been combined from the peptide level up to the protein level and across replicates. The development process of mzQuantML is ongoing, and we encourage further input (please see the group webpage for details http://www.psidev.info/mzQuantML).
Implementations
There is a growing list of implementations available for mzIdentML (http://www.psidev.info/tools-implementing-mzidentml). Results in mzIdentML format can be exported directly from Mascot (2) (export of version 1.0 available in version 2.3, version 1.1 exporter under development), and converters are currently available for Sequest (3) and Proteome Discoverer output (.msf and .protXML) (e.g. within ProCon: http://www.medizinisches-proteom-center.de/ProCon), OMSSA (4) and X!Tandem (5) (http://code.google.com/p/mzidentml-parsers/), and in the pipeline applications Scaffold (42) (import into Scaffold PTM and export of mzIdentML available in Scaffold version 3) and TPP (28) (results can be exported to mzIdentML via the ProteoWizard (53) converter). A beta exporter is also available for Phenyx (6). OpenMS (54) implements C++ code for reading (and as of release 1.9) writing mzIdentML. The OpenMS pipeline tools, TOPP (55), will fully support mzIdentML as of release 1.9 and can convert mzIdentML to and from various other identification formats. PeptideAtlas accepts mass spectrometer output files in a variety of formats, which are processed using standard parameters through the TPP, providing results for download in pepXML and protXML. The ProteoWizard converter can now be used to convert pepXML into mzIdentML, and the full integration of direct mzIdentML export using this mechanism is expected in PeptideAtlas in 2012. An open-source Java API for reading and writing mzIdentML has also been developed, available from http://code.google.com/p/jmzidentml/. PRIDE currently uses its own internal format called PRIDE XML for representing mass spectra and peptide and protein identifications, but is currently in the process of moving its internal pipeline and database schema over to support a complete import/export of mzIdentML (and the PSI standard for mass spectra, mzML). PRIDE can already take data submissions in mzIdentML version 1.1 by converting the files to PRIDE XML. As mentioned above, full import/export support for mzIdentML is under development and it is expected to finalize during 2012. In addition, work is ongoing to fully support the format by the PRIDE Inspector tool (http://code.google.com/p/pride-toolsuite/wiki/PRIDEInspector). It is expected that once mzIdentML becomes well established as a community format, tools will routinely use mzIdentML internally for data representation and processing.
The combination of the mzIdentML XML Schema plus the associated mapping file and semantic validation software define the minimum information required to create a “valid” file when converting from other identification formats used in proteomics. However, software is also under development to link the mzIdentML specifications formally to the corresponding MIAPE module to enable an automatic test for compliance. As such, an mzIdentML file could have several different states, depending on the user's requirements: (1) valid against the XML Schema but not semantically valid; (2) XML schema valid and semantically valid; and (3) XML schema valid, semantically valid and MIAPE compliant. For public database or tool import—levels (2) or (3) should be reached depending on the context. Level (1) should only be used in tools internally, and would not be considered suitable for transfer between tools or making data sets publicly available.
EXAMPLE FILES
All example files described in the text can be downloaded from: http://code.google.com/p/psi-pi/source/browse/trunk/examples/1_1examples/ • 55merge_mascot_full.mzid - example MS-MS search results including decoy matches from Mascot. • 55merge_omssa.mzid - example MS-MS search results including decoy matches from OMSSA. • 55merge_tandem.mzid - example MS-MS search results including decoy matches from X!Tandem. • MPC_example_Multiple_search_engines.mzid - an example of PSMs from different search engines, assembled into proteins using a third-party algorithm; false-discovery estimation using decoy database. • Mascot_NA_example.mzid - an example of a search against an EST database with Mascot. • Mascot_top_down_example.mzid - a single MS/MS spectra from an intact protein, searched with Mascot. • Sequest_example_ver1.1.mzid - a simple example derived from an “.out” file produced by SEQUEST. • mascot_pmf_example.mzid - example Peptide Mass Fingerprint search with Mascot. • spectraST.mzid - examples search against a spectral library using spectraST • Mascot_N15_example.mzid - an example of a search using two sets of residue masses, 14N and 15N with Mascot. • phenyx-example.mzid - a tandem MS example exported from the Phenyx software. • Mascot_MSMS_example.mzid - a further example of a tandem MS data file exported from Mascot.
DISCUSSION
The mzIdentML standard (and accompanying controlled vocabulary) has been developed over several years within the PSI's standardization process, which is open to all interested parties and transparent at each stage. As such, the format has had input from a wide range of stakeholders and represents the consensus view of academic research groups, industrial representatives and software vendors working in this area. The standard was fixed at version 1.1 in August 2011. Alterations to the schema that could affect software implementations cannot be made without re-entering the standardization process and no major changes, beyond minor bug fixes, are currently planned by the PSI. The PSI proteome informatics workgroup has a stable core of developers working on mzIdentML implementations and we are committed to providing documentation, help guides and support (via the mailing list) for external implementers in the coming years. We anticipate that the release of mzIdentML will greatly facilitate data sharing for proteomics, and its release will serve as the basis for informatics developments in quantitative proteomics. We encourage further input on the standard by joining the mailing list or attending a PSI meeting (see http://www.psidev.info/ for details).
Acknowledgments
We thank the steering committee and the editors (Norman Paton and Christian Stephan) of the Proteomics Standards Initiative for the provision of the document process and feedback on the specification documents. We also thank the following individuals who have contributed to the development of mzIdentML: Marc Sturm (Eberhard Karls University, Tübingen); Michael Kohl (Medizinisches Proteom-Center, Ruhr-Universitaet Bochum, Germany); Patrick Pedrioli (Swiss Federal Institute of Technology Zürich); Zsuzsanna Bencsath-Makkai (Biomedical Engineering, McGill University); Phil Jones, Lennart Martens, Richard Côté, David Ovelleiro and Luisa Montecchi-Palazzi (European Bioinformatics Institute); Jimmy Eng (Fred Hutchinson Cancer Research); Alexandre Masselot (GeneBio Geneva); David Horn (Agilent); Phillip Young (Waters); and Eugene Kapp (Ludwig Institute for Cancer Research).
In memory of our colleague Andreas Bertsch, who lost his life unexpectedly and far too early. We will all miss his contributions, arguments, and friendship.
Footnotes
We would also like to gratefully acknowledge past and current funding sources: ARJ [BBSRC: BB/H024654/1, BB/I00095X/1, BB/G010781/1] and EU FP7 grant ‘ProteomeXchange’ [grant number 260558]; JAS and SJH BBSRC: BB/F004605/1 to SJH; ME (total), DC (in part) and JAS (in part) were funded by the European Commission's ProDaC grant (6th Framework Programme, project number LSHG-CT-2006–036814). ME is currently funded by P.U.R.E. (Protein Unit for Research in Europe), a project of Nordrhein-Westfalen, a federal state of Germany. EWD was funded in part under NHLBI contract N01-HV-28179, NIGMS grant R01 GM087221, EU FP7 grant ‘ProteomeXchange’ [grant number 260558], P50 GM076547/Center for Systems Biology, and the Systems Biology Initiative of the State of Luxembourg. GM is funded by EU FP7 grant ‘ProteomeXchange’ [grant number 260558]. JAV was funded by the European Commission FP7 grants LipidomicNet [grant number 202272] and ProteomeXchange [grant number 260558]. FR and JG were funded by PRIDE Wellcome Trust grant [number WT085949MA]. OK acknowledges funding from BMBF [grant numbers 0313842A and 0315395F] and the European Union [Contract No. 262067- PRIME-XS] that contributed to this work.
 This article contains supplemental Tables S1 and S2.
This article contains supplemental Tables S1 and S2.
1 The abbreviations used are:
- PMF
- Peptide Mass Fingerprinting
- CV
- Controlled Vocabulary
- MIAPE
- Minimum Information About a Proteomics Experiment
- PSI
- Proteomics Standards Initiative
- PSM
- Peptide-Spectrum Match
- XML
- Extensible Markup Language.
REFERENCES
- 1. Zhang W., Chait B. T. (2000) ProFound: An Expert System for Protein Identification Using Mass Spectrometric Peptide Mapping Information. Anal. Chem. 72, 2482–2489 [DOI] [PubMed] [Google Scholar]
- 2. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 3. MacCoss M. J., Wu C. C., Yates J. R., 3rd (2002) Probability-Based Validation of Protein Identifications Using a Modified SEQUEST Algorithm. Anal. Chem. 74, 5593–5599 [DOI] [PubMed] [Google Scholar]
- 4. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open Mass Spectrometry Search Algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 5. Fenyö D., Beavis R. C. (2003) A Method for Assessing the Statistical Significance of Mass Spectrometry-Based Protein Identifications Using General Scoring Schemes. Anal. Chem. 75, 768–774 [DOI] [PubMed] [Google Scholar]
- 6. Colinge J., Masselot A., Giron M., Dessingy T., Magnin J. (2003) OLAV: Towards high-throughput tandem mass spectrometry data identification. Proteomics 3, 1454–1463 [DOI] [PubMed] [Google Scholar]
- 7. Ma B., Zhang K., Hendrie C., Liang C., Li M., Doherty-Kirby A., Lajoie G. (2003) PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 17, 2337–2342 [DOI] [PubMed] [Google Scholar]
- 8. Taylor J. A., Johnson R. S. (2001) Implementation and Uses of Automated de Novo Peptide Sequencing by Tandem Mass Spectrometry. Anal. Chem. 73, 2594–2604 [DOI] [PubMed] [Google Scholar]
- 9. Frank A., Pevzner P. (2005) PepNovo: De Novo Peptide Sequencing via Probabilistic Network Modeling. Anal. Chem. 77, 964–973 [DOI] [PubMed] [Google Scholar]
- 10. Mann M., Wilm M. (1994) Error-Tolerant Identification of Peptides in Sequence Databases by Peptide Sequence Tags. Anal. Chem. 66, 4390–4399 [DOI] [PubMed] [Google Scholar]
- 11. Tanner S., Shu H., Frank A., Wang L. C., Zandi E., Mumby M., Pevzner P. A., Bafna V. (2005) InsPecT: Identification of Posttranslationally Modified Peptides from Tandem Mass Spectra. Anal. Chem. 77, 4626–4639 [DOI] [PubMed] [Google Scholar]
- 12. Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) The Paragon Algorithm, a Next Generation Search Engine That Uses Sequence Temperature Values and Feature Probabilities to Identify Peptides from Tandem Mass Spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 13. Lam H., Deutsch E. W., Eddes J. S., Eng J. K., Stein S. E., Aebersold R. (2008) Building consensus spectral libraries for peptide identification in proteomics. Nat. Methods 5, 873–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Craig R., Cortens J. C., Fenyo D., Beavis R. C. (2006) Using Annotated Peptide Mass Spectrum Libraries for Protein Identification. J. Proteome Res. 5, 1843–1849 [DOI] [PubMed] [Google Scholar]
- 15. Frewen B., MacCoss M. J. (2007) Using BiblioSpec for creating and searching tandem MS peptide libraries. Curr. Protoc. Bioinformatics, p. Unit 13.17 [DOI] [PubMed] [Google Scholar]
- 16. Käll L., Storey J. D., MacCoss M. J., Noble W. S. (2008) Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res. 7, 29–34 [DOI] [PubMed] [Google Scholar]
- 17. Carr S., Aebersold R., Baldwin M., Burlingame A., Clauser K., Nesvizhskii A. (2004) the need for guidelines in publication of peptide and protein identification data: working group on publication guidelines for peptide and protein identification data. Mol. Cell. Proteomics 3, 531–533 [DOI] [PubMed] [Google Scholar]
- 18. Taylor C. F., Paton N. W., Lilley K. S., Binz P.-A., Julian R. K., Jones A. R., Zhu W., Apweiler R., Aebersold R., Deutsch E. W., Dunn M. J., Heck A. J. R., Leitner A., Macht M., Mann M., Martens L., Neubert T. A., Patterson S. D., Ping P., Seymour S. L., Souda P., Tsugita A., Vandekerckhove J., Vondriska T. M., Whitelegge J. P., Wilkins M. R., Xenarios I., Yates J. R., Hermjakob H. (2007) The minimum information about a proteomics experiment (MIAPE). Nat. Biotechnol. 25, 887–893 [DOI] [PubMed] [Google Scholar]
- 19. Desiere F., Deutsch E. W., King N. L., Nesvizhskii A. I., Mallick P., Eng J., Chen S., Eddes J., Loevenich S. N., Aebersold R. (2006) The PeptideAtlas project. Nucleic Acids Res. 34, D655–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones P., Côté R. G., Martens L., Quinn A. F., Taylor C. F., Derache W., Hermjakob H., Apweiler R. (2006) PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucleic Acids Res. 34, D659–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Craig R., Cortens J. P., Beavis R. C. (2004) Open Source System for Analyzing, Validating, and Storing Protein Identification Data. J. Proteome Res. 3, 1234–1242 [DOI] [PubMed] [Google Scholar]
- 22.(2009) Credit where credit is overdue. Nat. Biotechnol. 27, 579–579 [DOI] [PubMed] [Google Scholar]
- 23. Martens L., Chambers M., Sturm M., Kessner D., Levander F., Shofstahl J., Tang W. H., Rompp A., Neumann S., Pizarro A. D., Montecchi-Palazzi L., Tasman N., Coleman M., Reisinger F., Souda P., Hermjakob H., Binz P. A., Deutsch E. W. (2011) mzML–a community standard for mass spectrometry data. Mol. Cell. Proteomics 10, R110.000133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kerrien S., Orchard S., Montecchi-Palazzi L., Aranda B., Quinn A. F., Vinod N., Bader G. D., Xenarios I., Wojcik J., Sherman D., Tyers M., Salama J. J., Moore S., Ceol A., Chatr-Aryamontri A., Oesterheld M., Stümpflen V., Salwinski L., Nerothin J., Cerami E., Cusick M., Vidal M., Gilson M., Armstrong J., Woollard P., Hogue C., Eisenberg D., Cesareni G., Apweiler R., Hermjakob H. (2007) Broadening the horizon - level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 5, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kinsinger C. R., Apffel J., Baker M., Bian X., Borchers C. H., Bradshaw R., Brusniak M.-Y., Chan D. W., Deutsch E. W., Domon B., Gorman J., Grimm R., Hancock W., Hermjakob H., Horn D., Hunter C., Kolar P., Kraus H.-J., Langen H., Linding R., Moritz R. L., Omenn G. S., Orlando R., Pandey A., Ping P., Rahbar A., Rivers R., Seymour S. L., Simpson R. J., Slotta D., Smith R. D., Stein S. E., Tabb D. L., Tagle D., Yates J. R., Rodriguez H. (2012) Recommendations for mass spectrometry data quality metrics for open access data (corollary to the Amsterdam Principles). Proteomics 12, 11–20 [DOI] [PubMed] [Google Scholar]
- 26. Kinsinger C. R., Apffel J., Baker M., Bian X., Borchers C. H., Bradshaw R., Brusniak M.-Y., Chan D. W., Deutsch E. W., Domon B., Gorman J., Grimm R., Hancock W., Hermjakob H., Horn D., Hunter C., Kolar P., Kraus H.-J., Langen H., Linding R., Moritz R. L., Omenn G. S., Orlando R., Pandey A., Ping P., Rahbar A., Rivers R., Seymour S. L., Simpson R. J., Slotta D., Smith R. D., Stein S. E., Tabb D. L., Tagle D., Yates J. R., Rodriguez H. (2011) Recommendations for mass spectrometry data quality metrics for open access data (corollary to the Amsterdam Principles). Mol. Cell. Proteomics 10, 0111.015446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kinsinger C. R., Apffel J., Baker M., Bian X., Borchers C. H., Bradshaw R., Brusniak M.-Y., Chan D. W., Deutsch E. W., Domon B., Gorman J., Grimm R., Hancock W., Hermjakob H., Horn D., Hunter C., Kolar P., Kraus H.-J., Langen H., Linding R., Moritz R. L., Omenn G. S., Orlando R., Pandey A., Ping P., Rahbar A., Rivers R., Seymour S. L., Simpson R. J., Slotta D., Smith R. D., Stein S. E., Tabb D. L., Tagle D., Yates J. R., Rodriguez H. (2012) Recommendations for mass spectrometry data quality metrics for open access data (Corollary to the Amsterdam Principles). J. Proteome Res. 11, 1412–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keller A., Eng J., Zhang N., Li X.-j., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1:2005.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taylor C. F., Paton N. W., Garwood K. L., Kirby P. D., Stead D. A., Yin Z., Deutsch E. W., Selway L., Walker J., Riba-Garcia I., Mohammed S., Deery M. J., Howard J. A., Dunkley T., Aebersold R., Kell D. B., Lilley K. S., Roepstorff P., Yates J. R., Brass A., Brown A. J. P., Cash P., Gaskell S. J., Hubbard S. J., Oliver S. G. (2003) A systematic approach to modeling, capturing, and disseminating proteomics experimental data. Nat. Biotechnol. 21, 247–254 [DOI] [PubMed] [Google Scholar]
- 30. Orchard S., Albar J. P., Deutsch E. W., Binz P.-A., Jones A. R., Creasy D., Hermjakob H. (2008) Annual Spring Meeting of the Proteomics Standards Initiative 23–25 April 2008, Toledo, Spain. Proteomics 8, 4168–4172 [DOI] [PubMed] [Google Scholar]
- 31. Orchard S., Apweiler R., Barkovich R., Field D., Garavelli J. S., Horn D., Jones A., Jones P., Julian R., McNally R., Nerothin J., Paton N., Pizarro A., Seymour S., Taylor C., Wiemann S., Hermjakob H. (2006) Proteomics and Beyond A report on the 3rd Annual Spring Workshop of the HUPO-PSI 21–23 April 2006, San Francisco, CA, U.S.A. Proteomics 6, 4439–4443 [DOI] [PubMed] [Google Scholar]
- 32. Orchard S., Hermjakob H., Taylor C., Binz P. A., Hoogland C., Julian R., Garavelli J. S., Aebersold R., Apweiler R. (2006) Autumn 2005 Workshop of the Human Proteome Organisation Proteomics Standards Initiative (HUPO-PSI) Geneva, September, 4–6, 2005. Proteomics 6, 738–741 [DOI] [PubMed] [Google Scholar]
- 33. Orchard S., Jones A. R., Stephan C., Binz P.-A. (2007) The HUPO Pre-Congress Proteomics Standards Initiative Workshop HUPO 5th Annual World Congress Long Beach, CA, U.S.A. 28 October-1 November 2006. Proteomics 7, 1006–1008 [DOI] [PubMed] [Google Scholar]
- 34. Vizcaíno J. A., Martens L., Hermjakob H., Julian R. K., Paton N. W. (2007) The PSI formal document process and its implementation on the PSI website. Proteomics 7, 2355–2357 [DOI] [PubMed] [Google Scholar]
- 35. Jones A. R., Miller M., Aebersold R., Apweiler R., Ball C. A., Brazma A., DeGreef J., Hardy N., Hermjakob H., Hubbard S. J., Hussey P., Igra M., Jenkins H., Julian R. K., Laursen K., Oliver S. G., Paton N. W., Sansone S.-A., Sarkans U., Stoeckert C. J., Taylor C. F., Whetzel P. L., White J. A., Spellman P., Pizarro A. (2007) The Functional Genomics Experiment model (FuGE): an extensible framework for standards in functional genomics. Nat. Biotechnol. 25, 1127–1133 [DOI] [PubMed] [Google Scholar]
- 36. Deutsch E. (2008) mzML: A single, unifying data format for mass spectrometer output. Proteomics 8, 2776–2777 [DOI] [PubMed] [Google Scholar]
- 37. Creasy D. M., Cottrell J. S. (2004) Unimod: Protein modifications for mass spectrometry. Proteomics 4, 1534–1536 [DOI] [PubMed] [Google Scholar]
- 38. Montecchi-Palazzi L., Beavis R., Binz P.-A., Chalkley R. J., Cottrell J., Creasy D., Shofstahl J., Seymour S. L., Garavelli J. S. (2008) The PSI-MOD community standard for representation of protein modification data. Nat. Biotechnol. 26, 864–866 [DOI] [PubMed] [Google Scholar]
- 39. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 40. Jones A. R., Siepen J. A., Hubbard S. J., Paton N. W. (2009) Improving sensitivity in proteome studies by analysis of false discovery rates for multiple search engines. Proteomics 9, 1220–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nahnsen S., Bertsch A., Rahnenführer J., Nordheim A., Kohlbacher O. (2011) Probabilistic consensus scoring improves tandem mass spectrometry peptide identification. J. Proteome Res. 10, 3332–3343 [DOI] [PubMed] [Google Scholar]
- 42. Searle B. C., Turner M., Nesvizhskii A. I. (2008) Improving Sensitivity by Probabilistically Combining Results from Multiple MS/MS Search Methodologies. J. Proteome Res. 7, 245–253 [DOI] [PubMed] [Google Scholar]
- 43. Stephan C., Reidegeld K. A., Hamacher M., van Hall A., Marcus K., Taylor C., Jones P., Müller M., Apweiler R., Martens L., Körting G., Chamrad D. C., Thiele H., Blüggel M., Parkinson D., Binz P. A., Lyall A., Meyer H. E. (2006) Automated reprocessing pipeline for searching heterogeneous mass spectrometric data of the HUPO Brain Proteome Project pilot phase. Proteomics 6, 5015–5029 [DOI] [PubMed] [Google Scholar]
- 44. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Meth. 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 45. Montecchi-Palazzi L., Kerrien S., Reisinger F., Aranda B., Jones A. R., Martens L., Hermjakob H. (2009) The PSI semantic validator: A framework to check MIAPE compliance of proteomics data. Proteomics 9, 5112–5119 [DOI] [PubMed] [Google Scholar]
- 46. Gloriam D. E., Orchard S., Bertinetti D., Björling E., Bongcam-Rudloff E., Borrebaeck C. A. K., Bourbeillon J., Bradbury A. R., de Daruvar A., Dübel S., Frank R., Gibson T. J., Gold L., Haslam N., Herberg F. W., Hiltke T., Hoheisel J. D., Kerrien S., Koegl M., Konthur Z., Korn B., Landegren U., Montecchi-Palazzi L., Palcy S., Rodriguez H., Schweinsberg S., Sievert V., Stoevesandt O., Taussig M. J., Ueffing M., Uhlén M., van der Maarel S., Wingren C., Woollard P., Sherman D. J., Hermjakob H. (2010) A Community standard format for the representation of protein affinity reagents. Mol. Cell. Proteomics 9, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Binz P.-A., Barkovich R., Beavis R. C., Creasy D., Horn D. M., Julian R. K., Seymour S. L., Taylor C. F., Vandenbrouck Y. (2008) Guidelines for reporting the use of mass spectrometry informatics in proteomics. Nat. Biotechnol. 26, 862–862 [DOI] [PubMed] [Google Scholar]
- 48. Gibson F., Anderson L., Babnigg G., Baker M., Berth M., Binz P.-A., Borthwick A., Cash P., Day B. W., Friedman D. B., Garland D., Gutstein H. B., Hoogland C., Jones N. A., Khan A., Klose J., Lamond A. I., Lemkin P. F., Lilley K. S., Minden J., Morris N. J., Paton N. W., Pisano M. R., Prime J. E., Rabilloud T., Stead D. A., Taylor C. F., Voshol H., Wipat A., Jones A. R. (2008) Guidelines for reporting the use of gel electrophoresis in proteomics. Nat. Biotechnol. 26, 863–864 [DOI] [PubMed] [Google Scholar]
- 49. Orchard S., Salwinski L., Kerrien S., Montecchi-Palazzi L., Oesterheld M., Stumpflen V., Ceol A., Chatr-aryamontri A., Armstrong J., Woollard P., Salama J. J., Moore S., Wojcik J., Bader G. D., Vidal M., Cusick M. E., Gerstein M., Gavin A.-C., Superti-Furga G., Greenblatt J., Bader J., Uetz P., Tyers M., Legrain P., Fields S., Mulder N., Gilson M., Niepmann M., Burgoon L., Rivas J. D. L., Prieto C., Perreau V. M., Hogue C., Mewes H.-W., Apweiler R., Xenarios I., Eisenberg D., Cesareni G., Hermjakob H. (2007) The minimum information required for reporting a molecular interaction experiment (MIMIx). Nat. Biotechnol. 25, 894–898 [DOI] [PubMed] [Google Scholar]
- 50. Taylor C. F., Binz P.-A., Aebersold R., Affolter M., Barkovich R., Deutsch E. W., Horn D. M., Huhmer A., Kussmann M., Lilley K., Macht M., Mann M., Muller D., Neubert T. A., Nickson J., Patterson S. D., Raso R., Resing K., Seymour S. L., Tsugita A., Xenarios I., Zeng R., Julian R. K. (2008) Guidelines for reporting the use of mass spectrometry in proteomics. Nat. Biotechnol. 26, 860–861 [DOI] [PubMed] [Google Scholar]
- 51. Bradshaw R. A., Burlingame A. L., Carr S., Aebersold R. (2006) Reporting protein identification data. Mol. Cell. Proteomics 5, 787–788 [DOI] [PubMed] [Google Scholar]
- 52. Mueller L. N., Brusniak M. Y., Mani D. R., Aebersold R. (2008) An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J. Proteome Res. 7, 51–61 [DOI] [PubMed] [Google Scholar]
- 53. Kessner D., Chambers M., Burke R., Agus D., Mallick P. (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sturm M., Bertsch A., Gröpl C., Hildebrandt A., Hussong R., Lange E., Pfeifer N., Schulz-Trieglaff O., Zerck A., Reinert K., Kohlbacher O. (2008) OpenMS - An open-source software framework for mass spectrometry. BMC Bioinformatics 9, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kohlbacher O., Reinert K., Gröpl C., Lange E., Pfeifer N., Schulz-Trieglaff O., Sturm M. (2007) TOPP–the OpenMS proteomics pipeline. Bioinformatics 23, e191–197 [DOI] [PubMed] [Google Scholar]