Abstract
A decade of high-throughput screenings for intraviral and virus-host protein-protein interactions led to the accumulation of data and to the development of theories on laws governing interactome organization for many viruses. We present here a computational analysis of intraviral protein networks (EBV, FLUAV, HCV, HSV-1, KSHV, SARS-CoV, VACV, and VZV) and virus-host protein networks (DENV, EBV, FLUAV, HCV, and VACV) from up-to-date interaction data, using various mathematical approaches. If intraviral networks seem to behave similarly, they are clearly different from the human interactome. Viral proteins target highly central human proteins, which are precisely the Achilles' heel of the human interactome. The intrinsic structural disorder is a distinctive feature of viral hubs in virus-host interactomes. Overlaps between virus-host data sets identify a core of human proteins involved in the cellular response to viral infection and in the viral capacity to hijack the cell machinery for viral replication. Host proteins that are strongly targeted by a virus seem to be particularly attractive for other viruses. Such protein-protein interaction networks and their analysis represent a powerful resource from a therapeutic perspective.
From a systems biology perspective, a viral infection can be viewed at the cell level, as a combination of molecular perturbations allowing viral component production and assembly while generating minor or massive cellular dysfunctions. These perturbations are at least in part introduced into the host protein network through interactions of cellular proteins with viral proteins. Two systems are thus involved, namely the protein-protein interaction network of the virus, i.e. the intraviral interactome, and the protein-protein interaction network of the host, i.e. the host interactome. Their interplay creates a new system, the virus-host interactome, with emergent properties leading to viral replication and eventually to pathogenesis (1).
In a first attempt toward modeling of a viral infection at the cell level, protein interaction data for these three systems are required. Although far from being completed and elucidated, the human interactome and several intraviral and virus-host interactomes have been generated, using mainly the yeast two-hybrid system. High-throughput mapping of the intraviral protein interactions has been performed for eight human-infecting viruses, namely Epstein-Barr virus (EBV) (2), influenza virus (FLUAV) (3), hepatitis C virus (HCV) (4, 5), herpes simplex virus 1 (HSV-1) (6, 7, 8), Kaposi's sarcoma-associated herpesvirus (KSHV) (9, 10), SARS coronavirus (SARS-CoV) (11, 12), vaccinia virus (VACV) (13), and varicella zoster virus (VZV) (9, 14). Proteome-wide virus-human interactomes are also available for five human-infecting viruses, namely DENV1 (15), EBV (2), HCV (16), FLUAV (3), and VACV (17). Besides, numerous low-throughput studies have provided additional data that can be mined from the literature to complement networks construction. Several databases have been developed with the aim of integrating high-quality virus-virus and virus-host protein-protein interaction data such as VirHostNet (18) and VirusMint (19).
The study of complex networks is underpinned by the graph theory discipline coupled with bioinformatics analysis (20). Characterizing global statistical properties of a network, as well as local rules governing its individual nodes, can lead to fit to a network model in order to better understand and predict the system behavior and to guide experimental design (21). Eukaryotic protein interaction networks have been described as sharing some topological features with other complex systems, such as internet or social networks (22). Concerning the virology field, Uetz et al. and Fossum et al. performed two comparative analyses of the first versions of Herpes virus protein networks and began to find differences between the viral and host networks (6, 9). Dyer et al. reconstructed a host-pathogen interaction data set from literature mining and public databases (23). Although highly biased toward HIV-1 low-throughput interactions, this work still provided an interesting overview of virus preferential interactions with human proteins. Calderwood et al. and de Chassey et al. performed the first systematic screens of EBV and HCV proteins, respectively, against the human proteome and unraveled topological and functional signatures of viral targeting of the host (2, 16). The volume of interaction data reached today now motivates a deeper and up-to-date analysis of highly curated data sets gathering all aforementioned viruses.
Here we present an integrative and comparative computational analysis of all intraviral interactomes and virus-human interactomes integrating at least one large-scale mapping of interactions. High-quality and up-to-date protein-protein interaction networks were reconstructed. Scale-free, resilience, assortivity, and small-world signatures were examined, revealing that intraviral networks are very different from cellular networks. Analysis of virus-human interactomes revealed that viral proteins targeting a high number of host proteins are predicted to be more disordered than viral proteins targeting a single host protein. Analysis of viruses according to the human proteins they target identified common and specific viral strategies at a topological as well as functional level.
MATERIALS AND METHODS
Protein-Protein Interaction Network Reconstruction and Representation
Intraviral, virus-host and host-host protein-protein interaction networks were reconstructed from the VirHostNet database (18). Networks were visualized with Cytoscape (24) using a degree sorted circle layout (intraviral protein-protein interaction networks) or a force-directed layout (virus-human protein-protein interaction networks).
Statistical Analysis and Network Metric Computation
The R statistical environment was used to perform statistical analysis and the igraph R package to compute network topology measures (25, 26). MATLAB was also used, particularly for the statistical evaluation of power-law fit. Definitions and details concerning metrics are listed in supplemental Information.
Statistical Analysis of Degree Distribution
We used the protocol developed and implemented by Clauset et al. (27) (full description in supplemental Information). Briefly, for each degree distribution the best power-law model was first characterized. The scaling parameter and the lower bound of the power-law behavior were estimated using maximum likelihood methods. From this combination of parameters, the goodness-of-fit between the data and the power law was then computed using a Kolmogorov-Smirnov test. If the resulting p value is greater than 5.10−2, the power law is a plausible hypothesis for the data, otherwise it is rejected. The third step was to compare the power law with alternative hypotheses via a likelihood ratio test. For each alternative, if the likelihood ratio is significantly different from zero, then the sign indicates whether the alternative is favored over the power-law model or not.
Simulation of Random Failures and Deliberate Attacks
For each graph, nodes were removed one by one randomly (random failures) or by degree-decreasing order (deliberate attacks). At each step the single largest connected component was considered, its characteristic path length and size, i.e. number of nodes, were collected and plotted as a multiple or fraction of the original values.
Clustering Coefficient Enrichment
For each interactome, 1000 random graphs were built according to the edge-swapping model, which rewires edges while maintaining the degree distribution (28). For each random graph building, the number of rewiring trials was at least equal to three times the number of interactions in the real graph. The clustering coefficient was then computed in each random graph. The statistical evaluation of the clustering coefficient was obtained by comparing the value observed in the real network with the mean value collected from random networks.
Disorder-Degree Correlation Analysis
DisEMBL was used to predict disordered regions in the sequences of viral proteins targeting human proteins (29). The following procedure was derived from Haynes et al. (30). Sequence coordinates of predicted disordered regions were collected for viral ends (viral proteins interacting with one and only one human protein) and for viral hubs (viral proteins interacting with numerous human proteins). Various thresholds were chosen to define viral hubs: proteins connected to at least 15, 20, 25, 30, 50, 60, or 100 human proteins. Because the disorder content appeared partly dependent on the protein length, the sequences were examined through all possible segments of incrementing length. For each segment length, the percentages of residues predicted to be disordered within segments were collected and summed up.
Interconnectivity Analysis
Statistical significance for the interconnectivity of targeted proteins was assessed by a random resampling testing procedure (n = 10,000 permutations). For each permutation, we randomly extracted from the human interactome a number of proteins equivalent to the number of targeted proteins, and the number of shared interactions was determined. The randomization procedure was weighted and corrected according to the connectivity of proteins in order to prevent inspection bias on highly studied proteins. A theoretical distribution was computed for the 10,000 resampled values. From this distribution, an empirical p value was computed by counting the number of resampled values greater than the value observed for virus interactors.
Overlap Size Significance
Cellular interactors overlapping between each pair of viruses have been listed and the significance of the overlap size was assessed by random simulation using R (25). For each virus, we randomly drew as many proteins as interactors from the human proteome. The procedure was repeated 10,000 times and the overlap size was systematically measured. The p value was obtained by comparing the distribution of sizes with the observed number of overlapping interactors (significant overlap size: p value < 5.10−2). The significance of the overlap size between interactors targeted by at least one viral protein and interactors targeted by at least two viral proteins was assessed using a hypergeometric distribution and R.
RESULTS
Intraviral interactomes
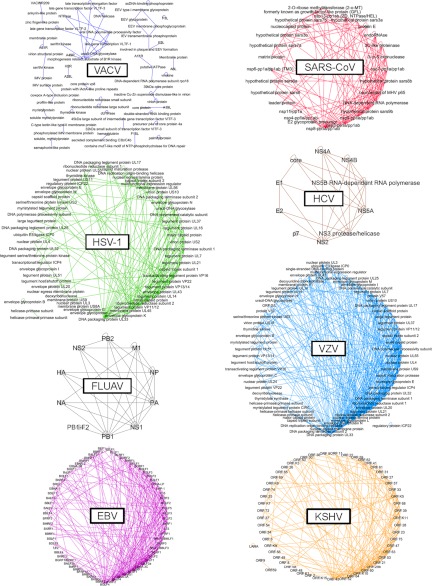
Eight human-infecting viruses, namely EBV (2, 6), FLUAV (3), HCV (4, 5), HSV-1 (6, 7, 8), KSHV (9, 10), SARS-CoV (11, 12), VACV (13), and VZV (9, 14) were introduced in this study because their interactome integrated at least one large-scale study (supplemental Fig. S1 and Table S1). In order to reconstruct high-quality and up-to-date interactomes for these viruses, high-throughput interactions were completed with low-throughput interactions from two different origins. First, interactions were extracted from VirHostNet, a public knowledge base specialized in the management and analysis of virus-virus and virus-host interactions offering today the largest high-confidence public data set (18). Second, interactions were accurately curated from the literature. Fig. 1 reveals that the eight interactomes are strikingly highly interconnected with a unique (or almost unique) single-connected component. VZV interactome is the densest, whereas VACV interactome stands apart from the others, most likely because of the lack of completeness (supplemental Table S1). The chosen layout shows the high heterogeneity in node degree for each interactome, ranging clockwise from low to high degree nodes, i.e. hubs.
Fig. 1.
Intraviral protein-protein interaction networks. VACV (dark blue), SARS-CoV (red), HSV-1 (green), HCV (brown), FLUAV (gray), VZV (light blue), EBV (magenta), and KSHV (orange). Node: viral protein. Edge: virus-virus protein-protein interaction. Nodes are ordered according to a degree sorted circle layout. Nodes are labeled according to the NCBI protein definition (for some imprecisely defined VACV proteins, the NCBI gene name or locus tag was used).
In order to examine more precisely the architectural features of these intraviral interactomes, key network metrics were computed (Table I, supplemental Information). The analysis was performed in parallel for the human interactome reconstructed from VirHostNet. Metric values for the human interactome were consistent with those already published (16, 31, 32), allowing comparison of intraviral and human interactomes. Mean degree values from intraviral interactomes confirm the conclusions drawn from the representations. VACV has the less connected proteins on average (mean degree = 3.64), in contrast to VZV whose proteins display the higher mean degree (mean degree = 11.10). Other intraviral interactomes have quite similar degree and betweenness values (mean degrees around 6, mean betweennesses around 0.01). One step further in the topological analysis was to investigate how the intraviral networks can compare with eukaryotic cellular networks. Indeed, most biomolecular networks have been found to be not random but rather commonly governed by a few simple principles (22). Among them, most cellular networks, and eukaryotic protein-protein interaction networks in particular, have been described as scale-free, diassortive and small-world.
Table I. Topological analysis of the intraviral and human interactomes. For each network are given: the network components, i.e. the number of nodes (proteins), the number of edges (interactions); the simple metrics, i.e. the mean degree, the mean betweenness, and the diameter computed from the single largest connected component; the scale-free information, i.e. the scale exponent and p value assessing whether the power-law can be rejected as a good statistical model of the data, the indication brought by the degree distribution comparison procedure and the indication brought by the structural comparison procedure; the assortativity coefficient; the small-world information, i.e. the characteristic path length and the clustering coefficient computed from the single largest connected component, and the enrichment of this coefficient over 1000 edge-swapping model-built networks.
| Network metrics | EBV | FLUAV | HCV | HSV-1 | KSHV | SARS-CoV | VACV | VZV | Human |
|---|---|---|---|---|---|---|---|---|---|
| Network components | |||||||||
| Nodes | 73 | 10 | 10 | 63 | 56 | 28 | 64 | 68 | 10707 |
| Edges | 266 | 34 | 32 | 158 | 162 | 114 | 61 | 373 | 55861 |
| Simple metrics | |||||||||
| Mean degree | 7.29 | 6.8 | 6.4 | 5.2 | 6.04 | 8.14 | 3.64 | 11.10 | 10.67 |
| Mean betweenness | 0.01051 | 0.01889 | 0.02333 | 0.01645 | 0.01444 | 0.01549 | 0.04545 | 0.00814 | 0.00014 |
| Diameter | 5 | 2 | 3 | 8 | 5 | 3 | 4 | 4 | 14 |
| Scale-free | |||||||||
| Scale exponent | 3.39 | 3.08 | 3.14 | 3.34 | 3.39 | 3.31 | 3.01 | 2.36 | 2.52 |
| p value | 0.579 | 0.244 | 0.445 | 0.364 | 0.931 | 0.731 | 0.554 | 0.003 | <10–4 |
| Degree distribution comparison | Moderate | Unlikely | Unlikely | Moderate | Moderate | Moderate | Moderate | Ambiguous | With cut-off |
| Structural comparison | Unlikely | Unlikely | Unlikely | Unlikely | Unlikely | Unlikely | Unlikely | Ambiguous | – |
| Assortativeness | |||||||||
| Assortativity coefficient | −0.106 | −0.340 | −0.090 | −0.034 | −0.077 | −0.045 | 0.366 | −0.155 | −0.066 |
| Small-world | |||||||||
| Characteristic path length | 2.53 | 1.38 | 1.47 | 2.97 | 2.53 | 1.87 | 2.00 | 2.09 | 3.94 |
| Clustering coefficient | 0.22 | 0.81 | 0.71 | 0.21 | 0.25 | 0.46 | 0.72 | 0.43 | 0.15 |
| Clustering coefficient enrichment | 0.99 | 0.97 | 0.99 | 1.02 | 1.23 | 0.95 | 1.68 | 0.88 | 6.53 |
The scale-free architecture has been proposed to be a representative model of many cellular networks (22, 33). A first method to assess whether interactomes are scale-free is to focus on their degree distributions. The degree distribution of a scale-free network is indeed expected to follow a power-law, at least asymptotically, with a scaling exponent typically ranging between 2 and 3 (33). We analyzed the degree distributions by applying the rigorous statistical method developed and implemented in Clauset et al. (27, 34) (supplemental Information). For each distribution, a maximum likelihood estimation followed by a Kolmogorov-Smirnov test were used to estimate the most probable power-law parameters and to compute a p value assessing whether we can reject the power law as a good statistical model for the data or not (Table I, supplemental Fig. S2 and Table S2). According to this test, the power law is not a plausible model for VZV as well as for human degree distributions. The power-law model cannot be rejected for VACV and to a lesser extent for HCV and FLUAV degree distributions, and exhibits suitable scale-free parameters. For EBV, HSV-1, KSHV, and SARS-CoV degree distributions the power-law model cannot be rejected either, however the scaling parameters above three suggest a weak role for hubs (22). To go a step further, the power-law model can be compared with several other competing distributions which might give a fit as good or better, or might conversely favor the power law. For each interactome, the power law was compared with several distributions via a likelihood ratio test according to the method described in Clauset et al. (27, 34). All results are given with a detailed interpretation in supplemental Table S3 and supplemental Information, and are summarized in Table I. The power law with exponential cutoff seems to be a more plausible model for the human degree distribution. Concerning the intraviral degree distributions, the pure power-law model never appears truly convincing. Several forms seem to fit better for FLUAV and HCV degree distributions. The power-law model cannot be completely rejected for EBV, HSV1, KSHV, SARS-CoV, and VACV degree distributions but is never preferred either. For VZV degree distributions, the results are ambiguous. Generally, none of the intraviral degree distributions seem to be well-characterized enough to be unequivocally assigned to any of the tested distributions.
A second method to address the scale-free question is to focus on network structure. This allows to better overcome noise and to capture local structures of the networks. We used the GraphCrunch 2 software tool to structurally compare each intraviral interactome to model-generated networks including several scale-free networks (35). Comparisons were performed using two constraining graphlet-based measures (supplemental Information). The results are described in details in supplemental Information and supplemental Fig. S3, and are summarized in Table I. Using this method, the scale-free model is not the best model for the intraviral interactomes, except for VZV for which the results remain ambiguous. The stickiness-index based model (36) is rather preferred, being the best-fitting model among those tested, in agreement with a previous case study (35).
A third method to assess the scale-free architecture concerns the network resilience. A scale-free architecture indeed leads a network to be highly tolerant to random failures but hardly resistant to deliberate attacks, because of its hublike core structure (37). Simulations of random failures by removing nodes randomly ordered one by one was first performed for intraviral and human interactomes (Fig. 2A and 2B). All characteristic path lengths globally increase slowly, except for VACV network because of the very small size of its largest connected component. Consistently, the sizes of the largest clusters of all networks slowly decrease. All the networks appeared rather robust to random failures. Simulation of deliberate attacks by removing the most connected nodes one by one was then performed for both intraviral and human interactomes (Fig. 2C and 2D). All intraviral networks maintain their characteristic path lengths during hub progressive removal, whereas human network clearly expands until a threshold at which the largest connected component is fragmented into an increasing number of small networks. The network size variations in response to attacks are consistent with characteristic path length behavior. Intraviral network sizes globally decrease slower than the human network size, which is particularly detracted as soon as the first hubs are removed. Therefore, intraviral networks do not seem very vulnerable to attacks, in agreement with the results obtained on the first herpesvirus interactome drafts (6, 9). Hubs, although present, do not seem to hold the whole network together, as it was already suggested by the scale exponent values (Table I). This resilience analysis discards a bit more the putative scale-free architecture for the intraviral interactomes.
Fig. 2.
Random failure and deliberate attack tolerance of intraviral and human interactomes. A, Changes in the characteristic path length (CPL) (as a multiple of the original value) during the progressive random removal of nodes in intraviral and human interactomes, simulating random failures. B, Changes in the network size, i.e. number of nodes, (as a fraction of the original value) during the progressive random removal of nodes in intraviral and human interactomes, simulating random failures. C, Changes in the characteristic path length (CPL) (as a multiple of the original value) during the progressive removal of nodes in degree-decreasing order in intraviral and human interactomes, simulating deliberate attacks. D, Changes in the network size, i.e. number of nodes, (as a fraction of the original value) during the progressive removal of nodes in degree-decreasing order in intraviral and human interactomes, simulating deliberate attacks.
The second frequently described signature of interactomes concerns the preferences of connections. Cellular networks are often qualified as diassortive, meaning that low-degree nodes are more likely to connect high-degree nodes, and reciprocally (28, 38). At first, correlation in node degrees was examined for each network (supplemental Fig. S4). Then, in order to statistically value these correlation profiles, the degree correlation of each network was compared with those of random networks built on the edge-swapping model, which rewires edges while maintaining the degree distribution (28) (supplemental Information, supplemental Fig. S4). HCV, FLUAV, and VACV small interactomes are not appropriate for rewiring, however in larger intraviral interactomes hubs do not seem to preferentially connect other hubs. We then applied a rigorous mathematical method by computing the assortivity coefficient, which is a measure indicating if a network shows diassortive, assertive, or neutral mixing (38). An assortive (resp. diassortive) mixing by degree means that nodes preferentially connect to others that have a similar (resp. different) degree. As their human counterpart, all intraviral interactomes, except VACV, display negative values of assortivity coefficient (Table I). It means that they are rather diassortive hence their proteins tend to pair up with proteins of different degrees.
The third popular signature assigned to cellular networks is the small-world architecture (22, 39). Such networks have a small characteristic path length and a large clustering coefficient compared with equivalent random networks. For each interactome, random counterparts were built on the edge-swapping model. The statistical evaluation of the clustering coefficient was obtained by comparing the value of the real network with the mean value computed from the random networks. There was no enrichment of clustering when intraviral interactomes were compared with their random counterparts. In contrast, the human interactome clustering coefficient was increased more than six-fold over random networks (Table I). When examining characteristic path lengths, the conclusions were similar (supplemental Table S4). Therefore intraviral interactomes can not be classified as small-world, in line with previous analysis (6, 9).
Overall, intraviral interactomes share a diassortive nature with the human interactome but strongly differ when regarding the scale-free and the small-world architectures. VACV sparse interactome stands apart whereas HCV and FLUAV small and compact interactomes are alike. Amazingly, VZV interactome analysis provides more ambiguous results.
Virus-Host Interactomes
Proteome-wide mapping of interactions between viral and human proteins has been reported for five human-infecting viruses, namely DENV (15), EBV (2), HCV (16), FLUAV (3), and VACV (17) (supplemental Fig. S1 and Table S5). The VirHostNet database and an extensive literature curation allowed the reconstruction of the corresponding virus-host interactomes with a high-quality data set (supplemental Table S6). For each virus, a virus-host interactome was built without a priori by using only high-throughput data (further referred as “without a priori” virus-host interactome). Indeed, by excluding small-scale studies, inspection biases are avoided, giving thus a more representative picture of the complete network although smaller.
The five virus-host interactomes reveal that the degree of viral protein is heterogeneous, some of them targeting a high number of human proteins (e.g. degree of EBV EBNA-LP = 139 and degree of HCV NS3 = 186) whereas the majority is targeting a small number (Fig. 3). These differences in connectivity led us to examine the structural nature of the viral proteins. Indeed, for eukaryotes, hubs (highly connected proteins) have been found to be more disordered than ends (proteins with only one partner) (30). The intrinsic structural disorder of viral ends (viral proteins connected to only one human protein) and of viral hubs (viral proteins highly connected to human proteins) was thus assessed. Disordered regions were predicted in viral hubs and then in viral ends using DisEMBL (29). Because a partial correlation was observed between length of disordered regions and length of proteins (data not shown), disorder measurement was normalized according to protein length. For all possible lengths of segment in the sequences of hubs and ends, the percentage of residues predicted to be disordered were collected and summed up. The proportion of predicted disorder was higher in hubs than in ends (Fig. 4), with differences in disorder content even higher than those observed between hubs and ends in eukaryotic interactomes (30). As the definition of a hub is not precise, the analysis was performed using increasing hub degree thresholds. Remarkably, the predicted disorder increased according to the degree threshold (Fig. 4), suggesting that the viral protein connectivity in virus-host interactomes may at least partly correlate to their structural disorder level. The tendency is conserved in the “without a priori” virus-host interactomes, indicating the robustness of the observation (supplemental Fig. S5).
Fig. 3.
Virus-host protein-protein interaction networks. DENV-human interactome (H-DENV), EBV-human interactome (H-EBV), FLUAV-human interactome (H-FLUAV), HCV-human interactome (H-HCV), and VACV-human interactome (H-VACV). Black node: viral protein; red node: human protein, red edge: virus-host protein-protein interaction; blue edge: host-host protein-protein interaction. Nodes are ordered according to a force directed layout. Viral protein nodes are labeled according to the NCBI protein definition (for some imprecisely defined VACV proteins, the NCBI gene name or locus tag was used).
Fig. 4.
Disorder-degree analysis of viral proteins in virus-host interactomes. Mean percentage of residues predicted to be disordered (y axis) within segments of various lengths (x axis) in sequences of viral proteins interacting with cellular proteins. Data from the five virus-host interactomes have been analyzed. Viral ends (black line) are viral proteins having only one human interacting protein. Viral hubs (colored lines) are viral proteins highly connected to human proteins. Various degree thresholds were used to define the set of hubs: at least 15 (violet line), at least 20 (blue line), at least 25 (turquoise line), at least 30 (green line), at least 50 (yellow line), at least 60 (orange line) and at least 100 human interactors (red line).
Disordered regions are known to be enriched with short linear motifs (SLMs) (40, 41). These SLMs, corresponding to short stretches of amino acid residues usually characterized by a simple sequence pattern, are involved in protein-protein interactions named “SLM-domain” interactions. According to that, when examining human proteins involved in virus-host interactomes, numerous domains were identified (supplemental Information, supplemental Fig. S6). These properties may allow viruses to target a large diversity of cellular proteins despite a small proteome. This is strikingly exemplified in supplemental Fig. S7 where proteins from small viral proteomes (FLUAV, DENV and HCV) target proportionally more distinct domains than proteins of viruses with larger proteomes (EBV and VACV).
To assess how viral proteins interplay with the human interactome, the virus-host interactomes were placed in the context of the human interactome. First, human proteins targeted by viral proteins were clearly overrepresented in the human interactome (Exact Fisher Test, all p values < 2.2 10−16). This suggests that viruses preferentially target host proteins already engaged in protein-protein interactions. Then, analysis of subnetworks of host proteins that are targeted by the viruses indicates that their interconnectivity appears significantly higher compared with the theoretical interconnectivity computed from resampled subnetworks (resampling tests, n = 10,000, all p values < 10−4, supplemental Fig. S8). Network descriptive metrics of each set of human proteins targeted by a virus were finally computed in the human interactome (Fig. 5A). For each virus, the degree distribution of targeted human proteins was significantly higher than the degree distribution in the human interactome (U test, all p values < 2.2 10−16), with average degrees of targeted proteins two to three times higher than the average degree of the human interactome (Figs. 5A and 5B). Therefore viral proteins appear to have a strong tendency to interact with highly connected cellular proteins. To go deeper into the analysis, the betweenness values and distributions were also examined (Figs. 5A and 5C). Similarly, for each virus the betweenness distribution of targeted human proteins was significantly higher than the betweenness distribution in the human interactome (U test, all p values < 2.2 10−16), with average betweennesses of targeted proteins four to five times higher than the average betweenness of the human interactome. In addition, for each virus the topological proximity of the targeted human proteins was examined through the path length between these proteins within the human interactome (Fig. 5A). For each virus, the path length distribution of targeted human proteins was significantly lower than the path length distribution in the human interactome (U test, all p values < 2.2 10−16), with characteristic path length between targeted proteins lower than in the human interactome. Thus, as a general hallmark, viruses target proteins that are strikingly interconnected, highly central, both locally (degree) and globally (betweenness), and that are relatively close to each other in the human interactome (2, 16, 23). The whole topological analysis was repeated using “without a priori” virus-host interactomes. All the aforementioned trends are conserved, indicating that they do not result from inspection bias (supplemental Figs. S9, S10, supplemental Table S7).
Fig. 5.
Topological analysis of the virus-host interactomes. A, Metrics of the human proteins and of the human proteins targeted by viruses in the human interactome. The number of nodes and edges, the mean degree, the mean adjusted betweenness and the characteristic path length are given first for all the human proteins, then for the human proteins targeted by DENV, EBV, FLUAV, HCV, and VACV. B, Degree distributions of human proteins and human proteins targeted by viruses in the human interactome. P(k) is the probability of a node to connect k other nodes in the network. Solid lines represent linear regression fits. Vertical dashed lines indicate the mean degree of each distribution. C, Betweenness distributions of human proteins and human proteins targeted by viruses in the human interactome. P(b) is the probability for a node to have a betweenness value of b in the network. Solid lines represent linear regression fits. Vertical dashed lines indicate the mean betweenness value for each distribution.
We next considered the overlaps of the lists of human proteins targeted by the five viruses in a pairwise manner (Table IIA). Overlaps range from 5 to 20% of each data set and are all significantly high (random simulation, all p values < 10−4, supplemental Fig. S11). The most striking example concerns HCV and EBV which commonly target 61 human proteins, corresponding to almost 20% of the EBV dataset. Two human proteins, LZTS2 and GOLGA2, have been identified in high-throughput screenings to interact with four viruses. LZTS2 has been studied for its role in cancer but its role in viral infections is unknown. In the human interactome LZTS2 interacts with TRAF2, suggesting that it may be involved in the functional modulation of the type I interferon pathway (42). We further assumed that, among the human proteins targeted by a given virus, those which are targeted by more than one viral protein are of particular importance, i.e. can be considered functionally essential because the virus targets them by different ways. These highly targeted proteins were also more frequently present in the lists of proteins targeted by other viruses in pairwise comparisons. For example, while 18.4% of the proteins targeted by EBV are also targeted by HCV (Table 2A), the percentage of overlap increased to 44.7% when proteins highly targeted by EBV were considered (Table 2B). This overlap is significantly high (hypergeometric distribution, p value = 5.2 10−6). The p value is less significant for other comparisons most likely because of the small size of the lists. Overall this suggests that proteins essential for a given virus tend to be also important for other viruses.
Table II. Protein overlaps between virus-host interactomes. A. Protein overlaps between full data sets. Overlaps are given as percentages of the number of human proteins targeted by DENV (column 3), EBV (column 4), FLUAV (column 5), HCV (column 6), or VACV (column 7). Stars indicate the level of significance as assessed by random simulation: *: < 5.10−2, **: < 10−3, ***: < 10−4. B. Protein overlaps between full data sets and data sets containing human proteins targeted by at least two proteins of the same virus. Overlaps are given as percentages of the number of human proteins targeted by at least two proteins of DENV (column 3), EBV (column 4), FLUAV (column 5), HCV (column 6), or VACV (column 7). Stars indicate the level of significance as assessed by the hypergeometric distribution: *: < 5.10−2, **: < 10−3, ***: < 10−4.
| A | % | Human proteins targeted by |
||||
|---|---|---|---|---|---|---|
| DENV | EBV | FLUAV | HCV | VACV | ||
| Human proteins targeted by | DENV | 100 | 4.2 (***) | 7.0 (***) | 6.5 (***) | 4.7 (***) |
| EBV | 7.4 (***) | 100 | 13.5 (***) | 14.6 (***) | 7.0 (***) | |
| FLUAV | 5.8 (***) | 6.3 (***) | 100 | 5.5 (***) | 4.7 (***) | |
| HCV | 14.4 (***) | 18.4 (***) | 14.7 (***) | 100 | 11.7 (***) | |
| VACV | 5.3 (***) | 4.5 (***) | 6.4 (***) | 6.0 (***) | 100 | |
| B | % | Human proteins targeted by at least two proteins of |
||||
|---|---|---|---|---|---|---|
| DENV | EBV | FLUAV | HCV | VACV | ||
| Human proteins targeted by | DENV | 100 | 8.5 | 8.8 | 15.8 (*) | 14.3 |
| EBV | 17.6 (*) | 100 | 17.6 | 47.4 (***) | 14.3 | |
| FLUAV | 5.9 | 14.9 (*) | 100 | 7.9 | 21.4 (*) | |
| HCV | 23.5 | 44.7 (***) | 14.7 | 100 | 28.6 | |
| VACV | 11.8 | 2.1 | 5.9 | 7.9 | 100 | |
The comparison of the five virus-host interactomes can also be done at an upper level, i.e. at the level of biological processes in which the targeted proteins are involved. Even if the lists of interactors are far from complete, a core of biological processes comprising shared aforementioned proteins and targeted by at least two viruses could be identified (supplemental Information, supplemental Fig. S12). These processes correspond to the cellular response to viral infection (such as immune response, cellular response to stress, positive regulation of cytokine production, apoptosis, cell cycle) and to the ability of the virus to hijack the cell machinery for its replication (such as nuclear transport, protein import to the nucleus, RNA processing, viral reproduction, cytoskeleton organization). Conversely, some targeted biological processes are more specifically related to the biology of a virus like for instance the targeting of the plasma lipoprotein particle assembly, of the cellular response to insulin stimulus, of the ER-nuclear signaling pathway and of the positive regulation of lipid metabolic process by HCV (supplemental Fig. S12). This suggests that comprehensive explorations of the virus-host interactomes can be very instrumental in the discovery and understanding of infection and induced-pathogenicity processes.
DISCUSSION
Using a high-quality data set of up-to-date curated interactions, this study is a comprehensive and comparative analysis of intraviral and virus-host interactomes. The criterion for the selection of a virus in this work is a large-scale exploration of protein-protein interactions. These data were combined with the low-throughput interaction data curated from the literature. In the virus-host interactomes, we have controlled that major structural and topological trends did not result from inspection biases. Because interactomes are far from complete and in constant refinement, lack some accuracy and may be subject to inherent variability, we do not expect this analysis to be definitive (20). Nevertheless, this work is gathering various methods from graph theory as well as structural and functional analysis and provides a common analytical framework allowing interactome comparisons.
It appears from this study that intraviral interactomes tend to be highly coupled modules. In contrast to what is currently reported for the human interactome, these networks do not present a scale-free nor a small-world signature and are hardly resistant to both random and deliberate attacks. None of the models examined here can fully describe the intraviral networks, even if the stickiness-based index may be an interesting hint. One intriguing possibility is that these networks cannot be well-characterized because they are only complete and functional in interaction with the host network, where they adopt its properties (6, 9, 43). We also confirmed that viruses target human proteins that are highly interconnected, central and close to each other in the human interactome. Structurally, viral proteins that are the most connected to human proteins display a higher content of predicted disorder. Significant overlaps in the human proteins targeted by viruses were observed in pairwise comparisons. Interestingly, cellular proteins targeted by at least two proteins of a same virus are more susceptible to be targeted by other viruses. Overlaps between data sets are also observed at the level of biological processes, reflecting common viral strategies.
Although popular and appealing, it is notable that the use of the scale-free model to describe networks in biology raises a number of reservations (20, 34, 44, 45). Methods often used to detect a power law from a degree distribution may lack rigor (27, 46, 47). Therefore we used here the statistical framework provided by Clauset et al. (27, 34). Another hurdle in this type of analysis is the power-law definition itself that can be criticized for its loose mathematical framework (20, 48, 49). In addition, as summarized by Han et al. (50), the apparent scale-free topology of an incomplete interactome may not be representative of the final topology of a full interactome. Overall, major efforts have to be made to overcome these difficulties, and results concerning the scale-free signature have to be taken cautiously. Here, to address the question of the scale-free architecture of intraviral interactomes, a degree distribution statistical analysis, a network structure comparison and a resilience analysis were combined.
Viruses seem to have evolved strategies to efficiently adapt to the scale-free and small-world architecture of the human interactome. By interacting with host hubs, viruses target the “Achilles' heel” of the host interactome. Because of the multi-functionality of these hubs, viruses manipulate a wide range of functions. In addition, some viral proteins have also evolved the ability to interact with numerous cellular proteins. This pleiotropy may be at least partly mediated by intrinsically disordered regions of these proteins, and it is striking to note that the most connected hubs tend to be the most intrinsically disordered. Indeed, this structural plasticity may allow viral hubs to adapt to the structures of a wide range of host protein interactors. Altogether, viruses are able to widely perturb the host protein network despite their relatively small genomes.
The first comparison of five virus-host interactomes reveals common cellular protein targets and biological processes. Viruses thus share a core of targets that take part in the cellular response to infection and in their capacity to hijack the cell machinery for their own replication. Viruses also target specific proteins and pathways, some of them being clearly related to their biology. The most striking is for HCV, which targets pathways associated with metabolism and clinical syndromes. Even if the list of targets is expected to increase, the data already represent a powerful resource for a therapeutic perspective. Common cellular targets as well as pathways could thus be used to identify broad-spectrum antiviral drugs (51). Overall, the possibility to target virus-host interactions considerably broadens the landscape of drugs that could be developed. In this way, SLMs are a great opportunity. Indeed, viruses use short linear motifs that mimic those of the human proteome to specifically target well-characterized domains of cellular proteins (52). They have also evolved original SLMs whose computational identification is in progress (53). These peptides could serve as scaffolds for peptide mimetic and structure-based drug design. Therefore, the list of virus-host interactors is an invaluable resource to derive new molecules especially for antiviral therapy.
The study of complex networks and their modeling are still in their infancy (21). We begin to apprehend the non-randomness and some statistical and topological signatures of most of biological networks, even if more sophisticated models are needed to go further. Accumulation of data is required to provide a more complete and accurate view of biological networks. In particular, the interaction maps will need to be filtered according to localization, expression profiles, time parameters and so on, and enriched by functional information to give a more dynamic picture of the networks. The systems biology challenge will be taken up with continuous and transdisciplinary effort involving experts from biology, mathematics, statistics, and computer science.
Acknowledgments
We thank particularly Hervé Gilquin for the very efficient assistance. This work strongly benefited from the ressources of PSMN (Pôle Scientifique de Modélisation Numérique) at the ENS Lyon. We also thank Maryam Meyniel for critical reading of the manuscript, Lilia Iakoucheva for technical details, Aaron Clauset and Adrián López García de Lomana for making their methods and codes publicly available as well as for helpful discussions.
This paper is dedicated to the memory of Chantal Rabourdin-Combe.
Footnotes
* This work was supported by Inserm and by the European Union's Seventh Program (FP7/2007–2013) under grant agreement no 267429 (SysPatho).
 This article contains supplemental Figs. S1 to S12 and Tables S1 to S7.
This article contains supplemental Figs. S1 to S12 and Tables S1 to S7.
1 The abbreviations used are:
- DENV
- dengue virus
- EBV
- Epstein-Barr virus
- FLUAV
- influenza virus
- HCV
- hepatitis C virus
- HSV-1
- herpes simplex virus 1
- KSHV
- Kaposi's sarcoma-associated herpesvirus
- SARS-CoV
- SARS coronavirus
- VACV
- vaccinia virus
- VZV
- varicella zoster virus
- SLM
- short linear motif
- CPL
- characteristic path length
- H-DENV
- DENV-human interactome
- H-EBV
- EBV-human interactome
- H-FLUAV
- FLUAV-human interactome
- H-HCV
- HCV-human interactome
- H-VACV
- VACV-human interactome.
REFERENCES
- 1. Tan S. L., Ganji G., Paeper B., Proll S., Katze M. G. (2007) Systems biology and the host response to viral infection. Nat. Biotechnol. 25, 1383–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Calderwood M. A., Venkatesan K., Xing L., Chase M. R., Vazquez A., Holthaus A. M., Ewence A. E., Li N., Hirozane-Kishikawa T., Hill D. E., Vidal M., Kieff E., Johannsen E. (2007) Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. U.S.A. 104, 7606–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shapira S. D., Gat-Viks I., Shum B. O., Dricot A., de Grace M. M., Wu L., Gupta P. B., Hao T., Silver S. J., Root D. E., Hill D. E., Regev A., Hacohen N. (2009) A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139, 1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Flajolet M., Rotondo G., Daviet L., Bergametti F., Inchauspé G., Tiollais P., Transy C., Legrain P. (2000) A genomic approach of the hepatitis C virus generates a protein interaction map. Gene 242, 369–379 [DOI] [PubMed] [Google Scholar]
- 5. Dimitrova M., Imbert I., Kieny M. P., Schuster C. (2003) Protein-protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 77, 5401–5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fossum E., Friedel C. C., Rajagopala S. V., Titz B., Baiker A., Schmidt T., Kraus T., Stellberger T., Rutenberg C., Suthram S., Bandyopadhyay S., Rose D., von Brunn A., Uhlmann M., Zeretzke C., Dong Y. A., Boulet H., Koegl M., Bailer S. M., Koszinowski U., Ideker T., Uetz P., Zimmer R., Haas J. (2009) Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog. 5, e1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vittone V., Diefenbach E., Triffett D., Douglas M. W., Cunningham A. L., Diefenbach R. J. (2005) Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79, 9566–9571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee J. H., Vittone V., Diefenbach E., Cunningham A. L., Diefenbach R. J. (2008) Identification of structural protein-protein interactions of herpes simplex virus type 1. Virology 378, 347–354 [DOI] [PubMed] [Google Scholar]
- 9. Uetz P., Dong Y. A., Zeretzke C., Atzler C., Baiker A., Berger B., Rajagopala S. V., Roupelieva M., Rose D., Fossum E., Haas J. (2006) Herpesviral protein networks and their interaction with the human proteome. Science 311, 239–242 [DOI] [PubMed] [Google Scholar]
- 10. Rozen R., Sathish N., Li Y., Yuan Y. (2008) Virion-wide protein interactions of Kaposi's sarcoma-associated herpesvirus. J. Virol. 82, 4742–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. von Brunn A., Teepe C., Simpson J. C., Pepperkok R., Friedel C. C., Zimmer R., Roberts R., Baric R., Haas J. (2007) Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One 2, e459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pan J., Peng X., Gao Y., Li Z., Lu X., Chen Y., Ishaq M., Liu D., Dediego M. L., Enjuanes L., Guo D. (2008) Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS One 3, e3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCraith S., Holtzman T., Moss B., Fields S. (2000) Genome-wide analysis of vaccinia virus protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A. 97, 4879–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stellberger T., Häuser R., Baiker A., Pothineni V. R., Haas J., Uetz P. (2010) Improving the yeast two-hybrid system with permutated fusions proteins: the Varicella Zoster Virus interactome. Proteome Sci 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Khadka S., Vangeloff A. D., Zhang C., Siddavatam P., Heaton N. S., Wang L., Sengupta R., Sahasrabudhe S., Randall G., Gribskov M., Kuhn R. J., Perera R., LaCount D. J. (2011) A physical interaction network of dengue virus and human proteins. Mol. Cell Proteomics. 10, M111.012187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Chassey B., Navratil V., Tafforeau L., Hiet M. S., Aublin-Gex A., Agaugue S., Meiffren G., Pradezynski F., Faria B. F., Chantier T., Le Breton M., Pellet J., Davoust N., Mangeot P. E., Chaboud A., Penin F., Jacob Y., Vidalain P. O., Vidal M., André P., Rabourdin-Combe C., Lotteau V. (2008) Hepatitis C virus infection protein network. Mol. Syst. Biol. 4, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang L., Villa N. Y., Rahman M. M., Smallwood S., Shattuck D., Neff C., Dufford M., Lanchbury J. S., Labaer J., McFadden G. (2009) Analysis of vaccinia virus-host protein-protein interactions: validations of yeast two-hybrid screenings. J. Proteome Res. 8, 4311–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Navratil V., de Chassey B., Meyniel L., Delmotte S., Gautier C., André P., Lotteau V., Rabourdin-Combe C. (2009) VirHostNet: a knowledge base for the management and the analysis of proteome-wide virus-host interaction networks. Nucleic Acids Res. 37, D661–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chatr-aryamontri A., Ceol A., Peluso D., Nardozza A., Panni S., Sacco F., Tinti M., Smolyar A., Castagnoli L., Vidal M., Cusick M. E., Cesareni G. (2009) VirusMINT: a viral protein interaction database. Nucleic Acids Res. 37, D669–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mason O., Verwoerd M. (2007) Graph theory and networks in Biology. IET Syst. Biol. 1, 89–119 [DOI] [PubMed] [Google Scholar]
- 21. Newman M. E. J. (2003) The structure and function of complex networks. SIAM Review 45, 167–256 [Google Scholar]
- 22. Barabási A. L., Oltvai Z. N. (2004) Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 5, 101–113 [DOI] [PubMed] [Google Scholar]
- 23. Dyer M. D., Murali T. M., Sobral B. W. (2008) The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 4, e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. R Development Core Team (2010) R: A Language and Environment for Statistical Computing. [Google Scholar]
- 26. Csardi G., Nepusz T. (2006) The igraph software package for complex network research. InterJournal Complex Systems 1695 [Google Scholar]
- 27. Clauset A., Shalizi C. R., Newman M. E. J. (2009) Power-law distributions in empirical data. SIAM Rev. 51, 661–703 [Google Scholar]
- 28. Maslov S., Sneppen K. (2002) Specificity and stability in topology of protein networks. Science 296, 910–913 [DOI] [PubMed] [Google Scholar]
- 29. Linding R., Jensen L. J., Diella F., Bork P., Gibson T. J., Russell R. B. (2003) Protein disorder prediction: implications for structural proteomics. Structure 11, 1453–1459 [DOI] [PubMed] [Google Scholar]
- 30. Haynes C., Oldfield C. J., Ji F., Klitgord N., Cusick M. E., Radivojac P., Uversky V. N., Vidal M., Iakoucheva L. M. (2006) Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput. Biol. 2, e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodrigues F. A., Costa Lda F., Barbieri A. L. (2011) Resilience of protein-protein interaction networks as determined by their large-scale topological features. Mol. Biosyst. 7, 1263–1269 [DOI] [PubMed] [Google Scholar]
- 32. Navratil V., de Chassey B., Combe C. R., Lotteau V. (2011) When the human viral infectome and diseasome networks collide: towards a systems biology platform for the aetiology of human diseases. BMC Syst. Biol. 5, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barabasi A. L., Albert R. (1999) Emergence of scaling in random networks. Science 286, 509–512 [DOI] [PubMed] [Google Scholar]
- 34. López Garcia De Lomana A., Beg Q. K., De Fabritiis G., Villà-Freixa J. (2010) Statistical analysis of global connectivity and activity distributions in cellular networks. J. Comput. Biol. 17, 869–878 [DOI] [PubMed] [Google Scholar]
- 35. Kuchaiev O., Stevanović A., Hayes W., Pržulj N. (2011) GraphCrunch 2: Software tool for network modeling, alignment and clustering. BMC Bioinformatics 12, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Przulj N., Higham D. J. (2006) Modelling protein-protein interaction networks via a stickiness index. J. R. Soc. Interface 3, 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Albert R., Jeong H., Barabasi A. L. (2000) Error and attack tolerance of complex networks. Nature 406, 378–382 [DOI] [PubMed] [Google Scholar]
- 38. Newman M. E. (2003) Mixing patterns in networks. Phys. Rev. E Stat Nonlin Soft Matter Phys. 67, 026126 [DOI] [PubMed] [Google Scholar]
- 39. Watts D. J., Strogatz S. H. (1998) Collective dynamics of ‘small-world’ networks. Nature 393, 440–442 [DOI] [PubMed] [Google Scholar]
- 40. Fuxreiter M., Tompa P., Simon I. (2007) Local structural disorder imparts plasticity on linear motifs. Bioinformatics 23, 950–956 [DOI] [PubMed] [Google Scholar]
- 41. Longhi S. (2010) Structural disorder in viral proteins. Protein Pept. Lett. 17, 930–931 [DOI] [PubMed] [Google Scholar]
- 42. Navratil V., de Chassey B., Meyniel L., Pradezynski F., André P., Rabourdin-Combe C., Lotteau V. (2010) System-level comparison of protein-protein interactions between viruses and the human type I interferon system network. J. Proteome Res. 9, 3527–3536 [DOI] [PubMed] [Google Scholar]
- 43. Bader S., Kühner S., Gavin A. C. (2008) Interaction networks for systems biology. FEBS Lett. 582, 1220–1224 [DOI] [PubMed] [Google Scholar]
- 44. Przulj N., Corneil D. G., Jurisica I. (2004) Modeling interactome: scale-free or geometric? Bioinformatics 20, 3508–3515 [DOI] [PubMed] [Google Scholar]
- 45. Khanin R., Wit E. (2006) How scale-free are biological networks. J. Comput. Biol. 13, 810–818 [DOI] [PubMed] [Google Scholar]
- 46. Li L., Alderson D., Doyle J. C., Willinger W. (2005) Towards a Theory of Scale-Free Graphs: Definition, Properties, and Implications. Internet Math. 2, 431–523 [Google Scholar]
- 47. Tanaka R., Yi T. M., Doyle J. (2005) Some protein interaction data do not exhibit power law statistics. FEBS Lett. 579, 5140–5144 [DOI] [PubMed] [Google Scholar]
- 48. Bollobas B., Riordan O. (2003) Robustness and Vulnerability of Scale-Free Random Graphs. Internet Math. 1, 1–35 [Google Scholar]
- 49. Wu J., Tan Y., Deng H., Zhu D. (2008) Relationship between degree-rank function and degree distribution of protein-protein interaction networks. Comput Biol. Chem. 32, 1–4 [DOI] [PubMed] [Google Scholar]
- 50. Han J. D., Dupuy D., Bertin N., Cusick M. E., Vidal M. (2005) Effect of sampling on topology predictions of protein-protein interaction networks. Nat. Biotechnol. 23, 839–844 [DOI] [PubMed] [Google Scholar]
- 51. de Chassey B., Meyniel-Schicklin L., Aublin-Gex A., Andre P., Lotteau V. (2012) Genetic screens for the control of influenza virus replication: from meta-analysis to drug dicovery. Mol. Biosyst. 8, 1297–1303 [DOI] [PubMed] [Google Scholar]
- 52. Davey N. E., Edwards R. J., Shields D. C. (2010) Computational identification and analysis of protein short linear motifs. Front. Biosci. 15, 801–825 [DOI] [PubMed] [Google Scholar]
- 53. Neduva V., Russell R. B. (2006) Peptides mediating interaction networks: new leads at last. Curr. Opin. Biotechnol. 17, 465–471 [DOI] [PubMed] [Google Scholar]