Abstract
FTH_0069 is a previously uncharacterized strongly immunoreactive protein that has been proposed to be a novel virulence factor in Francisella tularensis. Here, the glycan structure modifying two C-terminal peptides of FTH_0069 was identified utilizing high resolution, high mass accuracy mass spectrometry, combined with in-source CID tandem MS experiments. The glycan observed at m/z 1156 was determined to be a hexasaccharide, consisting of two hexoses, three N-acetylhexosamines, and an unknown monosaccharide containing a phosphate group. The monosaccharide sequence of the glycan is tentatively proposed as X-P-HexNAc-HexNAc-Hex-Hex-HexNAc, where X denotes the unknown monosaccharide. The glycan is identical to that of DsbA glycoprotein, as well as to one of the multiple glycan structures modifying the type IV pilin PilA, suggesting a common biosynthetic pathway for the protein modification. Here, we demonstrate that the glycosylation of FTH_0069, DsbA, and PilA was affected in an isogenic mutant with a disrupted wbtDEF gene cluster encoding O-antigen synthesis and in a mutant with a deleted pglA gene encoding pilin oligosaccharyltransferase PglA. Based on our findings, we propose that PglA is involved in both pilin and general F. tularensis protein glycosylation, and we further suggest an inter-relationship between the O-antigen and the glycan synthesis in the early steps in their biosynthetic pathways.
The investigation of protein glycosylation in bacteria has attracted a lot of attention during the last decade, because of an increased knowledge of the involvement of such a co-translational modification in the virulence of particular bacteria (1–3). In several pathogens, the loss of glycosylation was found to result in accumulation of unglycosylated flagellins in the cell, providing evidence that glycosylation is essential for the correct assembly of flagella and subsequent motility (4–6). On the other hand, it has been concluded that glycosylation does not play a major role in pilus-mediated adhesion in Neisseria meningitidis (7). Moreover, a general O-glycosylation system is present in the major intestinal symbiont Bacteroides fragilis, indicating that glycosylation is not always associated with virulence properties (8).
Francisella tularensis represents one of the six etiological agents under consideration as a potential biological threat because of its low infectious dose and the ease of its transmission (9). Two F. tularensis subspecies, holarctica and tularensis, are primarily infectious for humans (10). In the past few years, questions arose regarding the potential role of glycosylation in the virulence of F. tularensis. A previous report indicated that one potential pilin subunit encoded by pilA, which is required for virulence of both subspecies, is post-translationally modified (11). Since then, the glycosylation of PilA has been reported in other studies (12–14), and recently the O-linked carbohydrates associated with PilA have been characterized (14).
In our previous study, using both detection and enrichment methods in combination with liquid chromatography-mass spectrometry, we mapped the glycoproteome in the FSC200 strain of F. tularensis subsp. holarctica. In addition to PilA, we suggested that several additional candidate proteins could be targets for glycosylation including DsbA (FTH_1071), an uncharacterized protein FTH_0069, FopA, Tul4, and LemA (13). However, we were unable to elucidate the repertoire of glycans and their structures because of the failure of both enzymatic and chemical release techniques. Because their role in virulence may be associated with their carbohydrates, DsbA and FTH_0069 are of particular interest for an in-depth analysis of their glycosylation. The protein DsbA has recently been identified to be essential for intracellular survival and replication of F. tularensis in the mouse monocyte macrophage cell line J774.2 and for in vivo virulence in the mouse infection model of tularemia (15). FTL_1096, the DsbA homologue in the live vaccine strain (LVS), was recently found to be recruited into the membrane rafts of J774.2 macrophages upon F. tularensis invasion.1 The O-linked glycosylation of DsbA was recently elucidated in F. tularensis subsp. tularensis, holarctica LVS, and F. novicida (16). Here, we additionally confirmed that the DsbA homologue in the FSC200 strain is also glycosylated. Similarly, FTH_0069, an orthologue of FTT1676, which was recently identified as a novel virulence factor of F. tularensis subsp. tularensis SchuS4, is important for both intracellular survival and proliferation in mice (17). Previously, we demonstrated that FTT1676 induced a long term antibody response in a technician accidentally infected with SchuS4 (18). The glycosylation status of FTH_0069 has not been previously described, and here we provide the first evidence that FTH_0069 is glycosylated in the subspecies holarctica strain FSC200. Additionally, using bioinformatics, we identified proteins that are likely to be involved in a glycosylation pathway of F. tularensis subsp. holarctica FSC200. Three of these predicted proteins, FTT0791, FTT0798, and PglA (FTT0905), are known to be involved in DsbA and PilA glycosylation in the strains SchuS4 and FSC200, respectively (14, 16).
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The F. tularensis strains and plasmids employed in this study are listed in Table I. The bacteria for all the strains were grown, harvested, and lysed within a BioSafety Level 2 containment facility. F. tularensis was cultured on McLeod agar supplemented with bovine hemoglobin (Becton Dickinson) and IsoVitaleXTM (Becton Dickinson) at 36.8 °C for 24–48 h. The cultivation of F. tularensis in Chamberlain medium was performed as previously described (13).
Table I. F. tularensis strains and plasmids used in this study.
| Strain/plasmid | Characteristics/description | Reference/source |
|---|---|---|
| Strain | ||
| FSC200 | F. tularensis subsp. holarctica FSC200 | Ref. 47 |
| FSC679 (FSC200/ΔpglA) | FSC200/in-frame deletion of codons 10–469 of FTT_0905 | Ref. 14 |
| FSC699 (FCS200/Δ1103) | FSC200/in-frame deletion of codons 4–369 of FTT_1103 | Ref. 15 |
| FSC200/ΔwbtDEF:Cm | FSC200 with inactivated production of O-antigen | This study |
| E. coli S17-1 | thi thr leu tonA supE recA::RP4–2-Tc::Mu Kn::Tn7 | Ref. 20 |
| Plasmid | ||
| pSMP22-RT1 | Suicide plasmid used to inactivate O-antigen cluster of F. tularensis subsp. tularensis | Ref. 19 |
Escherichia coli S17-1 was grown on LB agar base or LB broth at 37 °C. When appropriate, antibiotics (all purchased from Sigma-Aldrich) were used at the following concentrations: for the cultivation of the transformed E. coli S17-1, 25 μg/ml of chloramphenicol was added to growth medium; the selection of transconjugants was performed on plates with 75 μg/ml polymyxin B and 2.5 μg/ml chloramphenicol; plates containing 5% sucrose were utilized for the sucrose selection of the mutated F. tularensis strains, and the clones of the mutated F. tularensis were cultured on agar supplemented with 2.5 μg/ml of chloramphenicol.
Construction of FSC200/ΔwbtDEF::Cm (O-antigen Mutant Strain)
A region of the F. tularensis subsp. holarctica FSC200 O-antigen cluster (wbtDEF) was targeted for deletion and insertion of antibiotic marker to disrupt the entire function of the cluster. For the inactivation of the O-antigen cluster, a suicide vector pSMP22-RT1 was generously provided by R. Thomas. pSMP22-RT1 was originally used for the deletion of the O-antigen encoding genes in F. tularensis subsp. tularensis (19). The cloned 2.5-kb deletion construct shows a 100% sequence homology with the sequence from FSC200. pSMP22-RT1 was transformed into E. coli S17-1 (20) and subsequently introduced into the FSC200 strain by conjugation using essentially the same method as published by Golovliov et al. (21). The F. tularensis subsp. holarctica strain FSC200 double recombinant strain was designated as FSC200/ΔwbtDEF::Cm.
Verification of FSC200/ΔwbtDEF::Cm Mutant Strain
The integration of the suicide plasmid and the subsequent generation of the ΔwbtDEF::Cm mutant in FSC200 was verified by PCR. The primers wbtD_1F and wbtF_1R (the primers were designed for the detection of the deletion construct cloned into pSMP22-RT1) resulted in an amplicon with a length of 1780 bp (the length of wild type amplicon is ∼2300 bp). ΔwbtDEF::Cm-positive clones were obtained after sucrose selection and identified using the primers wbtD_1F and wzy_1R (the primer was designed on a chromosomal sequence downstream from the wbtDEF cluster), providing a PCR product having 2470 bp (the length of an amplicon from the wild type is ∼3 kb long), and finally with a combination of the primers wbtD_1F and wbtE_1R (designed on a deleted region of wbtDEF) that does not give rise to any PCR product in FSC200/ΔwbtDEF::Cm mutant. The primer sequences are available on request.
Animal Infection Studies
Groups of five 6–8-week-old female BALB/c mice were infected subcutaneously with the FSC200/ΔwbtDEF::Cm mutant (using doses of 3 × 102 and 3 × 106 cfu/mouse) and the wild type FSC200 strain (dose 3 × 102 cfu/mouse). The control groups of mice were inoculated with a sterile saline only. The mice were housed under conventional conditions; food and water were given ad libitum, and the mice were allowed to acclimatize for at least 7 days before the beginning of the experiment. The infected mice were observed two or three times per day to properly monitor the progress of the infection.
Preparation of F. tularensis Membrane Protein-enriched Fraction
Cell lysis was performed using a French pressure cell as previously described (13). Fractions enriched in membrane proteins were collected by ultracentrifugation of the whole cell lysate at 115,000 × g for 1 h at 4 °C. The supernatant was discarded, and the membrane pellet was resuspended in ice-cold PBS and then collected by centrifugation at 115,000 × g for 30 min at 4 °C. The final membrane protein-containing pellet was resuspended in PBS, and the protein content in the sample was quantified by a bicinchoninic acid assay.
Lipopolysaccharide Purification
LPS2 was purified from F. tularensis subsp. holarctica FSC200, FSC200/ΔwbtDEF::Cm, and FSC200/ΔpglA mutant strains using the hot phenol-water extraction method of Westphal and Jann (22) with slight modifications. Briefly, a 1-mg amount of the crude membrane fraction was incubated with 1.4 ml of a 50%/50% phenol/water solution at 65 °C for 30 min. The samples were cooled on ice and centrifuged at 4 °C to separate organic and aqueous layer. The phenol layer was back-extracted three times with hot water. The three aqueous layers containing the LPS were centrifuged at high speed to remove any residual phenol.
Mini One-dimensional Gel Electrophoresis, Semi-dry Western Blot, and Immunodetection
SDS-PAGE was performed on a 30-μg sample of the membrane-proteins enriched fraction and a 10-μl aliquot of LPS from the first aqueous layer according to the protocol employed by Laemmli (23). After electrophoresis, the separated proteins and LPS were transferred onto PVDF membranes using a semi-dry Western blotting technique. The primary antibodies used for the immunodetection of DsbA and PilA were a rabbit anti-FTT1103 polyclonal antibody (Apronex, Vestec, Czech Republic) and rabbit anti-PilA serum (kindly provided by Kerstin Kuoppa at FOI, Swedish Defense Research Agency, Umea, Sweden), respectively. The detection of LPS was accomplished using a mouse monoclonal anti-F. tularensis LPS FB11 antibody (Abcam, Cambridge, UK). The secondary antibodies used were swine anti-rabbit IgG/HRP (Dako, Glostrup, Denmark) for both anti-FTT1103 and anti-PilA antibodies, whereas goat anti-mouse IgG/HRP (Dako) was used for anti-LPS. A chemiluminescence detection was performed using a BM chemiluminescence blotting substrate POD according to the manufacturer's instructions (Roche Applied Science).
Two-dimensional Gel Electrophoresis and Glycoprotein Detection
The membrane proteins were repeatedly precipitated with cold acetone prior to two-dimensional electrophoresis. The protein precipitate was resolubilized in a rehydration buffer containing 7 m urea, 2 m thiourea, 1% (w/v) ASB-14, 1% Triton X-100, 0.12% De Streak, 40 mm Tris base, 0.5% bromphenol blue, 1% ampholytes, pH 3–10, and 0.5% PharmalyteTM, pH 8–10.5. The isoelectric focusing, reduction, alkylation, and SDS-PAGE performed herein were described previously (13). Pro-Q Emerald staining was performed according to the manufacturer's protocol (Invitrogen), with slight modifications as described previously (13). The stained gels were visualized by illumination at 342 nm using a UV radiometer. After detection and excizing the glycoproteins, the gels were stained with a colloidal blue stain to detect all proteins as a control.
In-gel Digestion of Proteins
The protein spots detected on the two-dimensional gels were excised and subjected to an in-gel tryptic digestion according to the procedure described recently (13). The digestion was stopped by acidifying the samples with TFA, bringing the pH to 2–3.
MALDI-MS/MS Analysis and Protein Identification
Mass spectra acquired from the in-gel digestion of the proteins following Pro-Q Emerald staining were recorded in the positive reflectron mode on a 4800 MALDI-TOF/TOF mass spectrometer (AB Sciex, Foster City, CA) equipped with an Nd:YAG laser (355 nm) and operated in its delayed extraction mode. Internal calibration of mass spectra was conducted utilizing the tryptic autolytic peptides. Data acquisition and evaluation were performed as recently described (13). Acquired data were evaluated using GPS ExplorerTM software v.3.6 (AB Sciex), which integrates the Mascot search algorithm against F. tularensis OSU18 genome database (NC_008369.1, 1,555 sequence entries). Trypsin was selected as the proteolytic enzyme, and one missed cleavage was allowed. Carbamidomethylation of cysteine residues was set as a fixed modification, and oxidation of methionine was set as a variable modification. The mass tolerance values of the precursor and fragment ions were 100 ppm and 0.25 Da, respectively. The proteins were considered identified with confidence when the GPS protein score confidence interval (%) was equal to 100% and a minimum of two peptide sequences per protein were identified. PilA (FTH_0384) possesses the only tryptic peptide AQLGSDLSALGGAK with a theoretical mass of 1286.6830, which is measurable within the m/z range 800–4,000. Thus, the mass of 1287.6903 m/z [M+H]+ was added to the inclusion list of masses selected for fragmentation.
LC-ESI-MS/MS Analysis of Protein Glycosylation
A 1.0-μl aliquot of the in-gel digests obtained from the proteins DsbA and FTH_0069 was analyzed by C18 reversed phase liquid chromatography using either a Nanoflow 1100 HPLC (Agilent Technologies) coupled on-line to an LTQ-FT-ICR mass spectrometer (Thermo Finnigan) or an Ultimate 3000 (Dionex, Sunnyvale, CA) interfaced to an LTQ-Orbitrap (Thermo). The samples were desalted and preconcentrated on a C18 PepMap300 0.3 × 5-mm μ-precolumn containing 5-μm mean particle size silica, with a pore size of 300 Å (Dionex). After loading and washing the peptides for 10 min with mobile phase A (97/3/0.1, water/ACN/formic acid, v/v/v), the trapping column was switched in-line with the analytical column. The separation of the peptides was conducted at a flow rate of 250 nl/min with a column packed in-house (75-μm inner diameter × 150 mm) with Magic C18AQ (3-μm mean particle size with pores of 200 Å) from Michrom Bioresources, Inc. (Auburn, CA). A linear gradient from 3 to 55% phase B (99.9/0.1, ACN/formic acid, v/v) over a period of 45 min was followed by a steeper ramp from 55 to 80% phase B over 10 min. The column eluent was electrosprayed into the mass spectrometer using a 1.8 kV spray voltage. MS spectra were acquired in the mass range from 80 to 2,000 m/z at a low orifice potential, with fragmentation of the five most intense peaks in the linear ion trap. Collisional activation was performed using helium gas at normalized collision energy 35%. Diagnostic glycan oxonium ions were generated by an in-source CID at the orifice potential set at 100 V. The acquisition of the data was controlled using the Xcalibur software v2.0.7 (Thermo Finnigan).
The acquired RAW data were processed using Turbo RAW2MGF converter v1.0.7 (Indiana University). The processing parameters were as follows: minimum ion count in MS/MS was at 5, absolute total ion intensity threshold was set to 100, smoothing was not applied, the percentage peak height at which centroids were calculated was 100%, and charge states were calculated. Peak lists were subjected to MASCOT (v2.2) searching against the F. tularensis subsp. holarctica (OSU18) protein sequences database (NC_008369.1, 1,555 sequence entries) using the following searching criteria: trypsin was used as the protease, allowing for one missed cleavage, 2+ and 3+ charge state ions; carbamidomethylation of cysteine residues was set as a fixed modification, and oxidation of methionine was selected as a variable modification. The mass tolerance of the precursor and fragment ions was set to 5 ppm and 0.8 Da, respectively. The obtained data were further filtered through ProteinParser v2.1 (24). Thus, peptides with a Mowse probability score threshold less than 30; peptides containing internal KK, KR, RK, or RR motifs; and those containing less than six amino acids and/or having a mass lower than 600 Da were rejected. A minimum of two peptide spectrum matches fulfilling the above-mentioned criteria were required for protein identification. To determine glycosylation, the collected data were further analyzed manually using Xcalibur software and compared with in silico digests of the respective proteins generated by PeptideMass (http://expasy.org/tools/peptide-mass.html).
Bioinformatic Analysis of the F. tularensis Protein Glycosylation Pathway
Proteins involved in the glycosylation pathways of Campylobacter jejuni, Campylobacter coli, Caulobacter crescentus, Pseudomonas aeruginosa, N. meningitidis, Aeromonas punctata, and Helicobacter pylori, and the O-antigen synthesis of F. tularensis subsp. tularensis SchuS4 and F. novicida (supplemental Table 1) were used as queries for the initial PSI-BLAST v. 2.2.22+searches (25) against the nr database of NCBI (release on October 13, 2010) (26). Each PSI-BLAST run involved six iterations and was performed with the following criteria: (i) an expected threshold of 10−10 and a PSI-BLAST threshold of 10−15; (ii) an expected threshold of 1 and a PSI-BLAST threshold of 10−3 (less strict thresholds leading to a larger number of hits); and (iii) an expected threshold of 10 and a PSI-BLAST threshold of 1 (supplemental Table 1). All of the identified protein sequences from F. tularensis subsp. holarctica FSC200 were extracted and aligned with their respective queries and selected as top hits using MUSCLE (27). Multiple sequence alignments were inspected in BioEdit (28). Whenever needed, the domain compositions of the proteins were assigned using the Pfam database (29). For the target F. tularensis FSC200 strain, the best hit for a given PSI-BLAST run was assigned as follows. All of the proteins or individual domains from the target strain with at least 20% identity to the query protein sequence were selected: (i) if one of these sequences was significantly more similar to the query than others (a difference in sequence identity to query of >10%), then this sequence was assigned as the best hit; otherwise (ii) if there were more sequences with equal similarity to the query, then all these sequences were assigned as the best hits. The best hits from the individual runs were used as queries for the reciprocal PSI-BLAST searches for the homologues in the genome of the originally queried organism. The reciprocal PSI-BLAST searches were conducted, and their results were analyzed in an identical way to the initial runs of the PSI-BLAST searches. The results of the initial and reciprocal PSI-BLASTs were compared with distinguish any valid positive hits from potentially false positive hits. A hit was assumed to be valid if two proteins, each from a different genome, were each the best hit for the other. A reciprocal PSI-BLAST search could not be performed for hits of the P. aeruginosa queries because the query strain was not known, nor for the N. meningitidis c311#3, C. coli vc167, and A. punctata Sch3 queries, because the genome of these strains has not been sequenced.
RESULTS
PglA Is Responsible for Both Pilin and General Protein Glycosylation in F. tularensis
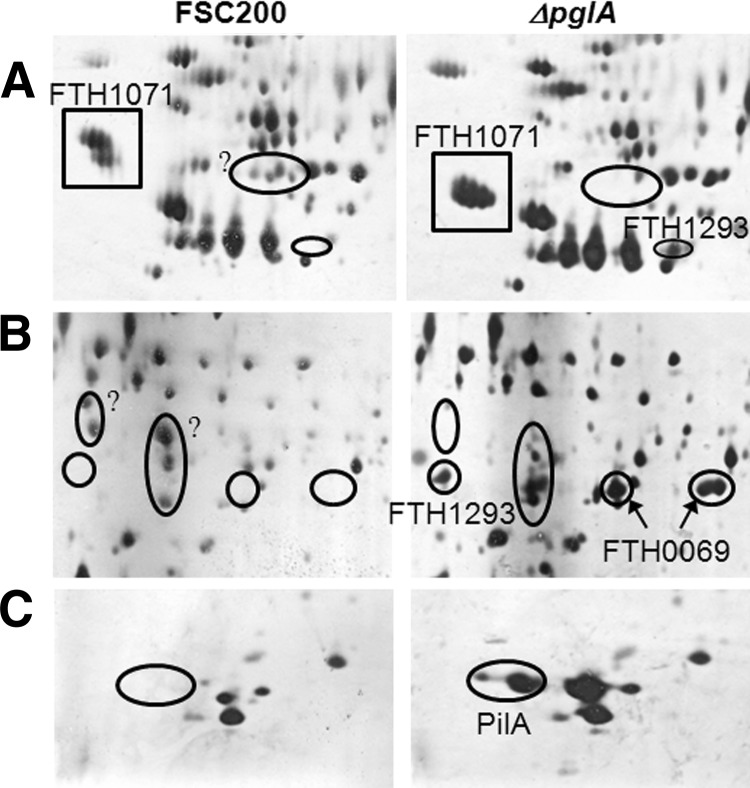
The bioinformatic analysis revealed a similarity in the middle regions of the F. tularensis protein FTT0905, annotated as a Type IV pilus glycosylation protein (recently designated PglA), and the glycosyltransferase PilO of P. aeruginosa. PilO transfers the synthesized oligosaccharide en bloc from the lipid carrier to the pilin protein and is thus indispensable for pilin O-glycosylation (30). PglA was recently shown to be directly involved in glycosylation as a protein-targeting oligosaccharyltransferase that was sufficient to glycosylate PilA in a reconstituted system in E. coli (14). We attempted to ascertain whether the function of PglA is limited exclusively to the glycosylation of pilin or whether it is the unique oligosaccharyltransferase used by F. tularensis for protein glycosylation. Therefore, we searched for additional glycosylated proteins that may undergo PglA-mediated glycosylation. The proteomes of the membrane-enriched fractions of a wild type FSC200 strain and a pglA mutant strain were compared using two-dimensional electrophoresis over a pH range of 3–10 to reveal any qualitative differences that would indicate glycosylation (i.e. a shift in molecular mass and/or pI). Because of the complexity of the proteome maps, only a few differences were able to be identified conclusively. Two spots of the proteins FTH_0069 (Fig. 1B, right panel) and FTH_1293 (Fig. 1A, right panel, and B, right panel) and a spot of PilA (Fig. 1C, right panel) were identified in the pglA mutant using MS. These proteins were also present in the wild type FSC200 strain, albeit in different regions of the gel. The exclusive position of the DsbA protein (FTH_1071) on the gel showed that this protein is shifted in both its molecular mass and pI in the pglA mutant when compared with the wild type strain of FSC200 (Fig. 1A, left and right panels).
Fig. 1.
Representative silver-stained two-dimensional gels for the proteomic comparison of parental FSC200 and a ΔpglA mutant strain. The enlarged regions A, B, and C with highlighted proteins that were altered in the mutant when compared with the FSC200 are shown. The proteins with altered locations on the gel are indicated with ovals; the proteins with obviously altered pI and MW are boxed. Question marks indicate detected protein spots that were not identified by mass spectrometry.
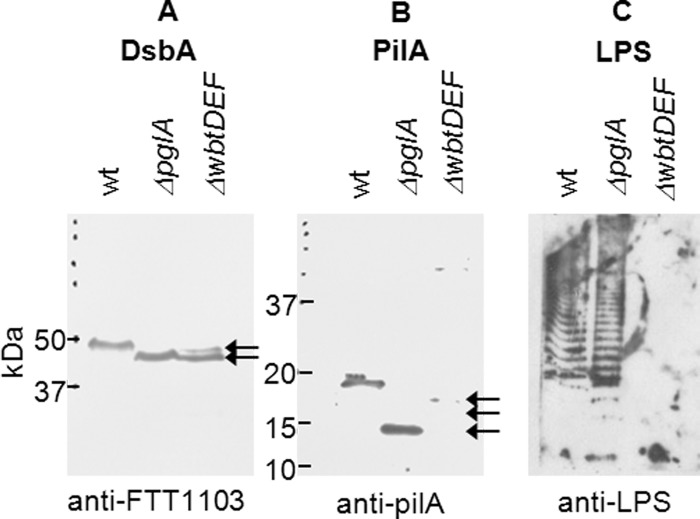
Using the carbohydrate-specific Pro-Q Emerald stain, the proteins FTH_0069, FTH_1293, PilA, and DsbA have recently been identified as being glycosylated in the strain FSC200, along with an additional seven potential glycoproteins (13). To verify the hypothesis that PglA is involved in their glycosylation, we employed carbohydrate-specific staining on the proteins expressed by the pglA mutant strain. In total, three biological replicates were analyzed. As demonstrated in the representative Fig. 2A, neither the pilin protein PilA nor the non-pilin proteins FTH_0069, FTH_1293, and DsbA were detected in the mutant strain when compared with the wild type FSC200 (13). This suggests the loss or truncation of the carbohydrate, potentially decreasing the signal intensity below the detectable limit for these particular proteins. Additionally, the influence of PglA on glycosylation of DsbA was demonstrated using an anti-DsbA antibody. An increase in the relative mobility of the protein in the pglA mutant strain was observed as a consequence of the truncation/loss of glycosylation (Fig. 3A, lane 2). This simple mobility shift detection clearly confirms that: (i) DsbA is glycosylated in FSC200 and that (ii) PglA is required for the glycosylation of DsbA.
Fig. 2.
Detection of glycoproteins using a carbohydrate-specific Pro-Q Emerald staining following two-dimensional SDS-PAGE of F. tularensis membrane-enriched proteins. A, detection of glycoproteins in FSC200/ΔpglA (left panel). The identical gel stained with colloidal blue (right panel). B, detection of glycoproteins in FSC200/ΔwbtDEF::Cm (left panel). Spot 1, FTH_1071; spot 2, FTH_1293; spots 3, 4, and 6–8, FTH_0384 (PilA); spots 5 and 6, FTH_1163. The identical gel stained with colloidal blue (right panel).
Fig. 3.
A and B, immunodetection of the proteins DsbA (A) and PilA (B) in FSC200 (wild type), FSC200/ΔpglA, and FSC200/ΔwbtDEF::Cm (lanes 1, 2, and 3, respectively) using their respective polyclonal antibodies. The molecular masses of both proteins originating from the wild type strain are higher in comparison with the masses of proteins isolated from both mutant strains. C, immunodetection of LPS in FSC200, ΔpglA, and FSC200/ΔwbtDEF::Cm (lanes 1, 2, and 3, respectively) using mouse monoclonal anti-LPS. LPS is not detected in FSC200/ΔwbtDEF::Cm.
DsbA naturally occurs in several charge and mass variants (31). To determine whether all these variants are glycosylated in FSC200, we employed an immunodetection of DsbA in FSC200, ΔpglA, and a mixture of both strains separated in the narrow pH range 3–6 by two-dimensional SDS-PAGE (Fig. 4). In total, eight DsbA spots were detected in two lanes in the wild type FSC200 strain (Fig. 4A), whereas in the pglA mutant, a lane with five spots of lower mass was observed (Fig. 4D), indicating that presumably all eight DsbA isoforms detected in the wild type strain are glycosylated. A definitive confirmation of PglA-mediated DsbA glycosylation was gained by the mass spectrometric analysis of DsbA from both the wild type and ΔpglA strains (data not shown). In summary, the above-mentioned observations suggest that PglA is indispensable for both pilin and general protein glycosylation in F. tularensis FSC200.
Fig. 4.
Immunoblot detection of DsbA isoforms in FSC200 (wild type), FSC200/ΔwbtDEF::Cm, FSC200/ΔpglA, and their mixtures after two-dimensional SDS-PAGE in a pH range from 3–6. A, eight DsbA spots were detected in two lanes (lanes 1 and 2) in the wild type strain. B, 12 isoforms of DsbA were detected in three lanes (lanes 1–3) in the FSC200/ΔwbtDEF::Cm. C, the mixture of the wild type and FSC200/ΔwbtDEF::Cm gave rise to the 13 DsbA spots in three lanes (lanes 1–3), indicating fully (lane 1), partially (lane 2), and nonglycosylated (lane 3) isoforms. D, a single lane (lane 3) with five spots of the lowest mass was observed in the pglA mutant when compared with a wild type. E, three lanes (lanes 1–3) with the 13 DsbA spots were detected in the mixture of wild type and the pglA mutant, indicating that all isoforms in the pglA mutant are not glycosylated.
O-antigen Encoding Gene Cluster Is Involved in the Glycosylation of F. tularensis Proteins
We have recently shown that O-antigen complicates the investigation of glycosylation by necessitating a β-elimination chemical cleavage rather than the enzymatic release of glycans (13). A mass spectrometric analysis of a β-eliminated FSC200 crude membrane protein-enriched fraction revealed the O-antigen repeating unit of FSC200 strain to be identical with those in F. tularensis subsp. holarctica strain 15 and subsp. tularensis (32, 33). Hence, we argued that the O-antigen gene cluster in subspecies holarctica is likely to be identical to that of F. tularensis subsp. tularensis. The sequences of the gene cluster encoding the O-antigen in F. tularensis subsp. tularensis and F. novicida have recently been determined by Prior et al. (33) and Thomas et al. (19), respectively. Thomas et al. (19) further generated wbtDEF mutants in both F. novicida and F. tularensis subsp. tularensis SchuS4, which failed to produce O-antigen. In addition, they recently reported that DsbA glycosylation remained unchanged in the wbtDEF::Cm mutant in SchuS4 using a carbohydrate-specific stain (16). Removal of O-antigen with a simultaneous preservation of the protein glycosylation would facilitate an analysis of the glycosylation via glycomic profiling. Therefore, based on a 100% identity of the O-antigen encoding gene cluster in SchuS4 and FSC200 strain, we decided to prepare a mutant strain of FSC200 in the wbtDEF gene cluster using the same strategy as Thomas et al. (19).
Similar to the observation of Thomas et al., the glycosylation of DsbA appeared to be retained in FSC200/ΔwbtDEF::Cm strain using carbohydrate-specific staining (Fig. 2B), although with a lower intensity than in the wild type strain (13). Several other spots were also identified as carbohydrate positive in the mutant analogue (Table II and supplemental Table 5). However, further investigation of the effect of wbtDEF disruption on glycosylation of DsbA using a mobility shift assay revealed bands of lower molecular mass in the O-antigen mutant (Fig. 3A, lane 3) when compared with a wild type FSC200 strain (Fig. 3A, lane 1). Immunodetection of DsbA in FSC200/ΔwbtDEF::Cm and a mixture of FSC200 + FSC200/ΔwbtDEF::Cm separated in the narrow pH range 3–6 by two-dimensional SDS-PAGE further revealed 12 isoforms of DsbA that were detected in three lanes in the O-antigen mutant (Fig. 4B), suggesting that two spots in the top lane are fully glycosylated, five spots in the middle lane are partially glycosylated, and five spots in the bottom lane are presumably lacking or truncated in their glycosylation, because the bottom lane was not detected in FSC200 (Fig. 4A). Notably, nonglycosylated and partially glycosylated DsbA isoforms are more acidic than the fully glycosylated protein variants.
Table II. Proteins of FSC200/ΔwbtDEF detected using Pro-Q Emerald stain.
| Spot | Gene locusa | Protein name | Molecular mass (kDa)b | pIb | PSORTbc | LipoPd | Protein scoree | Protein score CI (%)f | Peptide count (all/sequenced)g | Protein expected value |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | FTH_1071 | Probable thioredoxin family protein | 39.69 | 4.89 | ? | SPII | 765 | 100 | 13/5 | 4.9e−074 |
| 2 | FTH_1293 | Outer membrane protein FopA | 41.45 | 5.58 | OM | SPI | 541 | 100 | 13/5 | 1.2e−051 |
| 3 | FTH_0384 | Type IV pili fiber protein | 13.59 | 9.06 | ? | SPI | 100 | 100 | 1/1 | 1.6e−007 |
| 4 | dtto | 100 | 100 | 1/1 | 1.6e−007 | |||||
| 6 | dtto | 125 | 100 | 1/1 | 4.9e−10 | |||||
| 7 | dtto | 124 | 100 | 1/1 | 6.2e−10 | |||||
| 8 | dtto | 89 | 100 | 1/1 | 1.8e−006 | |||||
| 5 | FTH_1163 | Ribosomal protein L13 | 15.94 | 10.03 | cyt | cyt | 292 | 100 | 10/3 | 9.8e−027 |
| 6 | dtto | 189 | 100 | 9/2 | 2.0e−016 |
a The accession number in the genome sequence of F. tularensis subsp. holarctica OSU18 (NC_008369.1).
b Theoretical pI/molecular mass (average) calculated using the compute pI/molecular mass tool (http://web.expasy.org/compute_pi/).
c Prediction of the protein localization using the PSORTb program (http://psort.org/psortb). cyt, cytoplasmic; OM, outer membrane; ?, unknown localization.
d Prediction of lipoproteins (SPII cleavage site II) and SPI (cleavage site I) using LipoP algorithm (http://www.cbs.dtu.dk/services/LipoP/).
e The MOWSE score calculated by the Mascot search engine for each protein matched from the MS peak list; this score is based on the probability that peptide mass matches are nonrandom events.
f The confidence interval (CI) for the protein score.
g The number of peptides with unique sequences matching the selected protein.
The proteins with accession numbers written in bold type have previously been identified as carbohydrate-positive in the FSC200 strain (13).
An impairment of the O-antigen production in FSC200/ΔwbtDEF::Cm was confirmed using a monoclonal antibody against the O-antigen of F. tularensis LPS (Fig. 3C, lane 3). To further assess the effect of wbtDEF deletion on the virulence in FSC200, we infected BALB/c mice subcutaneously with the FSC200/ΔwbtDEF::Cm mutant using two doses of 3 × 102 and 3 × 106 cfu/mouse. Subsequently, we monitored the course of the disease for 3 weeks. We found that the inactivation of the genes encoding the O-antigen cluster in FSC200 resulted in a highly attenuated phenotype in the mice. The mice infected with the highly virulent FSC200 strain at the dose of 3 × 102 cfu/mouse succumbed at 5 days post-infection (supplemental Fig. 1). This is consistent with data reported by Thomas et al. (19), in which the inactivation of the wbtDEF gene cluster in SchuS4 resulted in virulence attenuation in vivo. Thus, it appears that, in the strain FSC200, the wbtDEF gene cluster is involved in both the production of the O-antigen repeating unit of LPS and in the synthesis of glycans.
Structural Characterization of the Glycosylation of FTH_0069
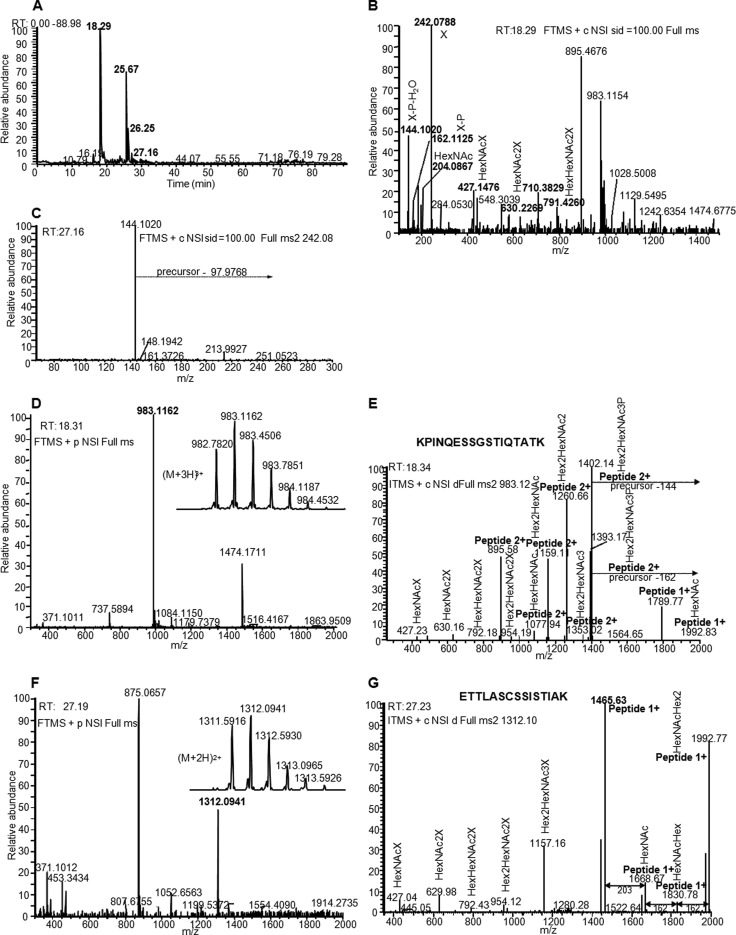
The uncharacterized protein FTH_0069 has recently been identified as carbohydrate-positive in FSC200 (13). For the analysis of its glycosylation, the protein was isolated by mini two-dimensional SDS-PAGE of the membrane protein-enriched fraction by excising the spots stained by Pro-Q Emerald. The two excised protein spots were in-gel-digested with trypsin, and the digest was analyzed by nano-scale HPLC coupled on-line with an LTQ-FT-ICR mass spectrometer using a stepped orifice voltage scan. The instrument was configured to allow the detection of diagnostic glycan oxonium ions from the cleavage of glycosidic bonds obtained from an in-source CID and from the product ion spectra of selected glycopeptide precursors. To acquire information on the glycan(s) and particular glycopeptides, eight scan events were defined in the instrument method. In the first scan event, the diagnostic glycan oxonium ions that were generated by an in-source CID at the orifice potential set at 100 V were monitored (i.e. a survey scan). The second scan event was set to fragment an unknown monosaccharide observed at an m/z value of 242.08 that was generated by the in-source CID. The third scan event recorded the entire mass range (spanning the m/z range from 80 to 2,000) to monitor the molecular ions of peptides and glycopeptides (i.e. a full scan) at a low orifice potential. The last five scan events were used to fragment and acquire MS/MS spectra by CID of the five most intense molecular ions from the full scan data (data-dependent analyses). These scan events were repeated over the course of the entire HPLC separation. Manual inspection of the acquired data led to the identification of a single glycan and a determination of two glycan-modified peptides. The glycan oxonium ions for hexose (Hex), N-acetylhexosamine (HexNAc), and the Hex-HexNAc disaccharide were observed in a survey scan at two retention times (RT), 18.29 and 25.67 min of an extracted ion chromatogram, indicating the presence of at least two glycopeptides (Fig. 5A). In addition, an unidentified monosaccharide observed at an m/z value of 242 (herein designated with an X) and further di- and trisaccharide fragments containing this unknown sugar were detected in this scan (Fig. 5B). An investigation of the MS/MS spectra near the given RT indicated several suspected glycopeptides with m/z values of 982.78 [M + 3H]3+, 1473.67 [M + 2H]2+, 874.73 [M + 3H]3+, 1311.59 [M + 2H]2+, 998.79 [M + 3H]3+, 1497.68 [M + 2H]2+, 899.16 [M + 4H]4+, and 1198.54 [M + 3H]3+. In these fragmentation spectra, glycan ions appearing at m/z values of 445, 630, 792, 954, and 1157 were observed. The identification of the glycopeptides was achieved through highly accurate FT-ICR mass measurement of the precursor ions. The masses of the corresponding unmodified peptides were derived from the mass difference between a particular glycopeptide and the glycan after both were deconvoluted (to the neutral molecule, M). Deconvolution of the triply charged glycopeptide species observed at an m/z value of 982.7824 and subtraction of the glycan (M = 1156.4024) resulted in a neutral peptide mass of 1788.9217, which differs from the theoretical mass of the peptide 288KPINQESSGSTIQTATK304 (M = 1788.9212) by only 0.29 ppm. A precursor ion appearing at an m/z value of 1473.6707 was a doubly charged species of this glycopeptide. Likewise, the doubly charged ion with a mass of 1311.5916, after deconvolution and substraction of the glycan (M = 1156.4024) yields a neutral mass of 1464.7677, which is 0.75 ppm more than 1464.7666 and corresponds to the theoretical mass of the peptide 273ETTLASGSSISTIAK287. The identity of the peptide was further unambiguously confirmed by the presence of peptide fragment ions in deconvoluted spectra (data not shown). Full MS scans, from which the two glycopeptides were selected for fragmentation, are shown in Fig. 5 (D and F) for the glycopeptide with m/z values of 982.7824 and 1311.5916, respectively. The interpreted ITMS spectra following the in-source CID of both identified glycopeptides with m/z values of 982.7824 and 1311.5916 are depicted in Fig. 5 (E and G), respectively. Additionally, glycopeptide fragments of various glycan lengths were observed in the MS/MS spectra of both glycopeptides, which allowed the monosaccharide sequence of the glycan to be determined as the difference between the two adjacent glycopeptide fragment peaks. Additionally, the loss of 98 Da was observed, suggesting the presence of either a phosphate or a sulfate group. Based on the slight difference in their mass (9.5 milli mass units), phosphorylated and sulfated compounds can be distinguished using a high resolution MS, such as FT-ICR. Thus, in-source CID fragmentation of the ion 242.07882 m/z followed by highly accurate mass analysis in the ion cyclotron generated the fragment with an m/z value of 144.10194 that most plausibly corresponds to the neutral loss of phosphoric acid (97.97688 Da), with a relative mass difference of 0.2 ppm. The glycan sequence was thus determined as follows: X-P-HexNAc-HexNAc-Hex-Hex-HexNAc where X denotes an unknown carbohydrate of a mass 144.1020 m/z (Fig. 5C). Using Xcalibur, we generated elemental composition of the mass 144.10203 m/z [M+H]+. Within 1 ppm mass tolerance, the only and thus most plausible chemical formula was determined as C7H14O2N (144.101905 m/z) with a relative mass difference of 0.866 ppm. Similarly, the precursor ion mass 242.07882 m/z corresponds most plausibly to C7H17O6NP (242.07880 m/z; a relative mass difference of 0.082 ppm). Additionally, an ion of the mass 162.11253 m/z [M+H]+, which was observed in the FT-MS spectra (Fig. 5B), was determined as having the formula C7H16O3N (162.11247 m/z), with a relative mass difference of 0.371 ppm, which corresponds to the loss of HPO3 (79.9663) from C7H17O6NP. Further confirmation of the presence of the phosphate group using other techniques would be useful because it is known that phosphorylation and sulfation are associated with significantly different biological functions.
Fig. 5.
FTH_0069 glycosylation. A, extracted ion chromatogram for the masses of the diagnostic glycan oxonium ions with m/z values of 162, 203, and 366 obtained from the LC-MS analysis of the FTH_0069 protein digest. The peptide 288KPINQESSGSTIQTATK304 eluted as a major peak at 18.29 min. The second major peak at 25.67 min corresponds to the glycosylated peptide 265ADEQREDKETTLASGSSISTIAK287 with two missed cleavages. The peak with a maximum at 26.25 min corresponds to the glycosylated peptide 270EDKETTLASGSSISTIAK287 with a single missed cleavage. The glycosylated peptide 273ETTLASGSSISTIAK287 eluted as a minor peak at 27.16 min. B, detection of glycan oxonium ions generated using in-source CID. An FT-MS spectrum obtained at RT 18.29 min of the LC-MS analysis of the FTH_0069 protein digest. C, the identification of the unknown monosaccharide with an m/z value of 242.08 in the glycan sequence of the FTH_0069 glycopeptide using the product ion spectrum of the in-source generated ion species with an m/z value of 242.08 at RT 27.16 min. The loss of phosphoric acid was observed. D, detection of the FTH_0069 glycopeptides in the full MS scan. The FT-MS spectrum obtained at RT 18.31 min. The inset presents a triply charged glycopeptide ion species that was selected by data-dependent analysis for MS/MS analysis. E, identification of a FTH_0069 glycopeptide with an m/z value of 982.78. The tandem mass spectrum of the triply charged glycopeptide ion at RT 18.34 min of the LC-MS analysis. F, detection of the FTH_0069 glycopeptides in the full MS scan at RT 27.19 min. The inset presents a doubly charged glycopeptide ion species that was selected by data-dependent analysis for MS/MS analysis. G, the identification of the FTH_0069 glycopeptide with a precursor m/z value of 1311.59 in the tandem mass spectrum of the doubly charged glycopeptide ion at RT 27.23 min.
The remaining glycopeptide with an m/z value 998.79 [M + 3H]3+ and its corresponding doubly charged ion (possessing an m/z value of 1497.68) has the sequence 270EDKETTLASGSSISTIAK287 and contains one missed cleavage. The peptide eluted at 26.25 min of the chromatographic separation. Similarly, the glycopeptide of the sequence 265ADEQREDKETTLASGSSISTIAK287, which was observed at m/z values of 899.16 [M+4H]4+ and 1198.54 [M + 3H]3+, contains two missed cleavages and eluted at 25.67 min of the LC run. These two sequences that contain missed cleavages include the above described glycosylated peptide, 273ETTLASGSSISTIAK287.
Overall, 27 peptide sequences of FTH_0069 have been identified within 5 ppm peptide mass tolerance, providing the protein sequence coverage of 43%. In addition to FTH_0069, four other proteins were identified. Further filtering the data (according to criteria described under “Experimental Procedures”) reduced the number of identified FTH_0069 peptides to 17, and the total number of identified proteins in the spot was reduced to four (supplemental Table 3). Nonetheless, to confirm the origin of the identified glycopeptides, the glycan residue mass of 1156.4047 Da was added to the MASCOT search engine as a variable modification (denoted as P-glycan1156 (ST)), and a peptide mass fingerprint search was performed against the F. tularensis OSU18 database (1,555 sequences). The MS data, which were required as an input for PMF search, were extracted from the original mgf file and converted to the [M+H]+ ion species using a home-made Perl script. This led to the unambiguous assignment of the glycan modification to the peptides derived exclusively from the protein FTH_0069 (supplemental Table 4). Specifically, sequences 273ETTLASGSSISTIAK287, 270EDKETTLASGSSISTIAK287, 265ADEQREDKETTLASGSSISTIAK287, and 288KPINQESSGSTIQTATK304 have been identified as each bearing a single glycan, which is consistent with the manual interpretation of the MS/MS data. Additionally, sequence coverage of the protein was increased to 52%. Moreover, the protein FTH_0069 has been identified as the most top-ranked protein when searching against NCBInr database (data not shown).
DISCUSSION
The first indication that a protein may be glycosylated is a discrepancy between its theoretical and experimentally determined molecular masses. This was shown in the first case for a possible post-translational modification in F. tularensis, where it was observed for a pilin subunit protein PilA, whose real molecular mass was around 4 kDa larger than expected from the size of the pilA gene (11). Similarly, in our investigation, the protein FTH_0069 showed an apparent molecular mass of around 50 kDa on the SDS-PAGE gel, which is significantly higher than the mass deduced from its amino acid sequence (37.5 kDa). Bioinformatic analysis assigned FTH_0069 as a lipoprotein (LipoP 1.0). This prediction was recently experimentally supported using a Triton X-114 phase partitioning (data not shown). Despite this, an increase in molecular mass could not be caused by palmitoylation alone, because the protein mass would be increased by only 570 or 810 Da in the case of modification by dipalmitoylglyceryl and tripalmitoylglyceryl, respectively. This indicates that, presumably, glycosylation must have contributed to the higher observed mass of the protein.
We previously discovered several candidate proteins as targets for glycosylation in F. tularensis, including the proteins DsbA, PilA, and FTH_0069 (13). The glycosylation status of DsbA has recently been described in F. tularensis strains SchuS4 and LVS and F. novicida (16), and therefore details of the glycosylation of the DsbA homologue in FSC200 strain (FTH_1071) are not included in the present study. However, confirmation of the glycosylation status of FTH_1071 by mass spectrometry was a prerequisite for further investigation of the changes of glycosylation in our prepared FSC200-derived mutant strains by the use of available anti-DsbA antibodies. (The mass spectrometric data are available upon request.) Recently, the glycan of the pilin protein PilA has also been determined in the strain FSC200 (14). Our intent here was to elucidate the glycosylation of the FTH_0069 protein in more detail. Using an LC-MS/MS approach, the glycan structure modifying two C-terminal peptides of FTH_0069 was characterized. The glycan with an m/z value of 1157 was determined to be a hexasaccharide consisting of two hexoses, three N-acetylhexosamine residues, and an unknown, phosphate group containing monosaccharide residue of chemical formula C7H17O6NP. The monosaccharide sequence of the glycan from FTH_0069 appears to be X-P-HexNAc-HexNAc-Hex-Hex-HexNAc. Interestingly, the glycan structure originating from the DsbA homologues in the F. tularensis strains LVS, FSC200, and SchuS4 was identical. Additionally, the same glycan structure was also recently identified as one of the multiple glycans modifying PilA (14). PilA glycans are composed of a common pentasaccharide, HexNAc-HexNAc-Hex-Hex-HexNAc, which is further extended with the unusual phosphate-linked moieties (14). Glycosylation sites were not revealed in FTH_0069; however, the amino acid sequences of both of the identified glycopeptides contain neither eukaryotic nor bacterial N-glycosylation motifs, suggesting that O-linked glycosylation is likely.
To have a sufficient amount of the protein for LC-MS analysis, it was necessary to pool the two pI isoforms of this low abundant protein into a single sample. Therefore, it is not clear whether (i) each identified glycopeptide originates from one of these two isoforms, (ii) both glycopeptides originate from both isoforms, or (iii) one of the two glycopeptides originates from one isoform and, at the same time, both glycopeptides originate from the second isoform. The first and second options would result in the same molecular mass of both protein isoforms, because the glycan associated with these isoforms has an identical mass. The third option would mean that the difference between the two isoforms is equal to the mass of the glycan. Considering the fact that both analyzed isoforms differ in their pI values and not in their molecular masses, one would argue that option (i) is correct. However, option (ii) cannot be entirely excluded because of the possibility of the existence of isobaric glycopeptides having a different monosaccharide sequence of the glycan (16). Indeed, the presence of the glycan oxonium ions observed at m/z values of 427, 648, and 731 in the tandem MS fragmentation spectra of the FTH_0069 glycopeptides is indicative of isobaric structures. In our study, more than two isoforms of the protein FTH_0069 were detected as being glycosylated using the Pro-Q Emerald stain, but they could not be excised from the gel before the fluorescence disappeared. Thus, a further characterization of each isoform of this protein is desirable.
It is particularly interesting that DsbA and FTH_0069 are both acylated and glycosylated. Such a dual post-translational modification has been observed in several proteins of various bacteria (34–36). Although it is the acyl moiety of lipoproteins that is recognized by Toll-like receptors TLR2 and is thus able to activate an innate immune response via TLR2-mediated signaling, the role of the glycan moiety of the glycosylated lipoproteins is unclear. The glycan moiety of mycobacterial lipoglycoprotein LprG seems to be directly involved in the stimulation of the innate immune responses via activation of MHC class II-restricted T cells (37). Moreover, glycosylation in conjunction with acylation appears indispensable for retaining the 19-kDa mycobacterial lipoglycoprotein within the cell (38). Thus, the function of the glycan(s) in the particular F. tularensis lipoproteins remains a subject of investigation.
Whereas TLR2 recognizes lipoproteins, TLR4 generally interacts with lipopolysaccharides of Gram-negative bacteria. TLR4 specifically recognizes a biologically active lipid A of the LPS, which is biphosphorylated and hexa-acylated. F. tularensis LPS, however, is not recognized by TLR4, because of its structural diversities. The general role of bacterial O-antigen, on the other hand, is to provide resistance to complement-mediated killing by serum (39). Thomas et al. (19) recently generated a knock-out mutant in the gene cluster wbtDEF in both F. novicida and F. tularensis subsp. tularensis SchuS4, which failed to produce O-antigen. SchuS4-derived mutant with abrogated production of O-antigen was completely attenuated in mice (19). Further studies showed that the disruption of the wbtDEF in SchuS4 did not affect the glycosylation of DsbA (16). We employed the same procedure to create a FSC200 strain with abrogated production of O-antigen. Similarly, we observed a completely attenuated phenotype of the FSC200/ΔwbtDEF::Cm mutant in a mouse model following infection. Our results, however, suggest that DsbA glycosylation was affected, although not completely lost in the FSC200/ΔwbtDEF::Cm variant. Similarly, we observed that glycosylation of PilA was affected in a similar manner in the mutant using immunoblotting (data not shown). The influence of the wbtDEF disruption on the glycosylation of FTH_0069 could not be tested because a specific anti-FTH_0069 antibody was not available. However, FTH_0069 was not detected using the carbohydrate-specific stain, which may suggest that its glycosylation was affected in the mutant strain. It is likely that the glycosylation was below the detection limit of the stain because of the splitting of this low abundance protein into several differentially glycosylated isoforms.
Studies of other bacteria have shown that the initial steps of O-antigen biosynthesis and glycan biosynthesis may involve the same genes (40, 41). It is thus possible that glycan and O-antigen biosynthesis are cross-linked at this point in the FSC200 strain, which might explain the affected glycosylation of the proteins in the FSC200/ΔwbtDEF::Cm strain. Nevertheless, glycosylated DsbA and PilA isoforms in FSC200 are still formed in the mutant, indicating the presence of an additional gene that would function as WbtD, WbtE, and/or WbtF in this strain.
A bioinformatic analysis of the genes/proteins likely to be involved in the glycosylation pathway of the FSC200 strain revealed that the protein WbtD (ZP_02274475) encoded by wbtD was predicted as a potential homologue of PglA of both N. meningitidis strain mc58 and C. jejuni strain 81-176 and both WlaC and WlaE of C. jejuni strain 81116 (supplemental Table 2). WbtD is required for the assembly of the O-antigen through the transfer of a GalNAcAN subunit to QuiNAc-Und-PP (33), which in particular resembles the function of the homologous PglA, WlaC, and WlaE proteins. These glycosyltransferases mediate the assembly of the respective glycans on a lipid carrier by transferring the second monosaccharide unit to the DATDH-Und-PP (42, 43). Two other genes of the O-antigen cluster: wbtE (encoding WbtE protein (ZP_02274474)) and wbtF (encoding WbtF (ZP_02274473)), possess only a slight similarity to the glycosyltransferases of the compared bacteria (supplemental Table 2). These data suggest that wbtD itself is responsible for affected glycosylation. Both the glycosylation of F. tularensis proteins (PilA, DsbA, FTH_0069, and possibly others) in the ΔwbtDEF::Cm mutant and the exact function of wbtDEF genes in the synthesis of glycans remain to be comprehensively defined.
It was demonstrated that the FTT0791 or FTT0798 mutants, defective in DsbA glycosylation in SchuS4 but not in the production of O-antigen, were not attenuated in the murine model of tularemia upon a subcutaneous route of infection (16). These observations are consistent with our bioinformatic data, in which the proteins FTT0791 and FTT0798 are potential homologues of the bacterial glycosyltransferases but are not similar to any known proteins in F. tularensis subsp. tularensis and F. novicida that would participate in the synthesis/production of O-antigen. Thus, it appears that the attenuated phenotype of ΔwbtDEF::Cm is solely the result of the loss of O-antigen and not the altered glycosylation in the mutant. Nevertheless, this needs to be verified in the strain FSC200 because it has been demonstrated that O-antigen from F. tularensis subsp. tularensis plays a relatively minor role (as compared with F. novicida) in the protection from serum killing, which presumably requires other surface structures. O-antigen is rather important for intracellular survival and replication in the subspecies tularensis (19).
Both flagellin and the general glycosylation pathway were found in C. jejuni. The flagellin and general glycosylation genes are located in distinct gene loci and give rise to distinct glycan structures. Thus, the product of a general glycosylation pathway is a heptasaccharide (44), whereas a flagellin-linked glycan is a pseudaminic acid or its derivatives (45). The same holds for the flagellin and pilin glycosylation in P. aeruginosa (30, 46). Therefore, the fact that F. tularensis pilin and non-pilin glycans possess the same pentasaccharide core may explain why the disruption of the wbtDEF impairs glycosylation in both PilA pilin protein and DsbA non-pilin protein, and, furthermore, why PglA acts as both a pilin and a general oligosaccharyltransferase. It appears that the role of WbtDEF in the protein glycosylation is to provide some of the monosaccharides for the synthesis of the glycan structures that are then translocated by PglA. The complete description of the function of each enzyme involved in the glycosylation pathway could eventually provide clarification of the possible role of protein glycosylation in the F. tularensis pathogenesis.
Acknowledgments
We thank Jitka Zakova and Alena Firychova for excellent technical assistance. The preparation of the FSC200/ΔwbtDEF mutant was facilitated by using a suicide vector pSMP22-RT1 that was kindly provided by Rebecca Thomas from Defense Science and Technology Laboratory (Porton Down, UK).
Footnotes
* This work was supported by Ministry of Education Grant ME08105, Czech Republic Ministry of Defense Grant FVZ0000604, Czech Science Foundation Grant GA203/09/0857, Ministry of Health of Czech Republic Project NS 9747, National Institutes of Health/Institute of General Medical Sciences Grant GM024349-25, and National Institutes of Health/National Center for Research Resources Grant RR018942.
 This article contains supplemental material. The data associated with this manuscript may be downloaded from PeptideAtlas.org, http://www.peptideatlas.org/repository/, under accession numbers PASS0043 and PASS00044.
This article contains supplemental material. The data associated with this manuscript may be downloaded from PeptideAtlas.org, http://www.peptideatlas.org/repository/, under accession numbers PASS0043 and PASS00044.
1 Härtlova, A., Balounova, J., Benesova, M., Straskova, A., Sobol, M., Krocova, Z., Hozak, P., Filipp, D., and Stulik, J., manuscript in preparation.
2 The abbreviations used are:
- LPS
- lipopolysaccharide
- Hex
- hexose
- HexNAc
- N-acetylhexosamine
- RT
- retention time.
REFERENCES
- 1. Arora S. K., Neely A. N., Blair B., Lory S., Ramphal R. (2005) Role of motility and flagelin glycosylation in the pathogenesis of Pseudomonas aeruginosa burn wound infections. Infect. Immun. 73, 4395–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Karlyshev A. V., Everest P., Linton D., Cawthraw S., Newell D. G., Wren B. W. (2004) The Campylobacter jejuni general glycosylation system is important for attachment to human epithelial cells and in the colonization of chicks. Microbiology 150, 1957–1964 [DOI] [PubMed] [Google Scholar]
- 3. Szymanski C. M., Burr D. H., Guerry P. (2002) Campylobacter protein glycosylation affects host cell interactions. Infect. Immun. 70, 2242–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goon S., Kelly J. F., Logan S. M., Ewing C. P., Guerry P. (2003) Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 50, 659–671 [DOI] [PubMed] [Google Scholar]
- 5. Josenhans C., Vossebein L., Friedrich S., Suerbaum S. (2002) The neuA/flmD gene cluster of Helicobacter pylori is involved in flagellar biosynthesis and flagellin glycosylation. FEMS Microbiol. Lett. 210, 165–172 [DOI] [PubMed] [Google Scholar]
- 6. Gryllos I., Shaw J. G., Gavín R., Merino S., Tomás J. M. (2001) Role of flm locus in mesophilic Aeromonas species adherence. Infect. Immun. 69, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marceau M., Forest K., Béretti J. L., Tainer J., Nassif X. (1998) Consequences of the loss of O-linked glycosylation of meningococcal type IV pilin on piliation and pilus-mediated adhesion. Mol. Microbiol. 27, 705–715 [DOI] [PubMed] [Google Scholar]
- 8. Fletcher C. M., Coyne M. J., Villa O. F., Chatzidaki-Livanis M., Comstock L. E. (2009) A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell 137, 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dennis D. T., Inglesby T. V., Henderson D. A., Bartlett J. G., Ascher M. S., Eitzen E., Fine A. D., Friedlander A. M., Hauer J., Layton M., Lillibridge S. R., McDade J. E., Osterholm M. T., O'Toole T., Parker G., Perl T. M., Russell P. K., Tonat K. (2001) Tularemia as a biological weapon: Medical and public health management. JAMA 285, 2763–2773 [DOI] [PubMed] [Google Scholar]
- 10. Svensson K., Larsson P., Johansson D., Byström M., Forsman M., Johansson A. (2005) Evolution of subspecies of Francisella tularensis. J. Bacteriol. 187, 3903–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Forslund A. L., Kuoppa K., Svensson K., Salomonsson E., Johansson A., Byström M., Oyston P. C., Michell S. L., Titball R. W., Noppa L., Frithz-Lindsten E., Forsman M., Forsberg A. (2006) Direct repeat-mediated deletion of a type IV pilin gene results in major virulence attenuation of Francisella tularensis. Mol. Microbiol. 59, 1818–1830 [DOI] [PubMed] [Google Scholar]
- 12. Salomonsson E., Forsberg A., Roos N., Holz C., Maier B., Koomey M., Winther-Larsen H. C. (2009) Functional analyses of pilin-like proteins from Francisella tularensis: Complementation of type IV pilus phenotypes in Neisseria gonorrhoeae. Microbiology 155, 2546–2559 [DOI] [PubMed] [Google Scholar]
- 13. Balonova L., Hernychova L., Mann B. F., Link M., Bilkova Z., Novotny M. V., Stulik J. (2010) Multimethodological approach to identification of glycoproteins from the proteome of Francisella tularensis, an intracellular microorganism. J. Proteome Res. 9, 1995–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Egge-Jacobsen W., Salomonsson E. N., Aas F. E., Forslund A. L., Winther-Larsen H. C., Maier J., Macellaro A., Kuoppa K., Oyston P. C., Titball R. W., Thomas R. M., Forsberg Å., Prior J. L., Koomey M. (2011) O-Linked glycosylation of the PilA pilin protein of Francisella tularensis: Identification of the endogenous protein-targeting oligosaccharyltransferase and characterization of the native oligosaccharide. J. Bacteriol. 193, 5487–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Straskova A., Pavkova I., Link M., Forslund A. L., Kuoppa K., Noppa L., Kroca M., Fucikova A., Klimentova J., Krocova Z., Forsberg A., Stulik J. (2009) Proteome analysis of an attenuated Francisella tularensis dsbA mutant: Identification of potential DsbA substrate proteins. J. Proteome Res. 8, 5336–5346 [DOI] [PubMed] [Google Scholar]
- 16. Thomas R. M., Twine S. M., Fulton K. M., Tessier L., Kilmury S. L., Ding W., Harmer N., Michell S. L., Oyston P. C., Titball R. W., Prior J. L. (2011) Glycosylation of DsbA in Francisella tularensis subspecies tularensis. J. Bacteriol. 193, 5498–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wehrly T. D., Chong A., Virtaneva K., Sturdevant D. E., Child R., Edwards J. A., Brouwer D., Nair V., Fischer E. R., Wicke L., Curda A. J., Kupko J. J., 3rd, Martens C., Crane D. D., Bosio C. M., Porcella S. F., Celli J. (2009) Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell. Microbiol. 11, 1128–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Janovská S., Pávková I., Reichelová M., Hubáleka M., Stulík J., Macela A. (2007) Proteomic analysis of antibody response in a case of laboratory-acquired infection with Francisella tularensis subsp. tularensis. Folia Microbiol. 52, 194–198 [DOI] [PubMed] [Google Scholar]
- 19. Thomas R. M., Titball R. W., Oyston P. C., Griffin K., Waters E., Hitchen P. G., Michell S. L., Grice I. D., Wilson J. C., Prior J. L. (2007) The immunologically distinct O antigens from Francisella tularensis subspecies tularensis and Francisella novicida are both virulence determinants and protective antigens. Infect. Immun. 75, 371–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Simon R., Priefer U., Pühler A. (1983) A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram-negative bacteria. Nat. Biotechnol. 1, 784–791 [Google Scholar]
- 21. Golovliov I., Sjöstedt A., Mokrievich A., Pavlov V. (2003) A method for allelic replacement in Francisella tularensis. FEMS Microbiol. Lett. 222, 273–280 [DOI] [PubMed] [Google Scholar]
- 22. Westphal O., Jann K. (1965) Bacterial lipopolysaccharides: Extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 5, 83–91 [Google Scholar]
- 23. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 24. Mann B., Madera M., Sheng Q., Tang H., Mechref Y., Novotny M. V. (2008) ProteinQuant Suite: A bundle of automated software tools for label-free quantitative proteomics. Rapid. Commun. Mass Spectrom 22, 3823–3834 [DOI] [PubMed] [Google Scholar]
- 25. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sayers E. W., Barrett T., Benson D. A., Bryant S. H., Canese K., Chetvernin V., Church D. M., DiCuccio M., Edgar R., Federhen S., Feolo M., Geer L. Y., Helmberg W., Kapustin Y., Landsman D., Lipman D. J., Madden T. L., Maglott D. R., Miller V., Mizrachi I., Ostell J., Pruitt K. D., Schuler G. D., Sequeira E., Sherry S. T., Shumway M., Sirotkin K., Souvorov A., Starchenko G., Tatusova T. A., Wagner L., Yaschenko E., Ye J. (2009) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 37, D5–D15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Edgar R. C. (2004) MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hall T. A. (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 [Google Scholar]
- 29. Finn R. D., Tate J., Mistry J., Coggill P. C., Sammut S. J., Hotz H. R., Ceric G., Forslund K., Eddy S. R., Sonnhammer E. L., Bateman A. (2008) The Pfam protein families database. Nucleic Acids Res. 36, D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Castric P. (1995) pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology 141, 1247–1254 [DOI] [PubMed] [Google Scholar]
- 31. Pavkova I., Reichelova M., Larsson P., Hubalek M., Vackova J., Forsberg A., Stulik J. (2006) Comparative proteome analysis of fractions enriched for membrane-associated proteins from Francisella tularensis subsp. tularensis and F. tularensis subsp. holarctica strains. J. Proteome Res. 5, 3125–3134 [DOI] [PubMed] [Google Scholar]
- 32. Vinogradov E. V., Shashkov A. S., Knirel Y. A., Kochetkov N. K., Tochtamysheva N. V., Averin S. F., Goncharova O. V., Khlebnikov V. S. (1991) Structure of the O-antigen of Francisella tularensis strain 15. Carbohydr. Res. 214, 289–297 [DOI] [PubMed] [Google Scholar]
- 33. Prior J. L., Prior R. G., Hitchen P. G., Diaper H., Griffin K. F., Morris H. R., Dell A., Titball R. W. (2003) Characterization of the O antigen gene cluster and structural analysis of the O antigen of Francisella tularensis subsp. tularensis. J. Med. Microbiol. 52, 845–851 [DOI] [PubMed] [Google Scholar]
- 34. Herrmann J. L., Delahay R., Gallagher A., Robertson B., Young D. (2000) Analysis of post-translational modification of mycobacterial proteins using a cassette expression system. FEBS Lett. 473, 358–362 [DOI] [PubMed] [Google Scholar]
- 35. Vik A., Aas F. E., Anonsen J. H., Bilsborough S., Schneider A., Egge-Jacobsen W., Koomey M. (2009) Broad spectrum O-linked protein glycosylation in the human pathogen. Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U.S.A. 106, 4447–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scott N. E., Bogema D. R., Connolly A. M., Falconer L., Djordjevic S. P., Cordwell S. J. (2009) Mass spectrometric characterization of the surface-associated 42 kDa lipoprotein JlpA as a glycosylated antigen in strains of Campylobacter jejuni. J. Proteome Res. 8, 4654–4664 [DOI] [PubMed] [Google Scholar]
- 37. Sieling P. A., Hill P. J., Dobos K. M., Brookman K., Kuhlman A. M., Fabri M., Krutzik S. R., Rea T. H., Heaslip D. G., Belisle J. T., Modlin R. L. (2008) Conserved mycobacterial lipoglycoproteins activate TLR2 but also require glycosylation for MHC class II-restricted T cell activation. J. Immunol. 180, 5833–5842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilkinson K. A., Newton S. M., Stewart G. R., Martineau A. R., Patel J., Sullivan S. M., Herrmann J. L., Neyrolles O., Young D. B., Wilkinson R. J. (2009) Genetic determination of the effect of post-translational modification on the innate immune response to the 19 kDa lipoprotein of Mycobacterium tuberculosis. BMC Microbiol. 9, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Clay C. D., Soni S., Gunn J. S., Schlesinger L. S. (2008) Evasion of complement-mediated lysis and complement C3 deposition are regulated by Francisella tularensis lipopolysaccharide O antigen. J. Immunol. 181, 5568–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. DiGiandomenico A., Matewish M. J., Bisaillon A., Stehle J. R., Lam J. S., Castric P. (2002) Glycosylation of Pseudomonas aeruginosa 1244 pilin: Glycan substrate specificity. Mol. Microbiol. 46, 519–530 [DOI] [PubMed] [Google Scholar]
- 41. Hug I., Feldman M. F. (2011) Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiology 21, 138–151 [DOI] [PubMed] [Google Scholar]
- 42. Szymanski C. M., Yao R., Ewing C. P., Trust T. J., Guerry P. (1999) Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32, 1022–1030 [DOI] [PubMed] [Google Scholar]
- 43. Linton D., Allan E., Karlyshev A. V., Cronshaw A. D., Wren B. W. (2002) Identification of N-acetylgalactosamine-containing glycoproteins PEB3 and CgpA in Campylobacter jejuni. Mol. Microbiol. 43, 497–508 [DOI] [PubMed] [Google Scholar]
- 44. Young N. M., Brisson J. R., Kelly J., Watson D. C., Tessier L., Lanthier P. H., Jarrell H. C., Cadotte N., St. Michael F., Aberg E., Szymanski C. M. (2002) Structure of the N-linked glycans present on multiple glycoproteins in the Gram-negative bacterium. Campylobacter jejuni. J. Biol. Chem. 277, 42530–42539 [DOI] [PubMed] [Google Scholar]
- 45. Thibault P., Logan S. M., Kelly J. F., Brisson J. R., Ewing C. P., Trust T. J., Guerry P. (2001) Identification of the carbohydrate moieities and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276, 34862–34870 [DOI] [PubMed] [Google Scholar]
- 46. Schirm M., Arora S. K., Verma A., Vinogradov E., Thibault P., Ramphal R., Logan S. M. (2004) Structural and genetic characterization of flycosylation of type a flagellin in Pseudomonas aeruginosa. J. Bacteriol 186, 2523–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Johansson A., Berglund L., Eriksson U., Göransson I., Wollin R., Forsman M., Tärnvik A., Sjöstedt A. (2000) Comparative analysis of PCR versus culture for diagnosis of ulceroglandular tularemia. J. Clin. Microbiol. 38, 22–26 [DOI] [PMC free article] [PubMed] [Google Scholar]