Abstract
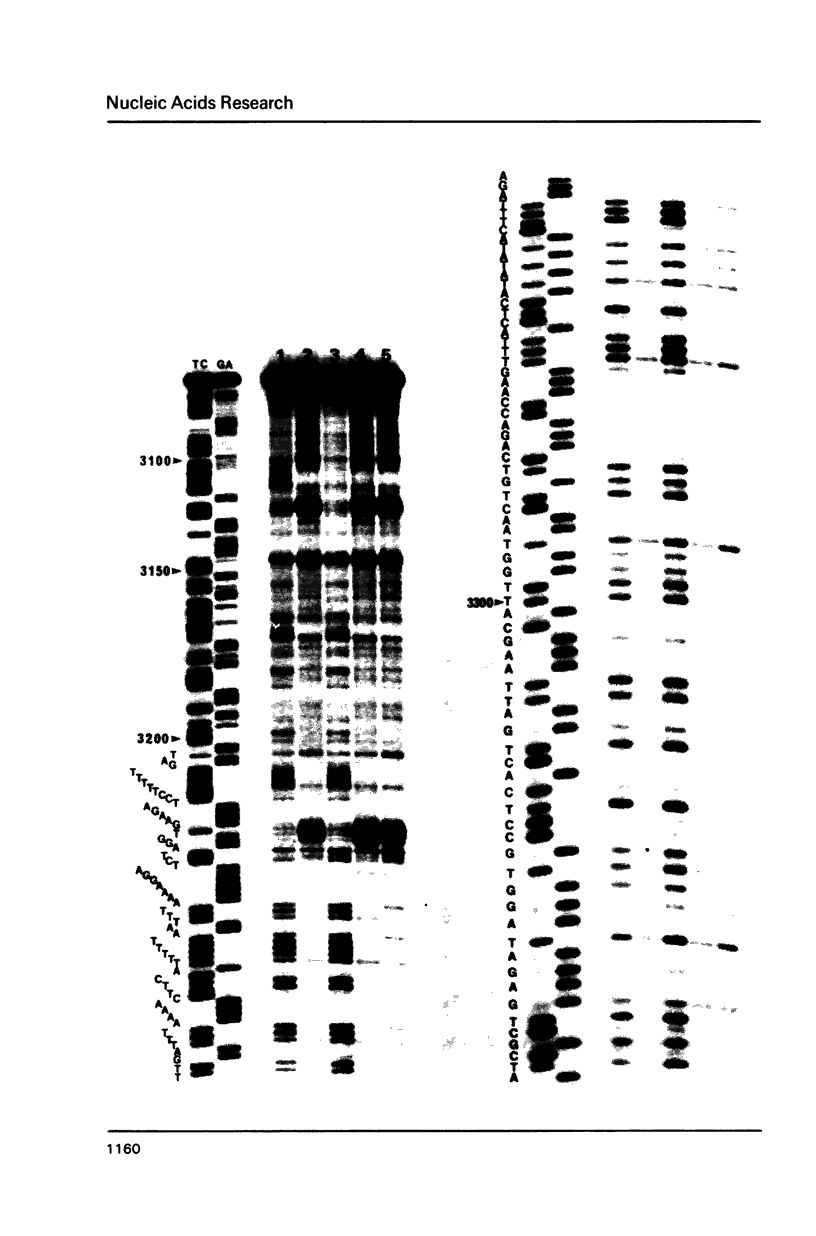
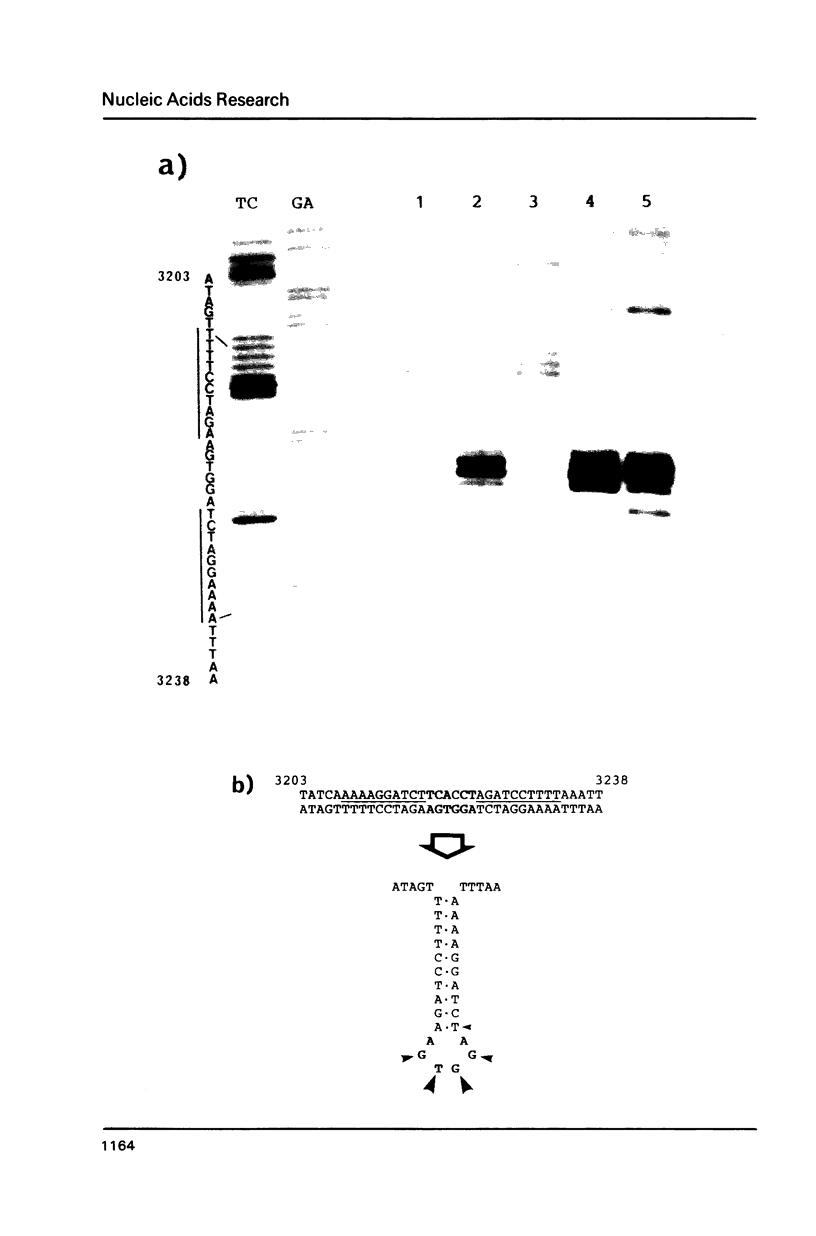
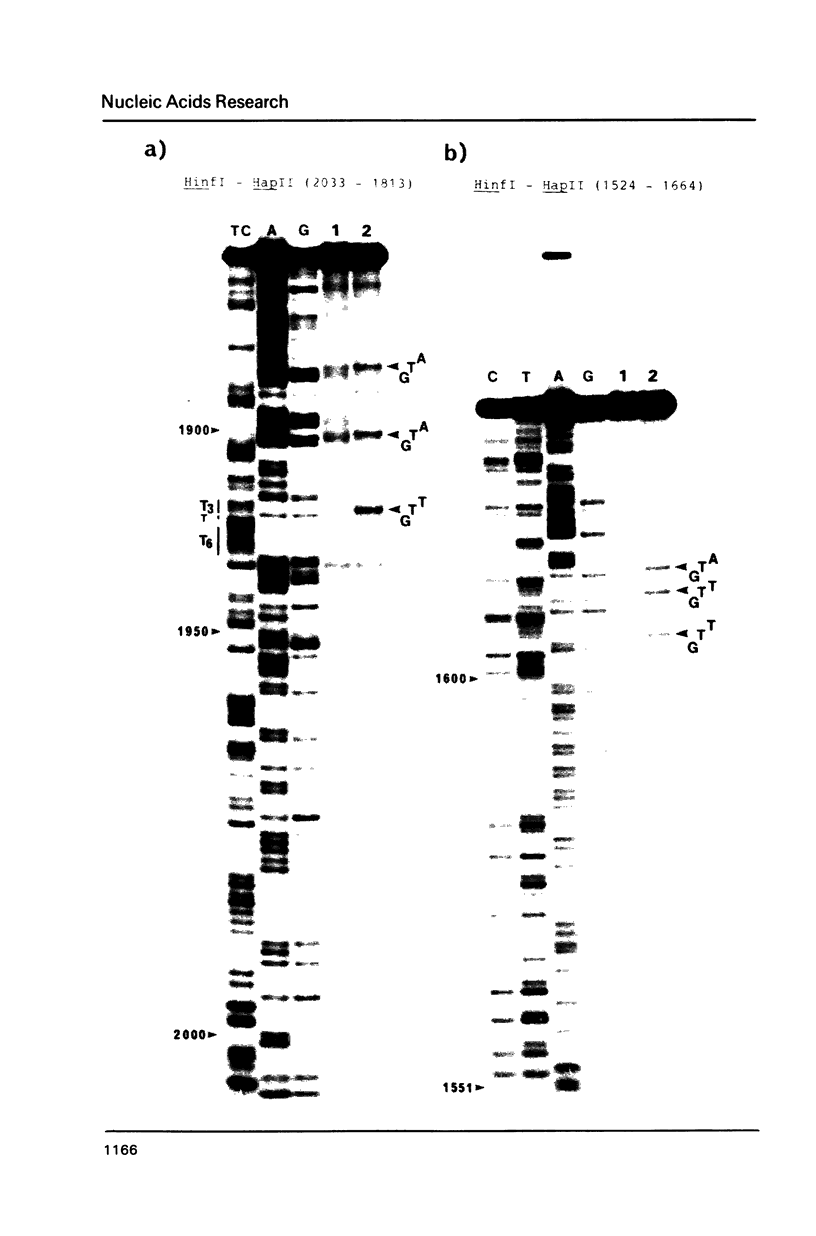
Ozone-reactive sites on the nucleobase moieties in supercoiled pBR322 DNA were investigated by using sequencing procedures. Ozonolysis in the absence of salt resulted in degradation of thymine residues in the A + T rich region located at 3100-3400bp. In the presence of salt, such as NaCl or MgCl2, a conformational change of plasmid DNA was induced. Subsequently the thymine and guanine residues in the loop of the cruciform located at 3120bp and 3220bp were degraded. In addition, central thymine residues present in sequences GTA, GTT and ATA were also degraded.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Gellert M., O'Dea M. H., Mizuuchi K. Slow cruciform transitions in palindromic DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5545–5549. doi: 10.1073/pnas.80.18.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K., Shinriki N., Ueda T. Degradation of nucleic acids with ozone. V. Mechanism of action of ozone on deoxyribonucleoside 5'-monophosphates. Chem Pharm Bull (Tokyo) 1984 Sep;32(9):3601–3606. doi: 10.1248/cpb.32.3601. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. Dynamic, sequence-dependent DNA structure as exemplified by cruciform extrusion from inverted repeats in negatively supercoiled DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):101–112. doi: 10.1101/sqb.1983.047.01.013. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. Hairpin-loop formation by inverted repeats in supercoiled DNA is a local and transmissible property. Nucleic Acids Res. 1981 Mar 25;9(6):1271–1289. doi: 10.1093/nar/9.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Hallam L. R. Thermodynamics of the ColE1 cruciform. Comparisons between probing and topological experiments using single topoisomers. J Mol Biol. 1984 Nov 25;180(1):179–200. doi: 10.1016/0022-2836(84)90436-4. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Kemper B. Cruciform-resolvase interactions in supercoiled DNA. Cell. 1984 Feb;36(2):413–422. doi: 10.1016/0092-8674(84)90234-4. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Palecek E. The supercoil-stabilised cruciform of ColE1 is hyper-reactive to osmium tetroxide. EMBO J. 1984 May;3(5):1187–1192. doi: 10.1002/j.1460-2075.1984.tb01949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. Structural perturbation in supercoiled DNA: hypersensitivity to modification by a single-strand-selective chemical reagent conferred by inverted repeat sequences. Nucleic Acids Res. 1983 May 25;11(10):3097–3112. doi: 10.1093/nar/11.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The kinetic properties of cruciform extrusion are determined by DNA base-sequence. Nucleic Acids Res. 1985 Mar 11;13(5):1443–1465. doi: 10.1093/nar/13.5.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miura K., Ueda T., Shinriki N., Ishizaki K., Harada F. Degradation of nucleic acids with ozone. IV. Specific internucleotidic bond-cleavage of ozone-treated transfer ribonucleic acids with aniline-acetate. Chem Pharm Bull (Tokyo) 1984 Feb;32(2):651–657. doi: 10.1248/cpb.32.651. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Shinriki N., Ishizaki K., Ikehata A., Yoshizaki T., Nomura A., Miura K., Mizuno Y. Degradation of nucleic acids with ozone. II. Degradation of yeast RNA, yeast phenylalanine tRNA and tobacco mosaic virus RNA. Biochim Biophys Acta. 1981 Oct 27;655(3):323–328. doi: 10.1016/0005-2787(81)90041-1. [DOI] [PubMed] [Google Scholar]
- Shinriki N., Ishizaki K., Sato S., Miura K., Sawadaishi K., Ueda T. Degradation of nucleic acids with ozone. VI. Labilization of the double-helical structure of calf thymus deoxyribonucleic acid. Chem Pharm Bull (Tokyo) 1984 Sep;32(9):3636–3640. doi: 10.1248/cpb.32.3636. [DOI] [PubMed] [Google Scholar]
- Sinden R. R., Pettijohn D. E. Cruciform transitions in DNA. J Biol Chem. 1984 May 25;259(10):6593–6600. [PubMed] [Google Scholar]
- Singleton C. K. Effects of salts, temperature, and stem length on supercoil-induced formation of cruciforms. J Biol Chem. 1983 Jun 25;258(12):7661–7668. [PubMed] [Google Scholar]
- Singleton C. K., Wells R. D. Relationship between superhelical density and cruciform formation in plasmid pVH51. J Biol Chem. 1982 Jun 10;257(11):6292–6295. [PubMed] [Google Scholar]