Abstract
The PapC usher is a β-barrel outer membrane protein essential for assembly and secretion of P pili that are required for adhesion of pathogenic E. coli, which cause the development of pyelonephritis. Multiple protein subunits form the P pilus, the highly specific assembly of which is coordinated by the usher. Despite a wealth of structural knowledge, how the usher catalyzes subunit polymerization and orchestrates a correct and functional order of subunit assembly remain unclear. Here, the ability of the soluble N-terminal (UsherN), C-terminal (UsherC2), and Plug (UsherP) domains of the usher to bind different chaperone-subunit (PapDPapX) complexes is investigated using noncovalent electrospray ionization mass spectrometry. The results reveal that each usher domain is able to bind all six PapDPapX complexes, consistent with an active role of all three usher domains in pilus biogenesis. Using collision induced dissociation, combined with competition binding experiments and dissection of the adhesin subunit, PapG, into separate pilin and adhesin domains, the results reveal why PapG has a uniquely high affinity for the usher, which is consistent with this subunit always being displayed at the pilus tip. In addition, we show how the different soluble usher domains cooperate to coordinate and control efficient pilus assembly at the usher platform. As well as providing new information about the protein-protein interactions that determine pilus biogenesis, the results highlight the power of noncovalent MS to interrogate biological mechanisms, especially in complex mixtures of species.
Gram-negative bacteria use pili, long non-covalently assembled protein polymers, as surface-exposed appendages to mediate attachment to host cells, thereby initiating infection (1). Pili assemble at the outer membrane of the host organisms via the chaperone-usher pathway (2). Two of the most comprehensively characterized pilus systems of this pathway include type 1 pili, which attach to receptors in the bladder eventually causing cystitis (3, 4), and P pili, which attach to Galα(1–4)Gal moieties in kidney cells causing inflammation and, in some cases, urinary tract infections (5). The chromosomal pyelonephritis-associated pilus (pap) gene cluster encodes six subunits (PapG, PapF, PapE, PapK, PapA, and PapH) that assemble to form P pili (Fig. 1A). The flexible fibrillum contains at its tip only one copy of the PapG subunit which, uniquely, contains both an adhesin and a pilin domain (Fig. 1A). This subunit is required to be displayed at the pilus tip for host adhesion (6). PapG is joined to one copy of PapF, which is then linked to 5–10 copies of PapE to form the flexible pilus tip. A single PapK subunit joins the flexible tip to the second segment, a right-handed, rigid, helical rod, consisting of thousands of PapA subunits (Fig. 1A). Pilus assembly is arrested by the incorporation of one copy of the PapH subunit which prevents further assembly and locks the fully assembled pilus into the usher complex in the outer membrane (7–10).
Fig. 1.
Schematic of subunits comprising a P pilus and structure of the usher platform. A, Organization of the P pilus, which includes one copy each of PapG (red), PapF (orange), PapK (green), and PapH (dark blue), five to ten copies of the PapE (yellow), and up to one thousand copies of PapA (cyan). The chaperone, PapD (D, brown), usher (C, pink), periplasm (P), extracellular space (Ex), and bacterial outer membrane (OM) are indicated. The PapG adhesin (left, pdb 1J8S (28)) and pilin (right, pdb 3MEO) domains are shown. B, Structure of the usher FimD protein in the apo (left, pdb 3OHN (15)) and activated (right, pdb 3RFZ (15)) forms. The soluble N-terminal (red), plug (magenta), and C-terminal (C1; lime, C2; dark green) domains are shown. Note that the plug domain is released from the usher lumen into the periplasm in the activated form. The structure of the activated form also contains a chaperone-subunit (brown-blue) complex bound to the C-terminal usher domains. C, Domain organization of the PapC usher colored as in (B).
Assembly of the different subunits in a precise order is required to form functional pili. Pilus biogenesis, thus, is a highly organized process which is mediated by a large β-barrel transmembrane protein known as the usher (PapC) (Fig. 1B). Subunits (PapX) enter the periplasm via the secYEG general secretary pathway (11) whereupon they each bind to a molecule of the same chaperone, PapD, forming a PapDPapX chaperone-subunit complex (supplemental Fig. S1A, S1C). Chaperone-subunit binding is required to prevent the subunit domain from misfolding, premature polymerization, and degradation (9, 12). Each PapDPapX complex is formed by the chaperone donating one of its β-strands (G1) to the pilin subunit, to complete its C-terminally truncated immunoglobulin (Ig) fold, through a process known as donor strand complementation (13) (supplemental Fig. S1A).
To bring about pilus assembly, PapDPapX complexes are targeted to the usher at the bacterial outer membrane (Fig. 1B). The usher is an 809 residue protein comprised of a 24-stranded β-barrel pore that, in the inactive state, is blocked by its plug domain (UsherP)1 (magenta domain Fig. 1B, left) (14–15). Chaperone-subunits are first bound by the soluble N-terminal domain of the usher (UsherN) (red domain Fig. 1B) (16). Binding of PapDPapG (the first complex recruited at the usher) to UsherN activates the usher and causes release of UsherP into the periplasm, opening the translocation pore (Fig. 1B, right) (14, 15). Subunits are released from the chaperone when the N-terminal extension (Nte) of the subunit next in assembly displaces the donated chaperone G1 β-strand in a process termed donor strand exchange (DSE) (9) (supplemental Fig. S1B). During, or after, DSE subunits are released from UsherN and passed (directly or indirectly) to a second pair of soluble domains located toward the C-terminal region of the usher protein (UsherC1 and UsherC2) (green domains Fig. 1B, right) (15), whereupon they emerge through the pore of the usher (15) creating the new, growing pilus. This process continues, (catalyzed by the usher) (17), until a PapDPapH complex reaches the usher and undergoes DSE, terminating pilus growth (9).
The relative rates of DSE between all 30 possible pairs of P pilus subunits and their Ntes have been investigated using electrospray ionization mass spectrometry (ESI-MS) (18, 19). These experiments revealed that cognate subunit-Nte pairs undergo more rapid rates of DSE than their noncognate counterparts, suggesting that the relative rates of DSE contribute toward subunit ordering in pilus biogenesis. However, the extent of subunit discrimination and the rates of DSE observed in vitro are too small or too slow, respectively, to explain the specificity and rate of pilus biogenesis in vivo (20–21). This is consistent with a role of the usher protein in catalysis of pilus biogenesis (17) and in subunit discrimination (22, 23). How this is accomplished, however, remains unclear, despite recent crystal structures of the usher complex both with and without chaperone-subunits bound (14, 15, 22–25) (Fig. 1B).
Here we describe the use of noncovalent ESI-MS to monitor interactions of the three soluble usher domains (UsherN, UsherC2, and UsherP) (Fig. 1C) individually with all six PapDPapX complexes (supplemental Fig. S1C) (UsherC1 of PapC could not be produced by recombinant expression). Combined with collision induced dissociation tandem mass spectrometry (CID-MS/MS) to assess the relative gas phase stability of the different complexes formed, and competition binding experiments, also monitored by ESI-MS, the results reveal the ability of all three soluble usher domains to bind all six chaperone-subunit complexes, suggesting their involvement in the movement of the chaperone-subunit complexes around the usher to bring about the correct ordering of subunits into the usher pore during pilus biogenesis.
EXPERIMENTAL PROCEDURES
Expression and purification of soluble usher domains and PapDPapX complexes
PapDhisPapGII (18, 26, 27), PapGad (28), PapDPapFG8N (18), PapDhisPapENtd (9), PapDhisPapK (9), PapDhisPapANtd1G15N (10) and PapDhisPapHNtd1 (10), and the UsherC2 domain (24) were cloned, expressed and purified as previously reported. Production of PapDhisPapGp, UsherN, UsherN24–131, and UsherP is described in the Supplementary Information.
Formation of Ternary PapDPapXUsherY Complexes
PapDPapX complexes and each soluble usher domain were dialyzed separately overnight into 5 mm ammonium acetate (pH 6.8) after which the usher domain (final concentration of 20 μm) (in the same buffer) was added to each chaperone-subunit complex (final concentration 10 μm) to a total volume of 30 μl in 5 mm ammonium acetate (pH 6.8). After 30 min at ambient temperature, 6 μl of sample was removed and analyzed by ESI-MS. The concentration and pH of ammonium acetate were selected based on previous optimization studies for buffer conditions for analysis of the chaperone-subunit complexes (18).
Competition Experiments
Competition experiments in which the UsherN domain was added to a mixture of PapDPapG and PapDPapA involved dialyzing all components overnight into 5 mm ammonium acetate at pH 6.8, followed by the analysis of the different protein complexes after mixing on the subsequent day. All of these experiments were performed in series on the same day. Increasing concentrations of UsherN (0–10 μm) were added to equimolar amounts (5 μm) of PapDPapG and PapDPapA. After 15 min, an aliquot was removed and analyzed by ESI-MS. Comparison of the relative ion intensity of different complexes was calculated as follows: relative ion intensity of PapDPapGUsherN = PapDPapGUsherNtotal intensity/(∑PapDPapGtotal intensity + PapDPapGUsherNtotal intensity). Likewise, relative intensities of PapDPapAUsherN were calculated by the following equation: relative ion intensity PapDPapAUsherN = PapDPapAUsherNtotal intensity/(∑PapDPapAtotal intensity + PapDPapAUsherNtotal intensity).
For competition experiments involving two usher domains and one chaperone-subunit complex, after overnight dialysis the first usher domain (10 μm) was added to each chaperone-subunit complex (10 μm) in a total volume of 20 μl in 5 mm ammonium acetate (pH 6.8). After 2 h at ambient temperature, 6 μl of sample were removed and analyzed by ESI-MS. The second usher domain or control protein (10 μm) was then added and after a further 2 h at ambient temperature, 6 μl of sample were removed and analyzed by ESI-MS.
Mass Spectrometry Data Acquisition and Interpretation
Ternary (chaperone-subunit-usher(N, C2, or P), and competitive (PapDPapG + PapDPapA + UsherN, or chaperone-subunit-usher(N, C2, or P) + usher(N, C2, or P)) complexes were identified using an LCT Premier (Micromass UK Ltd., Waters Corporation, Manchester, UK) time-of-flight mass spectrometer. ESI-CID-MS/MS experiments were performed with a Synapt HDMS, hybrid quadrupole-IMS-oa-TOF mass spectrometer (Micromass UK Ltd., Waters Corporation, Manchester, UK). Both mass spectrometers are interfaced with a NanoMate (Advion Biosystems Inc., Ithaca, NY) nanoESI autosampling device for sample introduction and ionization. Positive nano-ESI with a capillary voltage of 1.75 kV and a nitrogen nebulising gas pressure of 0.5 p.s.i. was used. A cone voltage of 70 V with a source temperature of 50 °C was applied. For identification of ternary and quaternary complexes, spectra were acquired over 1 min with the analyzer set to transmit m/z 1000–5000 for all complexes except those containing the PapDPapG subunit, in which case ions were acquired over the range of m/z 1000–7000. MassLynx v4.1 software was used to process raw data. ESI-CID-MS/MS experiments were performed by selecting and isolating ions representing the ternary complex in the trap collision cell that was filled with argon at a pressure of 1.18 × 10−2 mbar. Increasing collision energy was applied to the trap collision cell in 10 eV increments until the product ions were completely dissociated into the fragment ions. Profiles describing the fragmentation pathway were generated. These display the fraction of ternary ion intensity (as a percentage of total ion intensity) versus normalized collision energy
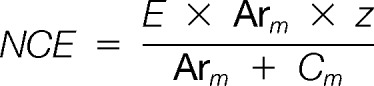 |
where E is the collision energy, Arm is the mass of Argon, Cm is the mass of the complex and z the charge of the precursor ions.
RESULTS
Chaperone-Subunit-Usher Ternary Complexes Identified by ESI-MS
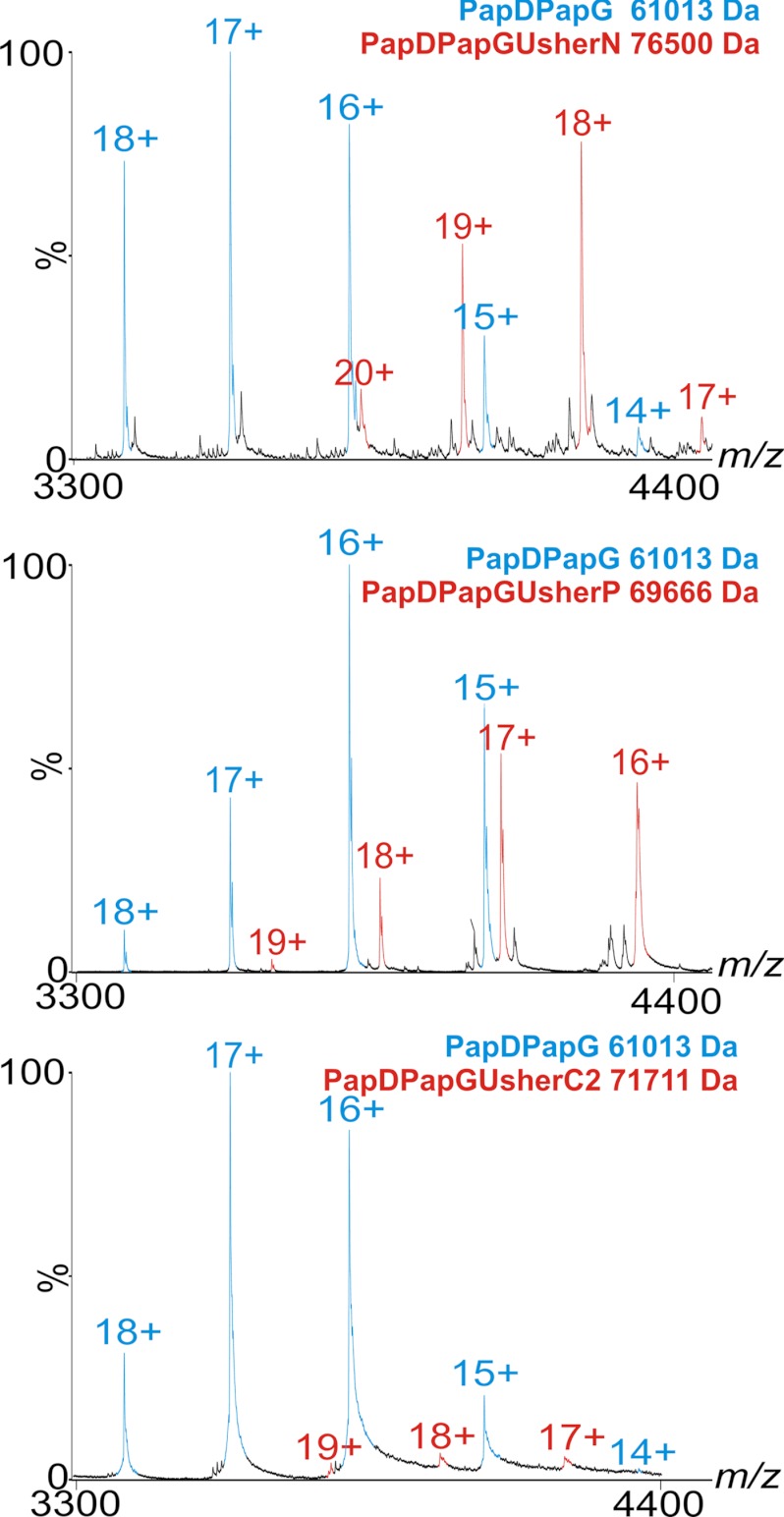
Structural analyses of the homologous usher proteins, PapC and FimD (from P pili and type I pili, respectively) have revealed that these proteins contain five functional domains: a membrane-embedded translocation domain and four soluble domains that reside in the periplasm in the activated state (Figs. 1B,C) (14, 15). The latter include the 131-residue N-terminal domain (UsherN), two C-terminal domains (UsherC1, 84 residues; UsherC2, 85 residues) and the 83-residue plug domain (UsherP) (Fig. 1C) (15). To determine whether the soluble Pap usher domains are able to bind different PapDPapX complexes, an ESI-MS in vitro assay was developed to detect interactions between the different soluble usher domains and different chaperone-subunit pairs. All six PapDPapX complexes (supplemental Fig. S1C) and three soluble usher domains (UsherN, UsherP, and UsherC2; Fig. 1C) were expressed and purified as previously reported (18) and their molecular masses confirmed by ESI-MS (supplemental Text and Fig. S2). Efforts to produce UsherC1 were not successful, presumably because this domain makes intimate interactions with the lumen of the usher translocation domain (Fig. 1B) and is therefore unstable in isolation. Each of the soluble usher domains was mixed separately with each PapDPapX complex and binding was monitored using noncovalent ESI-MS under ambient conditions (Experimental Procedures). Strikingly, ternary complexes of all six PapDPapX complexes with UsherN, UsherC2, or UsherP were identified using ESI-MS, resulting in ions indicative of complexes representing one usher domain bound to one chaperone-subunit in all cases (red peaks, Fig. 2, and supplemental Fig. S3). The ion series representing the ternary complex (red peaks, Fig. 2, and supplemental Fig. S3) appear interspersed with the ion series representing the free chaperone-subunit complex (cyan peaks, Fig. 2 and supplemental Fig. S3). The measured molecular masses of the ternary complex were in close agreement (within 0.03%) with the expected masses (supplemental Table S1).
Fig. 2.
Binding of PapDPapG to the three soluble usher domains. ESI-MS spectra of PapDPapGUsherN (top), PapDPapGUsherP (middle) and PapDPapGUsherC2 (bottom). PapDPapG ions are shown in blue and PapDPapGUsherX complexes are shown in red (unbound UsherN, UsherC2 and UsherP are not shown on this m/z scale). The differences in intensity of ions arising from the different complexes result from differential ionization potential and should not be interpreted as representative of the percent bound material (see also supplemental Figs. S4 and S5).
Despite clear evidence of binding between all of the PapDPapX complexes with each usher domain, different ion intensities are observed for the different complexes formed (supplemental Fig. S3). This likely reflects differences in binding affinity, complex stability and/or ionization efficiency. In accord with the latter proposition, control experiments showed that the three usher domains give very different MS responses; for example, the UsherC domain seemingly generates a lower ion count than either the UsherN or the UsherP domain when pairs of these proteins are mixed at equimolar concentrations in the absence of PapDPapX complexes (supplemental Fig. S4). Also, the various chaperone-subunits (in the absence of usher) do not ionize with identical efficiencies (supplemental Fig. S5). It is not unexpected, therefore, that the ternary complexes arising from different usher domains and different chaperone-subunit-usher domain complexes give rise to differing MS responses that will not reflect their relative abundances in solution quantitatively, and hence nor their KD values.
Previous reports have shown direct interactions of the chaperone-adhesin complex (PapDPapG) with the C-terminal domain (UsherC2) of the usher (15, 29), but no evidence has been reported for interactions of other chaperone-subunit complexes with this domain, nor for any PapDPapX complex with UsherP. The interactions observed here by ESI-MS appear to be specific, since there was no evidence for complexes with a molar ratio of >1 usher domain binding to each chaperone-subunit complex even when the usher domain is added in excess (supplemental Figs. S3 and S5). Furthermore, no binding was observed between UsherN, UsherP, and UsherC2 with the unrelated Ig domains I27 and β2-microglobulin, nor with free PapD or free PapX within the same sample (supplemental Text, Fig. S6).
In the crystal structure of UsherN bound to a chaperone-pilin complex, the N-terminal residues 2–23 of UsherN form a flexible tail that contacts the chaperone (22). Consistent with this, residues 2–11 of UsherN have been shown to be required for high affinity binding to chaperone-subunit complexes (16). To determine whether this is also the case in the experiments presented here, the ability of a truncated UsherN construct, UsherN24–131 (supplemental Text) to bind to all six PapDPapX complexes was also investigated. The ESI-MS results showed that UsherN24–131 is still able to form ternary complexes with all six PapDPapX complexes, indicating that the presence of residues 1–23 is not obligate for chaperone/subunit binding (red peaks, supplemental Fig. S7A). In addition to the known ability of UsherN to bind all six PapDPapX pairs (16, 22, 30), the results presented here reveal that UsherC2 and UsherP also recognize all six different chaperone-subunit complexes, suggesting that each of the soluble usher domains may play a role in the assembly of type P pili.
Differences in the Relative Stabilities of Chaperone-Usher-Subunit Complexes
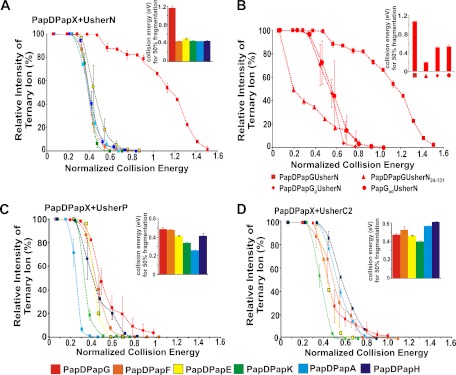
Because all soluble usher domains were found to be capable of forming stable interactions with each PapDPapX complex (red peaks, Fig. 2, and supplemental Fig. S3), the difference in the gas phase stability of the different PapDPapXUsherX ternary complexes was next determined using CID-MS/MS. These experiments are indicative of the relative gas phase stabilities of the different complexes formed (31). CID-MS/MS has been used widely to compare gas phase stabilities of protein complexes (32–35), including the stability effect of anions (34), mutations (36), and ligand binding (37–38) on protein complexes. In this study, this ubiquitous technique has been used to compare the behavior of differing chaperone-subunits complex bound to an usher domain in a qualitative context. In this experiment, ions representing the ternary complex were selected as the precursor ions. Accordingly, the collision energy applied to effect dissociation of different precursor ions was increased until complete dissociation of each ternary complex was observed. The fraction of the precursor ion intensity remaining compared with the total ion intensity was then plotted as a function of normalized collision energy (Fig. 3). Consistent with the known high affinity of PapDPapG for UsherN determined using Surface Plasmon Resonance (SPR) experiments (39), PapDPapGUsherN was also found to be the most stable ternary complex in vacuo (Fig. 3A), with the other five PapDPapXUsherN complexes showing substantially weaker, but nonetheless significant, gas phase stability relative to PapDPapGUsherN (Fig. 3A, inset).
Fig. 3.
Stability of different PapDPapXUsherX complexes determined using CID-MS/MS. A, CID-MS/MS fragmentation profiles of ternary complexes involving all six PapDPapX complexes bound to UsherN. B, Fragmentation profiles of PapDPapGUsherN (square), PapDPapGUsherN24–131 (triangle), PapDPapGpUsherN (diamond), and PapGadUsherN (circle). C, Fragmentation profiles of different PapDPapX complexes bound to UsherP and (D) as (C) but bound to UsherC2. Inset: bar graphs representing the normalized collision energy required to fragment each ternary ion to 50% of its original intensity. Error bars represent the S.E. of four replicate measurements.
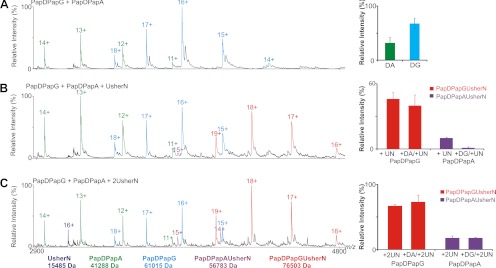
The ion intensities of the ternary complexes may not reflect in-solution abundances (31). To determine whether the relative gas phase stability is reflective of solution phase binding affinity for the noncovalent complexes investigated here, the interactions of PapDPapG and PapDPapA with UsherN were followed in more detail using ESI-MS. These two chaperone-subunit complexes were chosen specifically for this analysis since the binding of these complexes to UsherN has been determined previously using SPR (39). These solution phase binding data revealed that UsherN binds significantly more tightly to PapDPapG compared with PapDPapA (Kd values of 9.1 × 10−9 m and 1.8 × 10−7 m, respectively (39)), corroborating the CID-MS/MS data which show an enhanced gas phase stability for the PapDPapGUsherN complex compared with PapDPapAUsherN (Fig 3A). To demonstrate the different binding affinities of these complexes further using ESI-MS, PapDPapG and PapDPapA were mixed in an equimolar ratio and their relative ion abundances were determined (Fig. 4, top panel). ESI-MS analysis revealed that PapDPapG ionizes more efficiently than PapDPapA, as previously discussed. This equimolar mixture of PapDPapG and PapDPapA was then mixed with UsherN either in a 1:1:1 or 1:1:2 molar ratio of PapDPapG, PapDPapA, and UsherN, respectively, and the relative ion intensities of the different ternary complexes were measured and compared (Fig. 4, lower panels). The results revealed that when Usher N is added to PapDPapG and PapDPapA in a 1:1:1 ratio, UsherN is found predominantly bound to PapDPapG whereas very little, if any, binding was observed to PapDPapA (Fig. 4, central panel). By contrast, when UsherN was added in a 1:1 ratio to either PapDPapG or PapDPapA (without the competing chaperone-subunit complex), significant ion intensity corresponding to each ternary complex was observed (Figs. 4 and supplemental Fig. S3). Only when an excess of UsherN is added to PapDPapG/PapDPapA in a molar ratio of 1:1:2 is an observable ion intensity of the ternary PapDPapAUsherN complex detected (Figs. 4 and supplemental Fig. S5), thus reinforcing the ability of ESI-MS to reveal the relative binding affinity of different complexes, provided that suitable care is taken to measure the relative ionization efficiency of the different species formed, as well as to compare different species within carefully controlled experimental parameters. The data are thus consistent with the SPR and CID-MS/MS data, all of which indicate tighter binding of UsherN for PapDPapG.
Fig. 4.
Competition binding experiments with UsherN and mixtures of PapDPapG and PapDPapA. A, ESI-MS spectra of an equimolar mixture of PapDPapG + PapDPapA (PapDPapG (cyan), PapDPapA (green)). Bar charts showing the relative ion intensities of each complex within the mixture are shown alongside. B, ESI-MS spectra of equimolar PapDPapG + PapDPapA to which UsherN in a 1:1:1 molar ratio is added. Ions arising from the different complexes are colored: PapDPapG (cyan), PapDPapA (green), PapDPapAUsherN (purple), PapdPapGUsherN (red). Bar chart showing the relative ion intensities of ternary complexes PapDPapGUsherN (red) in samples that either lack (labeled PapDPapG + UN) or contain an equimolar concentration of competing PapDPapA (labeled PapDPapG + DA/UN) are shown alongside. Similarly, the ion intensity of PapDPapAUsherN (purple) without added PapDPapG (labeled PapDPapA +UN) or in the presence of the competing complex (labeled PapDPapA + DG/UN) are also shown (see Experimental Procedures). C, ESI-MS spectra of equimolar PapDPapG + PapDPapA to which UsherN in a 1:1:2 molar ratio is added, with relative ion intensities shown in the corresponding bar chart. The ions are colored and labeled as in (B). In all cases errors shown are the S.E. over several replicate measurements.
To determine the contribution of the unstructured N-terminal region of UsherN to the stability of the complexes formed between this usher domain and PapDPapG, ternary complexes were formed between PapDPapG and the truncated UsherN24–131 construct, and the gas phase stability of the resulting complex was determined using CID-MS/MS (Fig. 3B (triangles)). Consistent with the known role of the N-terminal region of UsherN in determining high affinity PapDPapG binding (16), the gas phase stability of the PapDPapGUsherN complex was reduced substantially when the unstructured N-terminal region was deleted (Fig. 3B). Interestingly, the stability of the other five chaperone-subunit complexes was not affected by the absence of the residues 1–23 in UsherN (supplemental Fig. S7A, S7B).
Although five of the six PapX subunits contain only a single pilin domain, PapG is unique among the pilin subunits in that it contains two domains, a pilin domain (PapGp) and an adhesin domain (PapGad) (Fig. 1A, supplemental Fig. S1C). Thus, we also tested complex formation and stability to gas phase dissociation of a PapDPapGpUsherN complex lacking the adhesin domain of PapG (Fig. 3B (diamonds)). Remarkably, complex stability is reduced substantially in this construct relative to its intact PapDPapG counterpart (Fig. 3B squares). These results reveal a hitherto unsuspected role of the PapG adhesin domain in enhancing the binding affinity of PapG for UsherN. Indeed, without the adhesin domain, the stability of the PapDPapGpUsherN complex is similar to that of other PapDPapX complexes (Fig. 3A). To confirm the ability of UsherN to bind to the adhesin domain alone, PapGadhesin (Ga) (residues 1–196 of PapG, Fig. 1A, supplemental Fig. S1C) was constructed (Experimental Procedures) (28) and the binding of this domain to UsherN was determined by ESI-MS (supplemental Fig. S8). The stability of the binary complex of PapGad with UsherN was also measured using CID-MS/MS (Fig. 3B (circles)), revealing that this complex has similar gas phase stability to PapDPapGp. The data strongly suggest, therefore, that a binding interface exists between the UsherN domain of PapC and the adhesin domain of PapG. This interaction rationalizes the remarkable stability of the PapDPapGUsherNternarycomplex that may facilitate activation of the usher with the PapDPapG complex, which is required to initiate the formation of a functional pilus (40).
In parallel with measurements of the gas phase stability of ternary complexes of different chaperone-subunits with UsherN, similar measurements were performed for all six PapDPapX complexes with UsherP and UsherC2. All of these complexes were stable to gas phase dissociation, resulting in measured stabilities similar to those formed between UsherN and PapDPapF/E/K/A/H, but weaker than the complex formed between UsherN and PapDPapG (compare bar graphs in Figs. 3A, 3C, and 3D). Small, but significant, differences in the stability of the ternary complexes involving UsherP and UsherC2 with the six PapDPapX complexes were observed (Fig. 3C, 3D), suggesting that the binding interface must involve at least some interactions with the subunit domain itself as the chaperone is the same across the PapDPapX complexes (supplemental Fig. S1C). Consistent with this, dissociation of the ternary complexes yielded ions representing the PapDPapX complex in addition to ions indicating the presence of free PapD, free PapX and free usher domains. Ions that were consistent with both PapDUsherY and PapXUsherY binary complexes were also detected. These findings support the proposal of a shared binding interface for all three usher domains with both the chaperone and subunit components of the complexes, consistent with recent crystallographic information of the equivalent subunits from type 1 pili (FimCFimH) binding to the N- and C-terminal domains of the Fim usher, FimD (15, 22). During the CID-MS/MS fragmentation of the ternary complexes, the UsherN domain was dissociated from the PapDPapX chaperone-subunit complex, following an expansion in its collision cross-sectional area, consistent with dissociation mechanisms reported elsewhere (35, 41–43).
Competition Between Different Usher Domains for Chaperone-Subunit Binding
Recent crystallographic analysis of the FimD usher in the act of DSE and subunit translocation through the pore of the usher has suggested that UsherN and UsherC2 compete for binding to each chaperone-subunit complex (15). Although UsherN has been proposed to be essential for receiving (16, 22) and discriminating (30, 39, 44) between the incoming chaperone-subunit complexes, usher domains C1 and C2 have been proposed to bind to the chaperone-subunit complex after DSE, releasing UsherN to accept the next incoming subunit (15). How handover of the chaperone-subunit complex between usher domains is conducted, however, remains unclear. Two scenarios can be envisioned: direct competition of UsherN and UsherC1/2 for a common binding site on the chaperone:subunit complex, or transfer between UsherN and UsherC1/2 via an intermediary domain or surface.
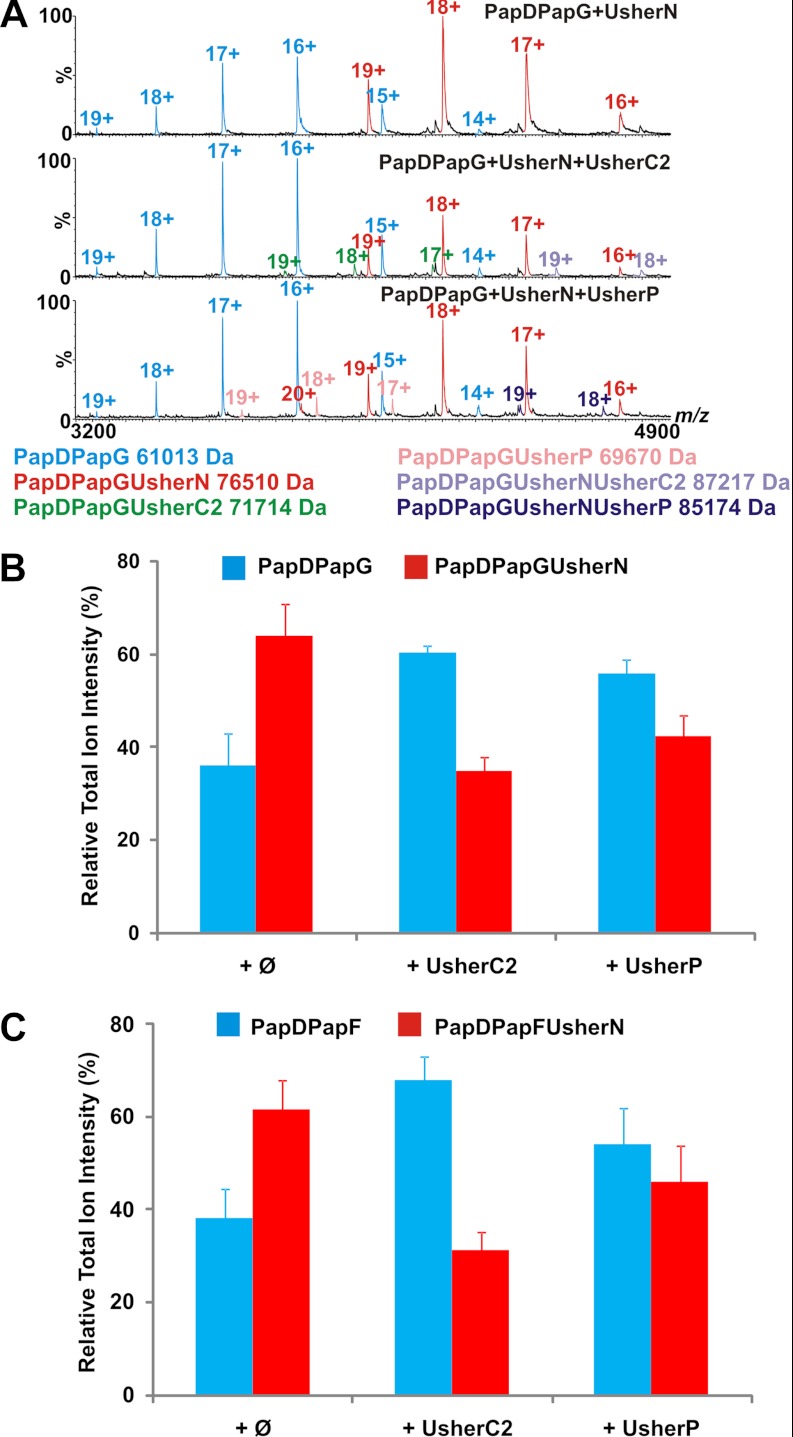
To determine whether UsherN, UsherP, and UsherC2 compete for a common binding site on each chaperone-subunit complex, the ability of each usher domain to bind simultaneously to PapDPapG or PapDPapF was investigated. Accordingly, equimolar concentrations of two different usher domains were added sequentially to PapDPapG or PapDPapF and the resulting mixtures were analyzed before, and after, addition of the second usher domain using ESI-MS. When equimolar amounts of UsherC2 or UsherP were added to complexes of PapDPapG or PapDPapF containing prebound UsherN (Fig. 5), the results revealed a significant depletion in the fraction of PapDPapGUsherN and PapDPapFUsherN ions compared with their unbound chaperone-subunit predecessors, suggesting that these domains can displace UsherN via direct competition for a common binding site or an allosteric shift. Importantly, when control proteins (UsherC2 of FimD or hen egg white lysozyme) were added, the fraction of PapDPapGUsherN to unbound PapDPapG was not depleted, suggesting the competition for UsherN is specific to the PapC usher domains (supplemental Fig. S9). Quaternary complexes were observed for PapDPapG/F+UsherN+UsherC2 and PapDPapG+UsherN+UsherP, but not for PapDPapG/F+UsherC2+UsherP (Fig. 5A). These potential intermediates may be “snapshots” of a possible displacement mechanism occurring between the two domains, or evidence of a handover process between UsherN, UsherP, and UsherC2.
Fig. 5.
Competition experiments with usher domains and different chaperone subunit complexes. ESI-MS spectra of (A) PapDPapGUsherN (top) to which UsherC2 (middle) or UsherP (bottom) was subsequently added. PapDPapG (cyan), PapDPapGUsherN (red), PapDPapGUsherC2 (green), PapDPapGUsherP (pink), PapDPapGUsherNUsherC2 (purple), and PapDPapGUsherNUsherP (dark blue) complexes are displayed. Histograms showing the relative ion intensities of (B) PapDPapGUsherN (red) and (C) PapDPapFUsherN (red) compared with the initial chaperone-subunit complexes (blue) before (Ø, left) or after addition of UsherC2 (middle) or UsherP (right). Note that although the relative intensity of ions arising from PapDPapXUsherN to free PapDPapX varies in different experiments, the loss of the ternary complex after addition of the second usher domain was consistently observed. Errors are the S.E. over four replicate measurements.
DISCUSSION
The Adhesin Domain is not an Innocent Bystander in Initiating Pilus Formation
The UsherN domain of PapC is known to be the first site of interaction of the usher with PapDPapG, the binding of which has to occur first in order to assemble a functional pilus (15, 16). In the case of the usher in the closely related Type 1 pilus, (supplemental Fig. S10) FimD, activation has been shown to require binding of the FimH subunit that contains the adhesin domain (supplemental Fig. S10), ensuring that this subunit is displayed at the pilus tip as required for functionality (45, 46). By contrast with the strict requirement of FimD for activation with FimH, PapC is less discriminating and may be activated by the binding of chaperone-subunit complexes other than PapDPapG (16, 40). Here we show using CID-MS/MS of the PapDPapXUsherNternary complexes that the PapDPapGUsherNcomplex is substantially more stable to gas phase induced dissociation than the other ternary complexes (Fig. 3A), consistent with the known high affinity of this complex determined using SPR (39, 44, 47). Dissection of PapG into its constituent adhesin and pilin domains, together with analysis of binding of different chaperone-subunit pairs to UsherN and its truncated variant UsherN24–131, revealed a dramatic reduction in stability when the UsherN24–131 construct was bound to PapDPapG (Fig. 3B), or when an adhesin-deleted PapDPapG complex was added to UsherN. These data indicate a hitherto unsuspected role of the adhesin domain in contributing to the high affinity binding of PapDPapG to UsherN. Interestingly, crystallographic analysis of the complex formed between the FimD UsherN domain bound to FimCFimHp (in which the adhesin domain of FimH is deleted) revealed that the first 23 residues of UsherN interact with the chaperone and not with the pilin domain (22). This may result from the lack of the adhesin domain in this complex, or could reflect differences in the manners by which the different UsherN domains of PapC and FimD recognize their initiating chaperone-subunit targets.
Coordinated Participation of UsherN, UsherP, and UsherC2 in Pilus Biogenesis
The usher plays numerous key roles in pilus biogenesis, including catalysis of DSE (17) and facilitation of subunit ordering (39, 40, 44). The usher is critical for pilus construction, as without this machinery pili are no longer assembled or secreted (20, 48, 49). Here, using noncovalent ESI-MS, we show that each soluble usher domain (UsherN, UsherC2, and UsherP) is able to bind each of the six PapDPapX complexes, suggestive of a potential role for all three soluble usher domains in pilus biogenesis (Fig. 2 and supplemental Fig. S3). To date, no interactions of UsherC or UsherP with chaperone-subunit complexes have been reported for the Pap system, highlighting the power of ESI-MS to observe these interactions. These observations are consistent with a recent crystal structure of the structurally and functionally homologous FimD usher in complex with the initiating FimCFimH chaperone-subunit complex, which reveals close contacts between FimCFimH with the UsherC1 and UsherC2 domains and less extended, but nevertheless significant, contacts with the plug domain (15). Furthermore, previous deletion experiments have shown that all three soluble usher domains are required for pilus assembly in vivo (14, 23, 29, 40, 50). Therefore, a well-orchestrated and specific series of interactions between the chaperone-subunits and all three soluble usher domains appears to be required to ensure effective pilus biogenesis.
A Refined Model for Usher Discrimination in Pilus Biogenesis
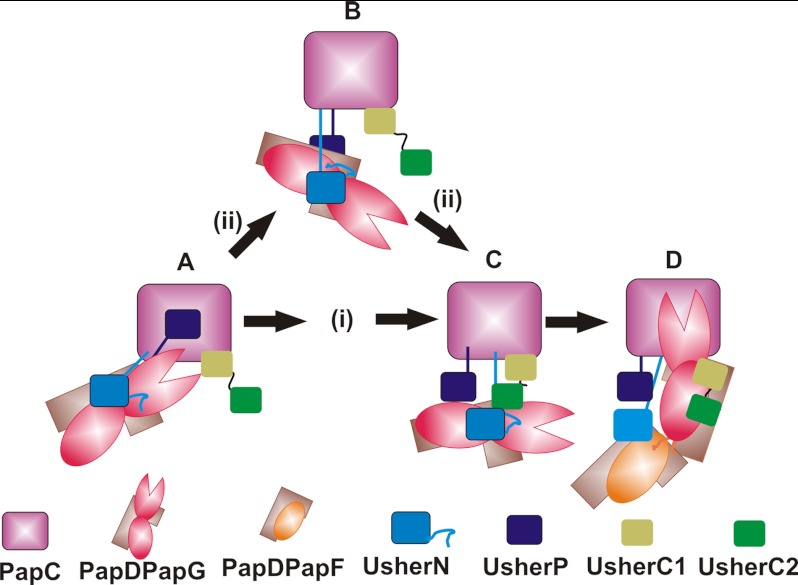
From the data presented, a model of the series of interactions between each PapDPapX complex with the usher domains during pilus biogenesis can be constructed (Fig. 6). Pilus assembly commences with the PapDPapG complex binding to the UsherN domain of PapC (Fig. 6 (step A)) (15, 16). Binding of PapDPapG causes activation of PapC and the consequent displacement of UsherP from the lumen of the usher into the periplasm (Fig. 6 (step B)) (15). The data presented here show that UsherP and UsherC2 are each able to bind all six PapDPapX complexes (Fig. 2, supplemental Fig. S3), indicating a potential role of these domains in pilus assembly. For UsherP this could involve facilitating the transfer of the chaperone-subunit complexes between the N- and C-terminal usher domains, consistent with the ability of this domain to bind all six chaperone-subunit complexes (supplemental Fig. S3), in addition to displacing the incoming subunit from UsherN (Fig. 5). Once the PapDPapXUsherN ternary complex is formed, the chaperone-subunit complex could be passed directly to the C-terminal usher domains (Fig. 6, pathway I, (Step C)), or this transfer could be mediated via UsherP (Fig. 6, pathway ii). In support of the latter, previous experiments have demonstrated a requirement of the plug domain for successful pilus formation, but not for the structure or stability of the usher itself (25, 51). Also, potential intermediates of these handover processes are observed by the detection of quaternary complexes involving the UsherC and UsherP domains binding simultaneously with ternary complexes involving UsherN (Fig. 5A). As a consequence of this transfer, UsherN would be released for recruitment of the next chaperone-subunit complex (Fig. 6 (step D)), whereas binding of the newly transferred chaperone-subunit complex to UsherC1/2 positions the Nte of the incoming subunit directly over the P5 pocket of PapG, allowing DSE to occur (Fig. 6 (step D)) (15).
Fig. 6.
Schematic of the proposed mechanism of pilus biogenesis. Pilus biogenesis commences with the PapDPapG complex (brown/red) binding to the UsherN domain (blue) of PapC, which involves the interaction of UsherN to the adhesin and pilin domains of PapG (A). PapDPapG binding causes the activation of PapC in which the UsherP domain (dark blue) leaves the lumen of the usher and moves into the periplasm (B,C). Once the PapDPapGUsherN ternary complex is formed, PapDPapG is passed onto the UsherC1/2 domains (lime green/dark green) (C) either directly (pathway i) or indirectly via the UsherP domain (pathway ii). At this point, the UsherC1/2 domains displace UsherN (D), releasing UsherN and allowing it to be in position for the arrival of the next chaperone-subunit complex: PapDPapF (brown/orange) (D). Once PapDPapF is bound to the UsherN domain, it is orientated in an ideal position for DSE to occur between its Nte and PapDPapG (15).
In summary, the data presented here add to the growing insights into the molecular mechanism of pilus biogenesis captured by crystallographic snapshots of different points in the assembly mechanism (14, 15, 23). Specifically, using ESI-MS we have revealed a new and hitherto unsuspected role of the adhesin domain in enhancing the affinity of PapG for UsherN, which may play a part in ensuring that this domain is used to initiate pilus formation by activating the usher. In addition, we reveal a potential role of the plug domain in orchestrating the transfer of each chaperone-subunit complex between the UsherN and UsherC domains, ensuring efficient transfer of subunits in the assembly line, and smooth handover between the UsherN and UsherC domains. Utilizing the sensitivity and ability of ESI-MS to monitor and analyze individual species within heterogeneous assemblies and to compare the relative strength of noncovalent interactions using both CID-MS/MS and competition experiments, binding interactions between usher domains and chaperone-subunits have been investigated. By extending this approach to interrogate interactions of chaperone-subunits with the whole membrane-embedded usher protein more insights into pilus biogenesis are sure to be revealed.
Acknowledgments
We thank Dr James Pullen for providing I27 and Dr David Smith for providing β2m. We thank Dr Sebastien Geibel for providing the UsherC2 of FimD. We dedicate this paper to the memory of Dr Denis Verger.
Footnotes
* This work was supported by grants from the Biotechnology and Biological Sciences Research Council (BB/F012284/1). The mass spectrometers were purchased with grants from the BBSRC (BB/E012558/1) (Synapt HDMS, used for CID-MS/MS experiments) and the Wellcome Trust (WT 075099/Z/04/Z) (LCT Premier, used for complex identification).
 This article contains supplemental Figs. S1 to S10, Table S1, and Experimental Procedures.
This article contains supplemental Figs. S1 to S10, Table S1, and Experimental Procedures.
1 The abbreviations used are:
- UsherP
- plug usher domain
- ESI-MS
- electrospray ionization mass spectrometry
- CID
- collision induced dissociation
- MS/MS
- tandem mass spectrometry
- Pap
- pyelonephritis associated pilus
- DSE
- donor strand exchange
- Nte
- N-terminal extension
- PapDPapX
- chaperone-subunitX
- where X = PapG
- PapF, PapK, PapE, PapA, or PapH
- UsherN
- N-terminal usher domain
- UsherC1/2
- C-terminal usher domains.
REFERENCES
- 1. Kuehn M. J., Heuser J., Normark S., Hultgren S. J. (1992) P pili in uropathogenic E. coli are composite fibres with distinct fibrillar adhesive tips. Nature 356, 252–255 [DOI] [PubMed] [Google Scholar]
- 2. Thanassi D. G., Saulino E. T., Hultgren S. J. (1998) The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr. Opin. Microbiol. 1, 223–231 [DOI] [PubMed] [Google Scholar]
- 3. Roberts J. A., Marklund B. I., Ilver D., Haslam D., Kaack M. B., Baskin G., Louis M., Möllby R., Winberg J., Normark S. (1994) The Gal(alpha 1–4)Gal-specific tip adhesin of Escherichia coli P-fimbriae is needed for pyelonephritis to occur in the normal urinary tract. Proc. Natl. Acad. Sci. U.S.A. 91, 11889–11893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wright K. J., Seed P. C., Hultgren S. J. (2007) Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol. 9, 2230–2241 [DOI] [PubMed] [Google Scholar]
- 5. Foxman B., Brown P. (2003) Epidemiology of urinary tract infections: transmission and risk factors, incidence, and costs. Infect. Dis. Clin. North Am. 17, 227–241 [DOI] [PubMed] [Google Scholar]
- 6. Lund B., Lindberg F., Marklund B. I., Normark S. (1987) The PapG protein is the alpha-D-galactopyranosyl-(1–4)-beta-D-galactopyranose-binding adhesin of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 84, 5898–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Båga M., Norgren M., Normark S. (1987) Biogenesis of E. coli Pap pili: papH, a minor pilin subunit involved in cell anchoring and length modulation. Cell 49, 241–251 [DOI] [PubMed] [Google Scholar]
- 8. Rêgo A. T., Chandran V., Waksman G. (2010) Two-step and one-step secretion mechanisms in Gram-negative bacteria: contrasting the type IV secretion system and the chaperone-usher pathway of pilus biogenesis. Biochem. J. 425, 475–488 [DOI] [PubMed] [Google Scholar]
- 9. Sauer F. G., Pinkner J. S., Waksman G., Hultgren S. J. (2002) Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell 111, 543–551 [DOI] [PubMed] [Google Scholar]
- 10. Verger D., Miller E., Remaut H., Waksman G., Hultgren S. (2006) Molecular mechanism of P pilus termination in uropathogenic Escherichia coli. EMBO Rep. 7, 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Driessen A. J., Nouwen N. (2008) Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77, 643–667 [DOI] [PubMed] [Google Scholar]
- 12. Hultgren S. J., Normark S., Abraham S. N. (1991) Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu. Rev. Microbiol. 45, 383–415 [DOI] [PubMed] [Google Scholar]
- 13. Sauer F. G., Fütterer K., Pinkner J. S., Dodson K. W., Hultgren S. J., Waksman G. (1999) Structural basis of chaperone function and pilus biogenesis. Science 285, 1058–1061 [DOI] [PubMed] [Google Scholar]
- 14. Remaut H., Tang C., Henderson N. S., Pinkner J. S., Wang T., Hultgren S. J., Thanassi D. G., Waksman G., Li H. (2008) Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell 133, 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Phan G., Remaut H., Wang T., Allen W. J., Pirker K. F., Lebedev A., Henderson N. S., Geibel S., Volkan E., Yan J., Kunze M. B., Pinkner J. S., Ford B., Kay C. W., Li H., Hultgren S. J., Thanassi D. G., Waksman G. (2011) Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate. Nature 474, 49–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ng T. W., Akman L., Osisami M., Thanassi D. G. (2004) The usher N terminus is the initial targeting site for chaperone-subunit complexes and participates in subsequent pilus biogenesis events. J. Bacteriol. 186, 5321–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nishiyama M., Ishikawa T., Rechsteiner H., Glockshuber R. (2008) Reconstitution of pilus assembly reveals a bacterial outer membrane catalyst. Science 320, 376–379 [DOI] [PubMed] [Google Scholar]
- 18. Rose R. J., Verger D., Daviter T., Remaut H., Paci E., Waksman G., Ashcroft A. E., Radford S. E. (2008) Unraveling the molecular basis of subunit specificity in P pilus assembly by mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 105, 12873–12878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rose R. J., Welsh T. S., Waksman G., Ashcroft A. E., Radford S. E., Paci E. (2008) Donor-strand exchange in chaperone-assisted pilus assembly revealed in atomic detail by molecular dynamics. J. Mol. Biol. 375, 908–919 [DOI] [PubMed] [Google Scholar]
- 20. Jacob-Dubuisson F., Striker R., Hultgren S. J. (1994) Chaperone-Assisted Self-Assembly of Pili Independent of Cellular-Energy. J. Biol. Chem. 269, 12447–12455 [PubMed] [Google Scholar]
- 21. Leney A.C., Phan G., Allen W., Verger D., Waksman G., Radford S.E., Ashcroft A.E. (2011) Second Order Rate Constants of Donor-Strand Exchange Reveal Individual Amino Acid Residues Important in Determining the Subunit Specificity of Pilus Biogenesis. J. Am. Soc. Mass Spectrom. 22, 1214–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nishiyama M., Horst R., Eidam O., Herrmann T., Ignatov O., Vetsch M., Bettendorff P., Jelesarov I., Grütter M. G., Wüthrich K., Glockshuber R., Capitani G. (2005) Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD. EMBO J. 24, 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eidam O., Dworkowski F. S., Glockshuber R., Grütter M. G., Capitani G. (2008) Crystal structure of the ternary FimC-FimF(t) -FimD(N) complex indicates conserved pilus chaperone-subunit complex recognition by the usher FimD. FEBS Lett. 582, 651–655 [DOI] [PubMed] [Google Scholar]
- 24. Ford B., Rêgo A. T., Ragan T. J., Pinkner J., Dodson K., Driscoll P. C., Hultgren S., Waksman G. (2010) Structural homology between the C-terminal domain of the PapC usher and its plug. J. Bacteriol. 192, 1824–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang Y., Smith B. S., Chen L. X., Baxter R. H., Deisenhofer J. (2009) Insights into pilus assembly and secretion from the structure and functional characterization of usher PapC. Proc. Natl. Acad. Sci. U.S.A. 106, 7403–7407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee Y. M., DiGiuseppe P. A., Silhavy T. J., Hultgren S. J. (2004) P pilus assembly motif necessary for activation of the CpxRA pathway by PapE in Escherichia coli. J. Bacteriol. 186, 4326–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lund B., Lindberg F., Normark S. (1988) Structure and antigenic properties of the tip-located P pilus proteins of uropathogenic Escherichia coli. J. Bacteriol. 170, 1887–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dodson K. W., Pinkner J. S., Rose T., Magnusson G., Hultgren S. J., Waksman G. (2001) Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor. Cell 105, 733–743 [DOI] [PubMed] [Google Scholar]
- 29. So S. S., Thanassi D. G. (2006) Analysis of the requirements for pilus biogenesis at the outer membrane usher and the function of the usher C-terminus. Mol Microbiol 60, 364–375 [DOI] [PubMed] [Google Scholar]
- 30. Nishiyama M., Vetsch M., Puorger C., Jelesarov I., Glockshuber R. (2003) Identification and characterization of the chaperone-subunit complex-binding domain from the type 1 pilus assembly platform FimD. J. Mol. Biol. 330, 513–525 [DOI] [PubMed] [Google Scholar]
- 31. Daniel J. M., Friess S. D., Rajagopalan S., Wendt S., Zenobi R. (2002) Quatitative determination of noncovalent binding interaction using soft ionization mass spectrometry. Int. J. Mass Spectrom. 216, 1–27 [Google Scholar]
- 32. Benesch J. L., Sobott F., Robinson C. V. (2003) Thermal dissociation of multimeric protein complexes by using nanoelectrospray mass spectrometry. Anal. Chem. 75, 2208–2214 [DOI] [PubMed] [Google Scholar]
- 33. Canon F., Paté F., Meudec E., Marlin T., Cheynier V., Giuliani A., Sarni-Manchado P. (2009) Characterization, stoichiometry, and stability of salivary protein-tannin complexes by ESI-MS and ESI-MS/MS. Anal. Bioanal. Chem. 395, 2535–2545 [DOI] [PubMed] [Google Scholar]
- 34. Han L., Hyung S. J., Mayers J. J., Ruotolo B. T. (2011) Bound anions differentially stabilize multiprotein complexes in the absence of bulk solvent. J. Am. Chem. Soc. 133, 11358–11367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dodds E. D., Blackwell A. E., Jones C. M., Holso K. L., O'Brien D. J., Cordes M. H., Wysocki V. H. (2011) Determinants of gas-phase disassembly behavior in homodimeric protein complexes with related yet divergent structures. Anal. Chem. 83, 3881–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hyung S. J., Robinson C. V., Ruotolo B. T. (2009) Gas-phase unfolding and disassembly reveals stability differences in ligand-bound multiprotein complexes. Chem. Biol. 16, 382–390 [DOI] [PubMed] [Google Scholar]
- 37. Hopper J. T., Oldham N. J. (2009) Collision induced unfolding of protein ions in the gas phase studied by ion mobility-mass spectrometry: the effect of ligand binding on conformational stability. J. Am. So.c Mass. Spectrom. 20, 1851–1858 [DOI] [PubMed] [Google Scholar]
- 38. van Duijn E., Simmons D. A., van den Heuvel R. H., Bakkes P. J., van Heerikhuizen H., Heeren R. M., Robinson C. V., van der Vies S. M., Heck A. J. (2006) Tandem mass spectrometry of intact GroEL-substrate complexes reveals substrate-specific conformational changes in the trans ring. J. Am. Chem. Soc. 128, 4694–4702 [DOI] [PubMed] [Google Scholar]
- 39. Saulino E. T., Thanassi D. G., Pinkner J. S., Hultgren S. J. (1998) Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. EMBO J. 17, 2177–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li Q., Ng T. W., Dodson K. W., So S. S., Bayle K. M., Pinkner J. S., Scarlata S., Hultgren S. J., Thanassi D. G. (2010) The differential affinity of the usher for chaperone-subunit complexes is required for assembly of complete pili. Mol. Microbiol. 76, 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Benesch J. L., Aquilina J. A., Ruotolo B. T., Sobott F., Robinson C. V. (2006) Tandem mass spectrometry reveals the quaternary organization of macromolecular assemblies. Chem. Biol. 13, 597–605 [DOI] [PubMed] [Google Scholar]
- 42. Ruotolo B. T., Hyung S. J., Robinson P. M., Giles K., Bateman R. H., Robinson C. V. (2007) Ion mobility-mass spectrometry reveals long-lived, unfolded intermediates in the dissociation of protein complexes. Angew. Chem. Int. Ed. Engl. 46, 8001–8004 [DOI] [PubMed] [Google Scholar]
- 43. Pagel K., Hyung S. J., Ruotolo B. T., Robinson C. V. (2010) Alternate dissociation pathways identified in charge-reduced protein complex ions. Anal. Chem. 82, 5363–5372 [DOI] [PubMed] [Google Scholar]
- 44. Dodson K. W., Jacob-Dubuisson F., Striker R. T., Hultgren S. J. (1993) Outer-membrane PapC molecular usher discriminately recognizes periplasmic chaperone-pilus subunit complexes. Proc. Natl. Acad. Sci. U.S.A. 90, 3670–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Connell I., Agace W., Klemm P., Schembri M., Mărild S., Svanborg C. (1996) Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. U.S.A. 93, 9827–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Langermann S., Palaszynski S., Barnhart M., Auguste G., Pinkner J. S., Burlein J., Barren P., Koenig S., Leath S., Jones C. H., Hultgren S. J. (1997) Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science 276, 607–611 [DOI] [PubMed] [Google Scholar]
- 47. Sauer F. G., Remaut H., Hultgren S. J., Waksman G. (2004) Fiber assembly by the chaperone-usher pathway. Biochim. Biophys. Acta 1694, 259–267 [DOI] [PubMed] [Google Scholar]
- 48. Norgren M., Båga M., Tennent J. M., Normark S. (1987) Nucleotide sequence, regulation and functional analysis of the papC gene required for cell surface localization of Pap pili of uropathogenic Escherichia coli. Mol. Microbiol. 1, 169–178 [DOI] [PubMed] [Google Scholar]
- 49. Klemm P., Christiansen G. (1990) The fimD gene required for cell surface localization of Escherichia coli type 1 fimbriae. Mol. Gen. Genet. 220, 334–338 [DOI] [PubMed] [Google Scholar]
- 50. Thanassi D. G., Stathopoulos C., Dodson K., Geiger D., Hultgren S. J. (2002) Bacterial outer membrane ushers contain distinct targeting and assembly domains for pilus biogenesis. J. Bacteriol. 184, 6260–6269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mapingire O. S., Henderson N. S., Duret G., Thanassi D. G., Delcour A. H. (2009) Modulating effects of the plug, helix, and N- and C-terminal domains on channel properties of the PapC usher. J. Biol. Chem. 284, 36324–36333 [DOI] [PMC free article] [PubMed] [Google Scholar]