Abstract
Ubiquitination controls the stability or function of many human proteins, thereby regulating a wide range of physiological processes. In most cases the combinatorial pattern of protein interactions that facilitate substrate recognition or modification remain unclear. Moreover, the efficiency of ubiquitination reactions can be altered by the formation of homo- and heterotypic E3-RING complexes. To establish the prevalence and nature of binary E3-RING/E3-RING interactions systematic yeast two-hybrid screens were performed to test 7269 potential interactions between 124 human E3-RING proteins. These studies identified 228 dimeric interactions between 100 E3-RINGs, of which 205 were novel. Complementary co-immunoprecipitation studies were performed to test predicted network interactions, showing a high correlation (64%) with primary yeast two-hybrid data. This data was integrated with known E3-RING interactions, tissue expression profiles and proteomic ubiquitination datasets to facilitate identification of subnetworks in which E3-RING dimerization events have the potential to alter network structure. These results reveal a widespread yet selective pattern of E3-RING dimerization events, which have the potential to confer further combinatorial complexity within human ubiquitination cascades.
Protein ubiquitination is the reversible conjugation of target proteins with a small, highly conserved protein tag, which in concert with an increasing variety of post-translational modifications confers high-fidelity regulation of many physiological processes. Although different forms of ubiquitination are known to elicit distinct physiological effects (1), the identity and range of E2/E3 ligase interactions that confer specific patterns of substrate modification remain unclear. Given the central role of these processes in protein homeostasis (2) it is not surprising that defects in protein ubiquitination are linked to the initiation or progression of disease, or degenerative conditions (3). For this reason it is imperative that we develop a better understanding of the combinatorial interactions that drive specific ubiquitination events in different cells.
The canonical ubiquitination cascade is mediated by three classes of protein that determine the specificity and architecture of substrate modification. These are the E1-activating enzymes, the E2-conjugating enzymes, and the E3-ligases. Currently there are eight ubiquitin or ubiquitin-like E1-activating enzymes, 42 E2-conjugating enzymes, and >600 predicted E3 “ligases” annotated in the human genome (4). These proteins work in a combinatorial manner to facilitate differential modification of specific protein substrates. In essence, E3 proteins can be divided into two subgroups; HECT domain ligases, which act as enzymatic intermediates in protein ubiquitination (5), and noncatalytic E3 proteins; the E3-RINGs and the cullin E3 multisubunit complexes (6). Of these, E3-RING proteins represent the largest single group with >300 members. In most cases, E3-RING proteins play a key role in controlling both substrate specificity and selective E2 recruitment.
Large numbers of E2/E3-RING interactions were recently characterized in two different high throughput yeast two-hybrid (Y2H)1 systems (7, 8). Data from these independent studies provide highly complementary information, which when integrated with data from numerous small-scale studies produce a high-density map of human E2/E3-RING combinatorial preferences. Despite the complexity of this network a further level of specificity may be imposed by the formation of homo- and heteromeric E3-RING complexes. Although only a limited number of multimeric E3-RING complexes have been reported (9) it is now clear that E3-RING multimerization events play an important allosteric or structural role in mediating ubiquitin transfer (10, 11). For example, heterodimeric RING1:BMI1 and BRCA1:BARD1 complexes have been both functionally and structurally defined (12, 13). Interestingly, in both of these cases one E3-RING recruits the E2 enzyme(s) whereas the second facilitates the efficient ubiquitination of selective substrates (12, 14). Furthermore, a subset of E3-RING proteins have been shown to form higher order multimeric complexes, which exhibit increased catalytic activity compared with their monomeric subunits (15–17). Therefore, selective induction, or perturbation of E3-RING dimers may offer new ways of regulating specific biological processes. Although previous reports provided invaluable insight into the structure and function of multimeric E3-RING complexes, the prevalence and specificity of homo- or heterotypic E3-RING interactions remains unclear. To address this important question two complementary Y2H studies were performed in order to generate a high-density binary network containing 228 dimeric E3-RING interactions. This data was integrated with previously known interactions to generate a functionally unbiased E3-RING centric network, which can inform studies in many different areas of ubiquitin biology. Finally, both experimental and literature derived evidence is provided to support the observation that large numbers of human E3-RING proteins form stable, enzymatically active complexes, which have the potential to alter the specificity, activity or form of target ubiquitination.
EXPERIMENTAL PROCEDURES
Construction of Y2H Bait and Prey Clones
Generation of the human E3-RING prey Y2H clone collection in pACTBD(/E)-B was described previously (7). The E3-RING prey Y2H clone collection in pGAD was extended from previously described (7) to include a subset of E3-RING proteins that had shown interactions in the pACT vector (supplemental File S1). Bait Y2H vectors pGBAD/E-B were generated previously (18). To generate the E3-RING bait Y2H clone collection in either pGBAD-B or pGBAE-B proof-read PCR products were generated from sequenced pDONR223 or pACTBD-B Y2H constructs by proof reading KOD polymerase PCR, according to manufacturer's instructions, before being transferred into bait vectors by in vivo gap repair cloning as previously described (18). This approach gave E3-RING collections in both bait and prey that were distributed across the entire E3-RING family. All constructs were sequence verified to confirm correct ORF insertion.
Yeast Two-Hybrid Matrix Screens
E3-RING bait clones were systematically mated against arrays of E3-RING prey clones on YPAD media for a period of 18 h. Diploids were selected following replication and growth on -Leu/-Trp plates for 48 h. Activation of the auxotrophic ADE2 and HIS3 reporters was independently assayed by replication to media lacking adenine or histidine (+2.5 mm 3AT). Growth on selective plates was scored over a period of 10 days. Activation of the enzymatic reporter gene LacZ was assayed after 5 days incubation on -Leu/-Trp plates. Briefly, yeast colonies were transferred to filter paper prior to repeated freeze/thaw fracturing in liquid nitrogen. Filter papers were then incubated in β-Gal substrate solution (6 mls Z buffer (60 mm Na2HPO4, 40 mm Na2H2PO4, 10 mm KCl, 1 mm MgSO4) 1.6 mg/ml X-Gal reagent (100 mg/ml in N,N-dimethylformamide), 11 μl β-mercaptoethanol) at 37 °C for up to 18 h to allow the development of a blue-white color change indicating the presence of an interaction. E3-RING screens in pACTBD/E-B were initially performed in pools, which were then deconvoluted to identify specific interactions. All pGAD screens were performed as direct pair-wise matings (See supplemental Experimental Procedures for more details).
Co-expression and Immunoprecipitation
Cells were transfected using GeneJuice® (Merck Biosciences, Germany) in Dulbecco's modified Eagle's medium with no additional supplements at a ratio of 3 μl transfection reagent to 1 μg total DNA according to the manufacturer's instructions. The amount of DNA used was adjusted to achieve optimal/equivalent levels of tagged protein expression in each experiment. Washed cells were lysed in 500 μl radioimmunoprecipitation assay buffer (10 mm Tris-HCl pH7.5, 150 mm NaCl, 1% w/v Triton X-100, 0.1% w/v SDS, 1% sodium deoxycholate) supplemented with mammalian protease inhibitor mixture (1:250 dilution) for 20 min on ice. Forty-eight microliters of the resulting supernatant was removed and added to 12 μl 5 × sample buffer (SB, Final concentrations: 62.5 mm Tris-HCl pH 6.8, 3% SDS, 10% glycerol, 3.3% 2-mercapotethanol, 0.01% bromphenol blue), prior to heating at 95 °C for 5 min. A 432 μl aliquot of the supernatant was then incubated with 30 μl protein-G agarose (Sigma Aldrich, Poole, UK) and 1.5 μl sheep anti-GFP (gift from Ian Prior, University of Liverpool) prior to mixing at 4 °C for 4 h. Agarose beads were then pelleted by centrifugation (13,000 rpm at 4 °C for 30 s), before being washed three times in ice cold IP wash buffer (0.1% Nonidet P-40, 25 mm Tris-HCl pH7.5 and 150 mm NaCl) and once in 10 mm Tris pH7.5, to remove remaining detergent. SDS-PAGE sample buffer was then added to the remaining beads prior to denaturing at 95 °C for 5 min and subsequent analysis by gel electrophoresis and Western blotting.
Flourescence Microscopy
Cells were transfected with EGFP tagged expression constructs as described for Co-IP studies above prior to fixation in 4% paraformaldehyde. Coverslips were then washed 3 × 5 min in PBS and rinsed in DH20, prior to being mounted onto a microscope slide facing downwards in Vectashield™ (Vector Laboratories, Peterborough, UK) containing DAPI for nuclear visualization.
GST Tagged Construct Generation, Production and Cleavage
GST conjugated E3-RING proteins for in vitro ubiquitination assays were prepared by inoculating 5 ml of 2XTY plus antibiotic with Rosetta™ 2(DE3) Singles™ Competent Cells (Invitrogren, UK) containing the required construct. Cultures were incubated at 37 °C overnight prior to addition of 45 ml of 2XTY broth plus antibiotic, supplemented with 100 μm ZnCl2 to aid correct folding of E3-RING domains. Cultures were then grown to an OD600 of 0.8–10 prior to addition of 100 μm IPTG and incubation for 16–18 h at 18 °C at 220 rpm. Cultures were then centrifuged at 4300rpm at 4 °C for 25 min before the pellet was resuspended in 1 ml sterile cold PBS and protease inhibitors were added (10 μl PMSF, 1 μm leupeptin hemisulphate, 1 μm pepstatin) prior to snap freezing in liquid nitrogen and over-night storage at −80 °C. Pellets were thawed on ice prior to addition of lysozyme to 1 mg/ml and incubation on ice for 30 min. Samples were then sonicated 3× 5s prior to addition of 1 μl DNAse1 and passage through a hypodermic needle to reduce viscosity. Samples were centrifuged at 33,000 rpm at 4 °C for 1 h prior to incubation with 50 μl glutathione cellulose beads for 2 h at 4 °C to bind the conjugated protein. Beads were then washed 3 × 1 ml in high salt buffer (NaCl 500 mm, Tris 25 mm pH 8.0) and 3 × 1 ml in high salt buffer (NaCl 150–200 mm, Tris 25 mm, pH 8.5) before addition of elution buffer (50 mm Tris, 50 mm NaF, 270 mm sucrose, 10 mm glycerol 2-phosphate disodium salt hydrate and 20 mm reduced glutathione, pH8.0). Samples were incubated at 50 rpm for 20 min at RT. Second and third eluates were collected by the same protocol. All elutions were analyzed using SDS-PAGE to estimate the protein concentration in comparison with a BSA standard and to verify the MW of protein product.
To produce cleaved E2 proteins for functional and biophysical assays, culture/inhibitor and bead volumes were all ratiometrically increased to give a 500 ml or 1 liter overnight culture prior to collection and freezing. Cells were then lysed using a French press prior to centrifugation at 4000 rpm for 5 min at 4 °C. Samples were incubated with 500 μl glutathione cellulose beads for 2 h at 4 °C to bind the conjugated protein. After binding beads were washed with 500 ml high and 500 ml low salt buffer prior to incubation with 5 ml low salt buffer plus 50/100 μl PreScission™ Protease (GElifesciences, UK) overnight at 4 °C. Cleaved protein was then eluted from the column prior to concentration using a Vivaspin 6 3kDa MWCO column (GE Healthcare) at 4,300rpm at 4 °C. Samples were snap frozen and stored at −80 °C until use.
In Vitro Ubiquitination Assays
His tagged E1-activating enzyme was from Boston BioChem (MA). Bovine ubiquitin and adenosine tri-phosphate (ATP) were from Sigma. Rabbit polyclonal anti-ubiquitin antibody was from Millipore (MA). Each reaction was undertaken as described previously (7).
Data Curation, Integration and Analysis
Protein interaction data was extracted from public databases HPRD, MINT, BioGrid and Intact as previously described (7). The E3-RINGs were also used to perform a further network analysis using the commercial MetaCore™ tool before exporting the interaction network into Cytoscape 2.6, integrating it with the public database sourced data sets and removing any redundant interactions. Molecular function enrichment was conducted using DAVID Functional annotation tool using the E3-RING ubiquitome as a background gene list and a p value cutoff of 0.05. Heatmaps were generated in R 2.10.1 using the heatmap.2 function. All basic interaction data analysis was performed using bespoke PERL programs generated using the PERL express text editor and run in PERL version 5.10.
E2 co-occurrence was calculated for E2s that contained ≥10 E3-RING interactions in the total E2/E3-RING data set (with the exception of UBE2V2 (Interactions = 8), which was included for comparison with the closely related UBE2V1). The total number of E3-RING subnetworks that both E2s were present in was divided by the total number of networks containing the E2 on the horizontal axis (i.e. its E3-RING degree).
The tissue expression data from 79 tissues was obtained and binarized at either a stringent cutoff used previously (250) (19), or a lower cutoff used as inferred expression in the original study (200) (20). This data was then mapped onto the node IDs represented in the physical interaction map to ascertain total coverage and co-expression.
Mass spectrometry identified ubiquitination sites (21–23) were converted from IPI accessions numbers to entrez gene IDs using the online tool bioDBnet, with only ubiquitination sites unambiguously assigned to a single gene ID taken forward for further analysis. Duplicated results were then removed taking the lower annotated site position to give a nonredundant list of annotated entrez gene IDs.
RESULTS
Construction, Benchmarking and Validation of a Binary E3-RING Dimerization Network
Systematic investigation of E3-RING dimers poses two main challenges. First, E3-RING dimers form by different modes of interaction (12, 17, 24–26), therefore the spread of binding affinities, or our ability to detect dimers, may be more variable than for E2/E3-RING interactions. Second, a high network density is required to reveal combinatorial patterns of homo and heterotypic interaction. To address these challenges we have applied the principle of “screen complementarity.” Three important studies (27–29) have demonstrated the fact that independent assay systems generate overlapping yet distinct data sets. With this principle in mind we reasoned that detection of E3-RING dimers could be enhanced by integrating data from two appropriately selected Y2H systems. Therefore, screen 1 was performed using the pACTBE(/D)-B medium expression prey vector (18) testing 10,049 potential interactions, whereas Screen 2 was performed using the low expression level pGAD prey vector (30) testing 4097 potential interactions. Initially a total of 477 interactions were detected in Screen 1, involving 72% of tested E3-RING clones. Fewer interactions (57) were detected in Screen 2, with only 41% of tested clones reporting positive hits (supplemental Fig. S1, supplemental Files S2 and S3). This raw data was then filtered to achieve a nonredundant set of 7269 tested interactions between 124 soluble E3-RINGs (supplemental Experimental Procedures). To combine interactions from Screens 1 and 2 into a format that reflects detection stringency, data was normalized for node promiscuity (supplemental Fig. S2). Data from Screen 2 and known literature derived interactions (LDIs) were used to establish a stringency filter for data derived from Screen 1. In this way network coverage was increased while eliminating a specific subset of interactions observed between promiscuous constructs that were only seen in Screen 1 (Supplemental Fig. S3). The resulting filtered data set contains 228 benchmarked dimeric interactions between 100 annotated E3-RINGs (Fig. 1A, supplemental File S4). A detailed description of the clone sets utilized and the Y2H experimental and analytical work-flow is provided in supplemental Experimental Procedures.
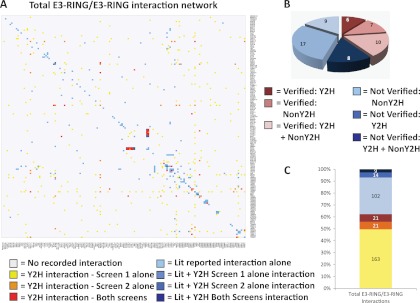
Fig. 1.
E3-RING/E3-RING network generation. A, Heatmap showing reported E3-RING/E3-RING interactions. Identical lists of E3-RINGs are ordered by sequence similarity on both axes. Colored boxes represent data source as indicated in the key. B, Reconfirmation rates for tested published interactions and the methodology used to observe them previously. C, Proportion of total interaction network derived from each data set described in (A), Lit + 1 Y2H screen data sets pooled into one group for bar chart.
To assess the efficacy of our benchmarking procedure and assign confidence levels to interactions observed in either or both screens, data from Screens 1 and 2 were analyzed to assess the proportion of known interactions detected in each data set. Of the 58 tested LDIs 40% were reconfirmed in our Y2H screens (Fig. 1B), indicating a comparable false negative rate to that reported in several previous publications (28, 29). Crucially, 14 of the 23 reconfirmed LDIs were only detected in one of the two screens (Fig. 1C). Also, seven of the LDIs tested in both screens were only detected in Screen 1, thus emphasizing the utility of dual screening methods for this particular class of binary interactions. Finally, a representative set of 32 positive (14% of total) and 10 negative Y2H interactions were retested in targeted Co-IP experiments using stringent assay conditions. The validation rate for interactions uniquely detected in Screen 2 was 67%. However, the verification rate for interactions detected in both Screens 1 and 2 was even higher at 78%, thereby demonstrating a very high correlation between primary Y2H data and our ability to detect the formation of corresponding E3-RING complexes in mammalian cells (Fig. 2 and supplemental File S4). Although the validation rate for interactions only detected in Screen 1 was lower (27%), 48% of reconfirmed LDIs were only detected in Screen 1. Therefore, it is highly likely that this data set contains a significant number of novel true-positive interactions that are not represented in data from Screen 2. Also, the degree of similarity observed between Y2H and Co-IP studies shows that ∼64% of tested interactions gave comparable results in both assays. This degree of similarity is consistent with trends observed in published comparisons between Y2H and Co-IP based methods (29).
Fig. 2.
E3-RING/E3-RING network validation. A, Classification of Y2H E3-RING/E3-RING interactions into low, medium or high stringency data sets. Percentage reconfirmation rates for tested Y2H interactions using Co-IP experiments are shown for each data set. B, Examples of Co-IP experiments. Positive and negative Y2H results are represented below pull down lanes. Dashed circles indicate reconfirmation of Y2H data.
LDIs not tested/observed in this study were integrated with our Y2H data to generate a final human E3-RING/E3-RING interaction network containing 59 homo- and 270 heterotypic dimers between 138 E3-RING proteins, of which 205 interactions are novel (Figs. 1A and 1C). Results from this study highlight the potential advantages of rationally applying the principle of “screen complementarity” to enhance network coverage and the selective yet widespread occurrence of homo- and heterotypic E3-RING complexes. The protein interactions from this publication have been submitted to the IMEx (http://www.imexconsortium.org/) consortium through IntAct (pmid: 17145710) and assigned the identifier IM-15696.
Constructing an Extended E3-RING Centric Network
To provide new insight into the spectrum of physiological processes that may be affected by E3-RING oligomerization events it was necessary to assemble an updated ubiquitome network, which enables the hierarchical relationship between different E2/E3-RING complexes and potential substrate proteins to be investigated. A list of 309 high confidence human E3-RING genes was generated from two previous Y2H studies (7, 8) and a bioinformatic analyses of annotated human E2 and E3-RING genes (4) (supplemental File S1). All available human E2/E3-RING interaction data was then extracted from literature and public databases to generate a combined high-density E2/E3-RING network containing 233 nodes and 966 binary interactions (supplemental File S5). This data not only re-emphasizes the combinatorial complexity that exists at this level of the human ubiquitome, but also reveals recurrent patterns of selective E2/E3-RING interaction; whereas global analysis shows that partner preference is not simply conferred by primary sequence similarity, subfamilies of E2 proteins do tend to have common E3-RING partners (supplemental Fig. S5A and B). This trend is most striking between members of the UBE2E family and UBE2W; all of which bind almost exclusively to a subset of E3-RINGs that also interact with members of the UBE2D family. Interestingly, both UBE2W and members of the UBE2D family are both known to mono-ubiquitinate target proteins in a priming reaction, prior to chain elongation by other E2 proteins, thus suggesting a potential for overlapping functionality (13, 31, 32). A similar binding pattern was also observed with the UBE2V inactive E2-like variants, as UBE2V2 interacts with a subset of E3-RINGs that also bind to UBE2V1, whereas both occur in a subset of D family binding E3-RINGs. Interestingly, the inactive TSG101 variant shows a completely distinct E3-RING binding profile in concordance with its divergent functions from other E2 variants.
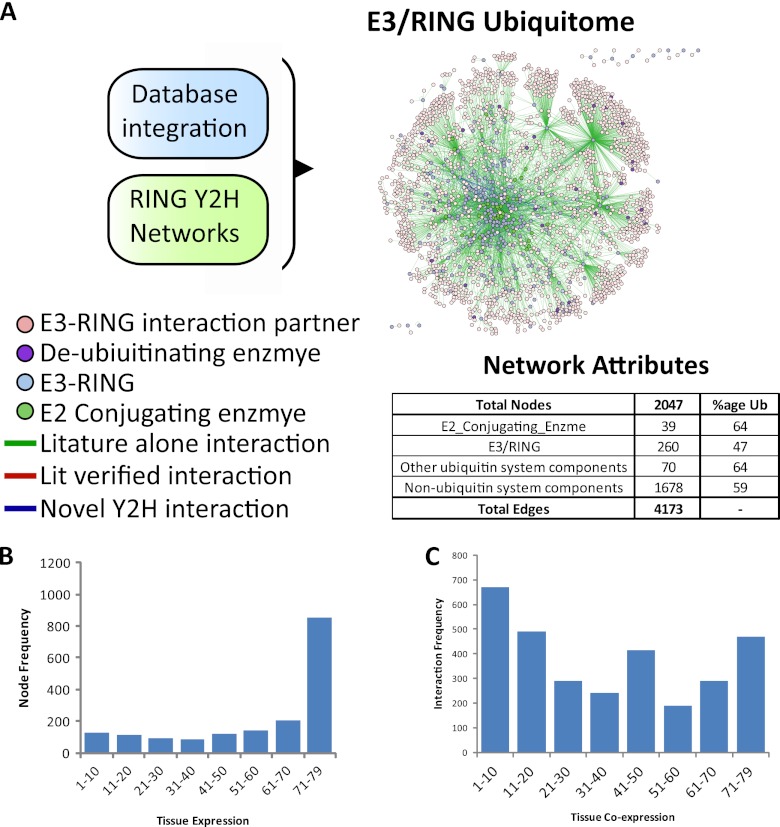
As E3-RING proteins mediate both recruitment of E2 proteins and substrate recognition, it is possible that a one-step extended E3-RING network may reveal subnetworks in which E3-RING dimerization events have the potential to change “information-flow” within human ubiquitination cascades. In addition, network structure and functional hierarchy may be dictated by cell-type specific expression of different network components. To investigate these possibilities data from public databases was combined with an extended network analysis in MetaCore™ (http://www.genego.com/metacore.php) prior to integration with novel E3-RING/E3-RING interactions observed in this study, in order to generate a final nonredundant human E3-RING centric network containing 4173 interactions involving 2047 different proteins (Fig. 3A, supplemental File S6).
Fig. 3.
E3-RING ubiquitome construction. A, Interaction dataset integration from curated databases or Y2H datasets. Entire network visualized in Cytoscape v2.6, proteins represented by colored nodes and physical interactions between them by connecting lines. Inset; a tabular breakdown of components of the E3-RING based ubiquitome and the percent that are ubiquitinated. B, Frequency of node expression across 79 tissues. C, Frequency of interactions between co-expressed nodes.
To investigate the potential for cell-type specific interactions this network was then integrated with gene expression data from 79 differing tissues (20). A previously established stringent cutoff of 250 (19) was utilized to define the presence or absence of protein expression. Interestingly, almost half (49%) of mapped nodes were expressed in almost all (≥71/79) tissues (Fig. 3B). However, interactions between these proteins account for only 15% (468) of total interactions within the E3-RING centric network (Fig. 3C). Therefore, although a core set of common interactions may drive fundamental ubiquitination processes in all cells, the majority of interactions appear to be cell-type specific. This trend is also highlighted by the fact that 38% (1162) of interactions involve nodes that are co-expressed in ≤20/79 tissues (Fig. 3C). Significantly, this trend persists when a lower cut-off value of 200 is used to infer protein expression patterns (Supplemental Fig. S6A, B, and C; supplemental File S7). It is therefore possible that cell-type specific variations in combinatorial preference may alter both network structure and function. Interestingly, transcriptional control processes are significantly enriched within the core co-expression network, suggesting that highly conserved ubiquitination cascades may regulate transcriptional networks in many differing cell types (supplemental File S7).
Finally, the E3-RING ubiquitome was integrated with three recent large-scale mass spectrometry studies that identify ubiquitin (or NEDD8) modification through a di-gly post tryptic signature (21–23). Together these studies identified 22,468 unique sites across 5812 Entrez gene IDs (supplemental File S8). Significantly, 1177 ubiquitination targets are represented within our E3-RING ubiquitome with ∼60% of E3-RING binding proteins (not including other ubiquitination related proteins) containing ubiquitinated residues (Table inset in Fig. 3A). Ubiquitinated proteins within this network show a wide range in their frequency of modified sites that is independent of the number of known E3-RING binding partners (supplemental Fig. S6D and E).
Together these data provide the first large-scale human E3-RING interaction network, which can be used to predict partner preferences within different ubiquitination cascades. Having generated this extended E3-RING centric network we were particularly interested to identify modules or subnetworks that contain examples of (1) non-E2 binding E3-RING proteins; (2) common nonubiquitin cascade interaction partners, (3) both homo- and heterotypic E3-RING interactions, or (4) heterodimers in which individual E3-RING proteins form active complexes with different sets of E2 proteins.
Subnetworks that Contain a Non-E2-Binding E3-RING Protein
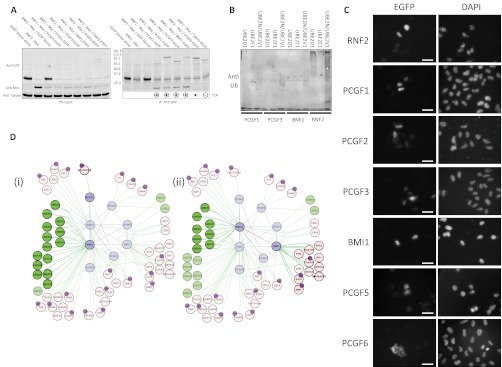
Despite multiple systematic screens and large numbers of focused biochemical studies, there is still a subset of E3-RING proteins that do not have any known E2 partners (9). Although in some cases this trend may reflect technical bias in available datasets, there is now convincing evidence for heteromeric E3-RING complexes (BRCA1:BARD1 and RNF2:BMI1) where only one of the E3-RING partners is functional with E2-proteins. It is highly probable that further examples of this form of “facilitated” E2/E3-RING interaction may occur within the human ubiquitome. In our studies, members of the PCGF subfamily were each found to interact with the closely related RING1 and RNF2 proteins in both Y2H and Co-IP experiments (Figs. 2B and 4A). Although some of these interactions have been observed in co-complex studies (33), this is the first evidence of specific binary interaction preferences within this subnetwork of functionally related proteins. Furthermore, all RNF2 Co-IP positive interaction partners were also found to localize to the nucleus (Fig. 4C). Auto-ubiquitination assays of four subnetwork components show that within these E3-RING pairs only RNF2 is capable of independently functioning with selected E2 proteins in vitro (Fig. 4B). Thus supporting the hypothesis that facilitated recruitment of E2s is necessary for members of the PCGF family to function as active dimeric ligases.
Fig. 4.
Verification of RNF2 and RING1 facilitated E2-recruitment subnetworks. A, Co-IP experiments were performed using myc-RNF2 and EGFP-tagged partners to test a selection of positive or negative Y2H interactions. Corresponding Y2H data is shown below Western blots. Circled symbols indicate reconfirmation of Y2H data. B, In vitro ubiquitination assay performed with RNF2 or selected PCGF family members in combination with specific E2s. A high MW black smear is indicative of in vitro ubiquitination activity. C, Flourescence microscopy of EGFP tagged RNF2 and PCGF family members in HeLa cells. White scale bar represents 50 μm. D, Integration of RNF2 and RING1 interaction subnetworks. (1) Subnetwork in which RNF2 would facilitate recruitment of specific E2 enzymes to PCGF1 substrates. (2) Subnetwork in which RING1 would facilitate recruitment of different E2 enzymes to alternate PCGF binding proteins. For simplicity, in network diagrams only edges relating to a subset of E3-RING dimerization events, E2 recruitment, and direct RING binding partners are shown. Ubiquintated noncascade proteins identified by mass spectroscopy are annotated.
RING1 and RNF2 both regulate epigenetic control through their involvement in the PRC1 complex, which represses transcription through mono-ubiquitination of histone H2A (33). This activity is significantly enhanced by BMI1, PCGF1, and PCGF2 in vivo (12, 34, 35), thus providing evidence for a functional relationship between select E3-RING complexes identified in this study. Furthermore, a recent report highlights the overlapping yet distinct functions of PCGF family members, showing that although PCGF3 and BMI1 are recruited to sites of DNA damage together with RNF2 but not RING1, they show differential dynamics of site occupancy (36). Analysis of the PCGF subnetwork shows that RNF2 has the potential to mediate a wide range of ubiquitination reactions through selective interaction with different members of the PCGF family (Fig. 4D1). Also, heterotypic interaction with RING1 would alter the range of E2 proteins that are recruited to PCGF specific substrates (Fig. 4D2). Thus, our data supports a model in which different E3-RING heterodimers could mediate modification of either the same (histone H2A) or distinct (cyclin D2) E3-RING specific substrates, thereby imposing different functional consequences.
Subnetworks of E3-RING Hetero-dimers That Share Common Nonubiquitin Cascade Partners
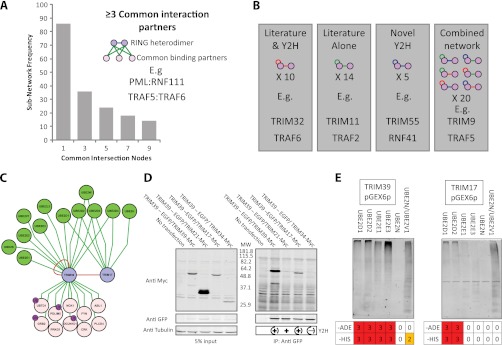
To further investigate combinatorial subnetworks involving heteromeric E3-RING complexes a “common intersection” analysis was performed to identify specific subnetworks in which two interacting E3-RING proteins share common non-E2 or E3-RING partners (Fig. 5A and supplementary File S6). From this analysis 36 E3-RING heterodimers were identified that share ≥3 common partners, including well studied examples such as RNF2:BMI1, BRCA1:BARD1, and MDM2:MDM4. However, the vast majority of subnetworks from this study were functionally uncharacterized and provide a resource for future experimental studies. For example, the novel TRAF5:TRAF6 and PML:RNF111 complexes identified in this study are implicated in signal transduction control and SMAD signaling respectively (For further description of these interactions see supplemental Experimental Procedures and supplemental Fig. S7).
Fig. 5.
E3-RING/E3-RING Subnetwork analysis. A, Graphical distribution of E3-RING heterodimeric interaction pairs that share common non-Ub related interaction partners. B, Number of RING proteins observed to homodimerize that also report heterodimerization in either literature databases, the Y2H network, or a combination of both. Color scheme of edges as in interaction network diagrams, below each graphic are examples of proteins shown to both homo- and heterodimerize in each data set. C, TRIM39:TRIM17 homo- and heterodimeric interaction subnetwork. For simplicity, in network diagrams only edges relating to E3-RING dimerization, E2 recruitment and common substrates are shown. Ubiquintated noncascade proteins identified by mass spectroscopy are annotated. D, Co-IP validation of Y2H interactions. Dashed circles in bottom right hand panel represent verification of both positive and negative Y2H data. E, Correlation of TRIM39 and TRIM17 in vitro ubiquitination activity with Y2H data obtained with these clones in our laboratory previously, number represents strength of interaction on growth selective agar. A high MW black smear is indicative of in vitro ubiquitination activity.
Homo- or Heterotypic E3-RING Interactions
As many E3-RING proteins exhibit both homo- and heterotypic interactions there is a strong potential for conditional changes in stiochiometry within multimeric E3-RING complexes (Fig. 1A). The Y2H network generated in this study verifies, or contributes to, 35 of the 49 known homo- and heterodimerising subnetworks (Fig. 5B). To test the hypothesis that nonproteolytic dimerization events may be prevalent among phylogenetically related E3-RING proteins, subnetworks were generated for the 19 E3-RINGs that form both homo- and heterodimers in our study. Further functional validation was then undertaken to investigate two subnetworks within the TRIM subfamily of E3-RINGs. TRIM39 and TRIM5 homo- and heterotypic interactions were verified by stringent Co-IP assays whereas no interactions could be detected between pairings not detected in our Y2H studies (Figs. 5C and 5D and supplemental Figs. 8A and C). No reduction in protein stability was observed during co-expression with interacting E3-RING proteins (TRIM5:TRIM32 or TRIM39:TRIM17 complexes), implying that the observed interactions represent stable multimeric complexes rather than E3-RING/substrate interactions. Interestingly, components of these heterodimeric pairs have also independently been implicated in common cellular functions; TRIM17 and TRIM39 in apoptosis (37, 38), TRIM5 and TRIM32 in the anti-HIV response (39–42), re-enforcing the potential physiological significance of these and other novel interactions observed in this study.
A variety of E3-RINGs are known to assemble into large homo-multimers, which have been shown to enhance ubiquitination activity (15, 17). In contrast to previously characterized heteromeric E3-RING complexes, where only one of the E3-RING proteins has been shown to function with E2 proteins, our verified TRIM protein subnetworks provides the first examples of stable E3-RING heteromeric complexes in which all interaction partners can independently bind E2 proteins to form functional ubiquitination complexes (Fig. 5E and supplemental Fig. S8B). Furthermore our data show that TRIM39 and TRIM17 function efficiently with an overlapping, but distinct, set of E2 proteins. As such, it is possible that a process of selective or conditional heterodimerization may control either the affinity of substrate binding or recruitment of different E2 proteins, in order to regulate the architecture or target of ubiquitination (Fig. 5C). It is highly likely that this kind of conditional subnetwork is a recurrent theme within human ubiquitination cascades as many more examples can be found within our extended E3-RING centric network (supplemental Fig. S9).
DISCUSSION
An emerging principle in network biology is the fact that different interaction techniques generate overlapping yet complementary data (29). Interestingly, this is equally true for results derived from different variants of the classic Y2H system (27). As a result, it is now clear that node coverage and network density can be significantly increased by the integration of data from complementary, or even parallel, studies performed in different vector/host-strain combinations. These observations are particularly pertinent for the analysis of focused biological networks in which combinatorial interactions have the potential to confer variations in network structure, information flow, and physiological consequence. In such cases, partner specificity and combinatorial preferences have been effectively revealed by systematic screens, which provide functionally unbiased high-density network coverage. In this study these principles have been used to generate a novel human E3-RING network that reveals a widespread potential for the formation of selective homo- and heteromeric E3-RING complexes. In addition, complementary Co-IP studies were performed to assign confidence levels to novel Y2H data and to verify predicted interactions that have the potential to confer differential, or conditional, information flow within human ubiquitination networks. By integrating this data into a network of all known E3-RING interactions it was possible to generate an E3-RING centric network, which provides a resource that can be used to inform hypothesis driven studies into a diverse range of E3-RING mediated ubiquitination events. Finally, we have analyzed this network to extract biologically interesting subnetworks and generated functional hypothesis that are supported by both literature and experimental observations.
The canonical E3-RING mediated ubiquitin cascade is often depicted as a simple linear process in which ubiquitin is transferred from an E1-activating enzyme, via an E2/E3-RING complex to a selective target. However, homo- and heterotypic E3-RING interactions within the human ubiquitome have the potential to confer an important level of combinatorial complexity within human ubiquitination cascades, a trend that is particularly prevalent among the TRIM, TRAF and PCGF subfamilies. Phylogenetically distinct E3-RINGs have been shown to form higher order structures, which have enhanced catalytic activity or altered partner preferences (15–17, 33, 43). Given the large numbers of heterotypic E3-RING interactions detected in this study it is possible that formation of multimeric E3-RING complexes may provide a general mechanism of controlling information flow within human ubiquitination cascades through differential E2 or substrate recruitment, thereby mediating different functional outcomes in a conditional manner. In support of this hypothesis, we observed ubiquitination activity for both heterodimeric E3-RING partners that were shown to form stable complexes in human cells. It seems reasonable to expect that some of the E3-RING pairs observed in our study may actually represent E3-RING/substrate interactions (9), which in vivo may drive the modification or degradation of one or both partners. Although our co-expression studies show no signs of partner mediated degradation, previous studies have shown that this can occur (44), we therefore expect that both forms of E3-RING dimerization will be represented and functionally important within the human ubiquitome.
As protein ubiquitination is implicated in a broad range of biological and pathological processes it is essential that we develop a better understanding of the multiple levels of combinatorial control that exist within this important system of post-translational regulation. The high-density binary E3-RING network generated in this study provides a new resource for studying the molecular basis of selective E3-RING interactions and the investigation of combinatorial modules that regulate specific physiological processes in different human cell types.
Acknowledgments
We thank Anja Bremm and David Kommander (LMB, Cambridge, UK) for their assistance with in vitro ubiquitination assays. We are also grateful to Kourosh Salehi-Ashtiani and Mark Vidal (Harvard, USA) for providing a proportion of E3-RING ORF Clones that were used in this and previous studies and to Josephine Worseck for critical reading of the manuscript.
Footnotes
* All research performed in this study was funded by the Wellcome Trust (GRANT Codes: 080911/2/06/2 and 083847/2/07/2).
 This article contains supplemental Experimental Procedures, Files S1 to S8 and Figs. S1 to S11.
This article contains supplemental Experimental Procedures, Files S1 to S8 and Figs. S1 to S11.
Author contributions: CS supervised all work described in this study. Experiments, image generation and computational analyses, with the exception of in vitro ubiquitination assays, were carried out by JW. In vitro ubiquitination assays were carried out by JW and RJ. JW and CS wrote the manuscript.
Conflict of Interest: We declare that they have no conflict of interest.
The protein interactions from this publication have been submitted to the IMEx (http://www.imexconsortium.org/) consortium through IntAct (pmid: 17145710) and assigned the identifier IM-15696.
1 The abbreviations used are:
- Y2H
- yeast two hybrid
- Co-IP
- Co-immuno precipitation
- LDI
- Literature Derived Interaction.
REFERENCES
- 1. Pickart C. M., Fushman D. (2004) Polyubiquitin chains: polymeric protein signals. Current Opinion Chem. Biol. 8, 610–616 [DOI] [PubMed] [Google Scholar]
- 2. Deshaies R. J., Joazeiro C. A. P. (2009) RING Domain E3 Ubiquitin Ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 3. Petroski M. D. (2008) The ubiquitin system, disease, and drug discovery. BMC Biochemistry 9, S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li W., Bengtson M. H., Ulbrich A., Matsuda A., Reddy V. A., Orth A., Chanda S. K., Batalov S., Joazeiro C. A. P. (2008) Genome-Wide and Functional Annotation of Human E3 Ubiquitin Ligases Identifies MULAN, a Mitochondrial E3 that Regulates the Organelle's Dynamics and Signaling. PLoS ONE 3, e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu J. Y., Lin Y. Y., Qian J., Tao S. C., Zhu J., Pickart C., Zhu H. (2008) Functional dissection of a HECT ubiquitin E3 ligase. Mol. Cell. Proteomics 7, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Petroski M. D., Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 7. Markson G., Kiel C., Hyde R., Brown S., Charalabous P., Bremm A., Semple J., Woodsmith J., Duley S., Salehi-Ashtiani K., Vidal M., Komander D., Serrano L., Lehner P., Sanderson C. M. (2009) Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome Res. 19, 1905–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van Wijk S. J. L., de Vries S. J., Kemmeren P., Huang A., Boelens R., Bonvin A. M. J. J., Timmers H. T. M. (2009) A comprehensive framework of E2-RING E3 interactions of the human ubiquitin-proteasome system. Mol. Syst. Biol. 5, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Bie P., Ciechanover A. (2011) Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ 18, 1393–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Budhidarmo R., Nakatani Y., Day C. L. (2012) RINGs hold the key to ubiquitin transfer. Trends in Biochemical Sciences 37, 58–65 [DOI] [PubMed] [Google Scholar]
- 11. Plechanovová A., Jaffray E. G., McMahon S. A., Johnson K. A., Navrátilova I., Naismith J. H., Hay R. T. (2011) Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat. Struct. Mol. Biol. 18, 1052–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buchwald G., Stoop P. v. d., Weichenrieder O., Perrakis A., Lohuizen M. v., Sixma T. K. (2006) Structure and E3-ligase activity of the Ring–Ring complex of Polycomb proteins Bmi1 and Ring1b. EMBO J. 25, 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Christensen D. E., Brzovic P. S., Klevit R. E. (2007) E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 14, 941–948 [DOI] [PubMed] [Google Scholar]
- 14. Hashizume R., Fukuda M., Maeda I., Nishikawa H., Oyake D., Yabuki Y., Ogata H., Ohta T. (2001) The RING Heterodimer BRCA1-BARD1 Is a Ubiquitin Ligase Inactivated by a Breast Cancer-derived Mutation. J. Biol. Chem. 276, 14537–14540 [DOI] [PubMed] [Google Scholar]
- 15. Kentsis A., Gordon R. E., Borden K. L. B. (2002) Control of biochemical reactions through supramolecular RING domain self-assembly. Proc. Natl. Acad. Sci. U. S. A. 99, 15404–15409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poyurovsky M. V., Priest C., Kentsis A., Borden K. L., Pan Z. Q., Pavletich N., Prives C. (2007) The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 26, 90–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., Guffanti A., Minucci S., Pelicci P. G., Ballabio A. (2001) The tripartite motif family identifies cell compartments. EMBO J. 20, 2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Semple J., Prime G., Wallis L., Sanderson C., Markie D. (2005) Two-hybrid reporter vectors for gap repair cloning. BioTechniques 38, 927–934 [DOI] [PubMed] [Google Scholar]
- 19. Bossi A., Lehner B. (2009) Tissue specificity and the human protein interaction network. Mol. Syst. Biol. 5, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Cooke M. P., Walker J. R., Hogenesch J. B. (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U. S. A. 101, 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., Choudhary C. (2011) A Proteome-wide, Quantitative Survey of In Vivo Ubiquitylation Sites Reveals Widespread Regulatory Roles. Mol. Cell. Proteomics 10, M111.013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., Gygi S. P. (2011) Systematic and Quantitative Assessment of the Ubiquitin-Modified Proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Danielsen J. M., Sylvestersen K. B., Bekker-Jensen S., Szklarczyk D., Poulsen J. W., Horn H., Jensen L. J., Mailand N., Nielsen M. L. (2011) Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteomics 10, M110.003590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yin Q., Lin S. C., Lamothe B., Lu M., Lo Y. C., Hura G., Zheng L., Rich R. L., Campos A. D., Myszka D. G., Lenardo M. J., Darnay B. G., Wu H. (2009) E2 interaction and dimerization in the crystal structure of TRAF6. Nat. Struct. Mol. Biol. 16, 658–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsuzawa S. I., Li C., Ni C. Z., Takayama S., Reed J. C., Ely K. R. (2003) Structural Analysis of Siah1 and Its Interactions with Siah-interacting Protein (SIP). J. Biol. Chem. 278, 1837–1840 [DOI] [PubMed] [Google Scholar]
- 26. Brzovic P., Keeffe J., Nishikawa H., Miyamoto K., Fox D., 3rd, Fukuda M., Ohta T., Klevit R. (2003) Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc. Natl. Acad. Sci. U.S.A. 100, 5646–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen Y. C., Rajagopala S. V., Stellberger T., Uetz P. (2010) Exhaustive benchmarking of the yeast two-hybrid system. Nat. Methods 7, 667–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rajagopala S. V., Hughes K. T., Uetz P. (2009) Benchmarking yeast two-hybrid systems using the interactions of bacterial motility proteins. Proteomics 9, 5296–5302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Braun P., Tasan M., Dreze M., Barrios-Rodiles M., Lemmens I., Yu H., Sahalie J. M., Murray R. R., Roncari L., de Smet A.-S., Venkatesan K., Rual J.-F., Vandenhaute J., Cusick M. E., Pawson T., Hill D. E., Tavernier J., Wrana J. L., Roth F. P., Vidal M. (2009) An experimentally derived confidence score for binary protein-protein interactions. Nat. Methods 6, 91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beningo K. A., Lillie S. H., Brown S. S. (2000) The Yeast Kinesin-related Protein Smy1p Exerts Its Effects on the Class V Myosin Myo2p via a Physical Interaction. Mol. Biol. Cell 11, 691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu K., Kovacev J., Pan Z. Q. (2009) Priming and Extending: A UbcH5/Cdc34 E2 Handoff Mechanism for Polyubiquitination on a SCF Substrate. Mol. Cell 37, 784–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garnett M. J., Mansfeld J., Godwin C., Matsusaka T., Wu J., Russell P., Pines J., Venkitaraman A. R. (2009) UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat. Cell Biol. 11, 1363–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vidal M. (2009) Role of polycomb proteins Ring1A and Ring1B in the epigenetic regulation of gene expression Int. J. Develo. Biol. 53, 355–370 [DOI] [PubMed] [Google Scholar]
- 34. Cao R., Tsukada Y. I., Zhang Y. (2005) Role of Bmi-1 and Ring1A in H2A Ubiquitylation and Hox Gene Silencing. Mol. Cell 20, 845–848 [DOI] [PubMed] [Google Scholar]
- 35. Sanchez C., Sanchez I., Demmers J. A. A., Rodriguez P., Strouboulis J., Vidal M. (2007) Proteomics analysis of ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol. Proteomics 6, 820–834 [DOI] [PubMed] [Google Scholar]
- 36. Pan M. R., Peng G., Hung W. C., Lin S. Y. (2011) Monoubiquitination of H2AX protein regulates DNA damage response signaling. J. Biol. Chem. 286, 28599–28607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lassot I., Robbins I., Kristiansen M., Rahmeh R., Jaudon F., Magiera M. M., Mora S., Vanhille L., Lipkin A., Pettmann B., Ham J., Desagher S. (2010) Trim17, a novel E3 ubiquitin-ligase, initiates neuronal apoptosis. Cell Death Differ. 17, 1928–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee S. S., Fu N. Y., Sukumaran S. K., Wan K. F., Wan Q., Yu V. C. (2009) TRIM39 is a MOAP-1-binding protein that stabilizes MOAP-1 through inhibition of its poly-ubiquitination process. Exp. Cell Res. 315, 1313–1325 [DOI] [PubMed] [Google Scholar]
- 39. Fridell R. A., Harding L. S., Bogerd H. P., Cullen B. R. (1995) Identification of a Novel Human Zinc Finger Protein That Specifically Interacts with the Activation Domain of Lentiviral Tat Proteins. Virology 209, 347–357 [DOI] [PubMed] [Google Scholar]
- 40. Hatziioannou T., Perez-Caballero D., Yang A., Cowan S., Bieniasz P. D. (2004) Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. U. S. A. 101, 10774–10779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Keckesova Z., Ylinen L. M., Towers G. J. (2004) The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. U. S. A. 101, 10780–10785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yap M. W., Nisole S., Lynch C., Stoye J. P. (2004) Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. U. S. A. 101, 10786–10791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zheng C., Kabaleeswaran V., Wang Y., Cheng G., Wu H. (2010) Crystal Structures of the TRAF2: cIAP2 and the TRAF1: TRAF2: cIAP2 Complexes: Affinity, Specificity, and Regulation. Mol. Cell 38, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Depaux A., Regnier-Ricard F., Germani A., Varin-Blank N. (2006) Dimerization of hSiah proteins regulates their stability. Biochem. Biophys. Res. Commun. 348, 857–863 [DOI] [PubMed] [Google Scholar]