Abstract
Phagosomes, by killing and degrading pathogens for antigen presentation, are organelles implicated in key aspects of innate and adaptive immunity. Although it has been well established that phagosomes consist of membranes from the plasma membrane, endosomes, and lysosomes, the notion that the endoplasmic reticulum (ER) membrane could play an important role in the formation of the phagosome is debated. However, a method to accurately estimate the contribution of potential source organelles and contaminants to the phagosome proteome has been lacking. Herein, we have developed a proteomic approach for objectively quantifying the contribution of various organelles to the early and late phagosomes by comparing these fractions to their total membrane and postnuclear supernatant of origin in the J774A.1 murine macrophage cell line. Using quantitative label-free mass spectrometry, the abundance of peptides corresponding to hundreds of proteins was estimated and attributed to one of five organelles (e.g. plasma membrane, endosomes/lysosomes, ER, Golgi, and mitochondria). These data in combination with a stable isotope labeling in cell culture method designed to detect potential contaminant sources revealed that the ER is part of the phagosomal membrane and contributes ∼20% of the early phagosome proteome. In addition, only a subset of ER proteins is recruited to the phagosome, suggesting that a specific subdomain(s) of the ER might be involved in phagocytosis. Western blotting and immunofluorescence substantially validated this conclusion; we were able to demonstrate that the fraction of the ER in which the ER marker GFP-KDEL accumulates is excluded from the phagosomes, whereas that containing the mVenus-Syntaxin 18 is recruited. These results highlight promising new avenues for the description of the pathogenic mechanisms used by Leishmania, Brucella, and Legionella spp., which thrive in ER-rich phagosomes.
Phagocytosis is the process that enables some cells, particularly professional phagocytes such as macrophage and dendritic cells, to engulf large particles (>0.5 μm). It can be triggered both by opsonin (e.g. Fc gamma Receptor II/III and Complement Receptor 3) and some pathogen-associated molecular pattern receptors (e.g. dectin-1). The organelle that is formed around the internalized particle is called a phagosome. One of its core functions is to link the destruction of pathogens with the processing of pathogen-derived antigens for presentation on major histocompatibility molecules class I and II to initiate an adaptive immune response. The phagosome being highly dynamic in nature, the description of the discrete steps toward maturation into phagolysosome is still actively pursued.
The notion inferred from the pioneering work of Elie Metchnikoff that phagosomes are made by the invagination of the cell surface has been extended in the last decade. Indeed, multiple endomembrane pools are now thought to be harnessed during phagosome formation and maturation (1). Briefly, activation of phagocytic receptors triggers the remodeling of the actin cytoskeleton, which forces the plasma membrane (PM)1 to enclose the external body. Thus, the PM clearly constitutes an important fraction of the nascent phagosome membrane (2). Nevertheless, although fractions of the PM are displaced or recycled (3), there is evidence that endomembranes of various origins are recruited to the nascent phagosomes. Indeed, it was shown that recycling endosomes are able to fuse at the phagocytic cup through the action of the soluble N-ethylmaleimide-associated receptor (SNARE) VAMP-3 (4). Similarly, fusion of late endosomes mediated by VAMP-7 was also suggested to be essential for optimal Fc and complement-mediated phagocytosis (5). These examples of focal exocytosis of endomembranes are in agreement with the absence of net reduction or even the extension of cell surface area that is observed during phagocytosis (6, 7). The maturation of the phagosome is further achieved by sequential fusion events with early endosomes, late endosomes, and lysosomes (1, 8). These sequential fusion events are likely triggered by a switch from Rab5 to Rab7, as shown for endosomes (9).
Early proteomics studies indicated that the endoplasmic reticulum (ER) might contribute to phagosome genesis as well (10). In our follow-up work, we provided compelling evidence of ER recruitment to phagosome by biochemical and morphological approaches (11). Furthermore, interaction between the ER and phagosomes led to the proposal that this process might favor the presentation of phagocytosed antigens on major histocompatibility molecule class I molecules, a phenomenon referred to as antigen cross-presentation, in macrophage and dendritic cells (12–14). In addition, phagosomes isolated from interferon-γ-treated macrophages were shown to display an increased level of ER resident proteins specifically implicated in cross-presentation (e.g. TAP1/2 and tapasin) and to mature at a lower rate toward phagolysosomes (15). Recently, a phylogenetic study based on large scale proteomics analyses performed on phagosomes isolated from various organisms has shown that the recruitment of the ER to phagosomes observed as early as in Amoeba is likely to have conferred novel functional properties to this organelle, including antigen cross-presentation, in animals displaying an acquired immune system (16).
Two ER resident soluble N-ethylmaleimide-associated receptors (SNARE), Sec22b and Syntaxin (Stx) 18, have been implicated in ER-PM/phagosome fusion (17–19). These two proteins have been shown to form a SNARE complex with Use1/D12 and BNIP1 and are best known for their participation in Golgi-ER retrograde transport (20, 21). However, Grinstein and co-workers (22) used selected heterologous ER markers such as GFP-KDEL and calnexin (Cnx)-GFP in real time fluorescence microscopy to provide evidence that the PM was the principal source of phagosome membrane, whereas the contribution of ER, if any, was minimal.
The relative contribution of the various organelles to the phagosome is still poorly characterized. Herein, we developed a large scale comparative proteomics approach to determine the contribution to the phagosome proteome of proteins annotated to five organelles. Remarkably, this approach highlighted the relative contribution of the cell membrane reservoirs and demonstrated that only a subset of the ER contributes to phagosome biogenesis. Interestingly, we demonstrate that GFP-KDEL is not found in the fraction of the ER contributing to phagocytosis in contrast with several endogenous ER markers.
EXPERIMENTAL PROCEDURES
Cell Lines and Plasmids
J774A.1 and RAW264.7 macrophage cell lines were grown in DMEM containing 10% heat-inactivated fetal bovine serum supplemented with glutamine and penicillin/streptomycin (Wisent, St. Bruno, Canada). J774A.1 and RAW264.7 stably expressing mVenus or mVenus-Stx18 and GFP-KDEL, respectively, were grown in the conditions previously reported (18, 22). GFP-KDEL-expressing plasmids and the derived RAW264.7 stable cell line were a kind gift from Dr. Sergio Grinstein.
Phagosomes Isolation by Floatation on Sucrose Gradient
Polystyrene bead (PB)-phagosomes were isolated from J774A.1 cells by floatation on sucrose gradient as described previously (23). Polystyrene microspheres of 0.79 μm (K080) were used (Merck-Estapor, Val de Fontenay, France). Internalization and chase were performed in phagocytosis medium (RPMI1640 supplemented with 25 mm HEPES, penicillin/streptomycin and glutamine; all of the components were obtained from Wisent). Lysis and gradient solutions were buffered with HEPES and contained 5 mm MgCl2. Phagosomes formed by internalization of PB for 15 min and chase for 0 or 45 min were obtained to monitor maturation. Postnuclear supernatant (PNS) and total membrane (TM) fractions were also obtained for the MS analysis and validation. All of the cell fractions were done in triplicate.
Expression Analyses Using SDS-PAGE and Mass Spectrometry
20 μg of biological triplicates of phagosomal/TM/PNS proteins were reduced with tris(2-carboxyethyl)phosphine (Pierce), alkylated by iodoacetamide (Sigma-Aldrich), and separated on a 4–12% precast NuPAGE gel (Invitrogen). The gel was Coomassie-stained, and the lanes were cut into 12 equal-sized pieces using an in-house cutting device. The gel pieces were digested by trypsin, and peptides were extracted three times with 90% ACN, 0.5 m urea. The combined extracts were dried and resuspended in 5% ACN, 0.1% TFA for mass spectrometry analyses.
Label-free Mass Spectrometry and Bioinformatics
Mass spectrometric analyses were performed as described in Ref. 15. The peptides were separated on a 150-μm-inner diameter, 10-cm reversed phase nano-LC column (Jupiter C18, 3 μm, 300 Å; Phenomex) with a loading buffer of 0.2% formic acid. Peptide elution was achieved by a gradient of 5–40% ACN in 85 min on an Eksigent 2D-nanoLC (Dublin, CA) operating at a flow rate of 600 nl/min. The nano-LC was coupled to an LTQ-Orbitrap mass spectrometer (Thermo-Electron, Bremen, Germany), and samples were injected in an interleaved manner. The mass spectrometer was operated in a data-dependent acquisition mode with a 1-s survey scan at a resolution of 60,000, followed by three product ion scans (MS/MS) of the most abundant precursors above a threshold of 10,000 counts in the LTQ part of the instrument.
Protein Identification
Peak detection of raw MS2 spectra was performed using Mascot Distiller v2.1.1 (Matrix Science) using the default LCQ and zoom scan parameters. The centroided data was merged into single peak list files and searched with the Mascot search engine v2.10 (Matrix Science) against the combined forward and reversed mouse International Protein Index database v3.37 containing 51,291 forward protein sequences. Search conditions included trypsin set as enzyme, one missed cleavage site, carbamidomethylation (Cys) as a fixed modification, and deamidation (Asn and Gln), oxidation (Met), and phosphorylation (Ser, Thr, and Tyr) as variable modifications. Precursor and fragment ion tolerances were set to 0.02 and 0.5 Da, respectively. For the identification of proteins, all of the assigned peptides with a MOWSE score >12 were considered. Proteins were used for further analysis if they had at least two different peptide identifications and the combined score of unique peptide identifications exceeded the score of the first reversed database hit reaching 1%. This resulted in a false positive rate of <1% on the protein level.
Abundance Analyses
Orbitrap raw data were transformed into peptide maps using the in-house software ProteoProfile using a minimal intensity of 10,000 counts. Peptide maps belonging to one experiment were clustered and aligned using clustering parameters of Δm/z = 0.02 and ±2 min (wide), ±1 min (narrow). Peptide clusters were aligned with Mascot identification files to assign sequence identity. Expression analyses were performed on proteins identified by at least two different peptide sequences. Expression values and relative standard deviation were gained by averaging the intensity differences and standard deviation of the four most intense peptide triplets after removing outlying peptide clusters. Significance was calculated using a two-tailed t test.
Localization Analysis
Proteins were mapped with their International Protein Index identifiers against UniProt 15.6 (as of 28.7.2009), and their subcellular locations were retrieved in Uniprot-curated general annotations. If a protein was attributed to more than one organelle, the location was manually assigned following this hierarchy: plasma membrane, endosome/lysosome, endoplasmic reticulum, Golgi, and mitochondria. For further analysis, protein ratios of Phago15/0 versus TM, Phago15/45 versus TM, and TM versus PNS were plotted according to their frequency in bins of 1 between −10 and 10.
To identify functional groups of ER proteins on the phagosome, log2 ratios of phagosome 15′ versus TM and phagosome 15′/45′ versus TM were clustered using MultiExperiment Viewer v4.5 using default settings. The keywords of these proteins were extracted from Uniprot 15.6 (as of July 28, 2009). Statistical analysis was performed on Uniprot keywords that had at least five occurrences within the list using a two-tailed Fisher exact test.
Estimation of the Relative Contribution of Each Source Organelle to the Phagosome
For the estimation of organelle contribution to phagosomes using the quantitative label-free data, we made the following assumptions: (i) the sum of the intensities of the three highest intense peptides of a given protein is a good indicator of its abundance (24), and (ii) the abundances of proteins give a good estimate of the contribution to the phagosome of their source organelles. Therefore, we determined the sum of the average intensities/ion counts of the three most abundant peptides for each protein obtained from the biological replicates. Proteins not identified on the phagosome were not considered in the estimation of the contribution of their respective organelle. We then summed these protein intensities over all protein members of the respective organelles, because this was statistically sounder and showed similar results compared with using just proteins with transmembrane domains. Because some organelles are more abundant and/or better assigned in databases, numbers of proteins differed considerably between the organelles, from ∼30 proteins for the Golgi to ∼170 proteins for mitochondria. We therefore averaged the organelle intensities over the number of proteins assigned to each of the organelles, based on Uniprot annotations as described above, to eliminate this bias. Finally, the intensities of all members of ER, PM, endosome/lysosome, mitochondria, and Golgi were set arbitrarily to 100% to yield the relative contribution of each source organelle to the phagosome proteome. Similar operations were performed for estimating the contribution of each organelle to the total membrane fraction.
SILAC Experiment to Identify Cellular Contaminants during Phagosome Isolation
RAW264.7 macrophages were grown in light medium (DMEM; Invitrogen) and exposed to polystyrene beads for 30 min. The cells were washed several times with PBS and then mixed with the same number of cells labeled with heavy arginine (Arg10) and lysine (Lys8) before cell lysis. Phagosomes were isolated as described above. Phagosomal extracts were denatured and reduced in 8 m urea, 25 mm Tris, pH 8.0, 5 mm tris(2-carboxyethyl)phosphine, alkylated with 5 mm iodoacetamide and digested with trypsin and three isolation replicates separated on an Ultimate 3000 RSLC nano system (Dionex/Thermo) on a 50-cm-long 75-μm-inner diameter C18 Pepmap column (Dionex) using a 6-h gradient from 5 to 35% B (A: H2O, 0.1% formic acid, B: 80% ACN, 0.08% formic acid). Eluting peptides were identified and quantified on an Orbitrap Velos Pro using a data-dependent “top 20” method, dynamically choosing the most abundant precursor ions from the survey scan (400–2000 Thomson, 60,000 resolution, AGC target value of 106). Precursors above the threshold of 500 counts were isolated within a 2-Thomson window and fragmented by Collision Induced Dissociation in the LTQ Velos using normalized collision energy of 35 and an activation time of 10 ms. Dynamic exclusion was defined by a repeat count of 1, list size of 500 features, and exclusion duration of 60 s. Lock mass was used and set to 445.120025 for ions of polydimethylcyclosiloxane. Mass spectrometric data was analyzed using MaxQuant 1.2.2.5 and the Andromeda search engine resulting in 1,428 proteins with at least two unique peptides at an false discovery rate of <1%. Subcellular localization information was retrieved from Uniprot (as of 20.11.2011) for 799 proteins and assigned 143 proteins to endosome/lysosome, 116 proteins to the plasma membrane, 38 proteins to the endoplasmic reticulum, 38 proteins to mitochondria, and 9 proteins to histones.
Validation by Western Blot and Antibodies
Three independent cell fractions of Phago15/0 and 15/45 and TM were obtained from J774A.1 cells specifically to perform the Western blot (WB). For each antibody tested, the samples were loaded on a 4–12% precast NuPAGE gel (Invitrogen). Proteins selected based on the MS data to represent various values in the fold change graph, ER functions, membrane, cytoplasmic, or luminal localization were probed by WB using the following antibodies: lysosome-associated membrane glycoprotein 1 (LAMP1) clone 1D4B (Developmental Studies Hybridoma bank, Iowa City, IA); cytosolic domain of Cnx (gift from Dr. John J. Bergeron), calreticulin (ab14234) (Abcam, Cambridge, UK); BIP/GRP78 (610978), CD51 (611012), early endosome antigen 1 (EEA1) (610456), α/β soluble N-ethylmaleimide-sensitive factor attachment protein (SNAP) (611898), and signal recognition particle 54 (610940) (BD Biosciences, Franklin Lakes, NJ); signal recognition particle receptor β (gift from Dr. Jacques Paiement); D12/USE1, Stx18, and Sec22b (18); Stx4 (hpa001330) and Stx18 (hpa003019) (Sigma-Aldrich); retinol dehydrogenase 11 (LS-C46836), protein-tyrosine phosphatase, nonreceptor type 1 (LS-C7486), and γSNAP (LS-C15085) (Lifespan Biosciences, Seattle, WA); Rab1a (11671-1-AP) and SPTLC2 (51012-2-AP) (Proteintech, Chicago, IL); Sar1 (07-692) (Millipore, Billerica, MA); neuropilin (GTX16786) (GeneTex, Irvine, CA); protein dissulfide isomerase (PDI) (SPA-891) (Stressgen, Ann Harbor, MI); and Na/K-ATPase (ma3–928) (Abgent, San Diego, CA). Horseradish peroxidase-coupled secondary antibodies directed against rabbit, mouse, rat, or chicken immunoglobulin G (IgG) (Jackson Immunoresearch Laboratories, West Grove, PA) were used with the appropriate primary antibody. The relative enrichment of Phago15/0 or 15/45 versus TM was obtained from the WB by densitometry using Quantity One (Bio-Rad). The log of the fold changes obtained from WB and MS was plotted, and a linear regression fitting was performed using Kaleidagraph 4.0 (Synergy Software, Reading, PA).
Immunofluorescence
J774A.1 were plated on coverslips coated with fibronectin (Sigma) and allowed to adhere for 3–4 h in their normal growth medium. Then the growth medium was removed and replaced with ice-cold phagocytosis medium (RPMI1640 with no fetal bovine serum), containing L1200 2.3 μm carboxyl PB (Estapor) covalently tethered to human IgG (Sigma-Aldrich). The beads were allowed to attach to cells for 30 min at 4 °C. Then PB-containing medium was removed from the cells and replaced by warm growth medium. Internalization was allowed to proceed for 6 min before washing, fixation in 4% paraformaldehyde solution and, permeabilization with 0.5% saponin. The cells were subsequently stained with the relevant variable primary antibodies and counterstained with phalloidin-BODIPY 558/568 (Invitrogen) and either one of two Cnx antibodies (see under previous subheading and below). Variable mouse or chicken monoclonal antibodies (see above) were revealed using anti-mouse and -chicken IgG Alexa 488-coupled secondary antibodies, respectively, and Cnx by combining the cytoplasmic epitope-targeted antibody and anti-rabbit Alexa 647. Variable polyclonal antibodies from rabbit were revealed using anti-rabbit IgG Alexa 647-coupled secondary antibody and Cnx directly using Cnx-FITC (BD Biosciences). All secondary antibodies were from Invitrogen. The images were acquired using a Leica TCS SP confocal microscope.
Recruitment of GFP-based ER Markers to Phagosomes
The cells were treated or not with interferon-γ (R & D Systems) at 100 units/ml. Activated cells spread better on the coverslips and thus displayed better imaging properties. The cells were fixed and stained with GFP antibody and Cnx antibody against a cytoplasmic epitope (see above), using Alexa 488- and 568-coupled secondary antibodies, respectively. The localization of GFP-KDEL and mVenus-Stx18 to the phagosome was monitored by immunofluorescence using the intrinsic fluorescence of the GFPs and by counterstaining with Cnx and phalloidin-BODIPY 558/568. Samples were observed and analyzed using a Leica TCS SP confocal microscope. Phagosomes were purified from stable cell lines expressing GFP-KDEL, mVenus, or mVenus-Stx18 and probed for the exogenous markers and Cnx, serving as an endogenous marker. Using densitometry, the ratio of enrichment on the phagosomes of the exogenous markers versus Cnx was determined.
RESULTS
To evaluate the contribution of various cellular organelles in the formation of phagosomes, we devised a large scale and unbiased strategy based on the assumption that the relative contribution of the putative source organelles to the phagosome can be determined using quantitative label-free MS. The rationale of our approach was the following (Fig. 1A). We prepared a PNS from resting J774A.1 cells and cells that had phagocytosed PB. We isolated phagosome and the TM fractions from the corresponding PNS crude lysates. To validate the capacity of our mass spectrometry approach to follow phagosomes maturation, we determined the protein composition of phagosomes following a 15-min pulse and either harvested immediately thereafter or chased for another 45 min (i.e. Phago15/0 and 15/45, respectively). The data sets for the TM versus PNS comprised 13,647 peptides corresponding to 1,764 proteins. Similarly, 16,021 peptides (1,969 proteins) and 16,003 peptides (1,955 proteins) were identified for Phago15/0 or 15/45 versus TM, respectively. Proteins retained in this analysis were reproducibly detected in at least two of three biological replicates at a false discovery rate <1% (Table I). The overlap between the different sets of Phago15/0 versus TM and 15/45 versus TM was 1,841 proteins or 93 and 94%, respectively. The identified proteins were mapped against the UniProt database (20), and annotations of function as well as subcellular localization were retrieved. Proteins attributed to PM, endosome/lysosome, ER, and the Golgi apparatus were selected for further analysis, because these organelles constitute the potential membrane pool of phagosomes; mitochondrial proteins were included in the analysis as a potential negative control (Table I and supplemental Tables S1–S3 and Fig. S1).
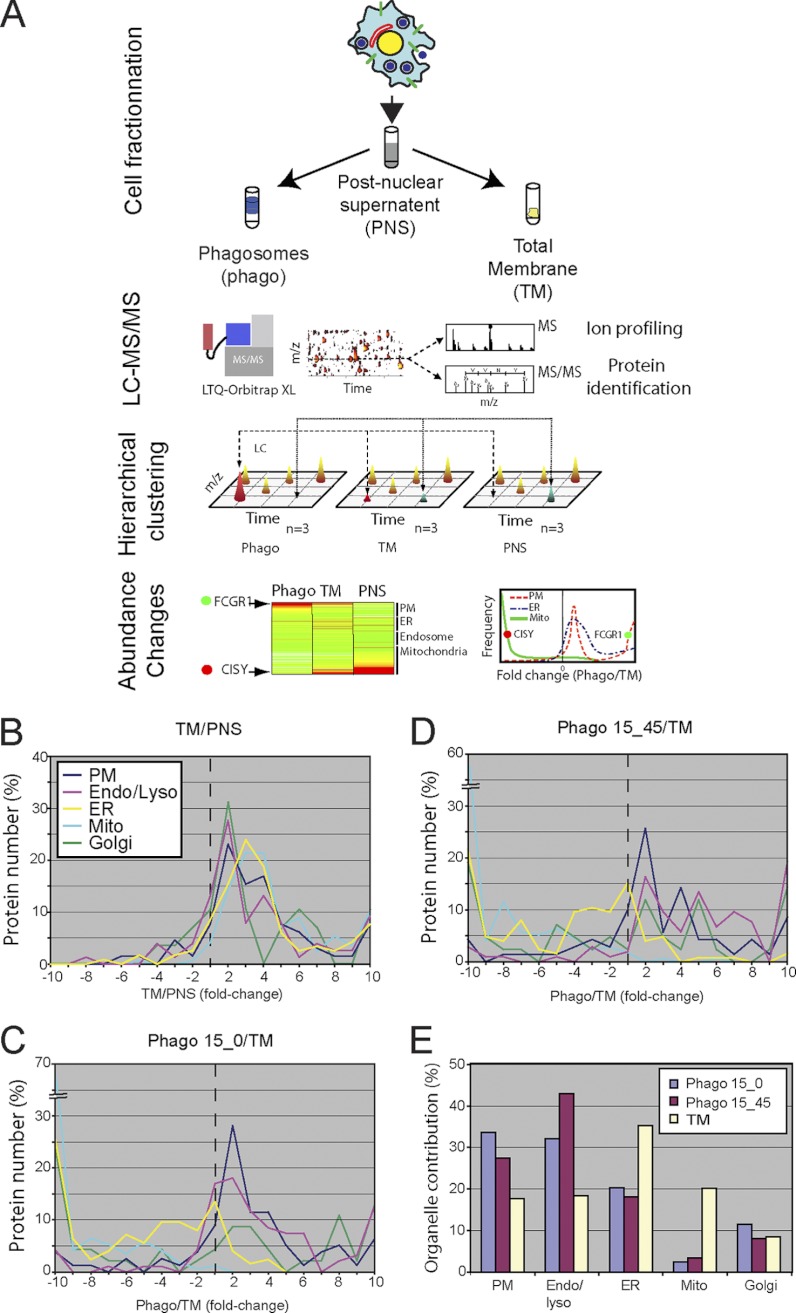
Fig. 1.
A proteomics approach reveals the contribution of the various source organelles to the phagosome proteome. The figure shows the rationale of the MS strategy designed to assess the relative contribution of diverse organelles to the phagosome membrane. PB phagosomes (Phago15/0 or 15/45) were purified from J774.1 cells on sucrose gradients in three independent trials. In parallel, TM and PNS were isolated. A, the protein composition of all four samples was characterized by MS, and their relative abundance within the TM, PNS, Phago15/0, and Phago15/45 fractions was determined. The proteins were assigned to five organelles (PM, endosome/lysosome, ER, Golgi apparatus, and mitochondria) according to the UniProt database. Protein fold changes were then plotted according to their frequency within −10 and 10 at bins of 1. B–D, the distribution of fold change for proteins of the TM versus PNS (B), Phago15/0 versus TM (C), and Phago15/45 versus TM (D) are shown. E, estimation of the contribution of each organelle to phagosome and TM proteomes. Mean peptide intensities for all assigned proteins within an organelle were averaged, yielding an estimate of the membrane contribution of the source organelle. The intensities of all members of ER, PM, endosome/lysosome, mitochondria, and Golgi were set arbitrarily to 100%, thus ignoring other endomembrane compartments.
Table I. Mass spectrometry statistics using Uniprot annotations.
| Peptides | Proteins identified |
||||||
|---|---|---|---|---|---|---|---|
| Total | PM | Endosome/lysosome | ER | Mitochondria | Golgi | ||
| TM/PNS | 13,647 | 1,764 | 65 | 76 | 117 | 168 | 29 |
| Phago 15_0/TM | 16,021 | 1,969 | 78 | 94 | 125 | 170 | 40 |
| Phago 15_45/TM | 16,003 | 1,955 | 70 | 104 | 125 | 174 | 36 |
When we used the TM/PNS ratio as a control, we found a homogenous distribution of proteins that peaks between 2 and 4 for all organelles scrutinized (Fig. 1B). Given that organelles are enriched in the TM, which is completely embedded in the PNS, this result was expected and demonstrates the validity of our approach. The comparison of the relative abundance of the proteins associated to the five potential “donor” organelles present in the phagosome samples versus the TM indicated that proteins from certain organelles were clearly enriched in the phagosome fractions, whereas others were significantly depleted (Fig. 1, C and D). Similar profiles were observed for early and more mature phagosomes (i.e. Phago15/0 and 15/45, respectively), with an exception in the fold change distribution that was especially obvious for endosome/lysosome and PM, as discussed below. Not surprisingly, most of the proteins associated to mitochondria were undetectable on phagosome fractions and therefore distributed mainly to the left side of the scale (Fig. 1, C and D). On the other hand, proteins from PM and endosome/lysosome are almost all found in the enriched half of the graph (i.e. on the right side of the scale). Remarkably, proteins from both the Golgi apparatus and the ER follow a multimodal distribution with a significant proportion of the proteins being depleted, whereas others are distributed close to the center of the scale or are clearly enriched (Fig. 1, C and D). This observation raises the possibility that only a subset of these organelles might contribute to the phagosome proteome. On the other hand, in the case of the Golgi, enriched proteins adopt a profile that is similar to the PM and endosome/lysosome, whereas the major peak of ER annotated proteins appears distinctively in the slightly de-enriched segment of the Phago/TM axis. To address these observations, we decided to perform a cluster analysis of the fold change of ER proteins that integrates subcellular location data extracted from Uniprot general annotations to estimate the relative contribution of each organelle to the phagosome proteome.
Heat maps of the relative abundance of proteins in phagosomes and TM highlighted the proteins from each of the potential organelle donors that are selectively enriched on phagosomes or depleted from this organelle compared with the TM (supplemental Figs. S1 and S2). Remarkably, using the STRING database, we identified specific protein complexes from the ER and Golgi that are enriched (e.g. ER chaperone complexes, ER, and Golgi-SNARE complexes) and depleted (e.g. coat protein complexes I and II components, reticulon complex, and ER signal peptidases), suggesting the translocation of specific ER and Golgi functions to the phagosome. To further characterize the functional properties of the ER subdomain associated to phagosomes, we performed a hierarchical clustering of 125 ER proteins identified in TM and phagosome fractions depending on the fold changes between Phago15/0, Phago15/45, and TM. These analyses allowed us to highlight three distinct clusters: cluster 1 contained 41 ER proteins present in the TM fraction and never detected on phagosomes; cluster 2 contained 77 proteins present on phagosomes, albeit at a slightly depleted level compared with the TM; and cluster 3 contained five highly phagosome-enriched proteins (supplemental Table S4). For the next analyses, we compared cluster 1 with clusters 2 and 3 combined. We extracted keywords from the Uniprot database for each member of these clusters and analyzed their occurrence (supplemental Table S5). For example, the ontology keywords “calcium” (1:10) and “chaperone” (0:11) were highly enriched in clusters 2 and 3 of ER proteins present on the phagosome. In contrast, the keyword “oxidoreductase” (9:6) was attributed to proteins present in cluster 1 of ER proteins not identified on the phagosome. Although the statistical power of the analysis was limited by the low samples size, the identified terms support the concept that some of the functional properties of phagosomes could arise from the specific contribution of a subset of the ER.
Next, we used a label-free proteomics approach to quantify the abundance of proteins assigned to each of these organelles to establish more precisely their relative contribution to phagosome genesis. For this, we determined the abundance of each protein in all fractions by determining the mean abundance of the three most abundant peptides (24). The protein mean abundances were then used to determine the relative proportion of each source organelle in a given fraction. This was done by obtaining the ratio of the sum of mean abundances from proteins assigned to a given organelle to that of all proteins found in each fraction analyzed (Fig. 1E). The resulting graph shows that proteins of the PM, endosome/lysosome, ER, and Golgi proteins constitute 34, 32, 20, and 12% of the Phago15/0 proteome, respectively. On the other hand, these organelles represent 27, 43, 18, and 8% for the Phago15/45, respectively. These observations recapitulate some known features of phagosome maturation obtained by tracking organelle markers by Western blotting, such as the decrease of PM and the increase of endosomal and lysosomal proteins during phagosome maturation (reviewed in Ref. 1) (see also supplemental Fig. S3). Similarly, ER markers were more recently shown to decline (11–14, 18). On the other hand, proteins assigned to mitochondria contribute only marginally (2–3%) to the phagosome proteome. Our results also indicate that the overall enrichment of the ER on the phagosome is rather poor. For one, the ER is very abundant in the TM. This is an obvious explanation for the overall lower fold change for Phago/TM ratios of the ER in comparison with the PM, endosome/lysosome, and Golgi (Fig. 1, C and D). It is also noteworthy that a significant portion (∼20–25%) of the ER contribution to the TM fraction in our estimation stems from the specific subset of ER proteins that does not contribute to the phagosome proteome. Nevertheless, ER annotated proteins contribute ∼20% of the early phagosome proteome (Fig. 1E).
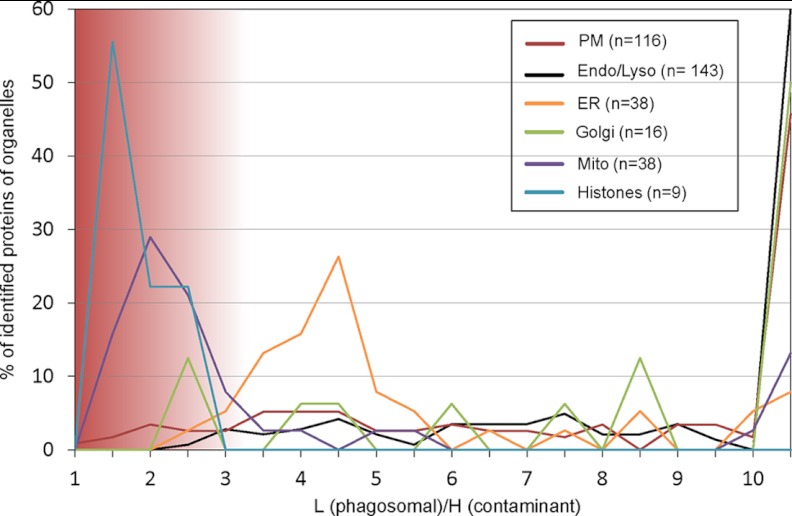
It has been suggested that phagosomes isolated by floatation in a sucrose gradient could be potentially contaminated by nonphagosomal proteins, such as ER-associated proteins (22). Previously we have shown that the amount of contaminants is rather small compared with other organelle purification methods using radioisotope labeling (11). Here we made use of a SILAC experiment to characterize contaminating proteins and their organelle of origin. First, we presented PB to RAW264.7 macrophages grown in normal, light DMEM for 30 min and mixed these cells prior to cell lysis with the same number of RAW264.7 cells grown in DMEM with heavy labeled lysine and arginine. In this scheme, we reasoned that contaminating proteins would show a light (phagosomes): heavy (contaminants) ratio of 1:1, whereas proteins translocated to the phagosome would show a ratio considerably higher than 1. To assess this, phagosomes were again isolated in triplicate using our standard protocol, and their tryptic peptides were analyzed on an Orbitrap Velos Pro. MaxQuant analysis and identification through the search engine Andromeda (25) resulted in identification of 1,428 proteins with at least two unique peptides at a false discovery rate of <1%. Similarly to before, we extracted subcellular localization from Uniprot and could assign 143 proteins to endosome/lysosome, 116 proteins to the cellular/plasma membrane, 38 proteins to the endoplasmic reticulum, 16 proteins to Golgi, 38 proteins to mitochondria, and 9 proteins to histones (Fig. 2 and supplemental Table S6). The data show clearly that mitochondrial proteins (median of 2.1) and histones (median of 1.5) appear at a low ratio of light:heavy, proving that these proteins are contaminants, albeit in rather low quantity (Fig. 1), which are most likely nonspecifically attaching to the phagosomes during isolation. Proteins of the endoplasmic reticulum (median of 4.1; average ratio of 5.2), as well as endosomal and lysosomal (median 13.0), plasma membrane proteins (median of 10.4), and Golgi (median of 9.9), appear noticeably separated from these contaminants, demonstrating that proteins from these organelles are truly located to the phagosome.
Fig. 2.
SILAC experiment to identify potential contaminations to the phagosome. RAW264.7 macrophages were grown in light DMEM, and phagocytosis was induced for 30 min. These cells were mixed with an equal number of cells grown in heavy labeled DMEM and lysed. Phagosomes were isolated and analyzed by quantitative MS. Subcellular localization of proteins was obtained from Uniprot, and organelles were plotted according to their ratio of light (L) to heavy (H) in bins of 0.5. A light to heavy ratio of ∼1 indicates a potential source of contamination such as mitochondria and histones, whereas most proteins associated to other organelles appear to be genuine phagosome components.
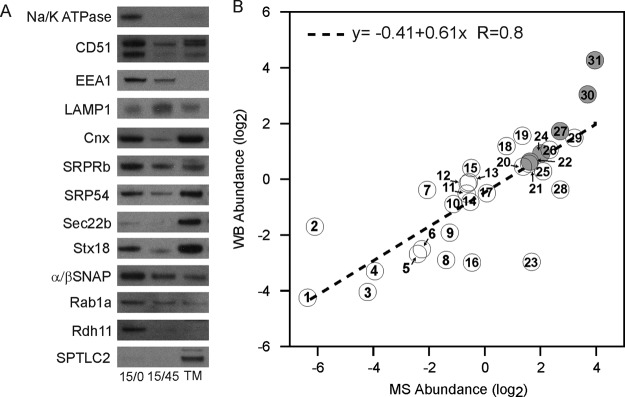
To validate the label-free MS data, WB were performed on biological triplicates of phagosome preparations (Fig. 3A and supplemental Fig. S4). A panel of ER markers were selected to monitor proteins with various Phago/TM fold change profiles, including proteins of interest not detected by MS (e.g. SRP54, Stx18, and D12). Phagosome maturation was monitored using antibodies against the early endosome and late endosome markers, EEA1 and LAMP1, as well as with proteins expected to be found at the PM, such as Na+/K+ ATPase and CD51. Remarkably, the relative abundances obtained from the WB by densitometry showed a very good correlation (y = −0.41 + 0.61x, r = 0.80) with the values determined from the label-free MS data (Fig. 3B and supplemental Table S7), thus validating our large scale approach. As shown by the latter, most ER proteins were found to be slightly less abundant on Phago15/0 versus TM. Exceptionally abundant proteins on the phagosome included retinol dehydrogenase 11 and SNAP proteins. In contrast, SPTLC2, protein-tyrosine phosphatase, nonreceptor type 1, and PDI were almost completely absent from phagosome fractions (Fig. 3 and supplemental Table S7 and Fig. S4). To complement the data obtained by WB and proteomics, we devised a fluorescent-assisted cell sorting (FACS) method to assess phagosome population throughout maturation. The FACS study of purified phagosomes indicates that the Cnx-enriched phagosomes and particularly a Cnx+/LAMP1− (e.g. C+/L−) subpopulation is more abundant on Phago15/0 versus 15/45 (supplemental Fig. S5). Interestingly, this subpopulation is depleted at the benefit of Cnx−/LAMP1+ and Cnx+/LAMP1+ as phagosomes mature.
Fig. 3.
Validation of the label-free MS results by Western blotting. A, similar amounts of protein extracts from Phago15/0 and 15/45 and TM were probed with antibodies against appropriate controls and ER annotated proteins that displayed various abundance levels according to MS. B, the fold changes obtained by MS were plotted against the relative enrichment determined by densitometry of WB triplicates, yielding a linear correlation with r = 0.80. ER and other organelle annotated proteins are represented by white and gray circles, respectively. The numbers provide a link to the proteins blotted and the early and late status of phagosomes probed in each instance (supplemental Table S7).
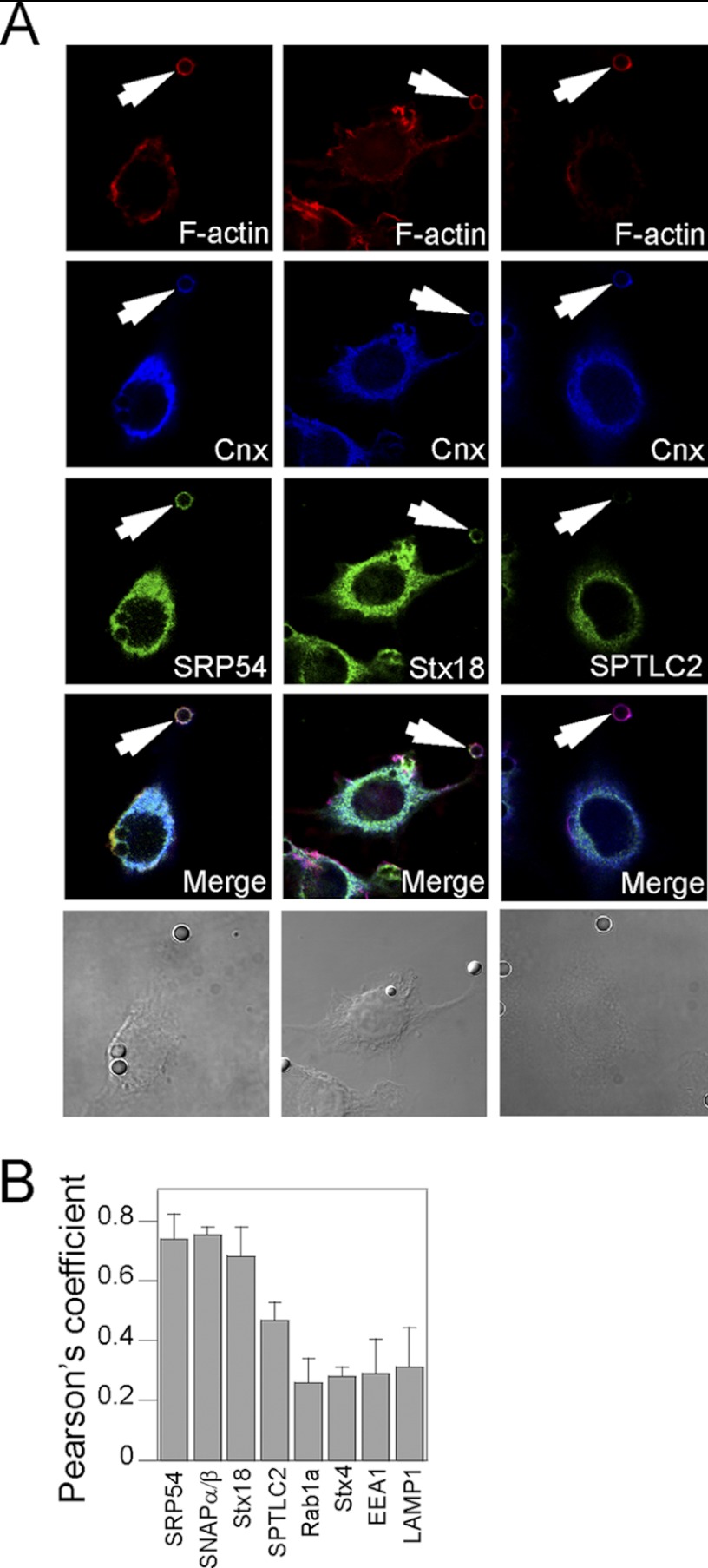
Furthermore, we used immunofluorescence microscopy to investigate whether selected ER proteins associated with phagosomes. The subcellular localization of the proteins studied was consistently compared with Cnx, the gold standard of ER resident proteins. In addition, F-actin staining with phalloidin-BODIPY was used to identify early phagosomes. The labeling for both SRP54 and Stx18 co-localized with that of Cnx throughout the cell cytoplasm, as well as in the phagosome vicinity (Fig. 4A). In contrast, SPTLC2, an enzyme involved in sphingolipid metabolism, displayed a typical ER labeling that was, nonetheless, co-localizing only partially with Cnx. Indeed, although Cnx staining extends to the cell periphery, SPTLC2 is mainly observed in the perinuclear region. Unlike Cnx, this protein was not detected in the phagosome vicinity, hence confirming the proteomics and WB data (Figs. 1 and 3). These observations were confirmed by obtaining the Pearson's coefficient for these staining and others on at least three independent fields (Fig. 4B). It is noteworthy that the quantification of the SPTLC2 and Cnx co-localization is intermediate between those of other ER markers (e.g. SRP54, α/βSNAP, and Stx18) and of endosome and PM markers (e.g. EEA1, LAMP1, and Stx4, respectively). Taken together, these data support the concept that the ER is made of subdomains in which all proteins are not equally distributed and that a subset of the ER interacts with phagosomes.
Fig. 4.
Validation of MS results by confocal microscopy indicates that only a subdomain of the ER is recruited to the phagosome. Early PB-IgG phagosomes were formed in J774A.1 cells that were plated on fibronectin-coated cyoverslips. After fixation and permeabilization, the cells were stained for various proteins detected on the phagosome (e.g. SRP54, Stx18, and SPTLC2) and counterstained with Cnx antibody and phalloidin-BODIPY to reveal nascent phagosomes (“Experimental Procedures”). A, the data indicate that the ER proteins SRP54 and Stx18 co-localize with Cnx on the phagosome, whereas SPTLC2 does not. B, quantification of the relative co-localization in whole cells of putative phagosome markers over Cnx using the mean Pearson's coefficients obtained by the analysis of at least three representative fields for each staining reveal significant differences in the distribution of several ER markers.
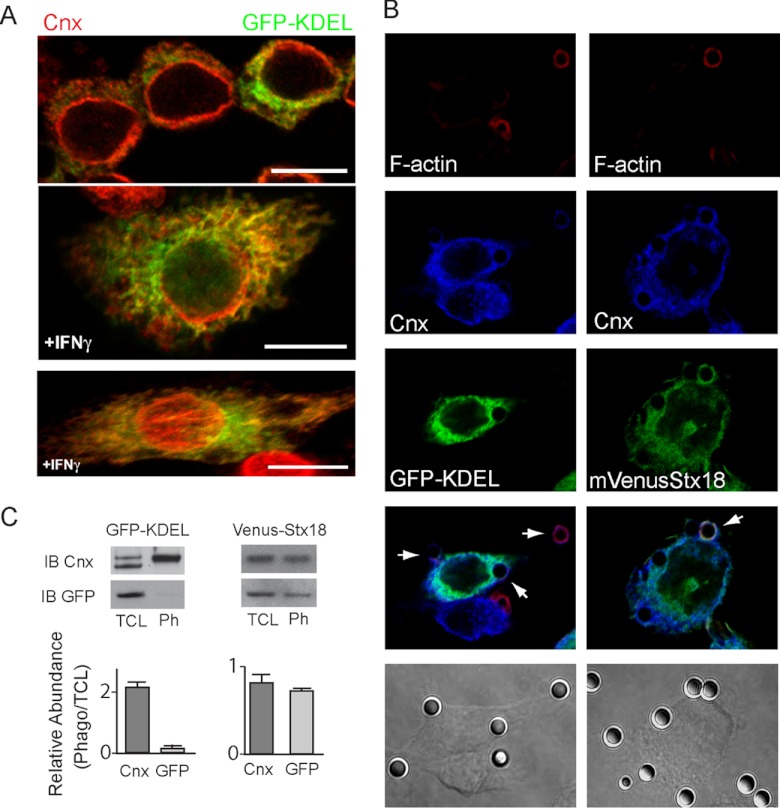
GFP constructs targeted to the ER and harboring a KDEL retention signal are popular ER markers. In earlier studies, this marker was shown not to be recruited to nascent phagosomes (22). To determine whether GFP-KDEL is a suitable marker to highlight the subdomain of the ER involved in phagocytosis, we compared its subcellular localization with Cnx by performing immunofluorescence on the Raw 264.7 GFP-KDEL stable cell line used in (22). As seen in Fig. 5A, Cnx and GFP-KDEL co-localized only partially. As observed for SPTLC2, GFP-KDEL was preferentially distributed in the perinuclear region of the ER and was less often observed at the cell periphery where Cnx was abundant. The formation of phagosomes in these cells did not lead to the recruitment of GFP-KDEL (Fig. 5B), as described previously (22). Interestingly, however, mVenus-Stx18, an ER marker used to study the fusion properties of this organelle (18), was clearly present in the vicinity of phagosomes, as well as at the cell periphery. WB confirmed that GFP-KDEL was poorly recruited to purified phagosomes, in comparison with the endogenous marker Cnx and mVenus-Stx18 (Fig. 5C).
Fig. 5.
mVenus-Stx18, but not GFP-KDEL, is localized to the subregion of ER implicated in phagocytosis. A, confocal microscopy of RAW264.7 stable cell line expressing GFP-KDEL in the absence (top panel) or presence (bottom two panels) of interferon-γ (IFNγ), which was used to flatten the cell to improve the spatial resolution; GFP and Cnx were detected by immunofluorescence. The bottom panel shows a flattened three-dimensional image rendered from multiple confocal sections obtained through the depth of the cell (the bar represents 10 μm). Note the discrepancy in the co-localization of GFP and Cnx, particularly in the perinuclear region and at the cell periphery. B, PB-IgG phagosomes were internalized by RAW264.7 GFP-KDEL and J774A.1 mVenus-Stx18. The cells were stained for Cnx, and F-actin was revealed by phalloidin-BODIPY to identify early phagosomes. C, WB using antibody against Cnx and GFP on phagosomes fraction obtained from the same cell line as described in B were performed to compare the recruitment of Cnx, GFP-KDEL, and mVenus-Stx18 to the phagosome fraction. TCL, total cell lysate; IB, immunoblot; Ph, phagosomes fraction.
DISCUSSION
The comparison of organelles with the cell fraction from which they are derived is a fundamental method in cell biology (26, 27). However, only a small set of markers is usually followed by WB, therefore limiting the ability to gain a global perspective of the interactions of a given organelle within its cellular environment. Quantitative proteomics circumvents this limitation by allowing the assessment of the relative distribution of hundreds of protein among various cellular fractions. We used a label-free proteomics approach to quantify the contribution of various membranes to the phagosome by comparing the abundance of proteins in three cellular fractions (e.g. phagosomes, TM, and PNS). To quantify each protein, we obtained a mean abundance based on the intensity of the three most abundant peptides for each protein, an approach that was previously used for “accurate absolute quantitation” in protein mixtures (24). Applying this approach to proteins of assigned organelles, we could estimate precisely the contribution of the PM, endosome/lysosome, ER, Golgi, and mitochondria to the phagosome and TM fractions. Although our approach required certain assumptions, including that membrane-bound proteins give a good estimate of the abundance of the putative membrane reservoir, we reasoned that it allows to measure with high accuracy the membrane composition of the phagosome. Because we used subcellular information from the well curated Uniprot database, this approach is unbiased and used hundreds of proteins to describe the origin of the phagosome proteome.
We tested our approach by determining the protein abundance and membrane composition of the PNS fraction, from which the TM fraction is derived. Indeed, all proteins annotated to specific organelles behaved similarly, displaying the expected increased abundance in the TM compared with the PNS. Although endosome/lysosome and PM proteins were found in a relatively narrow range of enrichment on the phagosome, indicating that the whole membranes of these organelles likely contribute to the formation of phagosomes, our data suggested that only a subgroup of Golgi and ER proteins contributes to the phagosomal membrane. We hypothesize that this phenomenon stems from the complex organization of these organelles in functional subdomains, as discussed below for the ER.
Our results show that the ER contributes ∼20% of the Phago15/0 proteome. This is significantly higher than the 2–3% contribution of mitochondrial proteins, which are most likely the result of a low level of contaminations stemming from unspecific attachment or co-sedimentation with PB phagosomes or of incorrect organelle assignment in the database, providing an approximate error for our analysis. Previously, Rogers and Foster (28) estimated that the ER contributed 0.3% to the phagosome proteome. Although the comparison with our study is complicated by the fact that their methods and the underlying mass spectrometric data were not fully released, there are clear analytical differences that could explain the discrepancy between the two approaches. One such difference is that the percentages reported in both studies are different. For example, Rogers et al. reported that 10% of the PM found in the total extract is recruited to the phagosome, whereas we report that the PM constitutes 34% of the Phago15/0 proteome (Fig. 1E). We think that our analysis provide a reasonable way of reporting the contribution of the organelles that can serve as a potential source of membrane to the phagosome proteome, whereas the former is highly dependent on the number of beads phagocytosed. Another factor that could lead to the underestimation of the ER contribution is the lower number of quantified phagosomal proteins (382 proteins used by Rogers et al. versus >1,700 in our study). In addition, they based their estimation of the contribution of the ER to the phagosome on the quantitative data of only five ER resident proteins, i.e. sec61, Cnx, protein disulfide isomerases PDIA3 and PDIA4, and calreticulin. Our data indicate that these proteins are indeed found in cluster 2 that regroups phagosome-recruited proteins but that their abundance ratio is rather low (−2.3, −2.6, −3.5, −3.3, and −5 for Phago15/0, respectively) (supplemental Table S4). Therefore, using these five proteins to estimate the ER contribution might not fully take into account the organizational complexity of the ER. In contrast, using the Uniprot database annotations, we quantified 125 ER proteins in phagosome 15/0 versus TM, including 82 found on the phagosome fraction (Table I and supplemental Table S4), allowing for the determination of their abundance and temporal distribution in very good agreement with WB data. Nevertheless, it is impossible to ascertain that the phagosome membrane composition is directly proportionate to the proteome contribution of each organelle. In the future, combined proteomic/lipidomics approaches might allow for directly estimating the membrane contribution.
The contribution of the ER to the phagosomal membrane has recently been a matter of debate, and it was argued that the ER could be a mere contaminant of the phagosome isolation method (22, 29). Herein, we have presented the results of a SILAC approach in which we mixed cells grown in light medium that had formed PB phagosomes with equal numbers of “contaminating” cells (i.e. without phagosomes) that were grown in isotopically labeled heavy medium to identify contaminants during phagosome isolation. Mitochondrial proteins and histones, which are present in low amounts in our subcellular isolation of phagosomes, are indeed displaying a low light (phagosome) to heavy (contaminant) ratio. This ratio is not 1:1, most probably because of the shorter distance, at the time of cell breakage, between phagosomes and their potential intracellular “light” contaminants than with their intercellular “heavy” counterparts. However, ER proteins with an average ratio of 5.2 (median of 4.1) separate clearly from the contaminating proteins of mitochondria and histones (median of 2.1 and 1.5, respectively), supporting the notion that ER proteins are indeed an integrative part of the phagosome. This is even more evident when we look at the ratios of a number of well accepted phagosomal markers such as Rab5a (light/heavy ratio = 4.1), Rab5b (light/heavy ratio = 4.7), Rab7b (light/heavy ratio = 4.5), and EEA1 (light/heavy ratio = 3.3), which are all in a similar range as classical ER markers such as Cnx, (median of 4.1), PDIA4 (median of 4.4), and calreticulin (median of 4.1). Taken together, the label-free and SILAC quantitative proteomics data presented above support the paradigm of the recruitment of the ER to the nascent phagosomes established in our initial study (11), while providing a more thorough estimation of the contribution of the ER and the other organelles to the phagosome proteome.
Furthermore, numerous WB validating our quantitative MS data confirmed the recruitment of ER markers to the phagosome. The decrease of several ER markers shown in this study and by others (11, 13, 14, 18), as well as our immunofluorescence and FACS experiments (Figs. 4 and 5 and supplemental Fig. S5), suggest a specific ER translocation to the very early phagosomes. In addition, a reduction of ER markers in the course of maturation is hardly reconcilable with the expected properties of impure phagosome preparations. Moreover, our FACS data indicate that a Cnx+/LAMP1− subpopulation is particularly abundant on early phagosome and that this subpopulation is eroded at the expense of the Cnx−/LAMP1+ and Cnx+/LAMP1+ as phagosome matures (supplemental Fig. S5). This result suggests: (i) ER recruitment is an early step in phagosome genesis that is observed across a significant proportion of the phagosomes population, and (ii) ER resident proteins transferred to the phagosomes are recycled or degraded during phagosome maturation. Because we used an antibody directed against the cytoplasmic epitope of Cnx in these experiments, the hypothesis of ER membrane recycling from the phagosome seems more likely. As yet, the mechanism by which the ER proteins are transported to the phagosome and recycled is still sketchy. For ER recruitment to the phagosome, a vesicular pathway depending on ER resident Stx18 and Sec22b interactions with PM-located syntaxins appears plausible (17–19, 30). In Ref. 30, Sec22b was also shown to be essential for cross-presentation. Nevertheless, more efforts will be necessary to identify the functional routes of ER in and out of the phagosome.
Apart from the classic smooth/rough ER dichotomy of this organelle anatomy (31), it has been argued by several groups that the ER is organized in several subdomains (32–36). Our analysis also suggests that only a subset of the ER fuses with the phagosome. Because of the limited knowledge on ER markers partition, we were not able to identify the exact subdomain fusing with the phagosome. Nevertheless, we have confirmed that mVenus-Stx18 is recruited to the phagosome, whereas GFP-KDEL is not (Fig. 5) (18, 22). Clearly there is a discrepancy between the conclusions that can be drawn from using these distinct constructs. A concern is that the GFP-KDEL is obviously a more artificial marker than any fluorescent protein fusion constructs with natural ER resident protein, such as mVenus-Stx18. In addition, we identified two functions that were significantly over-represented among the ER proteins present on the phagosome: chaperone activity and calcium binding/transporting. Among these proteins were Cnx and calreticulin that were identified on phagosomes from Dictyostelium discoideum, Drosophila melanogaster, Mus musculus, and Homo sapiens (10, 11, 13, 14, 16, 37–39). Apart from these chaperones, which share a role in calcium storage (40, 41), we identified the transmembrane proteins Stim1 and its binding partner Orai, which has been shown to be regulating calcium storage in the ER by allowing direct capture of extracellular calcium (42–45), in high amounts on the phagosome (supplemental Fig. S6). Stim1 was shown to locate to the cortical ER (34), a subdomain of the ER found in the vicinity of the PM. In addition, depletion of extracellular calcium store by metal chelators and ER calcium store by thapsigargin and siRNA against Stim-1 and Orai-1 in Caenorhabditis elegans was shown to affect phagocytosis of apoptotic cells (46). Taken together, those evidence suggest that the fraction of the ER harnessed by the phagosome might include a significant amount of cortical ER. It is suggested that this subcompartment of the ER could provide the local Ca2+ concentration spike important for phagocytic cup formation and several phagosome functions (reviewed in Ref. 47). Our results indicating that calcium function is one of the main functional annotations attributed to ER proteins associated with the phagosome and the early recruitment of the ER markers that we have observed are in agreement with this hypothesis (supplemental Table S5). Interestingly, GFP-KDEL was also shown to be excluded from cortical ER (34). Thus, exploiting GFP-KDEL as a marker of the whole ER might be inappropriate. On this basis, we argue that the poor localization of GFP-KDEL to the phagosome, first reported by Touret et al. (22) and reproduced here (Fig. 5), cannot be evoked to reject the contribution of the ER to phagosome genesis. Instead, we propose that these results support the notion that only a subset of the ER, which could include one or several of its subdomains, contributes to the phagosome proteome. However, specific experiments will have to be performed to characterize the role of specific ER subdomains, including cortical ER, in phagosomes formation, maturation, and immune functions.
Several pathogens, such as Brucella, Legionella, and Leishmania spp. utilize an ER-rich vacuole as their biological niche inside infected host cells. In particular, Brucella and Legionella spp. are pathogenic bacteria using protein effectors to manipulate phagosome maturation to form their ER-rich vacuoles (Refs. 48 and 49; reviewed in Ref. 50). The perversions of Rab1 and Sar1 functions, respectively, are apparently crucial events in this process. Recently, Leishmania parasitophorous vacuole were shown to accumulate continuously various ER markers until at least 24 h after infection, suggesting discrete hijacking of vesicular mechanism to subvert the regular phagocytic process (51). In light of our results, we hypothesize that these pathogens subvert a mechanism that is already at play in the phagocytosis of inert particles by favoring, for example, persistent ER recruitment or inhibiting the recycling of the ER as maturation proceeds. These prospects provide an exciting new avenue to tackle the role of the ER during phagocytosis of inert particles or pathogens and the ensuing immune response.
The raw data associated with this manuscript may be downloaded from the ProteomeCommons.org Tranche network using the following hash code: Om1ftT5XPIiGt5mKfNlmPq4+Ch6BTg6AXftEgf7iz294xENBgNEveDZK6jQWYpz17uR1WaJ3ExzpWg9mMnJSkLMY3SIAAAAAAAAPkg = =. The pass phrase is 7dFvhCoxZn1MiiHKcB9N.
Acknowledgments
We thank Olivier Caron-Lizotte for the clustering of proteomics data; Drs. Jacques Paiement, Isabelle Jutras, Marek Gierlinski, and Nicholas Schurch for helpful discussions; Dr. Moïse Bendayan and Irene Londono for electron microscopy; Michel Lauzon for confocal microscopy and electron microscopy; and Serge Sénéchal and Danièle Gagné for FACS and the sequencing platform of the Institute for Research in Immunology and Cancer.
Footnotes
* This work was supported by the Canadian Institutes of Health Research.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PM
- plasma membrane
- Cnx
- calnexin
- DMEM
- Dulbecco's modified Eagle's medium
- EEA1
- early endosome antigen 1
- ER
- endoplasmic reticulum
- FACS
- fluorescence-assisted cell sorting
- GFP
- green fluorescent protein
- IgG
- immunoglobulin G
- LAMP1
- lysosome-associated membrane glycoprotein 1
- PDI
- protein disulfide isomerase
- PB
- polystyrene bead
- PNS
- postnuclear supernatant
- SILAC
- stable isotope labeling in cell culture
- SNAP
- soluble N-ethylmaleimide-sensitive factor attachment protein
- SNARE
- soluble N-ethylmaleimide-associated receptor
- SPTLC2
- serine palmitoyltransferase, long chain base subunit 2
- SRP54
- signal recognition particle 54
- Stx
- syntaxin
- TM
- total membrane
- WB
- Western blot.
REFERENCES
- 1. Jutras I., Desjardins M. (2005) Phagocytosis: At the crossroads of innate and adaptive immunity. Annu. Rev. Cell Dev. Biol. 21, 511–527 [DOI] [PubMed] [Google Scholar]
- 2. Heine J. W., Schnaitman C. A. (1971) A method for the isolation of plasma membrane of animal cells. J. Cell Biol. 48, 703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pitt A., Mayorga L. S., Stahl P. D., Schwartz A. L. (1992) Alterations in the protein composition of maturing phagosomes. J. Clin. Invest. 90, 1978–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bajno L., Peng X. R., Schreiber A. D., Moore H. P., Trimble W. S., Grinstein S. (2000) Focal exocytosis of VAMP3-containing vesicles at sites of phagosome formation. J. Cell Biol. 149, 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braun V., Fraisier V., Raposo G., Hurbain I., Sibarita J. B., Chavrier P., Galli T., Niedergang F. (2004) TI-VAMP/VAMP7 is required for optimal phagocytosis of opsonised particles in macrophages. EMBO J. 23, 4166–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hackam D. J., Rotstein O. D., Sjolin C., Schreiber A. D., Trimble W. S., Grinstein S. (1998) v-SNARE-dependent secretion is required for phagocytosis. Proc. Natl. Acad. Sci. U.S.A. 95, 11691–11696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Holevinsky K. O., Nelson D. J. (1998) Membrane capacitance changes associated with particle uptake during phagocytosis in macrophages. Biophys. J. 75, 2577–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Desjardins M., Huber L. A., Parton R. G., Griffiths G. (1994) Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J. Cell Biol. 124, 677–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rink J., Ghigo E., Kalaidzidis Y., Zerial M. (2005) Rab conversion as a mechanism of progression from early to late endosomes. Cell 122, 735–749 [DOI] [PubMed] [Google Scholar]
- 10. Garin J., Diez R., Kieffer S., Dermine J. F., Duclos S., Gagnon E., Sadoul R., Rondeau C., Desjardins M. (2001) The phagosome proteome: Insight into phagosome functions. J. Cell Biol. 152, 165–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gagnon E., Duclos S., Rondeau C., Chevet E., Cameron P. H., Steele-Mortimer O., Paiement J., Bergeron J. J., Desjardins M. (2002) Endoplasmic reticulum-mediated phagocytosis is a mechanism of entry into macrophages. Cell 110, 119–131 [DOI] [PubMed] [Google Scholar]
- 12. Houde M., Bertholet S., Gagnon E., Brunet S., Goyette G., Laplante A., Princiotta M. F., Thibault P., Sacks D., Desjardins M. (2003) Phagosomes are competent organelles for antigen cross-presentation. Nature 425, 402–406 [DOI] [PubMed] [Google Scholar]
- 13. Guermonprez P., Saveanu L., Kleijmeer M., Davoust J., Van Endert P., Amigorena S. (2003) ER-phagosome fusion defines an MHC class I cross-presentation compartment in dendritic cells. Nature 425, 397–402 [DOI] [PubMed] [Google Scholar]
- 14. Ackerman A. L., Kyritsis C., Tampé R., Cresswell P. (2003) Early phagosomes in dendritic cells form a cellular compartment sufficient for cross presentation of exogenous antigens. Proc. Natl. Acad. Sci. U.S.A. 100, 12889–12894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Trost M., English L., Lemieux S., Courcelles M., Desjardins M., Thibault P. (2009) The phagosomal proteome in interferon-γ-activated macrophages. Immunity 30, 143–154 [DOI] [PubMed] [Google Scholar]
- 16. Boulais J., Trost M., Landry C. R., Dieckmann R., Levy E. D., Soldati T., Michnick S. W., Thibault P., Desjardins M. (2010) Molecular characterization of the evolution of phagosomes. Mol. Syst. Biol. 6, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Becker T., Volchuk A., Rothman J. E. (2005) Differential use of endoplasmic reticulum membrane for phagocytosis in J774 macrophages. Proc. Natl. Acad. Sci. U.S.A. 102, 4022–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hatsuzawa K., Tamura T., Hashimoto H., Hashimoto H., Yokoya S., Miura M., Nagaya H., Wada I. (2006) Involvement of syntaxin 18, an endoplasmic reticulum (ER)-localized SNARE protein, in ER-mediated phagocytosis. Mol. Biol. Cell 17, 3964–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hatsuzawa K., Hashimoto H., Hashimoto H., Arai S., Tamura T., Higa-Nishiyama A., Wada I. (2009) Sec22b is a negative regulator of phagocytosis in macrophages. Mol. Biol. Cell 20, 4435–4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lewis M. J., Pelham H. R. (1996) SNARE-mediated retrograde traffic from the Golgi complex to the endoplasmic reticulum. Cell 85, 205–215 [DOI] [PubMed] [Google Scholar]
- 21. Hatsuzawa K., Hirose H., Tani K., Yamamoto A., Scheller R. H., Tagaya M. (2000) Syntaxin 18, a SNAP receptor that functions in the endoplasmic reticulum, intermediate compartment, and cis-Golgi vesicle trafficking. J. Biol. Chem. 275, 13713–13720 [DOI] [PubMed] [Google Scholar]
- 22. Touret N., Paroutis P., Terebiznik M., Harrison R. E., Trombetta S., Pypaert M., Chow A., Jiang A., Shaw J., Yip C., Moore H. P., van der Wel N., Houben D., Peters P. J., de Chastellier C., Mellman I., Grinstein S. (2005) Quantitative and dynamic assessment of the contribution of the ER to phagosome formation. Cell 123, 157–170 [DOI] [PubMed] [Google Scholar]
- 23. Desjardins M., Celis J. E., van Meer G., Dieplinger H., Jahraus A., Griffiths G., Huber L. A. (1994) Molecular characterization of phagosomes. J. Biol. Chem. 269, 32194–32200 [PubMed] [Google Scholar]
- 24. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 25. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 26. De Duve C. (1971) Tissue fractionation: Past and present. J. Cell Biol. 50, 20d–55d [PubMed] [Google Scholar]
- 27. Van Der Sluijs P., Hull M., Zahraoui A., Tavitian A., Goud B., Mellman I. (1991) The small GTP-binding protein Rab4 is associated with early endosomes. Proc. Natl. Acad. Sci. U.S.A. 88, 6313–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rogers L. D., Foster L. J. (2007) The dynamic phagosomal proteome and the contribution of the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 104, 18520–18525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gagnon E., Bergeron J. J., Desjardins M. (2005) ER-mediated phagocytosis: Myth or reality? J. Leukocyte Biol. 77, 843–845 [DOI] [PubMed] [Google Scholar]
- 30. Cebrian I., Visentin G., Blanchard N., Jouve M., Bobard A., Moita C., Enninga J., Moita L. F., Amigorena S., Savina A. (2011) Sec22b regulates phagosomal maturation and antigen crosspresentation by dendritic cells. Cell 147, 1355–1368 [DOI] [PubMed] [Google Scholar]
- 31. Voeltz G. K., Rolls M. M., Rapoport T. A. (2002) Structural organization of the endoplasmic reticulum. EMBO Rep. 3, 944–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iinuma T., Aoki T., Arasaki K., Hirose H., Yamamoto A., Samata R., Hauri H. P., Arimitsu N., Tagaya M., Tani K. (2009) Role of syntaxin 18 in the organization of endoplasmic reticulum subdomains. J. Cell Sci. 122, 1680–1690 [DOI] [PubMed] [Google Scholar]
- 33. Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. (2009) A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433–1437 [DOI] [PubMed] [Google Scholar]
- 34. Orci L., Ravazzola M., Le Coadic M., Shen W. W., Demaurex N., Cosson P. (2009) From the cover: STIM1-induced precortical and cortical subdomains of the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 106, 19358–19362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Levine T., Rabouille C. (2005) Endoplasmic reticulum: One continuous network compartmentalized by extrinsic cues. Curr. Opin. Cell Biol. 17, 362–368 [DOI] [PubMed] [Google Scholar]
- 36. Borgese N., Francolini M., Snapp E. (2006) Endoplasmic reticulum architecture: Structures in flux. Curr. Opin. Cell Biol. 18, 358–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Müller-Taubenberger A., Lupas A. N., Li H., Ecke M., Simmeth E., Gerisch G. (2001) Calreticulin and calnexin in the endoplasmic reticulum are important for phagocytosis. EMBO J. 20, 6772–6782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gotthardt D., Blancheteau V., Bosserhoff A., Ruppert T., Delorenzi M., Soldati T. (2006) Proteomics fingerprinting of phagosome maturation and evidence for the role of a Gα during uptake. Mol. Cell. Proteomics 5, 2228–2243 [DOI] [PubMed] [Google Scholar]
- 39. Stuart L. M., Boulais J., Charriere G. M., Hennessy E. J., Brunet S., Jutras I., Goyette G., Rondeau C., Letarte S., Huang H., Ye P., Morales F., Kocks C., Bader J. S., Desjardins M., Ezekowitz R. A. (2007) A systems biology analysis of the Drosophila phagosome. Nature 445, 95–101 [DOI] [PubMed] [Google Scholar]
- 40. Camacho P., Lechleiter J. D. (1995) Calreticulin inhibits repetitive intracellular Ca2+ waves. Cell 82, 765–771 [DOI] [PubMed] [Google Scholar]
- 41. Roderick H. L., Lechleiter J. D., Camacho P. (2000) Cytosolic phosphorylation of calnexin controls intracellular Ca2+ oscillations via an interaction with SERCA2b. J. Cell Biol. 149, 1235–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roos J., DiGregorio P. J., Yeromin A. V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J. A., Wagner S. L., Cahalan M. D., Veliçelebi G., Stauderman K. A. (2005) STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 169, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liou J., Kim M. L., Heo W. D., Jones J. T., Myers J. W., Ferrell J. E., Jr., Meyer T. (2005) STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 15, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang S. L., Yu Y., Roos J., Kozak J. A., Deerinck T. J., Ellisman M. H., Stauderman K. A., Cahalan M. D. (2005) STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature 437, 902–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Prakriya M., Feske S., Gwack Y., Srikanth S., Rao A., Hogan P. G. (2006) Orai1 is an essential pore subunit of the CRAC channel. Nature 443, 230–233 [DOI] [PubMed] [Google Scholar]
- 46. Gronski M. A., Kinchen J. M., Juncadella I. J., Franc N. C., Ravichandran K. S. (2009) An essential role for calcium flux in phagocytes for apoptotic cell engulfment and the anti-inflammatory response. Cell Death Differ. 16, 1323–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nunes P., Demaurex N. (2010) The role of calcium signaling in phagocytosis. J. Leukocyte Biol. 88, 57–68 [DOI] [PubMed] [Google Scholar]
- 48. Ingmundson A., Delprato A., Lambright D. G., Roy C. R. (2007) Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450, 365–369 [DOI] [PubMed] [Google Scholar]
- 49. Celli J., Salcedo S. P., Gorvel J. P. (2005) Brucella coopts the small GTPase Sar1 for intracellular replication. Proc. Natl. Acad. Sci. U.S.A. 102, 1673–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roy C. R., Salcedo S. P., Gorvel J. P. (2006) Pathogen-endoplasmic-reticulum interactions: In through the out door. Nat. Rev. Immunol. 6, 136–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ndjamen B., Kang B. H., Hatsuzawa K., Kima P. E. (2010) Leishmania parasitophorous vacuoles interact continuously with the host cell's endoplasmic reticulum: Parasitophorous vacuoles are hybrid compartments. Cell Microbiol. 12, 1480–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]