Abstract
Bioactive molecules typically mediate their biological effects through direct physical association with one or more cellular proteins. The detection of drug-target interactions is therefore essential for the characterization of compound mechanism of action and off-target effects, but generic label-free approaches for detecting binding events in biological mixtures have remained elusive. Here, we report a method termed target identification by chromatographic co-elution (TICC) for routinely monitoring the interaction of drugs with cellular proteins under nearly physiological conditions in vitro based on simple liquid chromatographic separations of cell-free lysates. Correlative proteomic analysis of drug-bound protein fractions by shotgun sequencing is then performed to identify candidate target(s). The method is highly reproducible, does not require immobilization or derivatization of drug or protein, and is applicable to diverse natural products and synthetic compounds. The capability of TICC to detect known drug-protein target physical interactions (Kd range: micromolar to nanomolar) is demonstrated both qualitatively and quantitatively. We subsequently used TICC to uncover the sterol biosynthetic enzyme Erg6p as a novel putative anti-fungal target. Furthermore, TICC identified Asc1 and Dak1, a core 40 S ribosomal protein that represses gene expression, and dihydroxyacetone kinase involved in stress adaptation, respectively, as novel yeast targets of a dopamine receptor agonist.
Drugs often act as protein antagonists (inhibitors) or agonists (activators) through selective physical interactions with targets in disease-relevant pathways, yet many pharmaceuticals and chemical probes from cell-based phenotypic screens currently lack defined cellular targets (1). Although conventional “target-based” drug discovery pipelines emphasize functional characterization and in vitro inhibition/activation assays (2), unexpected side effects can occur when drugs interact with additional, unanticipated cellular proteins (3). Computational strategies often predict multiple off-target effects even for well known pharmaceuticals that are intended to be highly selective (4), and the ability of compounds to engage multiple targets can sometimes be clinically and biologically desirable (5, 6). Consequently, understanding drug action ultimately depends on an unbiased experimental validation of compound binding specificity in a physiologically relevant cellular context.
Although chemical genetic screening methods have been developed to identify drug-affected pathways (7–9), such approaches do not pinpoint the direct target(s) bound by a drug. Conversely, biochemical characterization of the protein targets of small molecules has traditionally been accomplished by immobilizing or labeling compounds for use as affinity ligands to probe cell lysates (10, 11), but the introduction of additional functional moieties (i.e. derivatization) can perturb a compound's bioactivity and is not amenable to high throughput screening of diverse compounds. To overcome these limitations, unbiased “label-free” chemical proteomics strategies coupling biochemical fractionation with mass spectrometry have recently been developed to identify drug targets. For example, a biochemical suppression approach identifies drug target by rescuing the activity of a drug-inhibited cell lysate by the addition of a biochemically fractionated cell extract (12), whereas drug affinity responsive target stability approach compares the proteomic profiles of fractionated drug-treated lysate before or after protease treatment to identify stabilized targets based on the premise that the drug-protein complex is less susceptible to digestion (13). Although these approaches do not require any chemical modification or labeling of either the compound or target, certain limitations hinder wider applicability. For instance, an assayable activity is required for biochemical suppression screening, whereas for drug affinity responsive target stability, drug binding may not affect or even increase target proteolysis, confounding data interpretation. Given the growing awareness of polypharmacy, increased interest in drug repositioning (14), and the rapidly increasing pace of cell-based phenotypic screens, novel label-free chemical proteomic methods are urgently needed to allow for the unbiased detection of the physical interactions of bioactive compounds with proteins in complex biological systems in a hypothesis-generating fashion.
Here, we introduce a potentially widely applicable drug target identification strategy based on nondenaturing high performance liquid chromatography coupled to LC-MS to monitor the interactions of small molecules and potentially other bioactive ligands with nearly native cellular proteins in complex biological mixtures. The procedure is based on a characteristic shift in the chromatographic retention time profile of a compound that occurs after binding to a protein target. High performance LC-MS/MS is then used to deconvolute the co-fractionating protein target(s). We demonstrate proof of principle with mechanistically diverse compounds, using TICC1 to demonstrate additional insights into compound affinity, target abundance, and binding-induced conformational change. Finally, we identify novel targets for one antifungal agent (4513-0042) and one dopamine-receptor agonist (A77636). We conclude that TICC can be used to identify previously unreported drug-protein interactions without the need for either compound or protein immobilization or labeling.
EXPERIMENTAL PROCEDURES
Reagents, Cell Extracts, and Recombinant Proteins
Stock solutions (10 mm) of methotrexate (Sigma-Aldrich), radicicol (Sigma-Aldrich), A77636 (Tocris Bioscience, Ellisville, MO), and 4513-0042 (Hit2Lead, San Diego, CA) were prepared in 50% methanol:water and stored at −20 °C. Sordarin (5 mg/ml in DMSO) was a gift from C. H. Ho (15), whereas trichostatin A was purchased from Sigma-Aldrich as 5 mm solution in DMSO. Working solutions of all drugs were prepared fresh by dilution prior to use. HPLC grade solvents were from Fisher. Recombinant polyhistidine-tagged DHFR fusion protein was generated in BL21(DE3)pLys Escherichia coli cells using the pST39-HIS-DHFR expression vector (courtesy of S. Tan) (16) (see supplemental “Methods” for details). HeLa cytosolic and nuclear (6.0 mg/ml) extracts were obtained from Paragon BioServices (Baltimore, MD). Soluble E. coli and Saccharomyces cerevisiae whole cell protein extracts were prepared by sonication and glass bead beating, respectively, as previously reported (17, 18). Dual N-terminal glutathione S-transferase (GST) and polyhistidine (HIS)-tagged Erg6p fusion protein was overexpressed in yeast as previously reported (19) and isolated using glutathione-Sepharose (GE Healthcare) using the manufacturer's protocol.
Yeast Cell Culture, Drug Treatment, and Gene Overexpression
For in vivo drug treatments, wild type haploid yeast (BY4741: MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0) were cultured overnight in yeast peptone dextrose medium at 30 °C, diluted to an A600 of 0.2 in fresh medium and then grown to an A600 of 0.5. To minimize drug consumption, the cells were pelleted and transferred to a sterile microtube, and radicicol was added to a final drug concentration of 50 μm in ∼1 ml of yeast peptone dextrose. After incubation at 30 °C for 20 min, the cells were washed three times with ice-cold water and harvested for protein extraction.
For the drug rescue experiments, yeast were transformed with a high copy 2-μm inducible pEG(KG) plasmid expressing either ERG6 or a control S. cerevisiae gene, Fra1, with dual N-terminal GST and HIS tags (courtesy of R. Sopko) (19) (see supplemental “Methods” for protocol). To evaluate resistance to 4513-0042, overnight cultures were diluted and grown in synthetic complete (SC) + 2% glucose (parental control) or SC-URA+ 2% sucrose (plasmid-bearing cells) media to an A600 of 0.5. The cultures were further diluted to an A600 of 0.025 in SC + 2% glucose (control) or SC-URA + 2% galactose (plasmid-bearing cells) and aliquoted into a 96-well microtiter plate (Sarstedt, Newton, NC). Vehicle (methanol) alone or compound (4513-0042 dissolved in methanol) was added to a final drug concentration of 300, 150, or 3 μm. Growth inhibition was measured by A600 (SpectraMax Plus384 spectrophotometer; Molecular Devices, Sunnyvale, CA) after incubation at 30 °C overnight.
HPLC Fractionation Method 1 (Low Resolution)
A HP 1100 HPLC system (Agilent Technologies, Mississauga, Canada) consisting of a binary pump with a vacuum degasser, a refrigerated (4 °C) autosampler with a 100- or 900-μl injector loop, a thermostatted (17 °C) column compartment, a refrigerated (4 °C) fraction collection module, and a multi-wavelength UV-visible detector was used. Dual ion exchange chromatography was performed by coupling a 200 × 2.1-mm LP anion column (1000 Å pore, 5-μm particle size) in series with a 200 × 2.1-mm LP cation column (1000 Å pore, 5-μm particle size) (PolyLC, Columbia, MD) using a previously optimized configuration (20).
The columns were loaded with ∼1.5 mg of yeast soluble protein lysate, 650 μg of E. coli lysate, 112 μg of purified recombinant HIS-DHFR, or either 50, 200, or 800 μg of HeLa cell-free extract, before or after preincubation with drug for 20 min on ice to minimize target denaturation and drug metabolism. Prior to loading, the samples were spin-filtered at 100 g through 0.45-μm disposable membrane cartridges (Millipore, Ottawa, Canada).
Mobile phase gradients were as follows: (i) MTX in vitro drug “dosing” assays: 0–20 min, 0–75% gradient of Buffer B (10 mm Tris-HCl, pH 7.8, 1 m NaCl); 20–24 min, 75% Buffer B; 24–25 min, 75–5% Buffer B; 25–35 min, 5–0% Buffer B; and 35–40 min, 100% Buffer A (10 mm Tris-HCl, pH 7.8); (ii) Radicicol in vivo yeast assays and all yeast and HeLa in vitro drug dosing experiments were as follows: 0–8 min, 100% Buffer A; 8–38 min, 0–45% gradient of Buffer B; 38–58 min, 45–100% Buffer B; 58–66 min, 100% Buffer B; 66–68 min, 100–0% Buffer B; and 68–86 min, 100% Buffer A. The fractions were collected every 1–2 min at a constant flow rate of 0.2–0.25 ml min−1. Protein UV absorbance was measured at 280 nm. Half of each fraction collected was used for drug quantification (SRM), and the remainder was used for protein identification (LC-MS/MS).
HPLC Fractionation Method 2 (High Resolution)
Dual column nondenaturing chromatography was performed by coupling a 75 × 7.5 mm (10 μm, 1000 Å) TSK gel heparin 5PW affinity column (Tosoh Bioscience, Grove City, OH) in series with a mixed mode (Poly LC, 200 × 4.6 mm, 5 μm, 1000 Å) column, both thermostatted at 17 °C. Mobile phase A consisted of 10 mm Tris-HCl buffer, pH 7.8, 3 mm sodium azide, 5% glycerol, 0.1 mm DTT, whereas mobile phase B contained 1.5 m NaCl in addition to all components of mobile phase A. Gradient elution was performed at 0.25 ml/min, using the following gradient: 0–34 min, hold at 90% A; 34–144 min, gradient 10–65% B; 144–148 min, gradient 65–100% B; hold for 34 min at 100% B, and re-equilibrate to starting conditions for 58 min. Fractions were automatically collected at 2.0-min intervals.
A second complementary weak cation exchange (WCX) method at pH 6.0 was developed to narrow down candidate lists and used for A77636 protein target identification. Nondenaturing chromatographic fractionation was performed using 250 × 4.6 mm BioWCX, NP5 column (Agilent, 5 μm, nonporous, proprietary COOH on polystyrene/divinylbenzene support). Mobile phase A consisted of 20 mm ammonium acetate buffer, pH 6.0, 3 mm sodium azide, and 5% glycerol, whereas mobile phase B contained 500 mm ammonium acetate buffer, pH 6.0, 3 mm sodium azide, and 5% glycerol. Gradient elution was performed at 0.7 ml/min, using the following gradient: 0–6 min, hold at 100% A; 6–30 min, gradient 0–100% B; hold for 50 min at 100% B, and re-equilibrate to starting conditions for 15 min. Fractions were automatically collected at 1.0-min intervals.
An aliquot of each fraction (0.1 ml) was subjected to drug assay by SRM, and the remainder was used for protein identification. The columns were loaded with ∼2.4 mg of yeast lysate (sordarin and A77636 experiments) or HeLa nuclear extract (TSA experiment). The instrument and the rest of conditions were the same as described for method 1.
Protein Processing
Protein fractions were thawed on ice and precipitated with 10% (v/v) trichloroacetic acid overnight at 4 °C. After centrifugation at 17,000 × g, the pellet was incubated with 0.3 ml of ice-cold acetone for 30 min at −20 °C and centrifuged, and the pellet was allowed to air dry for 10 min after removal of supernatant. Protein was solubilized in 50 μl of digestion buffer (50 mm ammonium bicarbonate, pH 8.0, 1 mm CaCl2). After reduction with 5 mm DTT for 30 min and carboxymethylation with 15 mm iodoacetamide (Sigma-Aldrich) in the dark, the proteins were digested overnight using sequencing grade trypsin (1 μg/fraction) (Roche Applied Science) at 37 °C with gentle shaking. After digestion, the samples were concentrated by SpeedVac and redissolved in 25 μl of purified water containing 1% formic acid. For sordarin (HPLC method 1) and 4513-0042, trichloroacetic acid step was omitted as described in the supplemental “Methods.”
LC-MS Drug Analyses by SRM
LC-MS analyses in SRM mode were performed using a Quantum Access or Vantage triple quadrupole tandem mass spectrometer (ThermoFisher Scientific, San Jose, CA), depending on sensitivity requirements. MS transitions and instrument parameters for each drug were optimized by directly infusing a 1–5 μm analyte calibration solution at a flow rate of 10 μl min−1. The main assay parameters for each drug are summarized in Table I. Dwell time was set to 50 ms (Quantum Access) or 100 ms (Vantage) for each transition, with the instrument operated at unit mass resolution (peak width at half-height set to 0.7 Da) for both Q1 and Q3 in all cases. For Quantum Access, the electrospray parameters were sheath gas pressure of 45 arbitrary units, spray voltage was 4000 V, and capillary temperature set to 383 °C. For Vantage, equipped with HESI probe, the electrospray parameters included vaporizer temperature of 500 °C, capillary temperature of 380 °C, voltage of 2800 V, and sheath and auxiliary gas at 20 and 15 units, respectively. The data were acquired and processed with XCalibur 2.0 (Quantum Access) or XCalibur 2.1 (Vantage) software (ThermoFisher). Capillary temperature and other spray parameters were selected to provide good electrospray stability and good desolvation at high flow rates. The drug compounds used in this study were not thermally unstable, so the use of such capillary temperatures is acceptable. For thermally labile compounds, the optimization of capillary temperatures needs to be performed, and lower capillary temperatures such as 200–250 °C are usually used.
Table I. Summary of fractionation and LC-MS drug assay conditions.
| Drug | Fractionation conditions | LC-MS system | LC drug assay conditions | Transition monitored | Ionization mode | Injection volume |
|---|---|---|---|---|---|---|
| MTX | Method 1 | Agilent 1200 binary HPLC TSQ Quantum Access triple quadrupole | C18 (400 μl min−1) | 455.0 → 307.6 | Positive ESI | 100 μl |
| Radicicol | Method 1 | Agilent 1200 binary HPLC TSQ Quantum Access triple quadrupole | C18 (400 μl min−1) | 363.1 → 183.2 | Negative ESI | 100 μl |
| 4513-0042 | Method 1 and 2 | Agilent 1200 binary HPLC TSQ Quantum Access triple quadrupole | C18 (400 μl min−1) | 353.0 → 285.0 | Positive ESI | 100 μl (Quantum) |
| Sordarin | Method 1 and 2 | Both | Diphenyl (200 μl min−1) | 493.6 → 333.4 | Positive ESI | 100 μl (Quantum) 10 μl (Vantage) |
| TSA | Method 2 | Refrigerated CTC Pal autosampler, Thermo MS Surveyor pumps TSQ Vantage triple quadrupole | PFP (300 μl min−1) | 303.0 → 148.1 | Positive ESI | 10 μl |
| A77636 | Method 2 | Refrigerated CTC Pal autosampler, Thermo MS Surveyor pumps TSQ Vantage triple quadrupole | PFP (300 μl min−1) | 330.2 → 135.2 | Positive ESI | 10 μl |
Mobile phases consisted of buffer A (HPLC grade water with 0.1% formic acid) and buffer B (HPLC grade acetonitrile with 0.1% formic acid) for all drug assays. Chromatographic separation consisted of a 1-min divert followed by either (i) a 10–90% B gradient over 3 min on a 20 × 2.0-mm Mercury MS Luna (3 μm C18) column (Phenomenex, Torrance, CA), (ii) 5–95% gradient over 3 min on a Pursuit Diphenyl 3 μm 50 × 2.0 mm (Varian, Palo Alto, CA) column, or (iii) a 5–90% A gradient over 3 min on a Kinetex PFP 2.6 μm 50 × 2.1 mm (Phenomenex) column, as summarized in Table I.
Protein Identification by LC-MS/MS
Peptide mixtures were loaded onto a 150-μm inner diameter fused silica microcapillary column (Polymicro Technologies, Phoenix, AZ) pulled to a fine tip using a laser puller (Sutter Instruments Inc., Novato, CA). For the radicicol and sordarin experiments, the column was packed with ∼10 cm of reversed phase (Zorbax Eclipse XDB-C18; Agilent). The microcolumn was placed in-line with a nanoLC- electrospray ion source (Proxeon, Odense, Denmark) and interfaced to an LTQ linear ion trap (ThermoFisher Scientific). The peptides were eluted at a flow rate of 200 nl min−1 using a 105-min gradient (A: 95% water, 5% ACN, 0.1% formic acid; B: 95% ACN, 5% water, 0.1% formic acid) consisting of 1 min 2% B, 2-min change to 6% B, 62-min gradient to 24% B, 26-min gradient to 90% B, followed by a 5-min hold on 90% B, 1-min gradient to 2%B, with a final hold at 2% B for 8 min. Precursor ions (400–2000 m/z) were subjected to data-dependent collision-induced dissociation as the instrument cycled through one full mass scan followed by three successive MS/MS scans targeting the most intense precursor ions, in centroid format with dynamic exclusion enabled (90 s with 1 repeat count in 30 s). For TSA and A77636 analyses, a high performance LTQ Orbitrap Velos hybrid instrument (ThermoFisher Scientific) was used with following modified conditions: 75-μm inner diameter fused silica column packed with 10 cm of Luna 3-μm C18(2) 100 Å reversed phase particles (Phenomenex), 300 nl min−1 elution using 105-min gradient as described above with two modifications: 72-min gradient to 24% B, followed by 16-min gradient to 90% B. Precursor ions (400–2000 m/z) were subjected to data-dependent collision-induced dissociation as the instrument cycled through one full mass scan at 60,000 full-width at half maximum followed by 10 successive MS/MS scans targeting the most intense precursor ions, with dynamic exclusion enabled (22.5 s with two repeat count in 22.5 s) and charge state selection enabled to select preferentially 2+ and 3+ ions.
For the multidimensional LC-MS/MS (i.e. MudPIT (21)), 10 μl of compound 4513-0042-containing protein fractions were analyzed using a 10-h-long multi-cycle procedure essentially as described previously (21, 22). Briefly, a 100-μm inner diameter microcolumn packed with 10 cm of 3-μm Luna-C18 (Phenomenex) was joined via a zero dead volume PEEK junction (Upchurch Scientific, Oak Harbor, WA) to a 250-μm inner diameter capillary packed with 3 cm of 5-μm PolySULFOETHYL A strong cation exchange resin (PolyLC) and 4 cm of Luna-C18 resin. Four sequential salt “bumps” (5 μl) consisting of 0, 125, 250, and 500 mm ammonium acetate, delivered from a 96-well plate by autosampler, were used to elute peptides from the SCX segment onto the reversed phase. Each bump was followed by a linear organic gradient over 105 min as described for LTQ method, but the flow rate was set to 300 nl min−1. The spectra were collected in data-dependent mode selecting the three most intense precursor ions, all in centroid format with the dynamic exclusion enabled (60 s with two repeat counts within 30 s).
Tandem mass spectra were extracted from .RAW files and searched using the SEQUEST database program (SEQUEST-PVM v.27 rev. 9) (23) against relevant protein database downloaded as FASTA-formatted sequences (yeast, http://downloads.yeastgenome.org/sequence/GenBank Feb 2007, human Swiss-Prot database Dec 2010) and containing reversed decoy sequences using a mass tolerance of 3 and 0 m/z for precursor and product ions, respectively, and fixed cysteine modification of 57 atomic mass units. In the current study, precursor mass tolerance was initially set on the basis of LTQ capabilities, so the same parameters were kept for Orbitrap data for consistency throughout the manuscript. However, we also confirmed that the use of 10 ppm high accuracy mass filter for precursor ion search of Orbitrap data did not affect our selection of the top hits for follow-up IP validation of A77636, identification of putative targets of A77636, or conclusions made in the manuscript. Match likelihoods were assigned a statistical confidence score using the STATQUEST model (22), and candidate peptide identifications were filtered using an estimate confidence score of >95%. Spectral counting was used to estimate the abundance of protein in adjacent drug-containing fractions. Candidate lists were filtered to include proteins present in all bound drug fractions. In the final step, elution profile of the bound drug was compared against all proteins identified in all of the bound fractions using simple correlation algorithm in MS Excel (correlation of normalized amount of bound of drug versus amount of protein present as estimated using spectral counting after normalization). Proteins with high correlation values represent putative drug targets, and correlation of ≥0.6 was used to generate putative protein target lists.
GST-Erg6p Drug Binding Assay
200 ng of either purified GST-HIS-tagged yeast Erg6p fusion protein (4 ng/μl), BSA (100 ng/μl), or recombinant GST (10 ng/μl; GenWay Biotech, San Diego, CA) were incubated with 20 μm 4513-0042 on ice for 20 min. Unbound compound was removed by size exclusion chromatography using 0.5-ml Zeba spin columns with a cut-off of 7000 Da (ThermoFisher). The eluant (void volume) was analyzed by SRM to measure the amount of bound drug.
Validation of A77736 Protein Targets
Immunoprecipitation experiments were performed for a prioritized list of candidate proteins: untagged wild type (as a negative control) DAK1, ERG20, FBA1, TSA1, PDC5, ASC1, GDI1, HOM6, and ALD6. (DAL80 was not available in current library, so tentative interaction of this protein with A77636 could not be investigated in this study.) All C-terminally tagged yeast strains were obtained from the TAP fusion library (27) in the BY4741 parental genetic background (MATa, ura3Δ0, leu2Δ0, his3Δ1, met15Δ0).
The cells were grown to mid-log phase in yeast peptone dextrose medium, collected by centrifugation, and lysed by vigorous vortexing with glass beads using ice-cold lysis buffer containing 10 mm Tris, pH 7.9, 150 mm NaCl, 0.1% Nonidet P-40, 10% glycerol, and protease inhibitor mixture (Roche Applied Science). Cellular debris was removed by high speed microcentrifugation for 10 min. The lysates were then adjusted by adding β-mercaptoethanol to a 10 μm final concentration, imidazole to 1 mm, magnesium acetate to 1 mm, and calcium chloride to 2 mm. Immobilized calmodulin beads (Amersham Biosciences) were added to the lysates and incubated with gentle mixing for 2 h at 4 °C. The beads were collected, washed two times with calmodulin-binding buffer-1 (10 mm Tris, pH 7.9, 150 mm NaCl, 0.1% Nonidet P-40, 10% glycerol, 10 mm β-mercaptoethanol, 1 mm imidazole, 1 mm magnesium acetate, and 2 mm calcium chloride) and two times with calmodulin binding buffer-2 (10 mm Tris, pH 7.9, 150 mm NaCl, 0.02% Nonidet P-40, 10% glycerol, 1 mm β-mercaptoethanol, 1 mm imidazole, 1 mm magnesium acetate, and 2 mm calcium chloride). Tagged proteins were eluted with buffer containing 10 mm Tris, 100 mm NaCl, 20 mm EGTA, 10 mm β-mercaptoethanol, and 10% glycerol.
To evaluate the pulldowns, 10 μl of 2× SDS sample buffer was added to 10 μl of each eluate and subsequently loaded on a 10% polyacrylamide SDS gel. After transfer to nitrocellulose, the blots were incubated with primary antibodies to calmodulin (Abgent, San Diego, CA) and then with secondary antibody followed by enhanced chemiluminescence detection. Confirmed lysates were then individually dosed with A77636 at a 2 μm final concentration, incubated for a minimum of 2 h, and subjected to HPLC WCX fractionation method 2, as described above.
RESULTS
Concept: Target Identification by Chromatographic Co-elution
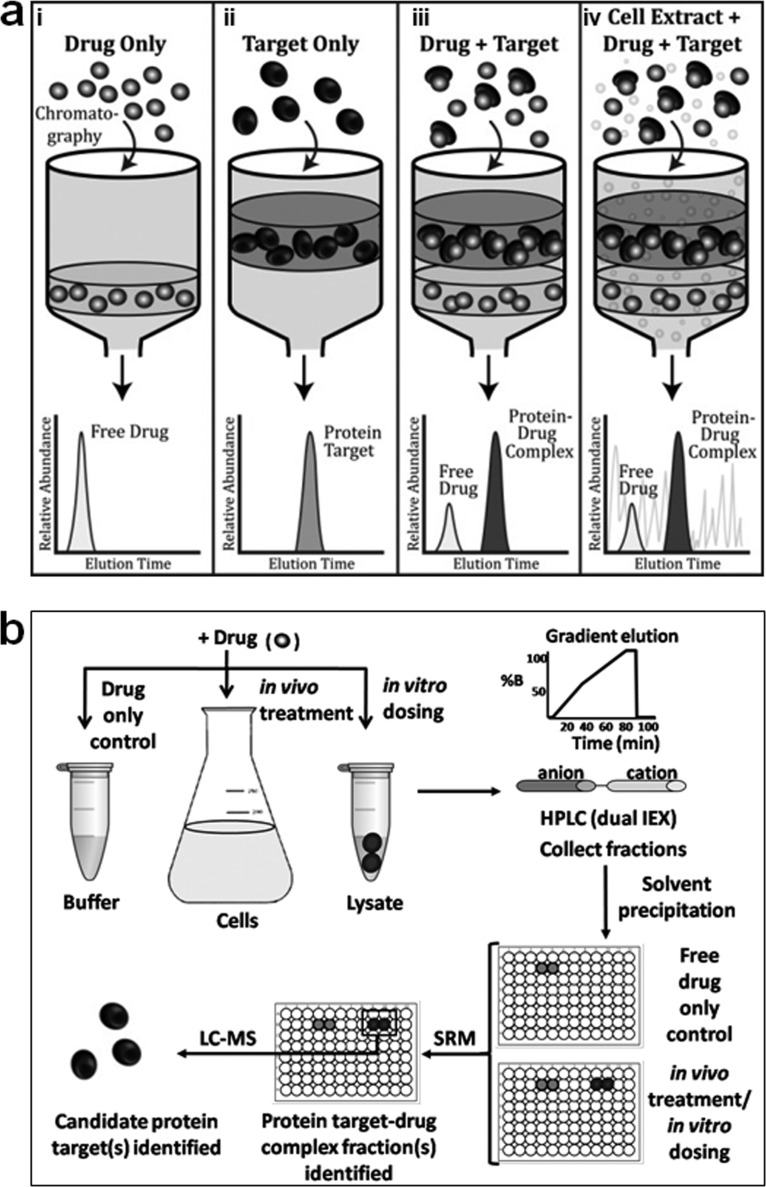
The TICC approach, illustrated schematically in Fig. 1a, is based on the co-fractionation of stable ligand-target complexes during nondenaturing HPLC. The main premise is that upon binding to one or more target proteins, a compound's chromatographic properties are altered with the ligand-target complex exhibiting a different characteristic elution profile relative to free (unbound) drug. That is, the compound's retention time is “shifted” to that of its interacting protein partner(s). In principle, the ligand can potentially represent any small molecule, drug, natural product, metabolite, or other analyte of interest with the requirement being sufficient stability during fractionation and subsequent detectability by mass spectrometry. Although size exclusion chromatography can be used to separate the free and bound compound, we have found that dual ion exchange (IEX) chromatography using a shallow salt gradient that is unlikely to perturb nonionic associations of compounds with proteins is far more effective at resolving compound-target complexes.
Fig. 1.
TICC method for monitoring ligand-protein interactions. a, schematic illustrating representative characteristic elution profiles of drug (panel i) or target alone (panel ii) during nondenaturing IEX-HPLC. Once bound to its direct target (panel iii), the retention time of the compound “shifts” to that of its interacting protein partner (i.e. ligand-protein complex), even in the presence (panel iv) of many irrelevant competitor proteins. b, typical TICC workflow: drug-treated (in vivo) or untreated cells (in vitro dosing) are lysed, and soluble protein extracts containing the ligand of interest are fractionated using dual IEX-HPLC. All of the fractions are collected and analyzed for the presence of drug by LC-MS/MS using SRM to characterize the profiles of free (unbound) and bound (shifted) ligand. Proteins present in the bound drug-containing (shifted) fractions are then analyzed by nanoflow LC-MS/MS to identify candidate co-eluting targets.
In the standard assay implementation (Fig. 1b), a compound of interest is incubated with a putative target in a biological mixture, for example by treating cultured cells (in vivo) or by dosing cell-free soluble protein extracts (in vitro), thus allowing the compound-target(s) interaction(s) to form. The ligand-target complex is then separated from irrelevant components by nondenaturing IEX-HPLC, with timed fractions collected for subsequent analysis by tandem mass spectrometry. To establish a control base-line reference profile, free ligand is first fractionated alone (i.e. in buffer only) using the same automated collection procedure. The elution profiles representing both free and bound ligand are then reconstructed using an appropriate analytical technique for the molecule of interest, such as UV absorption in certain cases but more generally using a highly sensitive and selective LC-MS assay, to document the amount of ligand present in each fraction. We typically perform LC-MS in SRM mode, which has become the gold standard technique for the detection of drugs in biological matrices within the past decade, because it enables the detection of small amounts of drug, which is particularly important when monitoring low abundance protein targets present in complex biological samples. In our standard SRM workflow, the ligand is ionized and injected into the first analyzer of a quadrupole mass spectrometer that only transmits a user-defined analyte of interest with the specified mass to pass through. This target precursor ion is then fragmented, and the resulting fragments are analyzed in a second analyzer, wherein only diagnostic fragment(s) of interest are allowed to pass through. By selectively monitoring the transitions describing the m/z values of desired precursor and reporter fragment ions in a single scan, high sensitivity and selectivity for most compounds of interest is readily achieved. The fast 3–7 min reversed phase LC method employed in our assays further separates the compound of interest from any isobaric interferences potentially present in complex biological samples to achieve accurate drug quantitation.
The fractions containing bound compound are identified by comparing the drug elution profile in drug control versus protein lysate sample, looking for a significant and reproducible shift (e.g. delay) in chromatographic retention time representing protein-bound compound. These fractions are then subjected to standard shotgun proteomic peptide sequencing to identify proteins co-eluting with the bound drug. In the final step, the resulting protein elution profiles are compared with the quantified bound drug elution profile after normalization of ion intensities, and statistical correlation calculation is used to identify the protein(s) that are most highly correlated with the bound drug elution profile, which present putative drug target(s).
Co-elution of MTX with Its Protein Target DHFR
As a first proof of principle, we monitored the co-elution of the folate antagonist MTX with its high affinity (Kd = 4.8 nm) enzyme target, DHFR (24). The MTX-DHFR complex represents a well characterized drug-target interaction supported by extensive biophysical and biological data addressing fundamental structure activity relationships (25, 26).
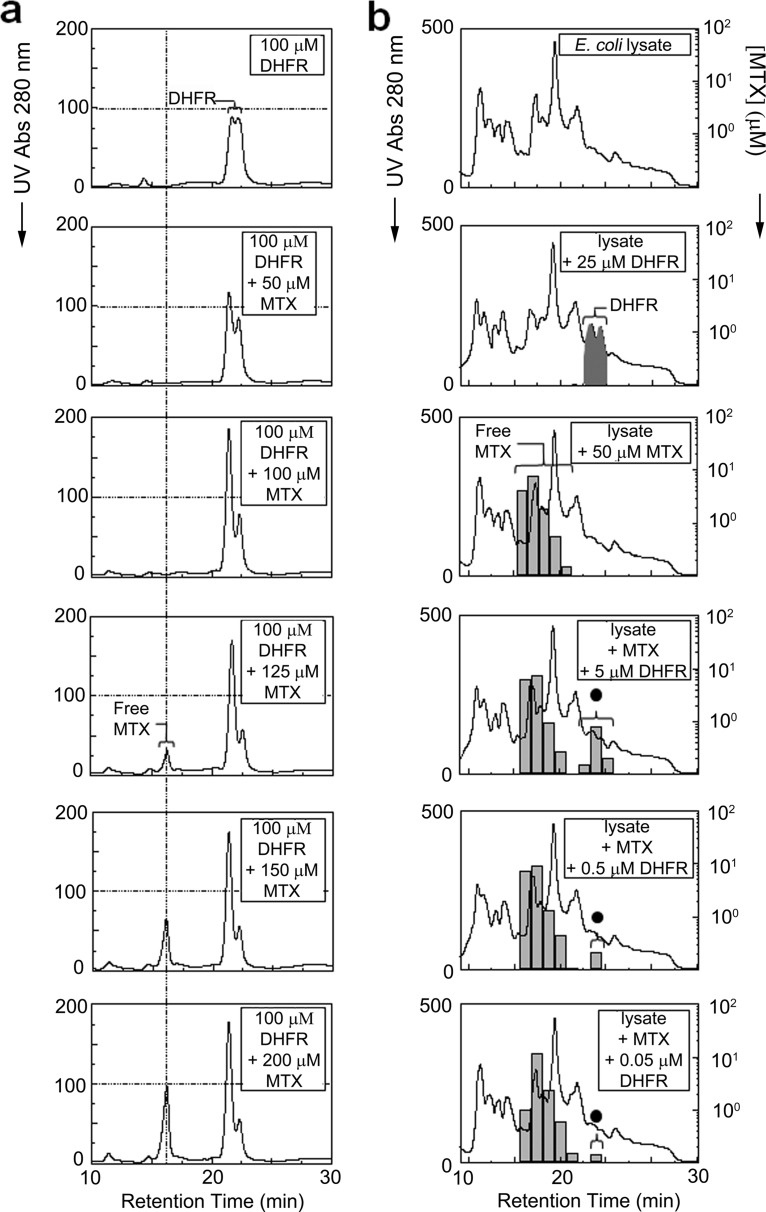
We compared the chromatographic profiles of purified recombinant DHFR (100 μm), in the absence or presence of varying amounts (50–200 μm) of MTX, by a dual IEX chromatography (method 1) using continuous UV absorption at 280 nm as a simple readout (Fig. 2a). Unbound drug, which showed an earlier retention time, was detectable only at a concentration (125 μm) sufficient to saturate its target, reflecting the previously documented one to one binding stoichiometry (27). Free drug peak intensity then increased proportionally at higher doses. Conversely, whereas DHFR alone eluted with a characteristic bimodal peak at 19–23 min, a pronounced shift in the relative abundance of these two conformational isoforms became evident upon MTX binding. The apparent increase in one of the isoforms is likely due to a more favorable conformation, consistent with the ligand-induced stabilization of a particular DHFR conformational state with MTX reported previously (24, 28–30). Hence, co-elution by TICC was both qualitatively distinct (i.e. shifted retention time) and quantitative (i.e. target saturation) and entirely consistent with expectations (27).
Fig. 2.
Stable interaction of methotrexate with its primary target dihydrofolate reductase. a, TICC of recombinant DHFR (100 μm) with various amounts (as indicated) of MTX by dual IEX-HPLC. The resulting UV absorbance (280 nm) traces for DHFR alone (top panel) or DHFR mixed with 50 (second panel), 100 (third panel), 125 (fourth panel), 150 (fifth panel), or 200 μm of MTX (bottom panel) are shown. A drug-induced conformational change is evident from the preferential detection of one of two prominent DHFR peaks as the MTX concentration is increased. Target saturation occurred at 125 μm MTX, whereas the peak intensity of free drug further increased proportionally to the amount of excess compound. b, selective DHFR-MTX co-elution in a complex mixture. The top three panels show dual IEX-HPLC elution profiles of E. coli cell lysate (650 μg of total protein) or extract dosed with either recombinant DHFR (25 μm) or MTX (50 μm) only. The presence of DHFR is indicated by black shading, whereas the black circle denotes DHFR-bound MTX. The bottom three panels show the corresponding elution profiles of extract dosed with a saturating amount (50 μm) of MTX and 5, 0.5, or 0.05 μm DHFR. Despite the presence of many other proteins, the specific interaction (co-elution) of MTX with DHFR was evident in each case by TICC, although the proportion of DHFR in the protein mixture was only 1, 0.1, or 0.01%, respectively.
Co-elution of MTX with DHFR in Complex Mixtures
To evaluate the specificity and sensitivity of TICC, we performed analogous fractionation experiments after introducing MTX alone or together with DHFR into an E. coli cell extract containing many irrelevant competitor proteins as a background mixture. In this case, timed fractions of eluant were collected, and the amount of MTX present was quantified by SRM, whereas the identity of proteins co-eluting with bound drug was subsequently established by shotgun LC-MS/MS.
The panels in Fig. 2b show the global protein chromatograms and corresponding MTX concentrations determined after IEX-HPLC fractionation of either cell-free extract alone, extract dosed with 25 μm DHFR only, extract dosed with 50 μm MTX only, or lysate dosed with both a fixed quantity (50 μm) of MTX and variable (5, 0.5, or 0.05 μm) amounts of DHFR. Despite the vast excess of nonspecific competitor proteins, no nonspecific binding of drug to E. coli proteins was evident, whereas specific and tight (i.e. quantitative) co-elution of the drug with DHFR was detectable even at the lowest target concentration (DHFR representing ∼0.01% total protein mass). The amount of bound (i.e. shifted) drug was proportional to target levels and showed a linear relationship between the amount of bound MTX in association with DHFR (supplemental Fig. 1). We conclude that interaction specificity, target abundance, and occupancy can be determined with good fidelity and sensitivity for high affinity compounds by TICC.
Co-elution of Radicicol with Human Hsp90
To establish the generality of the TICC approach, we next evaluated the binding profiles of another established anti-proliferative agent, radicicol, which is a potent selective inhibitor (Kd = 19 nm) of the ATPase activity of the conserved heat shock chaperone Hsp90 (31, 32) involved in selective protein stabilization.
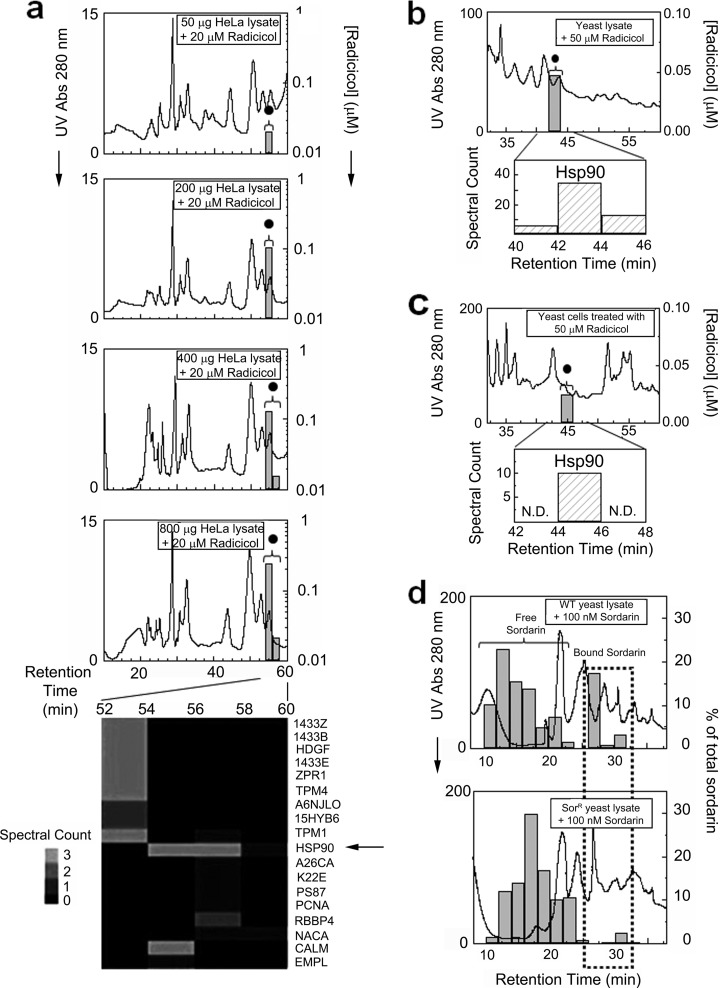
We again performed analogous co-fractionation experiments, after dosing radicicol (20 μm) into varying amounts (50–800 μg) of soluble protein lysate prepared from cultured human HeLa cells. Fig. 3a shows the global protein (based on UV absorption) and drug (measured by SRM analysis) elution profiles recorded after dual IEX-HPLC fractionation. No free drug peak was observed, whereas binding was detectable with as little as 50 μg of total protein and was proportional to the amount of HeLa extract loaded (supplemental Fig. 2). LC-MS/MS analysis of the radicicol-containing fractions confirmed the presence of Hsp90, which exhibited the highest profile correlation to drug of all the proteins identified (Fig. 3, a). Assuming the expected 1:1 stochiometry and 100% target occupancy, absolute quantification of radicicol signal by SRM indicates ready detection of ∼1.8 pmol of endogenous Hsp90 in a complex HeLa lysate. Moreover, a drug dilution series performed in parallel (supplemental Fig. 3) demonstrated Hsp90 saturation with 200 nm radicicol, consistent with the documented drug affinity, whereas no off-target binding was evident by TICC at even the highest (20 μm) drug concentration tested. Likewise, a stable radicicol Hsp90 complex was also detected after dosing yeast lysate with 50 μM radicicol (Fig 3b), or treating yeast cells to a growth-inhibiting dose (50 μm) of radicicol for 20 min (8, 31), from which a soluble extract was prepared and subsequently fractionated by dual IEX-HPLC (Fig. 3c). Again, Hsp90 was identified in the bound drug fraction with a high spectral count. Hence, target specificity, affinity, and abundance in a complex cellular context could be assayed by TICC after in vitro dosing or in vivo drug treatment.
Fig. 3.
Target detection and identification for radicicol and sordarin using TICC. a, in vitro drug dosing experiments using HeLa cell cytosolic protein extract. The top four chromatograms show dual IEX-HPLC elution profiles of radicicol (20 μm) mixed with 50, 200, 400, or 800 μg of lysate. A single peak representing protein-bound drug (black circle) was detected with increasing intensity by SRM in proportion to total protein load, whereas no column retention was observed for free drug. The heat map (bottom panel) shows the spectral counts of high confidence proteins identified by LC-MS/MS, including the known target Hsp90 (arrow) whose proteomic pattern correlated most closely with the radicicol profile. b and c, dual IEX-HPLC fractionation of radicicol (20 μm) dosing to a yeast whole cell extract (b) or after in vivo treatment of yeast for 20 min prior to cell lysis (c). Protein-bound radicicol detected by SRM (black circle), and the spectral counts obtained for Hsp90 in the same fractions by LC-MS/MS are reported. N.D., not determined. d, quantitative comparison of sordarin-binding in protein lysate prepared from wild type (WT) or sordarin-resistant (SorR) yeast strains. A marked reduction in protein-bound drug, denoted with a stippled box, was observed in the resistant strain, reflecting the lower affinity of mutant elf2 for sordarin. Excess free drug is indicated with a bracket.
Co-elution of Sordarin with Native elongation factor 2 (elf2) and Reduced Binding in a Drug-resistant elf2 Point Mutant
We next examined the utility of TICC to monitor the interactions of lower affinity drugs. We generated co-elution profiles and correctly identified the protein target of antifungal sordarin, a natural product that inhibits yeast translation elf2 (33) with low affinity (Kd = 1.26 μm) (34), after dosing protein lysates prepared from either wild type or sordarin-resistant yeast. The sordarin-resistant strain (15) used for this experiment had a single base pair substitution (S523Y) in the sordarin-binding pocket (34) in one of two paralogs (EFT2) encoding elf2. Because a functional allele of elf2 remains present, sordarin binding should be reduced, but not eliminated, in the sordarin-resistant lysate compared with wild type. Indeed, as seen in Fig. 3d, bound drug signal was markedly lowered in the mutant strain compared with control. This result demonstrates the potential of TICC to evaluate target affinity in different cellular contexts, providing insight into a drug resistance mechanism.
Although sordarin has a low affinity for elf2, elf2 was one of the 67 candidate targets initially identified by LC-MS/MS in the drug fractions (supplemental Table 1). However, the presence of other confounding proteins (i.e. chance co-elution) obscured target verification.
We addressed this issue by improving chromatographic resolution by adding a heparin HPLC precolumn while increasing the number of collected fractions from 36 (HPLC method 1) to 120 (method 2). We evaluated the performance of this modified fractionation method with wild type yeast lysate dosed with sordarin. Bound sordarin was reproducibly detected in fractions 52–55, and the corresponding proteins were identified using a more sensitive LTQ Orbitrap Velos instrument (supplemental Table 1). After filtering the candidates to only proteins detected in the drug-bound fractions using both HPLC methods 1 and 2, the bona fide target (elf2) showed the highest profile correlation (0.81) to sordarin (see supplemental Fig. 4d for overlaid drug-protein profiles), establishing the potential power of optimized TICC procedures to correctly identify the targets of even low affinity compounds. We also conclude that single column fractionation will generally not be sufficient for target identification, whereas the use of two or more complementary chromatographic methods can markedly improve target fidelity.
In addition, we examined the technical reproducibility of high resolution fractionation (method 2) by performing fractionations of yeast extract dosed with 1 μm of sordarin in triplicate. The results were highly reproducible, as shown by the overlaid profiles (supplemental Fig. 4). The drug binding response was linear (linear regression, r2 of 0.9997) over the concentration range tested (0.2–10 μm) (supplemental Fig. 4e). Nevertheless, we failed to detect an interaction using a low dose (0.02 μm) of sordarin, indicating the practical limit of detection of the assay.
Application of TICC to the Identification of Low Abundance Targets
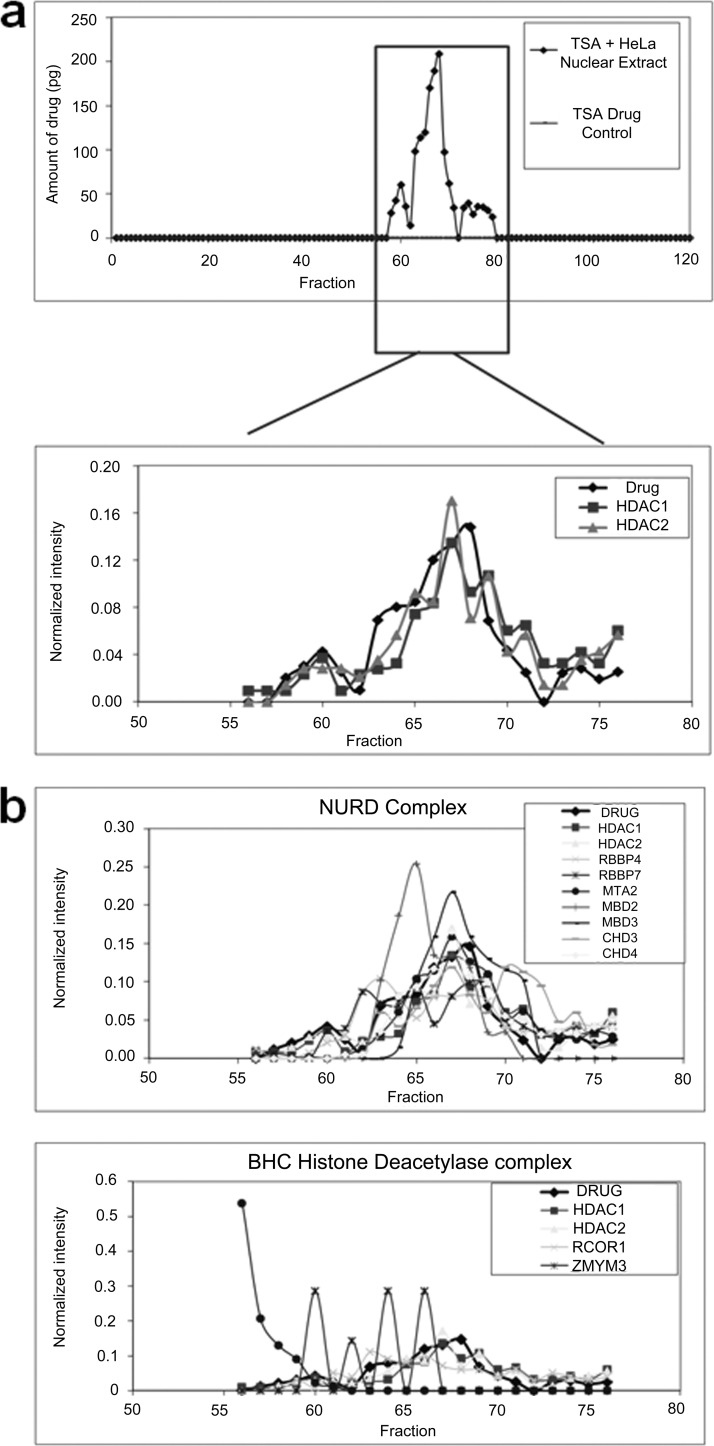
To test the performance of TICC to detect low abundance protein targets, we dosed a HeLa nuclear protein lysate with a 1 μm final concentration of TSA, a potent (Ki = 3.4 nm) inhibitor of human histone deacetylase such as the paralogs histone deacetylase 1 (HDAC) and HDAC2 (35, 36) and subjected the mixture to heparin dual ion exchange chromatography (HPLC fractionation method 2). To enhance detection, we used a high performance and sensitive LTQ Orbitrap Velos instrument for protein identification and an optimized SRM assay for TSA as described under “Experimental Procedures.” The results showed TSA binding in fractions 58–75. Consistent with expectation, HDAC1 and 2 were identified in the bound drug fractions (supplemental Table 2) and showed excellent overall agreement with the drug profile (Fig. 4a). Among these, nine other proteins also had high correlation (≥0.6) to TSA consistent with co-elution. Interestingly, two of these proteins are annotated subunits of either the nucleosome remodelling and histone deacetylation or other HDAC1/2-containing complexes, giving a preliminary indication that TICC may be able to detect not only direct physical interactors but also the actual endogenous macromolecular complex targeted by a drug (20) (Fig. 4b and supplemental Fig. 5). Proteins demonstrating similar matching profiles to a compound of interest by TICC may therefore provide additional insights into biological mechanisms.
Fig. 4.
Target identification for trichostatin A. a, The amount of TSA in each fraction is quantified after tandem heparin dual IEX-HPLC fractionation (method 2) of 2.4 mg of HeLa lysate dosed with 1 μm of TSA (top panel). The overlaid profiles of TSA with histone deacetylases HDAC1 and HDAC2 are shown in bottom panel, and HDAC1 and HDAC2 are known interactors of TSA. b, Drug elution profiles overlaid with two example histone deacetylase complexes shows good agreement of drug with nucleosome remodelling and histone deacetylation (NURD) complex (top panel) and poor agreement with BHC histone deacetylase complex (bottom panel) using the protein data collected in this study.
As shown in Fig. 4a, we did not observe any free drug peak during the fractionation of TSA standard solution, showing the free compound interacts nonspecifically with the column and/or is not eluted efficiently under chromatographic conditions employed. We investigated this issue further and determined the cause to be irreversible adsorption to the heparin column for certain compounds such as TSA. From an analytical perspective, this does not affect the performance of TICC for target detection and may even offer some advantages in improving signal to noise obtained for some bound compounds. Alternatively, heparin can be omitted for applications where drug recovery is problematic.
Application of TICC to Novel Target Identification for Anti-fungal Compound 4513-0042 Using MudPIT Strategy to Find Low Abundance Target
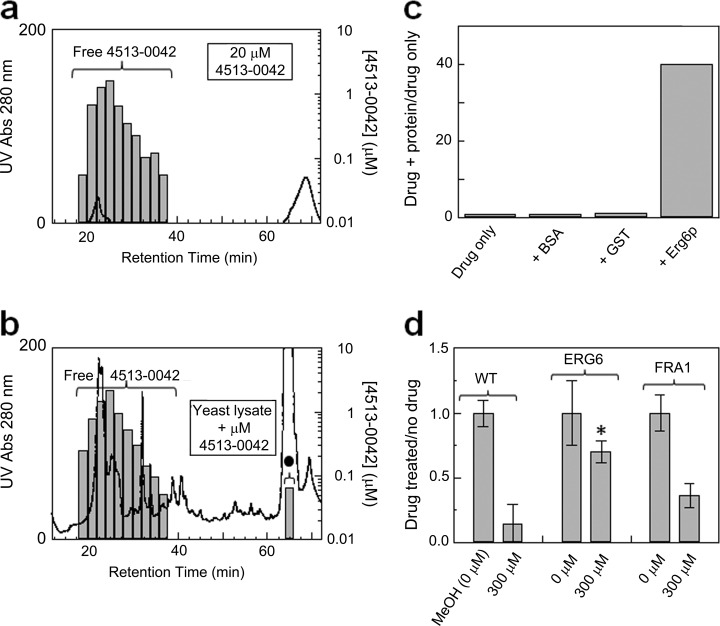
To evaluate the potential of TICC for unknown target identification, we first applied TICC to yeast lysates with the aim of characterizing the mechanism of action of an antifungal natural product, 4513-0042, which was recently proposed to disrupt ergosterol biosynthesis (37). 4513-0042 contains a hallmark azole ring common to drugs targeting the essential yeast membrane-associated protein Erg11p (38), and based on genetic perturbation criteria, Hoon et al. (37) previously reported Erg11p as a potential target of 4513-0042. We performed dual IEX-HPLC fractionation (method 1) of wild type cell extract after dosing of 20 μm of the compound.
As seen in Fig. 5, a single putative drug-target complex was evident by a shifted chromatographic fraction (cf. panels a and b). Because the bound drug fraction coincided within a prominent peak of abundant ribosomal proteins, both this and four adjacent fractions were analyzed by MudPIT to achieve deeper proteomic coverage (21). Although we did not detect Erg11p, likely because membrane-associated proteins were not efficiently solubilized by our extraction procedure, one candidate of the 69 proteins that had elution patterns similar to the drug (supplemental Table 3) was Erg6p, a cytoplasmic delta (24)-sterol C-methyltransferase in the same core ergosterol pathway. Of the 69 proteins identified, Erg6p is the only protein that participates in the ergosterol pathway, whereas the remaining 68 candidates are likely to be irrelevant proteins that co-fractionated with the drug as abundant components of a prominent co-eluting ribosomal peak or are involved in other process(es) that may not be directly relevant to ergosterol biosynthesis (supplemental Table 3).
Fig. 5.
Target identification for the anti-fungal compound 4513-0042. Dual IEX-HPLC elution profile of the antifungal compound 4513-0042 alone (a) or after in vitro dosing to a yeast whole cell extract (b). A new peak representing protein-bound drug (black circle) is evident at 64–66 min. c, plot showing the relative binding of 4513-0042 to purified GST-Erg6p, GST alone, or a BSA control. After incubation and separation by spin column gel filtration, the amount of compound in the protein-bound flow-through fraction (void volume) was quantified by SRM. d, plasmid-based overexpression of GST-Erg6p confers significant resistance to 4513-0042 as compared with a parental wild type (WT) strain or cells expressing an unrelated yeast factor (GST-Fra1p). Growth was recorded using triplicate cell culture readings at A600, and the ratio of drug treatment to no drug control was plotted. The error bars represent the coefficient of variation; a two-tailed Student's t test (equal variance) was applied. *, p < 0.01 compared with wild type 300 μm sample.
To confirm Erg6p as a bona fide target of 4513-0042, we affinity-purified endogenous Erg6p from wild type yeast and assessed direct binding to drug using ultrafiltration. As negative controls, we incubated compound with equivalent amounts of BSA and purified GST proteins in parallel. After incubation on ice, the amount of 4513-0042 present in the excluded volume in protein-bound form was quantified by SRM. Fig. 5c shows the normalized drug amounts measured in the respective void volumes. Consistent with the TICC data, GST-Erg6p showed pronounced drug binding compared with the controls, confirming that Erg6p can directly associate physically with 4513-0042.
To establish whether Erg6p is a physiologically relevant target, we examined whether overexpression of ERG6 confers resistance to 4513-0042 in vivo, as was reported previously for ERG11 (37). As a specificity control, we examined expression of Fra1, which encodes a regulator unrelated to ergosterol biosynthesis. As shown in Fig. 5d, elevated levels of ERG6 conferred a striking and highly significant (p < 0.01) resistance to 4513-0042 relative to wild type or Fra1-expressing yeast cells in the presence of compound. Taken together, these results suggest that Erg6p is a biologically relevant secondary target of 4513-0042.
Multiple Target Identification for Dopamine Receptor Agonist A77636
As a final application, we again applied TICC to yeast to investigate the off-target effects of the psychoactive drug A77636, a potent D1 dopamine receptor agonist that is reported to perturb protein glycoslyation, vesicle transport, and telomere biology in S. cerevisiae (39). After dosing 1 μm of the compound into soluble yeast extract, we detected three discrete drug binding peaks (fractions 13–18, 28–30, and 48–50) after fractionation by heparin dual ion exchange fractionation (HPLC method 2). Because the initial set of candidate proteins that closely co-eluted with drug (correlation, ≥0.6) representing putative targets was extensive (supplemental Table 4), we applied an alternate fractionation procedure based on weak cation exchange chromatography with acetate gradient elution at pH 6.0, which likewise produced three discrete bound drug peaks (fractions 17–31, 45–47, and 72–76), in excellent agreement with the heparin dual IEX results. (The overall workflow employed in this particular study, which is recommended as a general approach for TICC-based screening of complex samples, is summarized in detail in supplemental Fig. 7.)
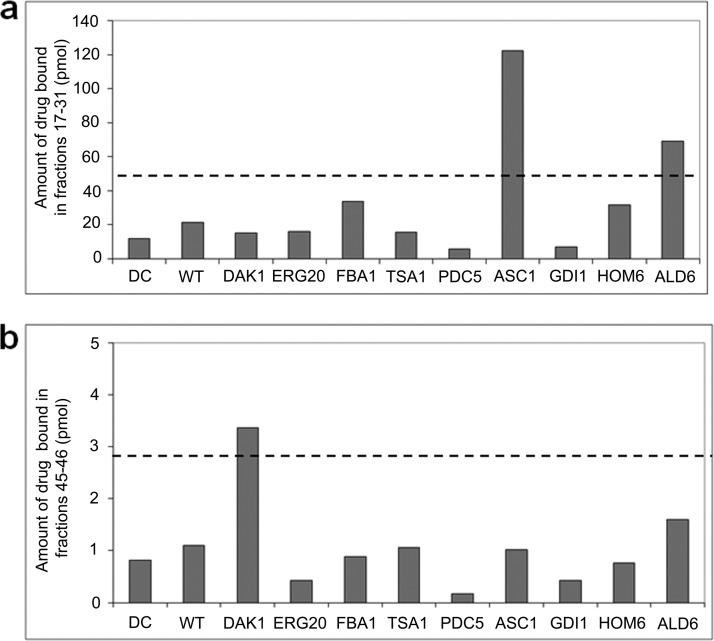
For WCX fractions 17–31, ASC1p, which is an ortholog of human RACK1, a regulator of adenylate cyclase that modulates D1 receptor internalization and the main pharmacological target of A77636, was the only protein candidate identified in all drug-bound fractions (40). For the remaining two peaks, we examined the nine proteins identified in common between the two complementary fractionation results (supplemental Table 4). Except for DAL80, which was the only putative target identified reproducibly in fractions 72–76 but was not available in tagged form, we evaluated the other eight candidates for direct physical binding with compound based on co-immunoprecipitation (Fig. 6) followed by weak cation exchange chromatography TICC. Consistent with our initial TICC predictions, we observed an elevated amount of bound drug in fractions 17–31 with purified ASC1p, confirming it is a true drug target consistent with our initial TICC predictions, whereas for fractions 45–47, DAK1, a kinase involved in detoxification and stress adaptation, was likewise validated.
Fig. 6.
Target confirmation for the psychoactive drug A77636 using TAP immunoprecipitation. Amount of drug bound (pmol) in fractions 17–31 (a) and fractions 45–47 (b) after subjecting individual IP lysates of each putative protein target dosed with 2 μm A 77636 to weak cation exchange fractionation pH 6.0 method. Using this method, A77636 interacted nonspecifically with the column, and a low leakage background level (around limit of quantitation, 1 pmol) was observed in all fractions. The results for drug control (DC) fractionation and control wild type (WT) fractionations are also included as references. Dashed lines illustrate significance thresholds calculated as three times the average amount of drug found in drug control and wild type. ALD6 exceeds significance threshold but is not considered to bind A77636 because it does not exhibit the expected peak shape and has poor correlation to bound drug profile. ASC1 and DAK1 are validated as bona fide interacting proteins for A77636.
Therefore, this example shows that TICC can be used to monitor drug interactions with multiple targets (i.e. polypharmacy). Although the detailed molecular mechanisms remain to be elucidated, the identification of possible “off-targets” by TICC may help to explain certain side effects observed with antipsychotic treatment (41).
DISCUSSION
Target identification or validation is essential for drug discovery, lead optimization, and exploration of the mechanism of action of drugs and chemical probes including unexpected off-target effects. A major advance in this field would be the development of generic protein-drug interaction screening technologies that are sensitive, accurate, and well suited to high throughput implementation with diverse compounds and biological samples.
In principle, TICC has the potential to allow the routine identification of the native physical interaction partners of bioactive compounds and other ligands in an unbiased manner in different cell types or biological contexts, i.e. not just annotated proteins involved in a known or predicted pathway. The method enables quantitative tracking of target-compound interactions in a physiologically relevant context without the need for labeling or immobilization of either the protein or compound. Although fractionation of ligand-bound proteins has been described before (12, 13), combining high resolution nondenaturing HPLC with sensitive tandem mass spectrometry-based drug and proteomic profiling together is novel. Furthermore, because the compound is not derivatized or modified prior to profiling, TICC minimizes artifacts by maintaining nearly native drug-target association parameters. We have established here that TICC is highly reproducible, scalable, automatable, and flexible in that one generically needs: (i) a ligand of interest that is detectable by MS; (ii) a nondisruptive fractionation procedure, such as IEX-HPLC; (iii) an accurate and sensitive readout for ligand detection and quantification, for which LC-MS/MS in selected reaction monitoring is well established; (iv) a procedure for protein identification, for which shotgun LC-MS/MS is optimal; and (v) a suitable biological source for target(s). In practice, target engagement can be examined using soluble protein lysates prepared from cultured cells (e.g. primary or transformed cell-lines), animal tissues, or model organisms (e.g. yeast) both after compound treatment in vivo or sample dosage in vitro prior to fractionation.
A key requirement for the success of the technique is the ability to separate free ligand versus protein-bound drug, which is likely when the compound of interest occupies a hydrophobic pocket or grove in the target. For complex mixtures, target deconvolution critically depends on resolving bona fide drug target from co-eluting “bystander” macromolecules exhibiting similar biophysical retention properties. To this end, we have exploited the concept of using multiple complementary/orthogonal fractionations to reduce the number of candidate targets prioritized for follow-up biochemical validation. This approach also allowed us to identify secondary targets of polypharmacological compounds such as A77636.
TICC is complementary to existing chemical genetic assays for drug target identification. Although in some cases chemical genetic assays can identify a list of genes that could be a mixture of true direct targets and proteins involved in the same or parallel biological pathway(s) as the target, the proteomic platform discussed here might be able to identify the targets that directly physically interact with the compounds of interest. The combined methods together can provide a common ground for target elucidation, validation, and characterization, while contributing to our understanding of biological pathways and networks affected by bioactive compounds.
Ultimately, the identification of low abundance or weakly bound targets is dictated by the combined effectiveness of minimizing irrelevant spurious (nontarget) proteins, the dynamic range of the target protein identification procedure, and the sensitivity of analytical LC-MS/MS drug assay. Examples we show in this study demonstrate strategies that can address these factors, such as the use of multiple fractionations to narrow down putative target lists, the use of high performance instrumentation to ensure detection of binding to low abundance and weak binding interactions, and the use of MudPIT to identify low abundance targets that may be missed with one-dimensional LC-MS/MS protein workflows. TICC performance for compounds that do not ionize well may require scaling the procedure to process larger amounts of biological material. Because the stability of the drug-target complex during the experiment is paramount, it is also imperative to use gentle nondenaturing buffers to preserve target-drug association. The development of more effective and rapid separations (e.g. ultrahigh pressure HPLC) combined with more accurate isotope label-based protein quantification procedures should improve overall assay performance.
TICC is applicable to both traditional target-based drug discovery pipelines and the characterization of chemical probes resulting from phenotypic screens and can potentially provide useful information regarding drug mechanism of action, selectivity, and off-target effects in a systematic, hypothesis-generating way. In principle, its applicability extends beyond small compounds, to include any monitor the interaction of other ligands, such as peptides or even antibodies, with proteins. Because data analysis is based on profile correlation, target specificity, affinity, and abundance can be systematically evaluated by titrating the ligand of interest across a physiologically relevant dose range, i.e. increasing drug concentration should drive target-ligand complex formation. For well behaved ligand-target combinations, TICC can provide both qualitative, i.e. identity of candidate target(s), and quantitative, i.e. target occupancy, stoichiometry and affinity information that are equally valuable for understanding the pharmacological properties of drugs. Furthermore, our results indicate that TICC may be useful for the characterization of protein complexes as drug targets.
Conversely, the study of membrane proteins, an important category for drug discovery, remains challenging and will require solubilization procedures and mild detergents compatible with HPLC and LC-MS/MS that do not disrupt the binding interaction between protein and ligand. Such efforts to modify TICC workflow to extend its applicability to membrane targets, are currently under way in our laboratory. In summary, the applicability of TICC is currently limited to noncovalent primarily hydrophobic protein-ligand interactions, to biological samples containing soluble proteins, and to protein-ligand interactions in the nanomolar to micromolar range, although future implementations of the technique may successfully address some of these challenges.
Acknowledgments
We thank E. Ericson and G. Giaever for advice and members of the Emili Laboratory for critical discussions and review of the manuscript. The pST39-HIS-DHFR expression vector was a kind gift from S. Tan. The pEG(KG) expression vector was a gift from R. Sopko. Sordarin-resistant yeast and sordarin were gifts from C. H. Ho.
J. N. Y. C. and D. V. designed and performed the fractionation experiments, analyzed and interpreted the data, and with A. E. co-wrote the manuscript. D. V. performed TSA, A77636, and sordarin (high resolution fractionation) SRM analyses and corresponding proteomic LC-MS/MS experiments. L. S. performed the methotrexate and radicicol SRM analyses and radicicol proteomic LC-MS/MS experiments. J. B. O. performed the SRM analyses and interpretation for 4513-0042. O. P. purified the TAP-tagged proteins for A77636 target validation. P. H. helped design fractionation experiments. N. B. performed the initial SRM analyses for sordarin. J. A. H. performed the proteomic LC-MS/MS procedure for the 4513-0042 and initial sordarin experiments. Y. W. purified DHFR and was involved in preliminary experiments. M. F. M. was involved in preliminary MTX SRM analyses. C. N. provided ongoing advice and target suggestions. A. E. devised the TICC method and directed the project from inception.
Footnotes
* This work was supported in part by operating funds from the Ontario Ministry of Innovation and the Council of Ontario Universities (to A.E.) and postdoctoral funding support from the Natural Sciences and Engineering Research Council of Canada (to D. V.).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- TICC
- target identification by chromatographic co-elution
- DHFR
- dihydrofolate reductase
- GST
- glutathione S-transferase
- HIS
- polyhistidine
- IEX
- ion exchange
- LTQ
- linear trap quadrupole
- MTX
- methorexate
- MudPIT
- multidimensional protein identification technology
- SRM
- selected reaction monitoring
- TSA
- trichostatin A
- SC
- synthetic complete
- WCX
- weak cation exchange
- elf
- elongation factor
- HDAC
- histone deacetylase.
REFERENCES
- 1. Chan J. N., Nislow C., Emili A. (2010) Recent advances and method development for drug target identification. Trends Pharmacol. Sci. 31, 82–88 [DOI] [PubMed] [Google Scholar]
- 2. Schreiber S. L. (2000) Target-oriented and diversity-oriented organic synthesis in drug discovery. Science 287, 1964–1969 [DOI] [PubMed] [Google Scholar]
- 3. Frantz S. (2005) Drug discovery: Playing dirty. Nature 437, 942–943 [DOI] [PubMed] [Google Scholar]
- 4. Keiser M. J., Setola V., Irwin J. J., Laggner C., Abbas A. I., Hufeisen S. J., Jensen N. H., Kuijer M. B., Matos R. C., Tran T. B., Whaley R., Glennon R. A., Hert J., Thomas K. L., Edwards D. D., Shoichet B. K., Roth B. L. (2009) Predicting new molecular targets for known drugs. Nature 462, 175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mestres J., Gregori-Puigjané E. (2009) Conciliating binding efficiency and polypharmacology. Trends Pharmacol. Sci. 30, 470–474 [DOI] [PubMed] [Google Scholar]
- 6. Hopkins A. L. (2008) Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 4, 682–690 [DOI] [PubMed] [Google Scholar]
- 7. Giaever G., Flaherty P., Kumm J., Proctor M., Nislow C., Jaramillo D. F., Chu A. M., Jordan M. I., Arkin A. P., Davis R. W. (2004) Chemogenomic profiling: Identifying the functional interactions of small molecules in yeast. Proc. Natl. Acad. Sci. U. S.A. 101, 793–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parsons A. B., Lopez A., Givoni I. E., Williams D. E., Gray C. A., Porter J., Chua G., Sopko R., Brost R. L., Ho C. H., Wang J., Ketela T., Brenner C., Brill J. A., Fernandez G. E., Lorenz T. C., Payne G. S., Ishihara S., Ohya Y., Andrews B., Hughes T. R., Frey B. J., Graham T. R., Andersen R. J., Boone C. (2006) Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell 126, 611–625 [DOI] [PubMed] [Google Scholar]
- 9. Lamb J., Crawford E. D., Peck D., Modell J. W., Blat I. C., Wrobel M. J., Lerner J., Brunet J. P., Subramanian A., Ross K. N., Reich M., Hieronymus H., Wei G., Armstrong S. A., Haggarty S. J., Clemons P. A., Wei R., Carr S. A., Lander E. S., Golub T. R. (2006) The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 [DOI] [PubMed] [Google Scholar]
- 10. Terstappen G. C., Schlüpen C., Raggiaschi R., Gaviraghi G. (2007) Target deconvolution strategies in drug discovery. Nat. Rev. Drug Discov. 6, 891–903 [DOI] [PubMed] [Google Scholar]
- 11. Rix U., Superti-Furga G. (2009) Target profiling of small molecules by chemical proteomics. Nat. Chem. Biol. 5, 616–624 [DOI] [PubMed] [Google Scholar]
- 12. Peterson J. R., Lebensohn A. M., Pelish H. E., Kirschner M. W. (2006) Biochemical suppression of small-molecule inhibitors: A strategy to identify inhibitor targets and signaling pathway components. Chem. Biol. 13, 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lomenick B., Hao R., Jonai N., Chin R. M., Aghajan M., Warburton S., Wang J., Wu R. P., Gomez F., Loo J. A., Wohlschlegel J. A., Vondriska T. M., Pelletier J., Herschman H. R., Clardy J., Clarke C. F., Huang J. (2009) Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. U. S.A. 106, 21984–21989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ashburn T. T., Thor K. B. (2004) Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 3, 673–683 [DOI] [PubMed] [Google Scholar]
- 15. Ho C. H., Magtanong L., Barker S. L., Gresham D., Nishimura S., Natarajan P., Koh J. L., Porter J., Gray C. A., Andersen R. J., Giaever G., Nislow C., Andrews B., Botstein D., Graham T. R., Yoshida M., Boone C. (2009) A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat. Biotechnol. 27, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lichty J. J., Malecki J. L., Agnew H. D., Michelson-Horowitz D. J., Tan S. (2005) Comparison of affinity tags for protein purification. Protein Expr. Purif. 41, 98–105 [DOI] [PubMed] [Google Scholar]
- 17. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St, Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt JF. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 18. Butland G., Peregrín-Alvarez J. M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Davey M., Parkinson J., Greenblatt J., Emili A. (2005) Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433, 531–537 [DOI] [PubMed] [Google Scholar]
- 19. Sopko R., Huang D., Preston N., Chua G., Papp B., Kafadar K., Snyder M., Oliver S. G., Cyert M., Hughes T. R., Boone C., Andrews B. (2006) Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 21, 319–330 [DOI] [PubMed] [Google Scholar]
- 20. Havugimana P. C., Wong P., Emili A. (2007) Improved proteomic discovery by sample pre-fractionation using dual-column ion-exchange high performance liquid chromatography. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 847, 54–61 [DOI] [PubMed] [Google Scholar]
- 21. Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 22. Kislinger T., Rahman K., Radulovic D., Cox B., Rossant J., Emili A. (2003) PRISM, a generic large scale proteomic investigation strategy for mammals. Mol. Cell. Proteomics 2, 96–106 [DOI] [PubMed] [Google Scholar]
- 23. Eng J. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 24. Subramanian S., Kaufman B. T. (1978) Interaction of methotrexate, folates, and pyridine nucleotides with dihydrofolate reductase: Calorimetric and spectroscopic binding studies. Proc. Natl. Acad. Sci. U. S.A. 75, 3201–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang T., Barclay B. J., Kalman T. I., von Borstel R. C., Hastings P. J. (1992) The phenotype of a dihydrofolate reductase mutant of Saccharomyces cerevisiae. Gene 121, 167–171 [DOI] [PubMed] [Google Scholar]
- 26. Wooden J. M., Hartwell L. H., Vasquez B., Sibley C. H. (1997) Analysis in yeast of antimalaria drugs that target the dihydrofolate reductase of Plasmodium falciparum. Mol. Biochem. Parasitol. 85, 25–40 [DOI] [PubMed] [Google Scholar]
- 27. Bolin J. T., Filman D. J., Matthews D. A., Hamlin R. C., Kraut J. (1982) Crystal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 A resolution: I. General features and binding of methotrexate. J. Biol. Chem. 257, 13650–13662 [PubMed] [Google Scholar]
- 28. Falzone C. J., Wright P. E., Benkovic S. J. (1991) Evidence for two interconverting protein isomers in the methotrexate complex of dihydrofolate reductase from Escherichia coli. Biochemistry 30, 2184–2191 [DOI] [PubMed] [Google Scholar]
- 29. Bennett B., Langan P., Coates L., Mustyakimov M., Schoenborn B., Howell E. E., Dealwis C. (2006) Neutron diffraction studies of Escherichia coli dihydrofolate reductase complexed with methotrexate. Proc. Natl. Acad. Sci. U. S.A. 103, 18493–18498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bystroff C., Kraut J. (1991) Crystal structure of unliganded Escherichia coli dihydrofolate reductase: Ligand-induced conformational changes and cooperativity in binding. Biochemistry 30, 2227–2239 [DOI] [PubMed] [Google Scholar]
- 31. Roe S. M., Prodromou C., O'Brien R., Ladbury J. E., Piper P. W., Pearl LH. (1999) Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J. Med. Chem. 42, 260–266 [DOI] [PubMed] [Google Scholar]
- 32. Schulte T. W., Akinaga S., Soga S., Sullivan W., Stensgard B., Toft D., Neckers LM. (1998) Antibiotic radicicol binds to the N-terminal domain of Hsp90 and shares important biologic activities with geldanamycin. Cell Stress Chaperones 3, 100–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Justice M. C., Hsu M. J., Tse B., Ku T., Balkovec J., Schmatz D., Nielsen J. (1998) Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J. Biol. Chem. 273, 3148–3151 [DOI] [PubMed] [Google Scholar]
- 34. Domínguez J. M., Martin J. J. (2001) Identification of a putative sordarin-binding site in Candida albicans elongation factor 2 by photoaffinity labeling. J. Biol. Chem. 276, 31402–31407 [DOI] [PubMed] [Google Scholar]
- 35. Yoshida M., Kijima M., Akita M., Beppu T. (1990) Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265, 17174–17179 [PubMed] [Google Scholar]
- 36. Kong X., Fang M., Li P., Fang F., Xu Y. (2009) HDAC2 deacetylates class II transactivator and suppresses its activity in macrophages and smooth muscle cells. J. Mol. Cell Cardiol. 46, 292–299 [DOI] [PubMed] [Google Scholar]
- 37. Hoon S., Smith A. M., Wallace I. M., Suresh S., Miranda M., Fung E., Proctor M., Shokat K. M., Zhang C., Davis R. W., Giaever G., St. Onge R. P., Nislow C. (2008) An integrated platform of genomic assays reveals small-molecule bioactivities. Nat. Chem. Biol. 4, 498–506 [DOI] [PubMed] [Google Scholar]
- 38. Tkacz J. S., DiDomenico B. (2001) Antifungals: What's in the pipeline. Curr. Opin. Microbiol. 4, 540–545 [DOI] [PubMed] [Google Scholar]
- 39. Ericson E., Gebbia M., Heisler L. E., Wildenhain J., Tyers M., Giaever G., Nislow C. (2008) Off-target effects of psychoactive drugs revealed by genome-wide assays in yeast. PLoS Genet. 4, e1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ryman-Rasmussen J. P., Nichols D. E., Mailman R. B. (2005) Differential activation of adenylate cyclase and receptor internalization by novel dopamine D1 receptor agonists. Mol. Pharmacol. 68, 1039–1048 [DOI] [PubMed] [Google Scholar]
- 41. Arana G. W. (2000) An overview of side effects caused by typical antipsychotics. J. Clin. Psychiatry 61, (Suppl. 8) 5–13 [PubMed] [Google Scholar]