Abstract
This study reports a global glycoproteomic analysis of pancreatic cancer cells that describes how flux through the sialic acid biosynthetic pathway selectively modulates a subset of N-glycosylation sites found within cellular proteins. These results provide evidence that sialoglycoprotein patterns are not determined exclusively by the transcription of biosynthetic enzymes or the availability of N-glycan sequons; instead, bulk metabolic flux through the sialic acid pathway has a remarkable ability to increase the abundance of certain sialoglycoproteins while having a minimal impact on others. Specifically, of 82 glycoproteins identified through a mass spectrometry and bioinformatics approach, ∼31% showed no change in sialylation, ∼29% exhibited a modest increase, whereas ∼40% experienced an increase of greater than twofold. Increased sialylation of specific glycoproteins resulted in changes to the adhesive properties of SW1990 pancreatic cancer cells (e.g. increased CD44-mediated adhesion to selectins under physiological flow and enhanced integrin-mediated cell mobility on collagen and fibronectin). These results indicate that cancer cells can become more aggressively malignant by controlling the sialylation of proteins implicated in metastatic transformation via metabolic flux.
The surfaces of mammalian cells are covered with a dense layer of carbohydrates, collectively known as the glycocalyx, that influence many aspects of the interaction between a cell and its microenvironment. To date, the biosynthesis of cell surface displayed glycans has been thought to be controlled largely by individual glycosyltransferases based on the assumption that flux through the metabolic pathways that supply activated nucleotide sugar donors (the substrates for these enzymes) is not a limiting factor. For example, this premise has been used in mathematical models of sialylation (1), where concentrations of CMP-Neu5Ac in the lumen of the Golgi were assumed to be much higher than the Km of sialyltransferases (2). In the past several years, the idea that nucleotide sugars, exemplified by CMP-Neu5Ac (shown in Fig. 1A), are not a limiting or controlling factor in glycosylation has garnered one major and unambiguous exception, notably that changes in flux through the hexosamine biosynthetic pathway (HBP)1 can alter UDP-GlcNAc levels with profound consequences on the branching of N-linked glycans (3). By changing the valence of these glycoconjugates, flux through the HBP can alter the galectin lattice and affect a host of downstream biological events including cancer progression (4); cell differentiation and proliferation (3); and autoimmunity, metabolic syndromes, and aging (5).
Fig. 1.
Overview of the sialic acid biosynthetic pathway and the selective sialylation of certain glycosites. A, Sialic acid biosynthesis begins with ManNAc, which is naturally supplied into the sialic acid biosynthetic pathway by conversion from UDP-GlcNAc by UDP-GlcNAc 2-epimerase (GNE). B, CMP-Neu5Ac produced from ManNAc is the substrate for a suite of sialyltransferases (STs) that install sialic acids into cell surface displayed protein- and lipid-bound glycoconjugates (in humans 20 STs exist that install either α2,3-, α2,6-, or α2,8-linked sialosides as described in detail elsewhere (44)). C, Using a “metabolic oligosaccharide engineering” approach (44, 45), increased flux was introduced into the pathway via 1,3,4-O-Bu3ManNAc (9). D, By increasing cellular levels of CMP-Neu5Ac, 1,3,4-O-Bu3ManNAc led to selectively enhanced sialylation of a subset of the glycans present on the cell surface; illustrative examples are provided by one of the potential N-glycans of CD44 (highlighted) and two of integrin α6 (please see references (17–19), Table I, and Fig. 7 for additional details).
In this report, we demonstrate that the HBP is not unique in its ability to control surface glycoproteins via bulk metabolic flux. In particular, counter to earlier assumptions that flux through the sialic acid pathway does not significantly alter the sialylation of individual glycans (2), analysis of two “high-demand” sialoglycans (i.e. polysialylated NCAM (6) and podocalyxin (7)) suggested that fluctuations in the intracellular concentrations of sialic acid and the corresponding supply of CMP-Neu5Ac critically affected their production. In the current report, we used a global cell level approach to investigate whether these two examples were outliers or whether metabolic flux controls the surface display of sialic acid with fine resolution across a wide range of N-linked glycoproteins. We found that the sialylation of certain N-linked glycoproteins increased dramatically (e.g. by five- to eightfold) when flux through the sialic acid pathway was enhanced by exogenously supplied substrate whereas there was negligible effect on other glycoproteins. Importantly, these changes altered the adhesive behavior of cancer cells in a manner consistent with the glycoproteomic analysis. Together, these findings expand the role of metabolic flux in controlling glycosylation beyond the HBP and provide a foundation to explore the hypothesis that cancer cells modulate their metastatic potential and malignant progression via changes to bulk metabolic flux through the sialic acid biosynthetic pathway.
EXPERIMENTAL PROCEDURES
Materials: Adhesion Molecules, Antibodies, and Reagents
E-selectin-IgG Fc (E-selectin) l-selectin-IgG Fc (l-selectin), P-selectin-IgG Fc (P-selectin), and unlabeled anti-CD44 mAbs (2C5) were purchased from R & D Systems (Minneapolis, MN). Alkaline phosphatase (AP)- and horseradish peroxidase (HRP)-conjugated anti-mouse IgG and AP-conjugated anti-rat IgM were from Southern Biotech (Birmingham, AL). Unlabeled anti-CD44 mAbs (515), and HECA 452, were purchased from Abcam (Cambridge, MA). Functional anti-integrin α6 mAbs (GoH3) was purchased from Novus Biologicals (Littleton, CO). Fluorescein labeled Sambucus nigra Lectin (SNA) and Ricinus communis agglutinin I (RCA) were purchased from Vector Labs, (Burlingame, CA) and the FITC-conjugated Maackia amurensis agglutinin (MAA) lectin was from EY Laboratories (San Mateo, CA). All other reagents were from Sigma-Aldrich (St. Louis, MO) unless otherwise stated.
Cell Culture
The human pancreatic carcinoma cell line SW1990 was obtained from the American Type Culture Collection (Manassas, VA) and cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 1.0% (v/v) of a 100 × stock solution of penicillin and streptomycin (Invitrogen, Carlsbad, CA). Prior to cell lysis, pancreatic cells were detached from culture flasks using Enzyme Free Cell Dissociation Media (20 min at 37 °C; Chemicon, Phillipsburg, NJ). CHO cells stably transfected with full-length E-selectin (CHO-E) or with phosphatidylinositol glycan-linked extracellular domain of P-selectin (CHO-P) were kindly donated by Affymax (Palo Alto, CA) and cultured as described previously (8). Synthesis of 1,3,4-O-Bu3ManNAc followed a previously reported procedure (9).
Periodate Resorcinal Assay
Total and bound sialic acid were quantified by the periodate resorcinol method. Briefly, treated and nontreated SW1990 cells were harvested with Enzyme Free Cell Dissociation Media and counted. The cells (2.25 × 106) were washed twice with 1.0 ml D-PBS (Dulbecco's phosphate-buffered saline), pelleted, resuspended in 200 μl D-PBS and divided into two 100 μl aliquots. Cell lysates were obtained by four freeze/thaw cycles; the lysates were then analyzed by the method of Jourdian and coworkers (10) adapted for a 96-well plate format (11). Oxidation of aliquots of each sample was performed in parallel at 0 °C and 37 °C to measure total sialic acid or glycoconjugate-bound sialic acid (note that CMP-sialic acid, although soluble, is detected in the what has conventionally been termed the “glycoconjugate-bound” fraction in this assay), respectively. Sialic acid concentrations were calculated by comparison with a standard curve (0–250 μm sialic acid) and expressed as the fold change from the control.
Lectin and Antibody Analysis of SW1990 Cells
Surface sialic acid was analyzed by labeling with fluorescein-conjugated RCA, FITC-conjugated MAA, and fluorescein-conjugated Sambucus nigra agglutinin (SNA) lectins. In brief, 500,000 SW1990 cells were harvested as described above, washed with D-PBS, and resuspended in 100 μl D-PBS. Lectin (4.0 μl at 0.1 mg/ml) was added and the cell-lectin suspension was incubated for 1.0 h. Excess lectin was removed by washes with 1.0 ml D-PBS and the cells were analyzed by flow cytometry as described previously (12). Monoclonal antibody binding to determine LeX, sLeX, and sLeA expression was performed by washing 500,000 cells with D-PBS and incubating them with anti-human CD15 FITC (eBioscience Inc., San Diego, CA), 10 μg/ml of mAb PE-labeled HECA 452 (PharMingen, San Diego, CA) in 100 μl D-PBS with 0.1% BSA, or anti-sialyl Lewis A (Millipore, Billerica, MA), respectively. Background fluorescence for each antibody was determined using isotype-matched Ig. The samples were measured in a flow cytometer (BD Biosciences, San Diego, CA) and the data were analyzed using the FlowJo software (Treestar, Inc., San Carlos, CA).
Western Analysis of CD44
Proteins (12 μg) were resolved by SDS-PAGE and transferred electrophoretically onto a nitrocellulose membrane. The membrane was blocked with 5% nonfat milk/0.1% TBS-Tween 20 at room temperature for 2.0 h and then probed with anti-human CD44 antibody (515) at 1:1000 at 4 °C overnight, followed by three washes with 0.1% TBS-Tween 20. HRP-conjugated anti-mouse IgG antibody was added at 1:2000 and incubated at room temperature for 1.0 h, followed by three washes with 0.1% TBS-Tween 20. The signal was visualized using SuperSignal Substrate (Pierce, Rockford, IL).
Mass Spectroscopy and Glyocopeptide Identification
Replicates of SW1990 cells (9 × 106) with or without treatment with 1,3,4-O-Bu3ManNAc were harvested and lysed using radioimmunoprecipitation assay buffer (Sigma-Aldrich) according to the manufacturer's instructions. The protein concentration was determined using the BCA assay (Pierce); 1.4 mg of protein was used in following experiment for each cell treatment condition. Urea (8.0 m) and tris(2-carboxyethyl) phosphine hydrochloride (5.0 mm) were added to each sample to the indicated final concentrations followed by incubation at 60 °C for 2.0 h. Iodoacetamide (10 mm final concentration) was added to each sample and they were incubated for 30 min at RT in the dark. Each solution was diluted eightfold with 100 mm KH2PO4 (pH 8.0). Trypsin (Promega, 30 μg) was added to each sample followed by incubation at 37 °C overnight with shaking. Silver staining was used to monitor the completeness of trypsin digestion as indicated by the disappearance of high MW protein bands and the appearance of lower MW peptide bands (<10 kDa). After digestion, the samples were centrifuged at 16,110 × g for 5.0 min to remove particulate matter.
The peptides were cleaned by elution through a C18 column and divided into two equal parts, N-linked total glycopeptides were then isolated from one sample using the solid phase extraction of glycopeptides method (13–15). This method was modified to extract N-linked sialoglycopeptides from the second aliquots of each sample. Briefly, the modified solid phase extraction of glycopeptidesmethod used 1.0 mm precooled sodium periodate at 0 °C for 15 min to oxidize sialic acid selectively. The resulting sialoglycopeptides were covalently conjugated to a solid support via hydrazide chemistry whereas nonsialylated glycopeptides were removed by washing prior to release of N-linked glycopeptides from solid support by PNGase F. The resulting sialoglycopeptides were concentrated by elution through C18 columns, dried, and resuspended in 20 μl of 0.4% acetic acid.
The peptides (5.0 μl/sample) were labeled with isobaric tag for relative and absolute quantitation (iTRAQ) 8plex (AB SCIEX) according to the manufacturer's instructions. The formerly N-linked total glycopeptides from control cells were labeled in duplicate by iTRAQ with 113 and 114 whereas those from 1,3,4-O-Bu3ManNAc-treated cells were labeled with 115 and 116. The formerly N-linked sialoglycopeptides from control cells were labeled in duplicate by iTRAQ 117 and 118, whereas those from1,3,4-O-Bu3ManNAc-treated cells were labeled in duplicate with 119 and 121 separately. The labeled peptides were then mixed, cleaned using strong cation exchange columns, and resuspended in 15 μl of 0.4% acetic acid.
iTRAQ-labeled peptides (6 μl) were analyzed by liquid chromatography-tandem MS (LC-MS/MS) using an LTQ-Orbitrap velos (ThermoFisher, Waltham, MA) coupled with a 15 cm × 75 μm C18 column (5 μm particles with 100 angstrom pore size). A nanoAquity UPLC at 300 nl/min with a 90-min linear acetonitrile gradient (from 5–32% B over 90 min; A = 0.1% formic acid in water, B = 0.1% formic acid in acetonitrile) was used. Top 10 data dependent with exclusion for 20 s was set. The samples were run with HCD fragmentation at a normalized collision energy of 45 and an isolation width of 1.2 Da. Monoisotopic precursor selection was enabled and the dynamic exclusion was set to 30 s with a repeat count of 1 and ± 10 ppm mass window. The source voltage was 2.0 kDa. A lock mass of the polysiloxane peak at 371.10123 was used to correct the mass in MS and MS/MS. Target values in MS were 1e6 ions at a resolution setting of 30,000 and in MS2 1e5 ions at a resolution setting of 7500.
MS/MS spectra were searched with MASCOT (version 2.2.0) using Proteome Discoverer (version 1.0) (Thermo Fisher) against human subdatabase of NCBI Reference Sequence (RefSeq) (version 40, released at April 16, 2010) containing 29,704 sequences. Integration window tolerance was set at 20 ppm for peak integration. The peptide cutoff score was set at 30. For this database search, the precursor mass tolerance and fragment mass tolerance was set at 15 ppm and 0.05 Da respectively, fixed modifications were set as iTRAQ 8plex labeling (N-term and K) and carbamidomethylation (C), and other database-searching parameters were set as flexible modification as follows: deamidation (NQ) and oxidation (M). Semi-tryptic end and 1 missed cleavage site was allowed. The False Discovery Rate was set at 0.01 to eliminate low-probability protein identifications. The data generated by mass spectrometry for the global N-glycopeptides and N-sialoglycopeptides analysis may be downloaded from ProteomeCommons.org Tranche using the following hash:
dcLmbIAQlBZvBYIgiJjGG5om/IK/fq7e+5TpB3TAg7PcWhk6CJzgTuePjUH7HJktURuizj6JD7Ack9TJDkqr8y9WGGQAAAAAAAACtg==. The hash may be used to prove exactly what files were published as part of this manuscript's data set, and the hash may also be used to check that the data has not changed since publication. For single peptide identification, the matched spectra can be found in supplemental Table S2.
Kyoto Encyclopedia of Genes and Genomes Pathway Analysis of Identified Glycoproteins
The DAVID Bioinformatics Resources 6.7 tool (National Institute of Allergy and Infectious Diseases, NIAID, NIH) was used to analyze the glycoproteins and sialoglycoproteins identified in the present study and assign them into categories compiled by the KEGG database. All analysis parameters were used in the default setting.
Evaluation of the Cut-off of the Protein Abundance Ratio for Protein Changes
To correct for any systematic errors of protein ratio introduced by sample handling and to determine the appropriate cutoff for protein changes, the distribution of abundance ratios of glycosylation changes was generated. Because the majority of proteins were not expressed differently in two cell states, we normalized the ratio based on the distribution of the protein abundance ratios from two cell states. Proteins that fell outside the normal distribution from the abundance ratio of two cell states were considered as altered proteins. The threshold to select protein changes was based on the ratio distribution of two cell states. The mean and standard deviation of the ratio from the two cell states were calculated, and the abundance of proteins with an abundance ratio outside of one standard deviation from the mean were flagged as altered.
Flow-based Adhesion Assays
To simulate the physiological shear environment of the vasculature, pancreatic carcinoma cells in D-PBS containing Ca+2/Mg+2, 0.1% bovine serum albumin were perfused over immobilized E-, or l-selectin-coated dishes at prescribed wall shear stresses using a parallel plate flow chamber (250-μm channel depth, 5.0-mm channel width). Adhesion was quantified by perfusing cells at 1.0 × 106/ml and recording for 2 or 5 min. Average rolling velocities were computed as the distance traveled by the centroid of the translating cell divided by the time interval at the given wall shear stress (16).
Blot Rolling Assays
SW1990 whole cell lysate was prepared by membrane disruption using 2.0% Nonidet P-40 followed by differential centrifugation. Blots of immunopurified CD44 from treated and untreated SW1990 whole cell lysate were stained with anti-CD44 (2C5) or anti-CD44 (515) or HECA-452 mAbs and rendered translucent by immersion in 90% D-PBS 10% glycerol. The blots were placed under a parallel plate flow chamber CHO transfectants expressing P- or E-selectin, resuspended at 1.0 × 106 cells/ml in 90% D-PBS/10% glycerol, and perfused at the shear stress of 0.5 dynes/cm2 (16). Molecular weight markers were used as guides to aid placement of the flow chamber over stained bands of interest. The number of interacting cells per lane was averaged over five 10× fields of view (0.55 mm2 each) within each stained region. Nonspecific adhesion was assessed by perfusing 5.0 mm EDTA in the flow medium.
Surface Plasmon Resonance Binding Assay
Surface plasmon resonance binding studies were performed using a BIAcore 3000 system (BIAcore Inc., Piscataway, NJ). Recombinant human E-, l-, and P-selectin-Fc chimera (500 μg/ml in 50 mm sodium acetate, pH 4.75; R&D Systems) were covalently immobilized via amine coupling to a sensor chip (CM5) to channels 2, 3, and 4 respectively as directed by the manufacturer (BIAcore). IgG was immobilized to Channel 1 and used as control. To protect the binding sites, CaCl2, MgCl2, fucose, and galactose were added to the immobilization buffer at a final concentration of 1.0 mm each. The immobilization of the selectins resulted in an average of 2500 resonance units. The binding assays were performed in PBS, with 1.0 mm CaCl2 and 1.0 mm MgCl2, pH 7.4 at 25 °C with a flow rate of 10 μl/min. The interactions of the same concentration of immunoprecipitated CD44 from 1,3,4-O-Bu3ManNAc treated and untreated control SW1990 cells were monitored and the signal response was recorded and corrected for nonspecific binding to the control channel. Data was analyzed using BIAevaluation software (V4.1, BIAcore).
Wound Healing Assays
Culture plates were coated with 20 μg/ml collagen-1 or 20 μg/ml fibronectin (BD Biosciences, Bedford, MA) in D-PBS. Cells were plated and 4.0 μl from a 50 mm stock solution of 1,3,4-O-Bu3ManNAc was added to treated samples (to give a final analog concentration of 100 μm) and 4.0 μl of 100% ethanol was added to the controls. Cells were incubated for 24 h at 37 °C to allow the formation of a confluent monolayer that was scratched by carefully dragging a p200 pipette tip across the cells. Fresh medium without FBS, and with or without 1,3,4-O-Bu3ManNAc was added, and the cells were incubated at 37 °C on a live-cell microscope where pictures of wound closure were taken every 20 min for 24 h. The wound area was measured using the Nikon Imaging Software (NIS) for each time point.
Modeled Representations of Identified Glycopeptides in the Overall Structures of CD44 and Integrin α6
For CD44, the identified glycan was modeled onto the NMR structure of CD44 (PDB ID:1UU), using the program GlyProt (http://www.glycosciences.de/glyprot/) (17). Because a corresponding structure is not available for integrin α6, the structure of the integrin αV (PBD ID: 3ije) was used to model the subunit repeat of the propeller where the identified glycopeptide was located using a comparative protein structural modeling approach (18). The GlycoProt program was used to N-glycosylate the model subunit with a simplified N-glycan structure. Visualization of structures was performed with the Chimera software (19).
Statistical Analysis
Data are expressed as the mean S.E. for at least three independent experiments. Statistical significance of differences between means was determined by analysis of variance.
RESULTS
1,3,4-O-Bu3ManNAc Increased Intracellular and Surface Sialic Acid
Pancreatic cancer SW1990 cells incubated with 1,3,4-O-Bu3ManNAc experienced an increase in total and glycoconjugate-bound sialic acid (Fig. 2A) consistent with previous results that cells incubated with ManNAc analogs have greatly increased levels of intracellular sialic acid and more modestly elevated surface display of this monosaccharide (12, 20, 21). Lectin binding analysis (Fig. 2B) confirmed that the surface display of sialic acid increased insofar as the number of “empty” penultimate galactose/GalNAc residues decreased (as measured by RCA binding) whereas a corresponding increase in the surface display of α2,6-linked (SNA) and α2,3-linked (MAA) sialic acids occurred. Similarly, antibody binding assays (Fig. 2C) showed that Lewis X (LeX) levels decreased in analog-treated cells whereas sialyl Lewis X (sLeX) and sialyl Lewis A (sLeA) levels increased. These results demonstrate that the overall level of glycans on the cell surface was not changing; instead the fraction of sialylated N-glycans was altered by analog-driven flux through the sialic acid pathway. It is noteworthy that the analysis of nonsialylated epitopes (e.g. CD15 (Fig. 2G)) indicated that analog-enhanced flux through the sialic pathway did not simply saturate all possible sites of sialic acid attachment because if it did, CD15 levels would be reduced to the levels observed in the isotype control. Once we established that bulk flux through the sialic acid pathway in SW1990 cells altered surface sialic acid display, we used mass spectrometry-based methods to identify the specific sites of glycosylation associated with these changes and then investigated whether these changes resulted in altered cell behavior, as described below.
Fig. 2.
Changes in intracellular and surface sialic acid in 1,3,4-O-Bu3ManNAc-treated SW1990 cells. A, Total (which includes all forms of sialic acid found within a cell, black bars) and “glycoconjugate-bound” (which also includes soluble CMP-sialic acid, white bars) sialic acid was measured by using the periodate resorcinol assay in cells incubated with the indicated concentrations of 1,3,4-O-Bu3ManNAc for 2 days. Cells treated with 100 μm 1,3,4-O-Bu3ManNAc for 2 days were analyzed by (B) lectin binding or (C) flow cytometry. In panels A, B, and C, the error bars represent the S.D. of three different experiments with representative flow cytometry data for lectins shown in (D) ricin agglutinin (RCA), (E) Sambucus nigra agglutinin (SNA), or (F) Macckia amurensis agglutinin (MAA) and for flow cytometry in (G) α-CD1, (H) α-CD15s (sLeX), and (I) sLeA.
Glycoproteomic Analysis of Metabolic Flux-driven Changes to N-Glycan Sialylation
Glycoproteomic methods were next used to perform a global analysis of sialylation changes that occurred in 1,3,4-O-Bu3ManNAc treated SW1990 cells. More specifically, to identify individual proteins whose sialylation status was impacted by analog-driven flux, the formerly N-linked total glycopeptides and sialoglycopeptides were isolated from cells that had been pretreated or not with 1,3,4-O-Bu3ManNAc, labeled with iTRAQ reagents, and analyzed by mass spectrometry (13–15, 22). Two proteins, CD44 and integrin α6, were used as examples to illustrate this process, as shown in Fig. 3. When conducted on a global scale to identify cell wide changes with a cutoff of a 1% false discovery rate, 105 unique glycopeptides (containing the NXS/T motif) were identified representing 82 unique glycoproteins; the ratio of the peptide level and protein level in each sample obtained from analog-treated cells to control cells was calculated and is given in supplemental Table S1. An important finding was that the relative levels of peptides isolated with or without pretreatment with 1,3,4-O-Bu3ManNAc was approximately the same when they were captured via their N-linked glycans. By contrast, the levels of certain peptides isolated after pretreatment with 1,3,4-O-Bu3ManNAc varied significantly from control samples when captured via the sialic acid specific strategy (Table I). Together, these results verified that this analog gave rise to sialic acid specific changes while leaving N-glycan levels unchanged.
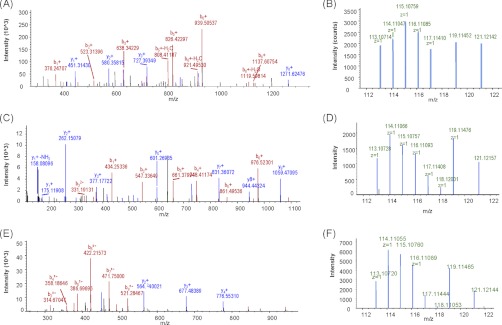
Fig. 3.
The identification and quantification of CD44 and integrin α6 by mass spectrometry. Glycopeptides were isolated from 1,3,4-O-Bu3ManNAc-treated and control SW1990 cells and identified as described in detail in the main text; to illustrate the identification process two examples (CD44 and integrin α6) are shown here. A, The matched fragments of CD44; B, the spectrum of iTRAQ reporter for CD44; C, the matched fragments of integrin α6 peptide, eINSLnLTESHnSR; D, the spectrum of iTRAQ reporter for integrin α6 peptide, eINSLnLTESHnSR; E, the matched fragments of integrin α6 peptide, anHSGAVVLLk; F, the spectrum of iTRAQ reporter for integrin α6 peptide, anHSGAVVLLk (amino acids indicated in lowercase at both ends of the sequence represent the iTRAQ labeled N termini and Lys; lowercase n in the nXT/S motif represents the formerly glycosylated Asp and deaminated after PNGase F release).
Table I. Sialoglycosylated protein changes after treatment of 1,3,4-O-Bu3ManNAc.
| Protein name | Identified sequencea | Pep avg TG S/Cb | Pep avg SG S/Cc | Pro avg TG S/Cd | Pro avg SG S/Ce |
|---|---|---|---|---|---|
| Integrin alpha chain, α6 | eINSLnLTESHnSR | 0.93 | 3.65 | 0.96 | 4.40 |
| Integrin alpha chain, α6 | anHSGAVVLLk | 1.13 | 4.22 | 0.96 | 4.40 |
| CD44 antigen | aFnSTLPTmAQmEk | 1.18 | 1.44 | 1.23 | 2.33 |
a Amino acids indicated in lower case at both ends of the sequence represent the iTRAQ labeled N-termini and Lys, lower case n in the nXT/S motif represents the formerly glycosylated Asp and delaminated after PNGase F release, lower case m represents the oxidation on M.
b Pep Avg TG S/C specifies the ratio of total glycopeptides of sample/control quantified at peptide level.
c Pep Avg SG S/C specifies the ratio of sialoglycopeptides of sample/control quantified at peptide level.
d Pro Avg TG S/C specifies the ratio of total glycopeptides of sample/control quantified at protein level.
e Pro Avg SG S/C specifies the ratio of sialoglycopeptides of sample/control quantified at protein level.
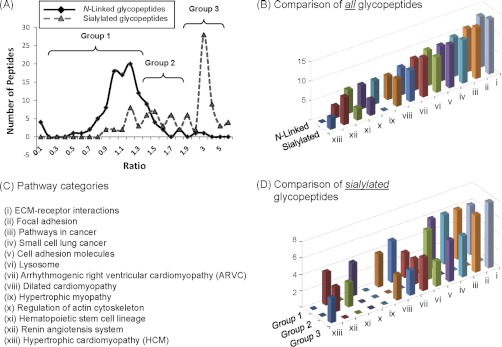
Several noteworthy trends observed from the glycoproteomic analyses are summarized in Fig. 4. First, histograms were used to compare the number of peptides with different abundance ratios. Because of errors introduced by analytical procedures, such as sample handling and quantification process, the ratios may be shifted slightly, therefore glycopeptides that were distributed within one S.D. (0.34) of the mean (1.07) were considered to be unchanged whereas those that fell beyond one S.D. of the normal distribution curve (<0.73 or >1.41) were considered to be changed. Based on these criteria, the ratio distribution of total N-linked glycoproteins identified from 1,3,4-O-Bu3ManNAc treated cells compared with untreated controls fit the curve of a normal distribution that almost exclusively fell within the “unchanged” group (i.e. between 0.73 and 1.41, Fig. 4A). By contrast, the ratio distribution of sialoglycopeptides was shifted to the right after treatment of 1,3,4-O-Bu3ManNAc indicating increased sialylation (Fig. 4A). Because not all sialoglycopeptides exhibited an increase and the ones that did had a distinct biomodal distribution, we divided these samples into three groups for further analysis: Group 1, statistically unchanged sialylation; Group 2, modestly increased sialylation; and Group 3, substantially increased sialylation.
Fig. 4.
Comparisons of changes to total N-glycans or sialoglycopeptides isolated and identified from SW1990 cells. A, Changes in the ratios of peptides captured by the total glycan or sialic acid specific methods from 1,3,4-O-Bu3ManNAc-treated and control cells (the ratio shown on the x axis represent the quantitative proportion of peptide isolated from analog treated to control cells, determined as shown in Fig. 3 for CD44 and integrin α6). B, Pathway classes of proteins represented by both of the classes of glycopeptides (e.g. analog-treated and control) shown in (A); the classes i to xiii are listed in (C). Because virtually all of the proteins identified from the total N-linked glycans fell within one S.D. of the mean (i.e. between 0.73 and 1.41) in a bell shaped distribution, pathway analysis was performed only for the entire group. D, Additional pathway analysis, however, was conducted for the sialoglycoproteins based on the groups of sialopeptides shown in A: specifically, Group 1 included proteins that fell within one S.D. of the mean; Group 2 included proteins that experienced a modest but statistically significant increase in sialylation upon analog treatment; and Group 3 included proteins with sialylation increases of greater than ∼twofold.
Pathway Analysis Revealed that Group 2 “Modestly Increased” Sialoglycoproteins Include Adhesion Molecules (e.g. CD44 and Integrin α6) Implicated in Metastasis
David pathway analysis was used to assign the glycopeptides identified from the mass spectrometry data to pathways delineated by the KEGG database. This analysis revealed that the total sets of proteins identified from both of the N-glycan and sialic acid-specific capture methods fit into 13 pathway classes that were distributed very similar to each other (Fig. 4B). The comparison of these two data sets revealed that each profile consisted of 12 categories (listed in Fig. 4C) with the main difference being a complete absence of proteins in the hypertrophic cardiomyopathy class (xiii) in the N-linked set and an absence of hypertrophic myopathy (ix) proteins in the sialylated set.
A more detailed analysis of the three groups of sialylated glycoproteins (i.e. Group 1, 2, and 3 from Fig. 4A) revealed that Groups 1 and 3 had a fairly broad distribution of pathway categories (Fig. 4D). By contrast, Group 2 proteins were restricted to 4 of the 13 categories with two of these categories (v, cell adhesion molecules; and xii, renin angiogensis system) completely absent from the unchanged (Group 1) and highly increased (Group 3) sialoglycoproteins.
Flux-driven Sialylation of CD44 and Integrin α6 Enhances Adhesion Associated with Metastasis
We reasoned that the “Group 2” proteins might be restricted to a modest, but real, increase in sialylation because they were involved in critical cellular processes very sensitive to changes in this PTM. The identification of CD44 and integrin α6 in this category (Table I) provided an opportunity to test whether any of the sialic acid-specific changes identified in the glycoproteomic data had a meaningful impact on cell behavior because of these molecules involvement in cancer progression and metastasis (23–25). Metastasis is a multistep process in which cancerous cells separate from the primary tissue and enter the circulatory system where they interact with various host cells before they migrate and lodge in the target organ and form secondary metastatic colonies. Mounting evidence suggests a role for selectins with ligands such as CD44 in the initial interactions between host cells and tumor cells in vasculature and integrins such as integrin α6 in tumor cell migration through the extracellular matrix. Accordingly, we used several in vitro assays that mimic pertinent aspects of the in vivo metastatic cascade to evaluate the effects of 1,3,4-O-Bu3ManNAc on the function of CD44 and integrin α6.
First, focusing on selectin-based adhesion relevant to CD44, a flow-based adhesion assay (16, 26) was used to compare the rolling velocities of 1,3,4-O-Bu3ManNAc-treated and control SW1990 cells to immobilized selectins under physiological flow conditions. The analog-treated cells adhered strongly to the E-selectin coated surface, and consequently rolled significantly slower than control cells at 0.5 dynes/cm2 (Fig. 5A). A smaller, but still statistically significant, decrease was noted for l-selectin-dependent rolling under the same shear conditions (Fig. 5B) consistent with prior work showing that CD44 possesses ligand activity for both of these selectins (27–29). Importantly, the site density of CD44 expressed in the analog-treated cells did not increase, as assessed by flow cytometry (Fig. 5C) and Western blot analysis (Fig. 5D) implicating the increased sialylation of these molecules as the sole factor contributing to the increased adhesion to selectins observed in the 1,3,4-O-Bu3ManNAc treated cells.
Fig. 5.
Selectin-mediated adhesion under flow. SW1990 cell rolling velocities on immobilized E-selectin (A) and L-selectin (B) after treatment with 100 μm 1,3,4-O-Bu3ManNAc for 2 days before perfusion through an in vitro flow chamber at the physiologic shear rate of 0.5 dyne/cm2 (p values are shown for n ≥ 3 independent experiments in comparison to untreated control samples). Verification that protein levels of CD44 did not change by (C) flow cytometry and (D) Western blot analysis. E, Selectin-dependent adhesion to SDS-PAGE resolved and blotted CD44 immunoprecipitated from 1,3,4-O-Bu3ManNAc-treated (or untreated control) SW1990 cells. CHO-E cells, or CHO-P cells were perfused at the wall shear stress level of 0.5 dynes/cm2 over SDS-PAGE immunoblots of immunopurified CD44. F, SPR sensorgram of the interaction between CD44 from 1,3,4-O-Bu3ManNAc treated and nontreated SW1990 cells and immobilized selectins.
Although sialylated epitopes on CD44 are linked firmly to cancer progression, additional factors nonetheless could have contributed to the increased adhesion of 1,3,4-O-Bu3ManNAc treated cells to E- and l-selectin. Therefore, to gain additional evidence that the observed increase in sialylation of CD44 was causatively linked to changes in the adhesive behavior of analog-treated SW1990 cells, a blot rolling assay was performed in which selectin-expressing CHO cells were perfused over SDS-PAGE-resolved immunoprecipitated CD44 from 1,3,4-O-Bu3ManNAc treated or untreated SW1990 cells. As shown in Fig. 5E, immunopurified CD44 from analog-treated cells exhibited a markedly increased capacity to capture E- and P-selectin-transfected CHO cells under dynamic flow conditions compared with control cells; this binding was completely ablated by sialidase treatment. In line with the flow-based rolling assays, a smaller difference was observed with l-selectin-expressing lymphocytes (data not shown). To further verify that CD44 from 1,3,4-O-Bu3ManNAc-treated SW1990 cells interacted more efficiently with E-selectin relative to controls, we used surface plasmon resonance to show that E-selectin rapidly and avidly bound to CD44 isolated from analog-treated SW1990 cells (Fig. 5F). In contrast, only modestly increased affinity of CD44 isolated from 1,3,4-O-Bu3ManNAc treated cells was observed for l- and P-selectins, indicating that sialylation of this glycoprotein preferentially enhanced adhesion to E-selectin. Taken together, this data indicates that changes in CD44 sialylation and not transcriptional control or other impacts to glycosylation were primarily responsible for the enhanced adhesion of analog-treated pancreatic cancer cells to E-selectin.
Moving to integrins, in general there have been several examples reported where hypersialylation of these cell adhesion molecules contributes to cancer progression by increasing cell motility (24, 25) through the extracellular matrix ECM after selectin-mediated extravasation from the vasculature. More specifically, however, although integrin α6 has long been known to play a role in the motility of pancreatic cancer cells (30), the precise involvement of sialic acids in this process remains unclear. To address this issue and test whether the increased sialylation of integrin α6 observed in 1,3,4-O-Bu3ManNAc treated cells influenced cell migration through the ECM, we used a wound-healing assay where the mobility of SW1990 cells on ECM substrates known to be integrin ligands was monitored. As shown in Fig. 6, the migration of analog-treated cells increased slightly on collagen type I (Fig. 6A), strongly on fibronectin (Fig. 6B), whereas no difference was seen on a control, BSA-coated surface (Fig. 6C). The function blocking GoH3 antibody to integrin α6 significantly impeded cell migration of 1,3,4-O-Bu3ManNAc-treated cells verifying that this integrin played a key role in the increasing the motility of the analog-treated cells. Finally, flow cytometry analysis with GoH3 showed that there was no difference in the protein levels of integrin α6 in the analog-treated cells (Fig. 6D), thereby verifying that the observed changes in adhesion were specific to changes in sialic acid display.
Fig. 6.
Wound healing, cell migration assays. SW1990 cells were treated (or not) with 100 μm 1,3,4-O-Bu3ManNAc (Cpd 1) and plated on culture plates pretreated with 20 μg/ml (A) collagen I, (B) fibronectin, or (C) BSA. After 24 h, wounds were created and new media without FBS but with (or without) fresh 1 was added. For each condition, 10 μg/ml of the integrin α6 functional blocking antibody (GoH3) was added to adjacent wells. The mobility of analog-treated cells was evaluated in comparison with control cells that had not been treated with analog by measuring the accumulated distance traveled per hour with the data given as a fold increase of analog-treated cells compared with control samples. D, Verification that protein levels of integrin α6 did not change was obtained by flow cytometry.
DISCUSSION
In the past, two methods have been used to alter metabolic flux through the sialic acid pathway (Fig. 1A) and both have had limitations. One has been the manipulation of GNE, an enzyme that regulates flux into the sialic acid pathway (31, 32). GNE, however, is a multifunctional protein that modulates the transcription of enzymes involved in glycosylation (33) and it also interacts directly with cell adhesion molecules (34) in multiple ways that could affect the endpoints evaluated in this study. Alternately, the sialic acid pathway can be supplemented with exogenous ManNAc but the high levels needed (often in the tens of millimolar (11)) are problematic whereas more efficiently utilized peracetylated analogs (35) have significant and often deleterious side effects (36). The current study avoided these problems by employing the “high flux” tributanoylated analog 1,3,4-O-Bu3ManNAc that allows intracellular sialic acid levels to be elevated to high levels (9) with negligible cytotoxicity or perturbation to gene regulation (37, 38). In this study, treatment of SW1990 cells with 1,3,4-O-Bu3ManNAc increased total sialic acid levels several fold and impacted glycoconjugate-bound and surface sialylation more modestly (e.g. by ≤ twofold) when measured using lectins or antibodies at cell-level resolution (Fig. 2).
Having confirmed that globally increased surface sialylation occurred in 1,3,4-O-Bu3ManNAc-treated SW1990 cells, we conducted a glycoproteomic analysis (Fig. 3) that revealed important insights into the global cellular impact of bulk flux through the sialic acid pathway. First, flux driven changes to intracellular sialic acid levels quite remarkably did not uniformly increase the sialylation of all surface glycans but instead selectively tuned the sialylation status of individual glycoproteins (Fig. 4A) with ∼31% of these molecules remaining statistically unchanged in 1,3,4,-O-Bu3ManNAc-treated cells while others (∼40%) experienced an increase of 200–800% or more. An even more interesting group was the subset of ∼30% of the glycoproteins that experienced a small (e.g. ∼1.5- to twofold) but statistically significant increase in sialylation after treatment with 1,3,4-O-Bu3ManNAc. David pathway analysis revealed that these proteins were limited to four categories (v, cell adhesion molecules; vi, lysosome; xi, hematopoietic stem cell lineages, and xii, renin angiogensis system, Fig. 4D).
An intriguing explanation for the fairly restricted subset of “Group 2” glycoproteins (Fig. 4D) that experienced tightly regulated increases in sialylation was that the activities of these proteins are unusually sensitive to sialylation. Accordingly, although cells are able to increase their sialylation thereby moving them out of “Group 1”, tight regulation is required to prevent their activity from being modified too severely, thus preventing them from moving into “Group 3.” Alternately, there may be no need to increase sialylation too greatly because even modest increases can modulate biological activities. We obtained experimental evidence for the latter premise by demonstrating that the sialylation status of CD44 and integrin α6, which are members of the cell adhesion category of tightly regulated proteins (i.e. “v” in the David pathway analysis, Fig. 4), contributed to changes in cancer cell mobility consistent with the known role of sialic acid in these processes. For example, increased sialylation of CD44 resulted in enhanced binding to selectins (Fig. 5); cancer cells exploit selectin-mediated adhesion to exit the vascular during metastasis. Another way sialylation assists metastasis is through integrin-mediated adhesion that facilitates cell migration through the ECM. We measured this end point via a wound healing assay wherein analog-treated SW1990 cells showed an increased ability to migrate across ECM components (Fig. 6). It is noteworthy that the increased mobility of analog-treated cells on ECM components complements enhanced selectin-mediated adhesion to facilitate two distinct facets of metastasis, namely the (1) tethering and rolling and (2) firm adhesion steps of the extravasation process.
Of the multiple potential sites of N-glycosylation for CD44 and integrin α6 that may have affected the activity of these molecules, we identified one site for CD44 and two for integrin α6 that were selectively hypersialated in cells treated with 1,3,4-O-Bu3ManNAc. Two of these three changes occurred in domains of the host proteins previously implicated in adhesion and provide the new information that sialic acid attached to a specific site of N-glycosylation is an important determinant of adhesion. These two sites are the hyaluronan binding domain of CD44 (Fig. 7A) (39) and the propeller domain of an integrin (Fig. 7B) (40). Although the impact of the increased sialylation observed for the third glycopeptide, located in the Calf-2 domain of integrin α6 (Fig. 7B) on adhesion is less clear, precedent that N-glycans present in the hinge region of an integrin can stabilize the open, activated form (41) suggests that allosteric effects of this third glycopeptide theoretically also could modulate integrin-mediated adhesion.
Fig. 7.
Representation of the identified glycopeptides aFnSTLPTmAQmEk in CD44 and eINSLnLTESHnSR and anHSGAVVLk in integrin α6. A, A cartoon representation of CD44 is shown with blue lollipops illustrating the positions of putative N-linked glycans. Successive “zoomed in” depictions show the location of the aFnSTLPTmZQEk glycopeptide within a computationally generated surface illustration of the HA binding domain of CD44. B, A cartoon representation of the integrin α6β1 complex (bottom) employs blue lollipops to illustrate the positions of putative N-linked glycans except for the red and green lollipops, which represent the actual N-glycans identified by mass spectrometry in this study. A modeled depiction of the integrin propeller subunit repeat containing the anHSGAVVLk glycopeptide is shown along with a zoomed in view of this glycopeptide with a representative N-glycan attached and shown using a sticks format. The green lollipop in the Calf-2 region represents the eINSLnLTESHnSR glycopeptide that also experienced selectively enhanced sialylation; sufficient structural information, however, is not available to further model this site.
In conclusion, this paper utilized 1,3,4-O-Bu3ManNAc, a recently developed molecular tool for manipulating flux through the sialic acid pathway (9, 42) that avoids pitfalls of previous genetic and small molecule-based approaches, to demonstrate that intracellular sialic acid levels selectively tune the sialylation status of individual surface glycans. This finding conclusively demonstrated that metabolic flux can determine the display and biological activity of sialic acid, an important cell surface carbohydrate. This work also supports the hypothesis that metabolic flux can alter the metastatic potential of cancer cells in a glycan-dependent manner reminiscent of how the activity of N-acetylgalactosyltransferases MGAT4/5 and the subsequent branching of N-glycans depends on flux through the HBP (3, 4, 43), which is the current exemplar of how metabolic flux-driven changes can modulate cell surface glycosylation.
Footnotes
* This work was supported by grants from the National Cancer Institute grant R01CA112314 for Y.T., E.T., R.B., K.J.Y., and Z.H.; grant R01CA101135 for R.T.A., S.-H.C., M.R.D., and K.K.; grant U01CA152813 for Y.T., L.C., Z.Z., and H.Z.; and grant P01HL107153-01 for R.B., K.J.Y., and H.Z.
 This article contains supplemental material.
This article contains supplemental material.
AUTHOR CONTRIBUTIONS: Y.T., L.C., Z.Z., and H.Z. performed the mass spectroscopy and bioinformatics experiments, R.T.A., S.-H.C., M.R.D., E.T., K.K., performed adhesion assays and characterization of sialic acid flux and display on analog treated cells, R.B. sysnthesized ManNAc analogs, and K.K. and K.J.Y. provided overall coordination of this project.
CONFLICT OF INTEREST: The authors have no competing financial interests.
1 The abbreviations used are:
- HBP
- hexosamine biosynthetic pathway
- SNA
- Sambucus nigra lectin
- RCA
- Ricinus communis agglutinin I lectin
- MAA
- Maackia amurensis agglutinin lectin
- CHO-E
- CHO cells (Chinese hamster ovary cells) stably transfected with full-length E-selectin
- CHO-P
- CHO cells stably transfected with full-length P-selectin
- D-PBS
- Dulbecco's phosphate-buffered saline
- 1,3,4-O-Bu3ManNAc
- 2-acetamido-1,3,4-tri-O-butanoyl-2-deoxy-α,β-d-mannopyranose
- GNE
- UDP-GlcNAc 2-epimerase
- sLeX
- Sialyl Lewis X
- sLeA
- Sialyl Lewis A.
REFERENCES
- 1. Krambeck F. J., Bennun S. V., Narang S., Choi S., Yarema K. J., Betenbaugh M. J. (2009) A mathematical model to derive N-glycan structures and cellular enzyme activities from mass spectrometric data. Glycobiology 19, 1163–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Monica T. J., Andersen D. C., Goochee C. F. (1997) A mathematical model of sialylation of N-linked oligosaccharides in the trans-Golgi network. Glycobiology 7, 515–521 [DOI] [PubMed] [Google Scholar]
- 3. Lau K. S., Partridge E. A., Grigorian A., Silvescu C. I., Reinhold V. N., Demetriou M., Dennis J. W. (2007) Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell 129, 123–134 [DOI] [PubMed] [Google Scholar]
- 4. Lau K. S., Dennis J. W. (2008) N-Glycans in cancer progression. Glycobiology 18, 750–760 [DOI] [PubMed] [Google Scholar]
- 5. Dennis J. W., Nabi I. R., Demetriou M. (2009) Metabolism, cell surface organization, and disease. Cell 139, 1229–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ricci E., Broccolini A., Gidaro T., Morosetti R., Gliubizzi C., Frusciante R., Di Lella G. M., Tonali P. A., Mirabella M. (2006) NCAM is hyposialylated in hereditary inclusion body myopathy due to GNE mutations. Neurology 66, 755–758 [DOI] [PubMed] [Google Scholar]
- 7. Galeano B., Klootwijk R., Manoli I., Sun M., Ciccone C., Darvish D., Starost M. F., Zerfas P. M., Hoffmann V. J., Hoogstraten-Miller S., Krasnewich D. M., Gahl W. A., Huizing M. (2007) Mutation in the key enzyme of sialic acid biosynthesis causes severe glomerular proteinuria and is rescued by N-acetylmannosamine. J. Clin. Invest. 117, 1585–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burdick M. M., Bochner B. S., Collins B. E., Schnaar R. L., Konstantopoulos K. (2001) Glycolipids support E-selectin-specific strong cell tethering under flow. Biochem. Biophys. Res. Commun. 284, 42–49 [DOI] [PubMed] [Google Scholar]
- 9. Aich U., Campbell C. T., Elmouelhi N., Weier C. A., Sampathkumar S. G., Choi S. S., Yarema K. J. (2008) Regioisomeric SCFA attachment to hexosamines separates metabolic flux from cytotoxicity and MUC1 suppression. ACS Chem. Biol. 3, 230–240 [DOI] [PubMed] [Google Scholar]
- 10. Jourdian G. W., Dean L., Roseman S. (1971) The sialic acids. XI. A periodate-resorcinol method for the quantitative estimation of free sialic acids and their glycosides. J. Biol. Chem. 246, 430–435 [PubMed] [Google Scholar]
- 11. Jones M. B., Teng H., Rhee J. K., Lahar N., Baskaran G., Yarema K. J. (2004) Characterization of the cellular uptake and metabolic conversion of acetylated N-acetylmannosamine (ManNAc) analogues to sialic acids. Biotechnol. Bioeng. 85, 394–405 [DOI] [PubMed] [Google Scholar]
- 12. Yarema K. J., Goon S., Bertozzi C. R. (2001) Metabolic selection of glycosylation defects in human cells. Nat. Biotechnol. 19, 553–558 [DOI] [PubMed] [Google Scholar]
- 13. Zhang H., Li X. J., Martin D. B., Aebersold R. (2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 [DOI] [PubMed] [Google Scholar]
- 14. Tian Y., Zhou Y., Elliott S., Aebersold R., Zhang H. (2007) Solid-phase extraction of N-linked glycopeptides. Nat. Protoc. 2, 334–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang H., Aebersold R. (2006) Isolation of glycoproteins and identification of their N-linked glycosylation sites. Methods Mol. Biol. 328, 177–185 [DOI] [PubMed] [Google Scholar]
- 16. Thomas S. N., Zhu F., Schnaar R. L., Alves C. S., Konstantopoulos K. (2008) Carcinoembryonic antigen and CD44 variant isoforms cooperate to mediate colon carcinoma cell adhesion to E- and L-selectin in shear flow. J. Biol. Chem. 283, 15647–15655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bohne-Lang A., von der Lieth C. W. (2005) GlyProt: in silico glycosylation of proteins. Nucleic Acids Res. 33, W214–W219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., Sali A. (2000) Comparative protein structure modeling of genes and genomes. Annu Rev. Biophys. Biomol. Struct. 29, 291–325 [DOI] [PubMed] [Google Scholar]
- 19. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 20. Almaraz R. T., Aich U., Khanna H. S., Tan E., Bhattacharya R., Shah S., Yarema K. J. (2012) Metabolic oligosaccharide engineering with N-acyl functionalized ManNAc analogues: cytotoxicity, metabolic flux, and glycan-display considerations. Biotechnol. Bioeng. 109, 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jacobs C. L., Goon S., Yarema K. J., Hinderlich S., Hang H. C., Chai D. H., Bertozzi C. R. (2001) Substrate specificity of the sialic acid biosynthetic pathway. Biochemistry 40, 12864–12874 [DOI] [PubMed] [Google Scholar]
- 22. Tian Y., Esteva F. J., Song J., Zhang H. (2012) Altered expression of sialylated glycoproteins in breast cancer using hydrazide chemistry and mass spectrometry. Mol. Cell Proteomics, Epub ahead of print: 10.1074/mcp.M1111.011403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hosono J., Narita T., Kimura N., Sato M., Nakashio T., Kasai Y., Nonami T., Nakao A., Takagi H., Kannagi R. (1998) Involvement of adhesion molecules in metastasis of SW1990, human pancreatic cancer cells. J. Surg. Oncol. 67, 77–84 [DOI] [PubMed] [Google Scholar]
- 24. Seales E. C., Jurado G. A., Brunson B. A., Wakefield J. K., Frost A. R., Bellis S. L. (2005) Hypersialylation of β1 integrins, observed in colon adenocarcinoma, may contribute to cancer progression by up-regulating cell motility. Cancer Res. 65, 4645–4652 [DOI] [PubMed] [Google Scholar]
- 25. Seales E. C., Jurado G. A., Brunson B. A., Wakefield J. K., Frost A. R., Bellis S. L. (2005) Hypersialylation of β1 integrins, observed in colon adenocarcinoma, may contribute to cancer progression by up-regulating cell motility. Cancer Res. 65, 4645–4652 [DOI] [PubMed] [Google Scholar]
- 26. Florey O., Haskard D. O. (2007) Analysis of flow-based adhesion in vitro. In: Cope A. P., ed. Methods in Molecular Medicine. Arthritis Research Methods and Protocols, pp. 323–332, Humana Press Inc., Totowa, New Jersey: [DOI] [PubMed] [Google Scholar]
- 27. Hanley W. D., Napier S. L., Burdick M. M., Schnaar R. L., Sackstein R., Konstantopoulos K. (2006) Variant isoforms of CD44 are P- and L-selectin ligands on colon carcinoma cells. FASEB J. 20, 337–339 [DOI] [PubMed] [Google Scholar]
- 28. Napier S. L., Healy Z. R., Schnaar R. L., Konstantopoulos K. (2007) Selectin ligand expression regulates the initial vascular interactions of colon carcinoma cells: the roles of CD44v and alternative sialofucosylated selectin ligands. J. Biol. Chem. 282, 3433–3441 [DOI] [PubMed] [Google Scholar]
- 29. Burdick M. M., Chu J. T., Godar S., Sackstein R. (2006) HCELL is the major E- and L-selectin ligand expressed on LS174T colon carcinoma cells. J. Biol. Chem. 281, 13899–13905 [DOI] [PubMed] [Google Scholar]
- 30. Weinel R. J., Rosendahl A., Pinschmidt E., Kisker O., Simon B., Santoso S. (1995) The α6-integrin receptor in pancreatic carcinoma. Gastroenterology 108, 523–532 [DOI] [PubMed] [Google Scholar]
- 31. Keppler O. T., Hinderlich S., Langner J., Schwartz-Albiez R., Reutter W., Pawlita M. (1999) UDP-GlcNAc 2-epimerase: A regulator of cell surface sialylation. Science 284, 1372–1376 [DOI] [PubMed] [Google Scholar]
- 32. Möller H., Böhrsch V., Lucka L., Hackenberger C. P., Hinderlich S. (2011) Efficient metabolic oligosaccharide engineering of glycoproteins by UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE) knock-down. Mol. Biosyst. 7, 2245–2251 [DOI] [PubMed] [Google Scholar]
- 33. Wang Z., Sun Z., Li A. V., Yarema K. J. (2006) Roles for GNE outside of sialic acid biosynthesis: modulation of sialyltransferase and BiP expression, GM3 and GD3 biosynthesis, proliferation and apoptosis, and ERK1/2 phosphorylation. J. Biol. Chem. 281, 27016–27028 [DOI] [PubMed] [Google Scholar]
- 34. Amsili S., Zer H., Hinderlich S., Krause S., Becker-Cohen M., MacArthur D. G., North K. N., Mitrani-Rosenbaum S. (2008) UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE) binds to alpha-actinin 1: novel pathways in skeletal muscle? PLoS ONE 3, e2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sarkar A. K., Fritz T. A., Taylor W. H., Esko J. D. (1995) Disaccharide uptake and priming in animal cells: inhibition of sialyl Lewis X by acetylated Gal β1,4GalcNAc β-onaphthalenemethanol. Proc. Natl. Acad. Sci. U.S.A. 92, 3323–3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim E. J., Sampathkumar S. G., Jones M. B., Rhee J. K., Baskaran G., Goon S., Yarema K. J. (2004) Characterization of the metabolic flux and apoptotic effects of O-hydroxyl- and N-acetylmannosamine (ManNAc) analogs in Jurkat (human T-lymphoma-derived) cells. J. Biol. Chem. 279, 18342–18352 [DOI] [PubMed] [Google Scholar]
- 37. Elmouelhi N., Aich U., Paruchuri V. D., Meledeo M. A., Campbell C. T., Wang J. J., Srinivas R., Khanna H. S., Yarema K. J. (2009) Hexosamine template. A platform for modulating gene expression and for sugar-based drug discovery. J. Med. Chem. 52, 2515–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Campbell C. T., Aich U., Weier C. A., Wang J. J., Choi S. S., Wen M. M., Maisel K., Sampathkumar S. G., Yarema K. J. (2008) Targeting pro-invasive oncogenes with short chain fatty acid-hexosamine analogues inhibits the mobility of metastatic MDA-MB-231 breast cancer cells. J. Med. Chem. 51, 8135–8147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Teriete P., Banerji S., Noble M., Blundell C. D., Wright A. J., Pickford A. R., Lowe E., Mahoney D. J., Tammi M. I., Kahmann J. D., Campbell I. D., Day A. J., Jackson D. G. (2004) Structure of the regulatory hyaluronan binding domain in the inflammatory leukocyte homing receptor CD44. Mol. Cell 13, 483–496 [DOI] [PubMed] [Google Scholar]
- 40. Isaji T., Sato Y., Zhao Y., Miyoshi E., Wada Y., Taniguchi N., Gu J. (2006) N-Glycosylation of the β-propeller domain of the integrin α5 subunit is essential for α5β1 heterodimerization, expression on the cell surface, and Its biological function. J. Biol. Chem. 281, 33258–33267 [DOI] [PubMed] [Google Scholar]
- 41. Luo B. H., Springer T. A., Takagi J. (2003) Stabilizing the open conformation of the integrin headpiece with a glycan wedge increases affinity for ligand. Proc. Natl. Acad. Sci. U.S.A. 100, 2403–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Z., Du J., Che P. L., Meledeo M. A., Yarema K. J. (2009) Hexosamine analogs: from metabolic glycoengineering to drug discovery. Curr Opin Chem Biol 13, 565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boscher C., Dennis J. W., Nabi I. R. (2011) Glycosylation, galectins and cellular signaling. Curr. Opin. Cell Biol. 23, 383–392 [DOI] [PubMed] [Google Scholar]
- 44. Du J., Meledeo M. A., Wang Z., Khanna H. S., Paruchuri V. D., Yarema K. J. (2009) Metabolic glycoengineering: sialic acid and beyond. Glycobiology 19, 1382–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Campbell C. T., Sampathkumar S. G., Weier C., Yarema K. J. (2007) Metabolic oligosaccharide engineering: perspectives, applications, and future directions. Mol. Biosyst. 3, 187–194 [DOI] [PubMed] [Google Scholar]