Abstract
Two different workflows were tested in order to develop methods that provide deeper insight into the secreted O-glycoproteome. Bovine serum samples were subjected to lectin affinity-chromatography both at the protein- and peptide-level in order to selectively isolate glycopeptides with the most common, mucin core-1 sugar. This enrichment step was implemented with either protein-level mixed-bed ion-exchange chromatography or with peptide-level electrostatic repulsion hydrophilic interaction chromatography. Both methods led to at least 65% of the identified products being glycopeptides, in comparison to ∼25% without the additional chromatography steps [Darula, Z., and Medzihradszky, K. F. (2009) Affinity enrichment and characterization of mucin core-1 type glycopeptides from bovine serum. Mol. Cell. Proteomics 8, 2515–2526]. In order to improve not only the isolation but also the characterization of the glycopeptides exoglycosidases were used to eliminate carbohydrate extensions from the directly peptide-bound GalNAc units. Consequent tandem MS analysis of the mixtures using higher-energy collision-dissociation and electron-transfer dissociation led to the identification of 124 glycosylation sites in 51 proteins. While the electron-transfer dissociation data provided the bulk of the information for both modified sequence and modification site assignment, the higher-energy collision-dissociation data frequently yielded confirmation of the peptide identity, and revealed the presence of some core-2 or core-3 oligosaccharides. More than two-thirds of the sites as well as the proteins have never been reported modified.
Glycosylation is one of the most frequent post-translational modifications of proteins. It is estimated that over 50% of all proteins undergo glycosylation during their lifespan (1). Apart from the regulatory O-GlcNAc modification, glycosylation occurs mostly on secreted proteins and extracellular domains of membrane proteins. Altered physiological conditions such as pregnancy (2) or disease including cancer (3, 4) may result in different glycosylation of target proteins involved. Hence, glycoprofiling is an indispensable part of biomarker research. Unfortunately, characterization of protein glycosylation of complex samples such as serum is a rather challenging task mainly because of two factors. First, bodily fluids usually feature a high background of nonglycosylated proteins. Moreover, modified sequences are frequently also present unmodified (heterogeneity), and when occupied, the same site may be modified with different carbohydrate structures (microheterogeneity). Second, up to now there is no single analytical approach that can readily identify both the glycosylation sites and the modifying sugar structures.
Glycosylation analysis of complex mixtures is usually restricted to N-glycosylation. This is because O-glycosylation lacks those features that facilitate N-glycosylation analysis; namely, a consensus sequence for modification and a single core structure for modification. Single sugar units as well as short or complex extended structures can modify Ser, Thr, and as recently reported Tyr residues (5, 6). For this reason there is no universal enzyme that can cleave all the O-linked carbohydrates (in the way e.g. PNGaseF can for N-linked glycopeptides), and those glycosidases that eliminate certain, well specified sugar structures leave an unmodified amino acid and thus no trace of the previous modification site. Sugar-elimination under basic conditions followed by Michael-addition has been used for the characterization of O-glycosylation (7), but its efficiency varies for the different sugar structures, definitely slower for Thr, and phosphorylated, sulfated, even unmodified Ser residues may also undergo the same reactions as well as alkylated Cys residues (7–9).
In the last two decades mass spectrometry has inevitably become the method of choice for protein characterization including post-translational modification analysis. However, MS characterization of O-glycopeptides by collision-induced dissociation (CID)1 activation is ineffective at identifying the peptide as sugar oxonium ions and fragment ions corresponding to carbohydrate fragmentation dominate MS/MS spectra. On the other hand, electron capture dissociation (10) and electron transfer dissociation (ETD) (11) analysis of glycopeptides is a more successful approach in this respect [ECD: 5,12; ETD: 6,13,14], despite the fact that these activation techniques are less efficient compared with CID, work considerably better on higher charge state peptide precursors and have significant precursor m/z limitations (15).
Currently, for successful ETD-based O-linked glycopeptide characterization one has to know either the protein(s) or the sugar structure to begin with. General glycopeptide enrichments as hydrophilic interaction liquid chromatography (16) or selective capture/release based on the unique properties of sialic acid (17) presently cannot be combined with large scale automated studies. Although CID data can provide information about sugar structure and ETD can characterize peptide sequence, there is currently no automated way to correlate these two types of data. Hence, only a fraction of glycopeptides enriched in a nonstructure-specific fashion can be characterized and it is done manually (17). Thus, either one can characterize glycosylation within a protein mixture of limited complexity (proteins identified from a strict database search can be subjected to a second search where undefined modifications over a mass range are considered (18)) or one has to apply some oligosaccharide-selective enrichment strategy for the glycopeptides, so that the database search could be restricted to a few sugar compositions.
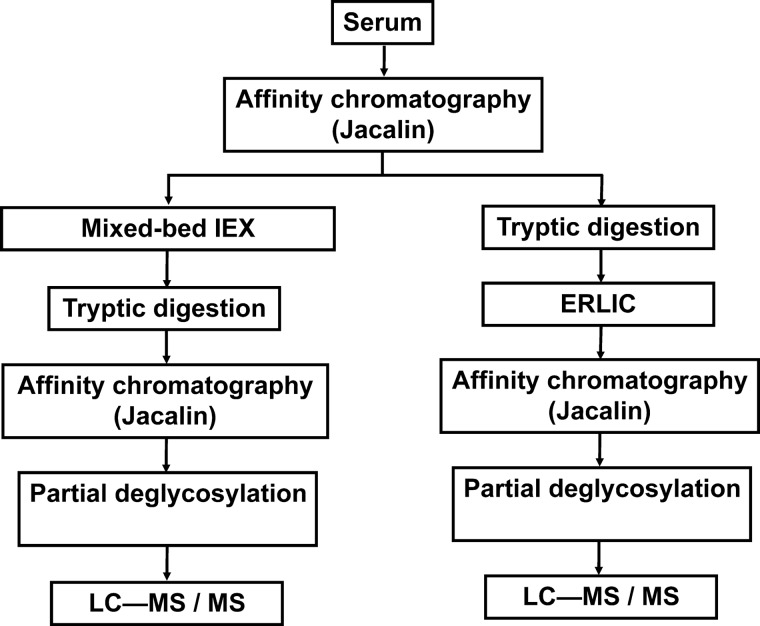
Jacalin, a lectin isolated from Artocarpus integrifolia has been reported binding GalNAcα1-modified glycopeptides in which C6-OH is free, but not recognizing such structures with substitution at the C6 position (19). Previously we have shown that Jacalin affinity-chromatography combined with MS-analysis by CID and ETD fragmentation is a viable experimental setup for characterization of the core-1 mucin-type glycoproteome of serum (14). However, our findings were restricted to the more abundant proteins of serum. In order to gain a deeper insight, we have now combined the affinity-enrichment with other protein- or peptide-level fractionations, and tested two different workflows.
In the protein-level fractionation approach, ion-exchange chromatography was implemented for fractionation of the glycoprotein mixture isolated by Jacalin lectin affinity-chromatography. Because of its high sample capacity, ion exchange is a popular method for separation of protein samples. Using a mixed-bed ion-exchange column that contains anion-exchange and cation-exchange material in equal amounts enables the retention and fractionation of proteins over the entire pI range (20). A further advantage of this separation step is that even abundant proteins are expected to be restricted to a few fractions, thus increasing the chances for the identification of less abundant glycoproteins.
In the peptide-level approach, the tryptic digest of the glycoprotein mixture isolated by Jacalin lectin affinity-chromatography was subjected to further separation applying the ERLIC (electrostatic repulsion hydrophilic interaction chromatography) principle (21). ERLIC is a mixed mode chromatography where the retention of any given compound depends on the combination of electrostatic repulsion from and hydrophylic interaction with the solid support (21). In the case of tryptic digests, unmodified peptides are expected to be protonated at pH 2 and therefore elute in the flow-through or early eluting fractions, whereas peptides modified by highly acidic groups such as phospho- and sulfopeptides, and sialylated glycopeptides are retained longer. As a result, sialylated glycopeptides can be selectively isolated from unmodified peptides. Although this workflow was expected to be limited to the selective isolation of sialylated glycopeptides, in our pilot studies the majority of the glycopeptides bore sialic acid residues. Therefore we did not consider this as a major limitation.
In this study glycopeptide enrichment results are compared from the two above described workflows. In order to ensure higher identification rates, i.e. to overcome the charge-density limits for successful ETD experiments, glycopeptides were treated with neuraminidase and β-galactosidase, and the sequences retaining only the core GalNAc units were subjected to MS/MS analysis using both HCD and ETD activation. We identified 124 glycosylation sites in 51 glycoproteins; an ∼6-fold improvement in comparison to our previous results, when only lectin affinity-chromatography was used. Thirty-five of the proteins were previously not known to be glycosylated. Similarly, more than half of the sites determined represent novel glycosylation sites.
EXPERIMENTAL PROCEDURES
Chromatography was performed on a Jasco semimicro HPLC system complete with a four-line degasser (DG-2080–54, Jasco), two pumps (PU2085, Jasco), a dynamic mixer (MX 2080–32, Jasco), a UV-VIS detector (Spectra-Flow 501, Sunchrom), and a fraction collector (CHF 122 SC, Advantec).
Glycoprotein Isolation by Jacalin Affinity Chromatography
Chromatography was performed as previously published (14), 2 ml of fetal calf serum was injected onto a 1 mm × 2000 mm (CV:1.57 ml) column packed with agarose-bound Jacalin (VectorLabs AL1153). After introducing the sample (flow rate: 50 μl/min), the column was washed with eight CV of solvent A (175 mm Tris.HCl, pH 7.5; flow rate:150 μl/min) then the species bound were eluted with five CV of solvent B (0.8 m galactose/175 mm Tris.HCl, pH 7.5; flow rate:150 μl/min) collecting 8-min fractions.
ERLIC Chromatography
The tryptic digest of the protein mixture isolated by Jacalin affinity-chromatography was fractionated on a weak anion-exchange column (PolyWAX LP, PolyLC Inc, 4.6 mm ID × 20 cm, 5 μm particle size, 300A pore size) applying the following gradient program (flow rate: 1 ml/min, UV-detection at 215 nm): 0–5 min: 0% B, 5–15 min: 0–10% B, 15–35 min: 10–60% B, 35–45 min: 60–100% B, 45–55 min:100% B (solvent A: 20 mm methyl-phosphonic acid pH:2/70% acetonitrile, solvent B: 200 mm TEAP (triethylammonium phosphate) pH 2/60% ACN; the pH of solvent A and solvent B were adjusted using 10 m aqueous NaOH and triethylamine, respectively). 1-min fractions were collected, dried down to ∼200 μl and desalted on 100 μl C-18 tips (Omix, Varian) and concentrated.
Mixed-bed Ion Exchange Chromatography
Protein mixture isolated by Jacalin affinity-chromatography from 2 ml fetal calf serum was fractionated on a mixed-bed ion exchanger column (PolyCATWAX, PolyLC Inc, 4.6 mm ID × 20 cm, 5 μm particle size, 1000 Å pore size) applying the following gradient program (flow rate: 0.5 ml/min, UV-detection at 275 nm): 0–5 min: 0% B, 5–15 min: 0–10% B, 15–35 min: 10–60% B, 35–45 min: 60–100% B, 45–55 min:100% B (solvent A: 20 mm ammonium acetate pH:7, solvent B: 800 mm ammonium acetate pH:7). 2-min fractions were collected and dried down before further treatment.
Tryptic Digestion
Samples were supplemented with guanidine hydrochloride to give a final concentration of 6 m. Disulfide bridges were reduced using dithiothreitol (56 °C for 30 min) and the resultant free sulfhydryl groups were derivatized using iodoacetamide (1.1x equivalent to dithiothreitol, 30 min in the dark at room temperature). Samples were then diluted eightfold with 100 mm ammonium bicarbonate to reduce the guanidine hydrochloride concentration, and incubated with porcine trypsin (Fluka 93614; 1% (w/w) of the estimated protein content) at 37 °C for 4 h. Digestion was stopped by adding trifluoroacetic acid (final pH ≤3). The resulting peptide mixtures were desalted on C18 reversed phase and concentrated.
Glycopeptide Isolation by Jacalin Affinity Chromatography
Chromatography was performed as previously described (14). The tryptic digest of a glycoprotein mixture was injected onto a 1 mm × 200 mm (CV:0.157 ml) column packed with agarose-bound Jacalin. After introducing the sample (flow rate: 50 μl/min), the column was washed with 20 CV of solvent A (175 mm Tris HCl, pH 7.5; flow rate:150 μl/min) then the species bound were eluted with 20 CV of solvent B (0.8 m galactose/175 mm Tris.HCl, pH:7.5; flow rate:150 μl/min) collecting 4-min fractions. Fractions of interest were acidified and desalted on 100 μl C-18 tips (Omix, Varian) prior to further treatment. The fractions to be purified were pulled up onto pipette-tips pretreated following the manufacturer's instructions, the galactose and salt were removed with 0.1% formic acid in water (5 × 200 μl). Peptides and glycopeptides were eluted with 200 μl 0.1% formic acid/50% acetonitrile/water. Samples were concentrated in a vacuum centrifuge.
Partial Deglycosylation of O-Glycopeptides (14)
Sialic acid and β-galactose units of glycopeptides were removed by incubation with neuraminidase (5–10 U/sample, New England Biolabs P0720; in 100 mm sodium citrate, pH 6.0) for 1 h at 37 °C followed by overnight treatment with β-galactosidase (10 U/sample, New England Biolabs P0726; in 100 mm sodium citrate, pH 4.5) at 37 °C. Enzymatic deglycosylation was stopped by acidification to pH ≤3 with 10% trifluoroacetic acid solution, and the resulting peptide mixtures were desalted on 10 μl C-18 tips (Millipore ZTC18S960).
Mass Spectrometry
Glycopeptide mixtures were separated on nanoflow reversed phase HPLC (nanoAcquity, Waters, Milford, MA) directing the eluent to nanospray sources of a linear ion trap-Orbitrap (Velos-Orbitrap, Thermo Fisher Scientific) mass spectrometer operating in positive ion mode.
Samples were injected onto a UPLC trapping column (Symmetry, C18 5 μm, 180 μm × 20 mm; Waters) (15 μl/min with 3% solvent B) followed by a linear gradient of solvent B (5 to 35% in 35 min, followed by a short wash at 50% solvent B, before returning to starting conditions; flow rate: 400 nl/min; nanoACQUITY UPLC BEH C18 Column, 1.7 μm, 75 μm × 200 mm; solvent A: 0.1% formic acid in water, solvent B: 0.1% formic acid in acetonitrile).
MS data acquisition was carried out in data-dependent fashion acquiring sequential HCD and ETD spectra of the three most intense, multiply charged precursor ions identified from each MS survey scan. ETD experiments were performed in the linear trap, whereas HCD activation was carried out in the collision cell. MS and HCD spectra were acquired in the Orbitrap, and ETD spectra in the linear ion trap. Ion populations within the trapping instruments were controlled by integrated automatic gain control. For HCD, the AGC target was set to 50,000, with dissociation at 35% of normalized collision energy, activation time: 0.1 ms. For ETD, the automatic gain control target values were set to 10,000 and 200,000 for the isolated precursor cations and fluoranthene anions, respectively, and allowing 100 ms of ion/ion reaction time. Supplemental activation for the ETD experiments was enabled (supplemental activation energy: 15). Dynamic exclusion was also enabled (mass width low: 0.5 Th, mass width high 1.5 Th), exclusion time: 45 s.
Some glycopeptides fractions were combined and analyzed on an LTQ-Orbitrap Elite (courtesy of Thermo Scientific, San Jose, CA). A single spectrum from this analysis that enabled unambiguous site assignment for E1BB91 was included in the supplementary Figs.
Data Interpretation
Peaklists from LTQ-Orbitrap raw data files were created by using the UCSF in-house peak-picking program PAVA (22). The software generates separate HCD and ETD peaklists.
From the above ETD peaklists “glycopeptide-only” versions were also prepared after HCD-based filtering. An in-house script (supplemental File S1) was used to screen HCD data for the HexNAc specific carbohydrate ion m/z = 204.087 with a mass accuracy of 0.01 Da. Whenever such a fragment was not found, the ETD spectrum of the corresponding precursor ion was deleted from the ETD peaklist. Similar ETD peaklists screened for 204.087 and 366.14; and 204.087 and 407.167 (mass accuracy: 0.01 Da) were also prepared.
Database searching was performed by ProteinProspector v.5.8.1 against the UniProt database (07.06.2011), supplemented with a random sequence for each entry, and species specified as Bos taurus (66914/33089872 entries searched). Search parameters were as follows: trypsin was selected as the enzyme, two missed cleavages were permitted, and nonspecific cleavages were also permitted at one of the peptide termini. Mass accuracies of 15 ppm for precursor ions, 20 ppm for HCD fragment ions, and 0.8 Da for ETD fragment ions were considered. Fixed modification was carbamidomethylation of Cys residues. Variable modifications were the acetylation of protein N termini; Met oxidation; and the cyclization of N-terminal Gln residues; plus HexNAc modification on Thr and Ser residues. A maximum of three modifications per peptide were permitted. Search parameters for HCD data also included HexNAc as a variable modification subject to neutral loss; i.e. fragments were assumed to be unmodified. Acceptance criteria were as follows: minimum peptide score: 22, minimum protein score: 22; maximum peptide E-value: 0.1, maximum protein E-value: 0.1; minimum best discriminant score: 1. SLIP score as a measure of reliability of site assignments was set to six (23). Only the best identification is reported for each unique sequence (considering differently modified sequences as unique).
Data was also searched permitting nonspecific cleavages at both termini, which identified a few new glycopeptides that after careful inspection were included in the data set (supplemental Figs.). An additional database search was performed on the subset of identified proteins allowing up to 4 variable modifications per peptide applying the same acceptance criteria as above with manual validation of data providing additional glycosylation information to the original database search results.
With the 204 and 366 and 204 and 407-filtered peaklists separate searches were performed. Search parameters were as above, except HexHexNAc and HexHexNAcSA or HexNAc2 on Ser/Thr residues were also permitted as variable modifications. Acceptance criteria reporting those modifications were the same.
Novelty of the glycosylation site assignments is based on information available in the UniProt database in November 2011.
RESULTS AND DISCUSSION
Previously we have shown that affinity chromatography with agarose-bound Jacalin is a useful but limited approach for the isolation of mucin core-1 type O-glycosylated structures. Although we were able to determine 34 O-glycosylation sites in 16 different proteins, evidently there are a much higher number of O-glycoproteins in serum. In order to lower nonspecific background and enable detection of less abundant glycoproteins and/or glycoforms, after the first protein-level affinity enrichment, an additional fractionation step was included into our enrichment protocol either at the protein- or the peptide-level (Fig. 1). Although the analysis of intact glycopeptides would be desirable, our previous experience showed that ETD frequently does not yield sufficient information because of the low charge-density of these molecules (15). We showed that this situation could be somewhat improved by partial deglycosylation, i.e. retaining only the core GalNAc units (14). Thus, this was the final step in both of our sample preparation protocols.
Fig. 1.
Enrichment strategy for mucin-type core-1 O-glycopeptides.
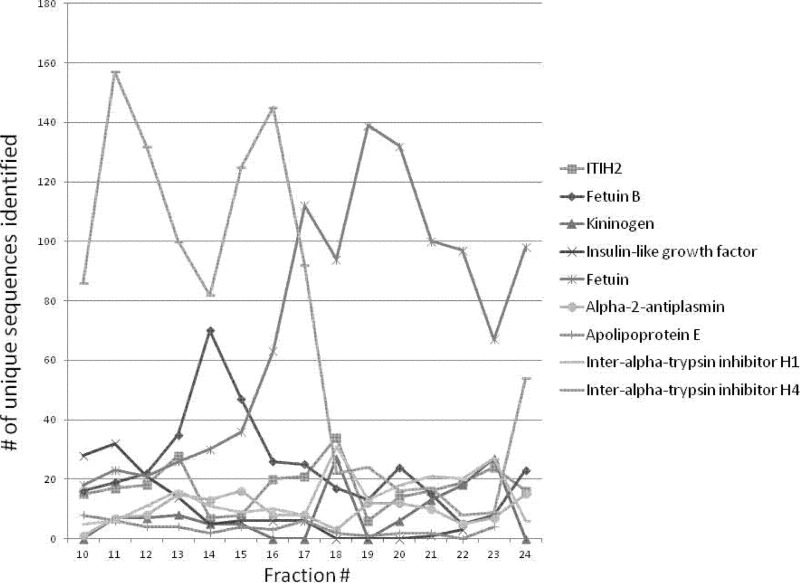
In the protein-level approach mixed-bed ion-exchange chromatography was used. It was expected that this fractionation would make the less abundant glycoproteins more “visible” once they were separated from the major components. Each fraction collected was digested with trypsin and subjected to peptide-level affinity-chromatography, and then to partial deglycosylation as described in the Experimental section. MS characterization of these samples revealed that our enrichment strategy was successful in significantly eliminating nonglycosylated background—65% (400 out of 614 peptides identified, supplemental Table S1) of the peptides identified represented glycosylated sequences compared with <25% achieved with affinity chromatography alone (14). As expected, new glycoproteins and glycosylation sites were identified (Table I, carefully inspected spectra corresponding to novel glycosylation sites are presented in the supplementary Figs.). At the same time the data also reflected poor separation efficiency of the mixed-bed ion-exchange chromatography: as deduced from the distribution of unique sequences representing nine abundant proteins (Fig. 2), the glycoproteins apparently spread out covering the whole chromatographic run. The most likely explanation for this phenomenon is that these proteins exist in multiple glycoforms that are retained differently. As a result the overwhelming majority of the glycopeptides still represented a few abundant glycoproteins. Heterogeneity in site occupancy as well as rampant proteolytic activity in serum resulting in nontryptic cleavages caused the number of glycopeptides representing the same proteins to multiply (supplemental Tables S1–S4).
Table I. O-glycosylation sites identified in the present study (See supplemental Figs.). Site assignments are given for the Uniprot entry corresponding to the best characterized bovine sequence. Uniprot IDs in parantheses refer to the protein identified by database search. Names with asterisk refer to proteins described as “uncharacterized protein”, the name of the closest well characterized (human) homolog is given. Legend for modification sites: new; discovered by us earlier (14, 30); known confirmed now; known found earlier; known in human found now, known in human found earlier. Reference data for known sites is given in supplemental File S2.
| Uniprot ID | Protein name | Modification site | Enrichment protocol |
|---|---|---|---|
| A2I7N2 | SERPINA3–6 | 31-gT | Both |
| A4IFA5 | VASN protein | 455-gS, 460-gT | Both |
| A5D7R6 | ITIH2 protein | 673-gS, 691-gT | Both |
| A5PK77 | SERPINA11 protein | 387-gT | mIEX |
| A5PKA3 | CCDC80 protein | 89-gT | ERLIC |
| A6QLD8 | ADAMTSL4 protein | 605-gT | Both |
| E1BB91 | Collagen alpha-3(VI) chain* | 2929-gT | ERLIC |
| E1BCW0 | Hepatocyte growth factor activator* | 355-gT, 360-gT, 365-gT | Both |
| E1BI67 | Interleukin-18-binding protein* | 50-gS | ERLIC |
| E1BKQ9 | Polypeptide N-acetylgalactosaminyltransferase 5* | 429-gT | Both |
| F1MER7 | Basement membrane-specific heparan sulfate proteoglycan core protein* | 3374-gT | ERLIC |
| F1MMK9 | Protein AMBP* | 198-gT | Both |
| F1N1I6 | Gelsolin* | 27-gT, 34-gT, 44-gT | Both |
| O18977 (F1MPK6) | Tenascin-X | 3146-gT or 3147-gT(682-gT or 683-gT) | ERLIC |
| P00735 | Prothrombin | 193-gT, 205-gT, 206-gS | ERLIC |
| P00743 | Coagulation factor X | 485-gT | ERLIC |
| P00744 | Vitamin K-dependent protein Z | 388-gT | Both |
| P01030 | Complement C4 (fragments) | 420-gT | Both |
| P01044 (F1MNV4) | Kininogen-1 | 136-gT, 149-gS or 150-gT, 399-gT, 400-gT, 406-gS, 581-gS, 586-gT, 605-gT | Both |
| P02672 | Fibrinogen alpha chain | 464-gT, 525-gT | ERLIC |
| P02676 | Fibrinogen beta chain | 4-gT | Both |
| P06868 | Plasminogen | 366-gT, 378-gT | Both |
| P07224 | Vitamin K-dependent protein S | 104-gT | ERLIC |
| P07456 (B8QGI3) | Insulin-like growth factor 2 | 99-gT, 106-gT, 154-gS, 163-gT, 168-gT, 173-gS; 174-gS | Both |
| P07589 | Fibronectin | 280-gT, 2156-gT, (2157-gT) | Both |
| P12763 | Alpha-2-HS-glycoprotein | 217-gT, 271-gS, 280-gT, 282-gS, 296-gS, 314-gT, 320-gS, 324-gS, 325-gS, 334-gT, 341-gS | Both |
| P17690 | Beta-2-glycoprotein 1 | 32-gS, 33-gT | mIEX |
| P19035 | Apolipoprotein C-III | 90-gS or 92-gT | ERLIC |
| P28800 | Alpha-2-antiplasmin | 24-gS/27-gS/28-gT, 398-gT, 400-gT, 489-gS | 24/27/28 mIEX; 398&400 ERLIC 489 both |
| P50448 | Factor XIIa inhibitor | 74-gT | mIEX |
| P81187 | Complement factor B | 26-gT | ERLIC |
| P81644 | Apolipoprotein A-II | 40-gT | Both |
| Q03247 | Apolipoprotein E | 31-gT, 32-gT, 211-gT, 307-gS, 309-gT, 310-gS | Both |
| Q05717 | Insulin-like growth factor-binding protein 5 | 171-gT | ERLIC |
| Q0VCM5 | Inter-alpha-trypsin inhibitor heavy chain H1 | 643-gS, 648-gT | Both |
| Q28083 | Collagen alpha-1(XI) chain (Fragment) | 86-gT | ERLIC |
| Q28107 | Coagulation factor V | 1151-gS, 1154-gT, 1171-gT | Both |
| Q29RQ1 | Complement component C7 | 696-gT | mIEX |
| Q2KIU3 | Protein HP-25 homolog 2 | 72-gT | Both |
| Q32KM8 | Augurin | 47-gT | ERLIC |
| Q32PI4 (F1N4M7) | Complement factor I | 57-gT | mIEX |
| Q3MHN2 | Complement component C9 | 24-gT, 26-gS | ERLIC(mIEX) |
| Q3SWW8 | Thrombospondin-4 | 270-gT, 282-gS, 284-gT | ERLIC |
| Q3SYR5 | Apolipoprotein C-IV | 35-gT | ERLIC |
| Q3T052 Q5EA67 | Inter-alpha-trypsin inhibitor heavy chain H4 | 629-gS, 635-gS, 677-gS, 683-gS, 686-gS, 688-gT, 689-gS, 695-gS, 698-gT, 705-gT, 706-gT, 708-gT, (719-gS) | Both |
| Q3ZBS7 | Vitronectin | 63-gT, 97-gT, 98-gT, 107-gT, 142-gT or 143-gS | Both |
| Q58CQ9 | Pantetheinase | 504-gT | Both |
| Q58D34 | Peptidase inhibitor 16 | 408-gT | ERLIC |
| Q58D62 | Fetuin-B | 19-gT, 20-gS, 157-gS, 173-gT, 262-gS, 273-gT, 292-gT, 295-gT, 299-gT | Both |
| Q95121 | Pigment epithelium-derived factor | 34-gT | Both |
| Q9N2I2 | Plasma serine protease inhibitor | 35-gT, 36-gT | Both |
Fig. 2.
Distribution of glycopeptides from nine abundant proteins in the mixed-bed ion exchange separation fractions. Different site assignments count as unique. The x-axis in represents the elution time: 2 min fractions were collected. (The color version of this Fig. can be seen in supplemental Table S3.) In order to illustrate the complexity of the system more liberal acceptance criteria were applied for the data used in this Figure: discriminative score ≥ 0; peptide score ≥15; E ≤ 0.1; mass error ≤ 10 ppm.
In the other workflow tested, after the protein level affinity-chromatography, the Jacalin-bound fraction was digested with trypsin, then ERLIC chromatography was used for the fractionation of the resulting peptide mixture. In our pilot studies it was found that ERLIC fractionation in itself was insufficient for the selective isolation of sialylated glycopeptides (unpublished results). In order to overcome this problem, an affinity-enrichment was applied with Jacalin on the collected ERLIC fractions. This step again was followed by partial deglycosylation. Affinity purification combined with ERLIC also proved to be very effective for glycopeptide isolation, 76% (473 out of 619 total peptide hits, supplemental Table S2) of the identified sequences belonged to glycopeptides.
Identification of glycopeptides first was carried out by database searching of HCD and ETD MS/MS data (acquired on the isolated chromatographic fractions), independently. The majority of the glycopeptides was identified from ETD data (supplemental Tables S1 and S2). Although ETD spectra provided unambiguous site assignments (See supplemental Figs.), the presence of isomeric structures, i.e. the same sequences modified at different positions cannot be excluded. The supplementary Tables clearly indicate the complexity of the mixture and the frequent ambiguity of the site assignments. HCD-based search results frequently provided confirmation for the identity of the modified sequences, even if the site could not be localized from the spectra (Shown in supplemental Tables S3 and S4). Although HCD data frequently do not yield sufficient information for glycopeptide identification, they almost always display characteristic carbohydrate fragmentation. In order to exploit this feature for our advantage, HCD data were used to screen for the presence of glycopeptides.
An in-house script was created that searches for reporter ions in the HCD spectra that indicate the presence of certain carbohydrate structures (this same script can be used for searching for other diagnostic ions, such as m/z = 216 for phosphotyrosine). Individual ions, or ion combinations, and their relative or absolute mass accuracy can be specified, as well as a relative intensity threshold (within the “n” most intense fragment ions in the spectrum) (supplemental File S1). The HCD peaklists generated from LC/MS/MS experiments were filtered with this program. Whenever the specified ions were absent in an HCD spectrum, the corresponding precursor ion and its fragments were removed from the appropriate ETD data set. In our case, the HexNAc oxonium ion at m/z = 204.087, within 0.01 Da was specified as an essential glycopeptide-identifying fragment, and no limit was specified for the fragment ions screened. Database searches were repeated with the filtered peaklists and results obtained with these ETD-peaklists are reported in supplemental Tables S3 and S4. In addition to more confident data interpretation, the screening also accelerates database searching. This simple filtering procedure removed 90% of data corresponding to nonglycosylated peptides with less than 1% or 3% of glycopeptide data loss in the mixedIEX and ERLIC data sets discussed here, respectively.
In order to check the efficiency of exoglycosidase treatment and also to search for carbohydrate structures other than mucin core-1, HCD data were also screened for the presence of carbohydrate oxonium ion combinations, such as m/z 204.087 and 366.140, and m/z 204.087 and 407.167 (during this screening the ions of interest had to be among the 60 most abundant HCD fragment). Combined filtering for m/z 204 and 366 identifies GalNAcGal structures present because of incomplete deglycosylation by beta-galactosidase. The results confirmed that the enzymatic sugar removal was reasonably efficient: ∼7.5% of the glycopeptide IDs belong to HexNAcHex or HexNAcHexSA containing structures (supplemental Table S5). In addition, the filtering protocol enabled us to identify glycopeptides with carbohydrate structures featuring a different core: m/z = 407.167 indicating a HexNAc2 structure was detected in several HCD spectra representing four different proteins, ITI H4 (Q5EA67), insulin-like growth factor II (P07456), kininogen-1 isoform (F1MNV4), and hepatocyte growth factor activator (E1BCW0) (supplemental Table S6).
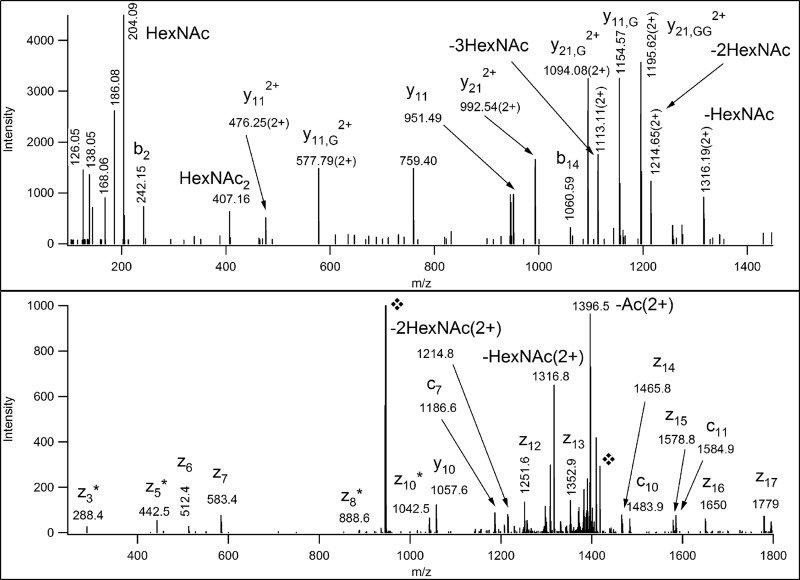
Although the corresponding carbohydrate oxonium ion indicates the presence of a HexNAc2 structure on these peptides, the exact sites of modification usually cannot be determined from the corresponding ETD spectra due to incomplete fragmentation. Moreover, if the right number of HexNAc modifications were permitted most of them would be assigned as modified with single sugar units at different positions (see supplemental Table S6). There were some exceptions, when the site assignment was unambiguous as shown in Fig. 3. The corresponding sugar structure evidently cannot be derived from the core-1 Galβ1–3GalNAcα structure. The presence of directly linked HexNAc units suggests core-2 and core-3 structures if one does not consider rare core structures. Although only core-3 structures bind to Jacalin we cannot exclude the presence of core-2 oligosaccharides. All peptides carrying the 407.2 Da modifications were multiply glycosylated and hence it is reasonable that Jacalin, specific for the core-1 structure captured them. These differently glycosylated peptides beautifully illustrate the need for carbohydrate structural information prior to the database search with ETD data. Without the HCD fragmentation information a significant portion of these glycopeptides would be incorrectly assigned or not assigned at all.
Fig. 3.
HCD (upper panel) and ETD (lower panel) spectra of the E1BCW0 glycopeptide, IQPPPT(HexNAc2)EALLTLPGPT(HexNAc)AAGPAGR. The precursor ion was at m/z 945.4967 (3+). In the HCD spectrum the Gs indicate the number of sugar units on the peptide fragment. In the ETD spectrum the fragments are fully glycosylated. However, sugar loss from the precursor ion was detected as indicated.  labels the original and the charge-reduced precursor ions.
labels the original and the charge-reduced precursor ions.
Jacalin has been reported to display binding specificity toward mannose residues (24). N-linked structures featuring multiple mannose residues should yield an oxonium ion at m/z 163.0606 (25). Thus, the HCD data were screened looking for this diagnostic fragment. However, we did not find any proof for the presence of N-linked glycopeptides.
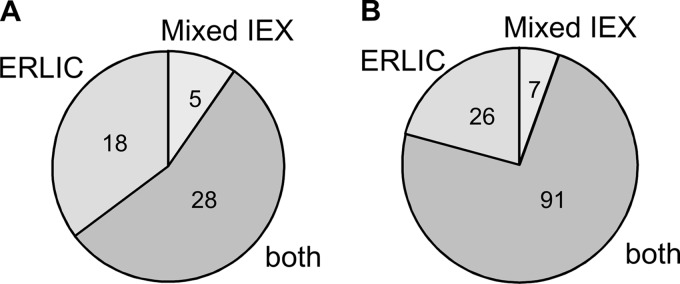
Summarizing the database search results for the two enrichment approaches (detailed in supplemental Tables S3 and S4), in the present study we have identified 124 O-glycosylation sites in 51 different proteins (Table I, supplementary Figs.). According to the current Uniprot database, 76 of these sites are novel (six of these sites were reported glycosylated in the corresponding human homolog) and 35 of the 51 glycoproteins have not been reported O-glycosylated previously. These results represent an ∼sixfold improvement compared with sample preparation by Jacalin enrichment alone, where 21 sites were identified (14). All the previously identified 21 sites were found with the present protocols. Of the 51 glycoproteins identified, 28 were found with both enrichment methods, whereas 18 and five additional glycoproteins were identified by the peptide-level ERLIC and the protein-level mixed-bed ion-exchange fractionation, respectively (Fig. 4A). In terms of glycosylation sites, 92 were identified in both experiments and an additional 26 and seven glycosylation sites were identified by the ERLIC and mIEX methodology, respectively (Fig. 4B). 73% of the total identified O-glycosylation sites were found by both approaches, providing a level of validation to the results. The ERLIC-based enrichment performed slightly better both in terms of the number of identified glycoproteins and sites of modification.
Fig. 4.
Distribution of O-glycoproteins (A) and O-glycosylation sites (B) identified by the Jacalin-mixed-bed ion-exchange and the Jacalin-ERLIC experiment.
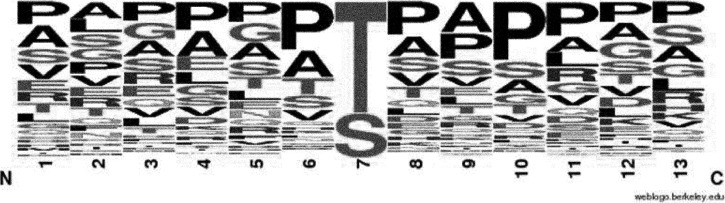
We carefully inspected the glycosylation sites identified as to whether these would reveal any tendencies about their localization. There are several GalNAc-transferases responsible for initiation of O-glycosylation with distinct but overlapping substrate specificity (26). Consequently, it is unlikely that a universal consensus sequence exists for this modification. However, frequency of different amino acids flanking the modification site might be of use for further studies including prediction algorithms. Residues for the six positions both “upstream and downstream” were considered, and only unambiguous site-assignments were included (supplemental Table S7). Interestingly, more than two-thirds of the modified sequences contain glycosylated Thr residues versus Ser modifications: 85 and 34, respectively. The distribution of the flanking amino acids (Fig. 5) confirms that secreted O-glycosylation primarily occurs in sequence stretches that are rich in Pro, Ala, Gly residues (27, 28). Additional potential modification sites, i.e. Ser and Thr residues also relative frequently occur in close vicinity (26). If we display distributions unique to Ser and Thr residues some differences start to emerge (supplemental Table S8). However, there are no sufficient data points to draw conclusions from these differences.
Fig. 5.
Amino acid distribution around the O-glycosylation sites determined (supplemental Table S7, color version also is presented there). Ser- and Thr-specific distributions are presented in supplemental Table S8. This Figure was generated by http://weblogo.berkeley.edu/. “The height of symbols within the stack indicates the relative frequency of each amino at that position.”
Regarding site localization within the protein sequences, the majority of the sites are located near protein termini (59% of the novel glycosylation sites) or near domain boundaries (an additional 13%). We have already reported this phenomenon in our pilot studies when affinity enrichment alone was used (14). We still do not know whether this is a bias of the affinity enrichment protocol (i.e. the first protein-level enrichment) or indicates that O-glycosylation is preferential to protein termini. However, O-glycosylation has been implicated in protein processing (29), and such a role would explain the glycosylations detected close to processing sites. In the present study we also have identified glycopeptides that show great variation in site occupancy (there is also some indication of different oligosaccharide structures, supplemental Tables S3–S6), yielding a wide variety of coexisting multiply modified sequences. As an example, insulin-like growth factor II (P07456) was found to be multiply glycosylated on its C-terminal region (Table II) with Ser-154, Thr-163, Thr-168, Ser-173, and Ser-174 being glycosylated either alone or in different combinations. HCD data of glycopeptides representing this sequence stretch revealed the presence of HexNAc2, i.e. not only mucin core-1, but perhaps core-2 or core-3 glycan. Such observations may suggest that for some proteins the modification of a given region not an individual residue has biological significance.
Table II. Glycopeptides representing the same region, Ser154-Lys175 with variable glycosylation, identified from insulin-like growth factor II.
| m/z | z | ppm | DB peptide | Protein mods | Expect |
|---|---|---|---|---|---|
| 764.1268 | 4 | −2 | SHRPLIALPTQDPATHGGASSK | HexNAc@154=5; HexNAc2@163=5; HexNAc@174=5 manual evaluation | 0.0029 |
| 570.8846 | 5 | −6 | SHRPLIALPTQDPATHGGASSK | HexNAc@154=48; HexNAc@163=36; HexNAc@168=19 | 0.033 |
| 713.3555 | 4 | −4 | SHRPLIALPTQDPATHGGASSK | HexNAc@154=57; HexNAc@163=15; HexNAc@173 174 | 4.9e-8 |
| 840.4152 | 3 | −3 | SHRPLIALPTQDPATHGGASS | HexNAc@154=60; HexNAc@163=27 | 1.8e-6 |
| 611.8168 | 4 | −3 | SHRPLIALPTQDPATHGGASSK | HexNAc@163=27 | 1.9e-5 |
| 788.3960 | 3 | 0 | SHRPLIALPTQDPATH | HexNAc2@154=42; HexNAc@163=18 | 0.019 |
| 724.0330 | 3 | −1 | LIALPTQDPATHGGASSK | HexNAc@163=26; HexNAc@174=11 | 8.4e-9 |
| 686.3384 | 3 | −1 | IALPTQDPATHGGASSK | HexNAc@163=50; HexNAc@168=26 | 0.0015 |
| 754.0312 | 3 | −2 | IALPTQDPATHGGASSK | HexNAc@163=29; HexNAc@173=24; HexNAc@174=28 | 1.2e-6 |
| 686.3372 | 3 | −3 | IALPTQDPATHGGASSK | HexNAc@163=22; HexNAc@173=6 | 2.7e-8 |
| 825.9274 | 2 | 2 | IALPTQDPATHGGASSK | 6.4e-5 |
In conclusion, we have developed two selective sample preparation methods that combined with partial deglycosylation and HCD/ETD MS/MS analyses provide a better insight into the secreted O-glycoproteome, albeit only such GalNAcα1-containing structures are enriched that are not modified in position C6. In addition, structural information is lost due to partial deglycosylation. The single GalNAc identified could derive from a T-antigen (Core 1), from its sialylated version or may represent the original Tn-antigen present. Similarly, the HexNAc2 structures detected could derive from Core-3 structures that bind to Jacalin, or Core-2 structures that do not bind to the lectin, but happened to be on glycopeptides modified also with other sugar structures of Jacalin-specific affinity.
These methods can be readily applied to urine or CSF samples providing useful tools for glycosylation analysis. However, for the characterization of O-glycosylation in membrane proteins because of the limitations in protein level fractionation, a combination of ERLIC and lectin affinity-chromatography is more promising.
Acknowledgments
We are grateful to Dr. Andrew Alpert for the generous gift of the chromatographic columns used for this study. We would like to thank Dave Maltby and Agnes Arva for their technical assistance. We appreciate the help of Dr. Robert Chalkley in editing our manuscript.
Footnotes
* This work was supported by NIH grant NCRR P41RR001614 and by the Howard Hughes Medical Institute (to the UCSF Mass Spectrometry Facility, Director A.L. Burlingame), and the Hungarian National Office for Research and Technology BAROSS-DA07-DA-ESZK-07-2008-0036.
 This article contains supplemental Tables S1 to S8, Supplemental files S1 and S2 and Figure.
This article contains supplemental Tables S1 to S8, Supplemental files S1 and S2 and Figure.
1 The abbreviations used are:
- CID
- collision-induced dissociation
- AGC
- automatic gain control
- CV
- column volume
- ECD
- electron capture dissociation
- ERLIC
- electrostatic repulsion hydrophilic interaction chromatography
- ETD
- electron transfer dissociation
- GlcNAc
- N-acetyl glucosamine
- GalNAc
- N-acetyl galactosamine
- HCD
- higher-energy collision-dissociation
- Hex
- hexose
- HexNAc
- N-acetyl-hexosamine
- mixedIEX
- ion exchange on a mixed-bed column
- SA
- sialic acid
- TEAP
- triethylammonium phosphate.
REFERENCES
- 1. Apweiler R., Hermjakob H., Sharon N. (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 [DOI] [PubMed] [Google Scholar]
- 2. Kenan N., Larsson A., Axelsson O., Helander A. (2011) Changes in transferrin glycosylation during pregnancy may lead to false-positive carbohydrate-deficient transferrin (CDT) results in testing for riskful alcohol consumption. Clin. Chim. Acta 412, 129–133 [DOI] [PubMed] [Google Scholar]
- 3. Brockhausen I. (2006) Mucin-type O-glycans in human colon and breast cancer: glycodynamics and functions. EMBO Rep. 7, 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arnold J. N., Saldova R., Hamid U. M., Rudd P. M. (2008) Evaluation of the serum N-linked glycome for the diagnosis of cancer and chronic inflammation. Review. Proteomics 8, 3284–3293 [DOI] [PubMed] [Google Scholar]
- 5. Halim A., Brinkmalm G., Rüetschi U., Westman-Brinkmalm A., Portelius E., Zetterberg H., Blennow K., Larson G., Nilsson J. (2011) Site-specific characterization of threonine, serine, and tyrosine glycosylations of amyloid precursor protein/amyloid beta-peptides in human cerebrospinal fluid. Proc. Natl. Acad. Sci. U.S.A. 108, 11848–11853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Steentoft C., Vakhrushev S. Y., Vester-Christensen M. B., Schjoldager K. T., Kong Y., Bennett E. P., Mandel U., Wandall H., Levery S. B., Clausen H. (2011) Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat. Methods 8, 977–982 [DOI] [PubMed] [Google Scholar]
- 7. Wells L., Vosseller K., Cole R. N., Cronshaw J. M., Matunis M. J., Hart G. W. (2002) Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell. Proteomics 1, 791–804 [DOI] [PubMed] [Google Scholar]
- 8. McLachlin D. T., Chait B. T. (2003) Improved beta-elimination-based affinity purification strategy for enrichment of phosphopeptides. Anal. Chem. 75, 6826–6836 [DOI] [PubMed] [Google Scholar]
- 9. Medzihradszky K. F., Darula Z., Perlson E., Fainzilber M., Chalkley R. J., Ball H., Greenbaum D., Bogyo M., Tyson D. R., Bradshaw R. A., Burlingame A. L. (2004) O-sulfonation of serine and threonine: mass spectrometric detection and characterization of a new posttranslational modification in diverse proteins throughout the eukaryotes. Mol. Cell. Proteomics 3, 429–440 [DOI] [PubMed] [Google Scholar]
- 10. Zubarev R. A., Horn D. M., Fridriksson E. K., Kelleher N. L., Kruger N. A., Lewis M. A., Carpenter B. K., McLafferty F. W. (2000) Electron capture dissociation for structural characterization of multiply charged protein cations. Anal. Chem. 72, 563–573 [DOI] [PubMed] [Google Scholar]
- 11. Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takahashi K., Wall S. B., Suzuki H., Smith A. D., 4th, Hall S., Poulsen K., Kilian M., Mobley J. A., Julian B. A., Mestecky J., Novak J., Renfrow M. B. (2010) Clustered O-glycans of IgA1: defining macro- and microheterogeneity by use of electron capture/transfer dissociation. Mol. Cell. Proteomics 9, 2545–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scott N. E., Parker B. L., Connolly A. M., Paulech J., Edwards A. V., Crossett B., Falconer L., Kolarich D., Djordjevic S. P., Højrup P., Packer N. H., Larsen M. R., Cordwell S. J. (2011) Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 10, M000031-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Darula Z., Medzihradszky K. F. (2009) Affinity enrichment and characterization of mucin core-1 type glycopeptides from bovine serum. Mol. Cell. Proteomics 8, 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Good D. M., Wirtala M., McAlister G. C., Coon J. J. (2007) Performance characteristics of electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics 6, 1942–1951 [DOI] [PubMed] [Google Scholar]
- 16. Christiansen M. N., Kolarich D., Nevalainen H., Packer N. H., Jensen P. H. (2010) Challenges of determining O-glycopeptide heterogeneity: a fungal glucanase model system. Anal. Chem. 82, 3500–3509 [DOI] [PubMed] [Google Scholar]
- 17. Halim A., Nilsson J., Rüetschi U., Hesse C., Larson G. (2012) Human urinary glycoproteomics; attachment site specific analysis of N-and O-linked glycosylations by CID and ECD. Mol Cell Proteomics. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chalkley R. J., Baker P. R., Medzihradszky K. F., Lynn A. J., Burlingame A. L. (2008) In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol. Cell. Proteomics. 7, 2386–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tachibana K., Nakamura S., Wang H., Iwasaki H., Tachibana K., Maebara K., Cheng L., Hirabayashi J., Narimatsu H. (2006) Elucidation of binding specificity of Jacalin toward O-glycosylated peptides: quantitative analysis by frontal affinity chromatography. Glycobiology 16, 46–53 [DOI] [PubMed] [Google Scholar]
- 20. Lee S., Chen Y., Luo H., Wu A. A., Wilde M., Schumacker P. T., Zhao Y. (2010) The first global screening of protein substrates bearing protein-bound 3,4-Dihydroxyphenylalanine in Escherichia coli and human mitochondria. J. Proteome Res. 9, 5705–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alpert A. J. (2008) Electrostatic repulsion hydrophilic interaction chromatography for isocratic separation of charged solutes and selective isolation of phosphopeptides. Anal Chem. 80, 62–76 [DOI] [PubMed] [Google Scholar]
- 22. Medzihradszky K. F., Chalkley R. J., Trinidad J. C., Michaelevski A., Burlingame A. L. (2008) The utilization of Orbitrap higher collision decomposition device for PTM analysis and iTRAQ-based quantitation. 56th ASMS Conference on Mass Spectrometry, Denver, CO [Google Scholar]
- 23. Baker P. R., Trinidad J. C., Chalkley R. J. (2011) Modification site localization scoring integrated into a search engine. Mol. Cell. Proteomics 10, M111.008078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bourne Y., Astoul C. H., Zamboni V., Peumans W. J., Menu-Bouaouiche L., Van Damme E. J. M., Barre A., Rougé P. (2002) Structural basis for the unusual carbohydrate-specificity of jacalin towards galactose and mannose. Biochem. J. 364, 173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Medzihradszky K. F., (2005) Characterization of protein N-glycosylation. Methods Enzymol. 405, 116–138 [DOI] [PubMed] [Google Scholar]
- 26. Gill D. J., Clausen H., Bard F. (2011) Location, location, location: new insights into O-GalNAc protein glycosylation. Trends Cell Biol. 21, 149–158 [DOI] [PubMed] [Google Scholar]
- 27. Wilson I. B., Gavel Y., von Heijne G. (1991) Amino acid distributions around O-linked glycosylation sites. Biochem. J. 275, (Pt 2), 529–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Christlet T. H., Veluraja K. (2001) Database analysis of O-glycosylation sites in proteins. Biophys. J. 80(2), 952–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gram Schjoldager K. T., Vester-Christensen M. B., Goth C. K., Petersen T. N., Brunak S., Bennett E. P., Levery S. B., Clausen H. (2011) A systematic study of site-specific GalNAc-type O-glycosylation modulating proprotein convertase processing. J. Biol. Chem. 286, 40122–40132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Darula Z., Chalkley R. J., Lynn A., Baker P. R., Medzihradszky K. F. (2011) Improved identification of O-linked glycopeptides from ETD data with optimized scoring for different charge states and cleavage specificities. Amino Acids 41, 321–328 [DOI] [PMC free article] [PubMed] [Google Scholar]