Abstract
Background:
Hepatocellular carcinoma (HCC) is the third common cause of cancer-related deaths and its prognostication is still suboptimal. The aim of this study was to establish a new prognostication algorithm for HCC.
Methods:
In all, 13 biomarkers related to the etiopathogenesis of HCC were evaluated by immunohistochemistry using tissue microarrays containing 121 primary HCC resection cases, and validated in subsequent cohort of 85 HCC cases. The results were compared with Affymetrix Gene Chip Human Genome U133Plus microarray data in a separate cohort of 228 HCC patients.
Results:
On immunohistochemical evaluation and multivariate Cox regression analysis p53, alpha fetaprotein (AFP), CD44 and CD31, tumour size and vascular invasion, were significant predictors for worse survival in HCC patients. A morpho-molecular prognostic model (MMPM) was constructed and it was a significant independent predictor for overall survival (OS) and relapse-free survival (RFS) (P<0.000). The OS and RFS of HCClow was higher (104 and 78 months) as compared with HCChigh (73 and 43 months) (P<0.0001for OS and RFS). Hepatocellular carcinoma patients with higher stage (III+IV), >5 cm tumour size, positive vascular invasion and satellitosis belonged to HCChigh group. The validation group reproduced the same findings. Gene expression analysis confirmed that 7 of the 12 biomarkers were overexpressed in >50% of tumour samples and significant overexpression in tumour samples was observed in AFP, CD31, CD117 and Ki-67 genes.
Conclusion:
The MMPM, based on the expression of selected proteins and clinicopathological parameters, can be used to classify HCC patients between good vs poor prognosis and high vs low risk of recurrence following hepatic resection.
Keywords: p53, microvessel density, prognosis, liver cancer, survival
Hepatocellular carcinoma (HCC) is one of the most common cancers in men and the third most common cause of cancer-related death worldwide (Parkin, 2000). Regions of high incidence are areas of subSaharan Africa and South-east Asia (World Health Organization, http://www.who.int/whois), mainly linked to the presence of risk factors such as chronic Hepatitis B and/or Hepatitis C infection (Lai et al, 2003). Surgical resection followed by liver transplant is the mainstay treatment; however, this treatment is available only for a subset of patients, and even though hepatic resection is curative, the long-term prognosis is still poor (Chen et al, 2006). Conventionally, Barcelona Clinic Liver Cancer (BCLC) (Llovet et al, 1999) and Tumour-Node-Metastasis (TNM) (Vauthey et al, 2002) staging systems, serum alpha fetoprotein (AFP) level and tumour size are used for the prognostication of HCC patients. These staging systems incorporate histopathological features of HCC tumours such as tumour size, number of tumours, vascular invasion and satellitosis. Recently, a significant number of tissue-based markers have been studied in relation to prognosis (survival and tumour recurrence). However, none of these biomarkers, alone or in combination with other clinicopathological conventional features, are used in the routine clinical practice. We selected a panel of tissue-based molecular markers on the basis of their role in hepatocarcinogenesis and following the recent published reports on molecular/genomic classification of HCC. Briefly, we selected p53 (TP53), Ki-67 (PCNA), cyclin D1 (CCND1), (related to proliferation and cell cycling, G3 (Boyault et al, 2007) cluster A (Lee et al, 2004); β-catenin, E-cadherin (Wnt signalling pathway, S1, G5, G6) (Hoshida et al, 2009; Boyault et al, 2007); CD44 (HB subtype); cancer stem cell-related (CD133, CD117); angiogenesis-related (CD31) and hepatocyte functional markers (AFP, Hepar, CD10) (Yang et al, 2010). We performed immunohistochemical analysis on 121 pairs of human HCC tissues and their corresponding non-tumour hepatic tissues followed by confirmation of immunoexpression on 50 full sections. We then constructed a morpho-molecular prognostic model (MMPM) based on the prognostic power of the histological parameters and the relative expression of the immunohistochemical markers. The resulted MMPM predicted patient outcome (death/relapse) more powerfully than any molecular markers. The robustness of MMPM was corroborated and reproducible on a separate cohort of 85 HCC cases.
Materials and methods
Three independent cohorts of patients were included in the study. For immunohistochemical analysis, formalin-fixed paraffin-embedded tissues from surgically resected specimens of HCC patients who had undergone curative hepatectomy between 1990 and 2003 (cohort 1, n=121) and from 2004 to 2009 (cohort 2, n=85) at National University Hospital, Singapore were taken. For gene expression analysis, an independent cohort of 228 patients with HCC (cohort 3) was recruited from Queen Mary Hospital, Hong Kong, between 1993 and 2007 as described previously (Luk et al, 2006). For the latter, the tumour and adjacent non-tumourous tissues were collected after hepatectomy, and were immediately snap frozen and stored at −80 °C prior to analysis. Cohort 1 and 2 were analysed for immunohistochemistry in a tissue microarray (TMA) format as described previously (Das et al, 2008). Clinicopathological information was obtained from the medical records and included ethnicity, age, gender, tumour number, tumour size, stage, histological grading, vascular invasion, satellitosis and preoperative serum AFP (Table 1). Tumour differentiation was defined according to the Edmondson grading system (Edmondson and Steiner, 1954). Tumour staging was defined according to the sixth edition of the TNM classification of the International Union against Cancer (Sobin and Wittekind, 2002). Patients were followed up for death/relapse. Overall survival (OS) was defined as the interval between surgery and death or date of last observation. The death data was censored at the last follow-up for living patients. Patients of both cohorts were followed up until May 2010. Relapse-free survival (RFS) was defined from the date of surgery until the detection of recurrent tumour or the date of last follow-up. The RFS data was censored for patients without tumour recurrence. Ethics approval for this study was obtained from National University Singapore-Institutional Review Board (NUS-IRB; 10–133).
Table 1. Clinicopathological features of two cohorts with hepatocellular carcinoma.
| Clinical and pathological features | Cohort1, n (%) | Cohort 2, n (%) |
|---|---|---|
| Age (years) | ||
| <50 | 27 (22.3) | 19 (22.4) |
| >50 | 70 (57.9) | 66 (77.6) |
| Sex | ||
| Male | 104 (86) | 64 (75) |
| Female | 17 (14) | 21 (25) |
| Ethnicity | ||
| Chinese | 99 (81.8) | 63 (74.1) |
| Others | 22 (18.2) | 22 (25.9) |
| Serum AFP (ng dl−1) | ||
| <20 | 35 (28.9) | 33 (38.8) |
| >20 | 59 (48.8) | 37 (43.5) |
| HbsAg | ||
| Yes | 68 (56.2) | 52 (61.2) |
| No | 33 (27.3) | 28 (32.9) |
| Alcoholic a | ||
| Yes | 32 (26.4) | 27 (31.8) |
| No | 55 (45.5) | 45 (52.9) |
| Tumour differentiation | ||
| I+II | 104 (86) | 71 (83.5) |
| III+IV | 17 (14) | 14 (16.5) |
| TNM stage | ||
| I+II | 85 (71.2) | 68 (80) |
| III+IV | 21 (17.4) | 17 (20) |
| Tumour number | ||
| Solitary | 75 (62) | 64 (75.3) |
| Multiple | 46 (38) | 21 (24.7) |
| Size (cm) | ||
| <5 | 46 (38) | 46 (54.1) |
| >5 | 75 (62) | 38 (44.7) |
| Vascular invasion b | ||
| Yes | 52 (43) | 17 (20) |
| No | 41 (33.9) | 60 (70.6) |
| Satellitosis | ||
| Yes | 15 (12.4) | 12 (14.1) |
| No | 106 (87.6) | 73 (85.9) |
Abbreviations: AFP=α-fetoprotein; HbsAg=hepatitis B surface antigen; TNM=tumour-node-metastasis.
Alcoholic intake of approximately >60 mg per day for prolonged period.
Cases with complete clinical information were included in the analysis.
Immunoreactivity for each marker was assessed semi quantatively by evaluating the extent and intensity of the staining; extent was recorded by the percentage of positive tumour cells in relation to the total number of tumour cells; intensity was recorded as absent, weak, moderate or strong. No or weak staining was considered as negative and moderate or strong staining was considered as positive. Immunohistochemical staining was assessed by two independent and trained viewers (SS and OCW). The cut-off percentage for determining positive expression of each protein was determined by receiver-operating characteristics (ROC) analysis against the OS as described previously (Heagerty et al, 2000). Using values derived from the area under the ROC, values above 0.5 indicate significant discriminatory power for survival (Zlobec et al, 2007). Gene expression profiling was performed by cDNA microarray using AffyU133Plus assay (Affymetrix, Santa Clara, CA, USA) containing 47 000 probes. All procedures for hybridisation, labelling and scanning of gene chips were as described previously in accordance with the manufacturer’s recommendations (Burchard et al, 2010). Raw gene expression profiling data were deposited to GEO database with the accession number of GSE25097.
Statistical analysis was performed using SPSS v 15.0 for Windows (SPSS, Armonk, NY, USA). Cumulative OS and RFS was calculated by the Kaplan–Meier method and analysed by the log-rank test followed by multivariate analyses using Cox proportional hazard regression model for statistically significant factors. A P-value ⩽0.05 was considered statistically significant.
Results
Immunohistochemical expression of the markers
All the samples were assessed for the immunohistochemical expression of the 13 protein markers (Figure 1). The subcellular localisation of the expression (cytoplasmic/membranous/nuclear) along with the cut-off values and frequency of positive expression of the markers are given in Supplementary Table 1 (S1). To establish the reliability of TMAs for this analysis, we further analysed the expression in full sections of these markers in 50 HCC cases randomly chosen from cohort 1. The results were concordant in 96% (48/50) of the cases. Two cases showed nuclear staining of p53 (50, 30%) in the TMA, but weak staining (less than 10%) in the corresponding full tumour sections. These cases were taken as p53 negative according to the results on full tumour sections.
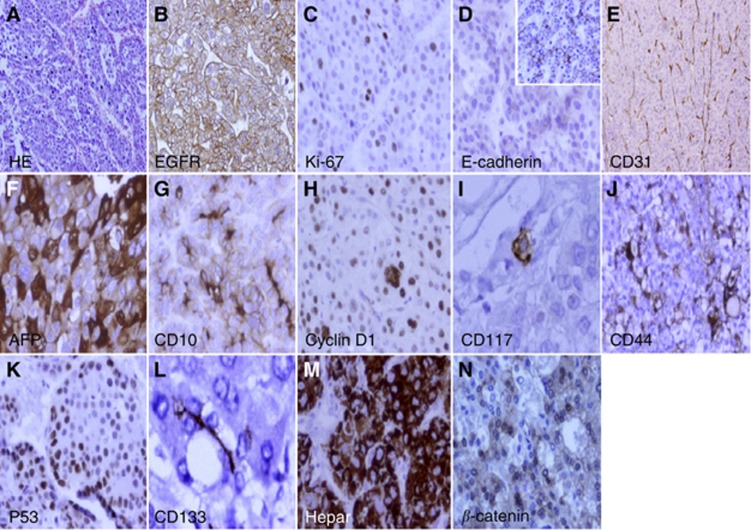
Figure 1.
Representative positive expression of protein markers: (A) HE; (B) epidermal growth factor receptor (EGFR); (C) Ki-67; (D) E-cadherin; (E) CD31; (F) AFP; (G) CD10; (H) cyclin D1; (I) CD117; (J) CD44; (K) P53; (L) CD133; (M) Hepar; (N) β-catenin by immunohistochemistry in tumour tissue microarrays (original magnification × 400).
Prognostic significance of 13 protein markers expression and clinicopathological characteristics
The mean OS was 93.51±9.88 months. The mean RFS was 60.11±8.05 months. The OS for 1-, 3-, 5-year were 81%, 65% and 50%, respectively. The RFS for 1-, 3-, 5-year were 61%, 44% and 29%, respectively. On univariate analysis p53, CD44, AFP, CD31, Ki-67, E-cadherin and cyclin D1 were unfavourable predictors of OS and RFS. Among the clinicopathological parameters higher TNM stage, more than 5 cm tumour size, positive satellitosis and vascular invasion were poor prognostic factors for OS or RFS (Table 2). Multivariate Cox regression model showed that p53, AFP, CD31 and CD44, vascular invasion and tumour size are statistically significant, independent factors for prognosis (Table 3).
Table 2. Univariate overall and recurrence-free survival analysis for clinicopathological features and protein markers.
|
Overall survival
|
Recurrence-free survival
|
|||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | P -value | HR | 95% CI | P -value |
| Clinicopathological | ||||||
| Gender (male vs female) | 1.44 | 0.81–2.55 | NS | 0.90 | 0.54–1.52 | NS |
| Age, years (>50 vs <50) | 1.19 | 0.65–216 | NS | 1.00 | 0.62–1.62 | NS |
| Ethnicity (Chinese vs non-Chinese) | 0.70 | 0.33–1.49 | NS | 0.88 | 0.50–1.53 | NS |
| Serum AFP, ng dl−1 (>20 vs <20 | 1.66 | 0.96–2.85 | 0.066 | 1.62 | 1.02–2.55 | 0.057 |
| Grade (I+II vs III+IV) | 0.70 | 0.30–1.64 | NS | 0.91 | 0.50–1.64 | NS |
| TNM stage (I+II vs III+IV) | 2.42 | 1.38–4.22 | 0.002 | 2.52 | 1.56–4.06 | 0.000 |
| Tumour size, cm (>5 vs <5) | 1.68 | 1.00–2.83 | 0.048 | 2.20 | 1.44–3.37 | 0.000 |
| Number (solitary vs multiple) | 1.21 | 0.70–2.68 | NS | 1.49 | 0.96–2.31 | NS |
| Satellitosis (yes vs no) | 3.01 | 1.58–5.72 | 0.001 | 3.34 | 1.93–5.79 | 0.000 |
| Vascular invasion (yes vs no) | 3.39 | 1.91–6.01 | 0.000 | 2.89 | 1.82–4.58 | 0.000 |
| Protein expression | ||||||
| P53 (positive vs negative) | 2.38 | 1.42–4.0 | 0.001 | 1.83 | 1.18–2.84 | 0.006 |
| Cyclin D1 (positive vs negative) | 1.67 | 0.99–2.82 | 0.049 | 1.04 | 0.68–1.60 | NS |
| CD31 (positive vs negative) | 1.85 | 1.04–3.29 | 0.033 | 1.67 | 1.06–2.63 | 0.026 |
| EGFR (positive vs negative) | 1.08 | 0.63–1.84 | NS | 0.97 | 0.63–1.51 | NS |
| Ki-67 (positive vs negative) | 3.16 | 1.11–8.99 | 0.031 | 2.05 | 0.74–5.65 | NS |
| β-catenin (positive vs negative) | 1.12 | 0.65–1.90 | NS | 1.10 | 0.72–1.68 | NS |
| E-cadherin (positive vs negative) | 1.99 | 1.00–3.97 | 0.045 | 1.37 | 0.83–2.26 | NS |
| AFP (positive vs negative) | 2.21 | 1.26–3.87 | 0.005 | 2.13 | 1.35–3.36 | 0.001 |
| Hepar-1 (positive vs negative) | 0.58 | 0.34–1.00 | NS | 0.71 | 0.46–1.09 | NS |
| CD10 (positive vs negative) | 1.13 | 0.66–1.94 | NS | 1.28 | 0.82–1.98 | NS |
| CD117 (positive vs negative) | 1.82 | 0.66–5.0 | NS | 1.16 | 0.74–3.48 | NS |
| CD133 (positive vs negative) | 1.41 | 0.56–3.55 | NS | 0.47 | 0.15–1.5 | NS |
| CD44 (positive vs negative) | 2.21 | 1.28–3.83 | 0.004 | 1.34 | 0.88–2.04 | NS |
Abbreviations: AFP=α-fetoprotein; CI=confidence interval; EGFR=epidermal growth factor receptor; HR=hazard risk ratio; NS=not significant; TNM=tumour-node-metastasis.
Univariate analysis, Cox proportional hazards regression model.
Table 3. Multivariate analysis for the protein markers and clinicopathological features for OS and RFS.
|
OS
|
RFS
|
|||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | P -value | HR | 95% CI | P -value |
| Clinicopathological | ||||||
| TNM stage (I+II vs III+IV) | 1.92 | 0.68–5.42 | NS | 0.98 | 0.38–2.53 | NS |
| Tumour size, cm (>5 vs <5) | 2.38 | 1.18–4.81 | 0.013 | 2.98 | 1.70–5.23 | 0.000 |
| Satellitosis (yes vs no) | 0.77 | 0.25–2.41 | NS | 2.25 | 0.81–6.23 | NS |
| Vascular invasion (yes vs no) | 2.98 | 1.38–6.41 | 0.004 | 1.89 | 1.06–3.362 | 0.030 |
| Protein expression | ||||||
| P53 (pos vs neg) | 1.93 | 1.03–3.63 | 0.039 | 1.93 | 1.03–3.63 | 0.030 |
| Cyclin D1 (pos vs neg) | 1.82 | 0.88–3.76 | NS | 1.11 | 0.56–2.19 | NS |
| CD31(pos vs neg) | 2.41 | 1.10–5.31 | 0.028 | 1.70 | 0.97–3.00 | 0.060 |
| Ki-67 (pos vs neg) | 3.16 | 1.11–8.99 | NS | 1.50 | 0.49–4.56 | NS |
| E-cadherin (pos vs neg) | 1.99 | 1.00–3.97 | NS | 0.64 | 0.32–1.26 | NS |
| AFP (pos vs neg) | 2.22 | 1.03–4.78 | 0.041 | 1.68 | 0.81–3.46 | NS |
| CD44 (pos vs neg) | 2.12 | 1.02–4.41 | 0.044 | 0.94 | 0.49–1.82 | NS |
Abbreviations: AFP=α-fetoprotein; CI=confidence interval; HR=hazard ratio; neg=negative; OS=overall survival; pos=positive; RFS=recurrence-free survival; TNM=tumour-node-metastasis.
Multivariate analysis, Cox proportional hazard regression model. Variables were adopted for their prognostic significance by univariate analysis.
Morpho-molecular prognostic model
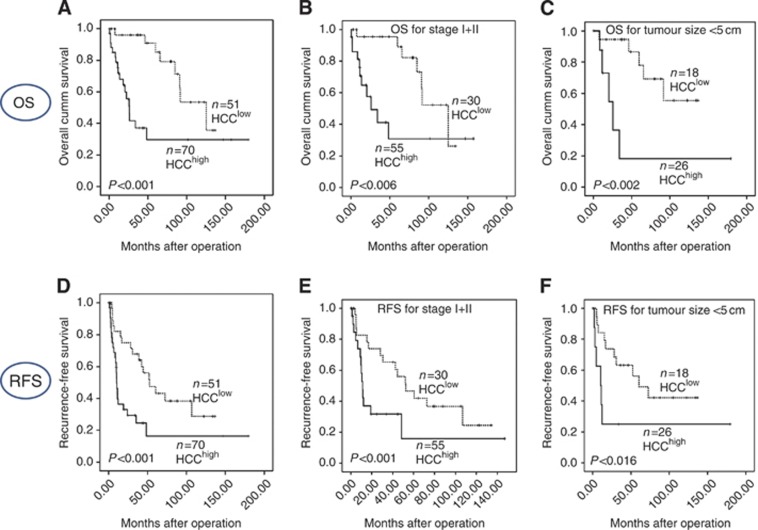
The risk scores for MMPM were calculated using Cox regression model for multivariate analysis, and it was as follows: (0.800 × CD31)+(0.597 × p53)+(0.662 × AFP)+(0.485 × CD44)+(0.583 × size)+(1.001 × vascular invasion). The protein marker represents the expression level (positive=1, negative=0), and the histological features can be present (=1) or absent (=0). The median of the final score was 3.240. Accordingly, the 121 cases were dichotomised in two groups, HCChigh (score>3.240) and HCClow (score<3.240). The OS and RFS in HCClow was significantly higher (104 and 78 months) than in HCChigh (73 and 43 months) (P<0.0001 for OS and RFS, respectively) (Figure 2). The HCChigh also expressed higher serum AFP (ng dl−1) (3706±9199 vs 346±1625; P=0.006) and higher MVD (21.10±12.65 vs 15.95±13.33; P=0.015) as compared with HCClow. Patients with higher stage (III+IV), >5 cm tumour size, positive vascular invasion and satellitosis belonged to HCChigh group as compared with HCClow group (P<0.001, P<0.001, P<0.001 and P=0.022, respectively). On multivariate analysis, the MMPM was an independent prognostic factor for OS (P=0.008) against clinicopathological factors, however for RFS (P=0.074) it was not significant. The prognostic power of the MMPM was higher than the individual markers (p53, CD44, AFP and CD31) and the clinicopathological features alone as shown in the ROC curve in Supplementary Figure 1 (SF1). Further, when stratified by tumour size and TNM stage the MMPM could be a robust predictor of OS (P-value 0.002 and 0.006, respectively) and RFS (P-value 0.016 and 0.000) (Figure 2).
Figure 2.
Kaplan–Meier survival analysis of low- and high-risk HCC patients by MMPM in cohort 1: overall survival (A), stratification of OS of cohort 1 for early TNM stage (I+II) (B), and tumour size <5 cm (C), recurrence-free survival (D), stratification of RFS for early TNM stage (I+II) (E) and tumour size <5 cm (F).
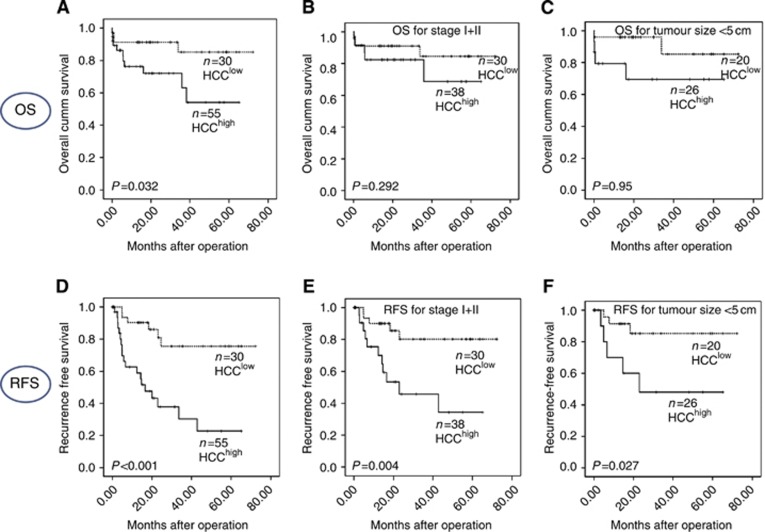
Validation of the MMPM
Validation for the predictive power of the MMPM was done in another cohort of 85 HCC patients. Patients classified as HCChigh had a significant shorter OS and RFS (43.2 and 26.3 months) as compared with HCClow (63.7 and 58.5 months) (P=0.032 and 0.000 for OS and RFS, respectively) (Figure 3). Similar to cohort 1, we observed that the patients with higher TNM stage (III+IV) (14/16; P<0.001), >5 cm tumour size (23/35; P=0.009), positive vascular invasion (17/17; P<0.001) and satellitosis (9/11; P=0.048) belonged to the HCChigh group. Higher MVD was also observed in high-risk group as compared with low-risk group (20±2 vs 18±2). However, it was not significant (P=0.551). Similarly, the serum level of AFP was of borderline significance (P=0.060). The P-values of early TNM stage (I+II) and tumour size less than 5 cm were 0.004 and 0.027, respectively, for RFS. For OS, early TNM stage (I+II) and tumour size <5 cm were not significant, P=0.29 and 0.095, respectively, which is likely to be owing to small sample size.
Figure 3.
Kaplan–Meier survival analysis of low- and high-risk HCC patients by MMPM in cohort 2: overall survival (A), stratification of OS of cohort 2 for early TNM stage (I+II) (B), and tumour size <5 cm (C), recurrence-free survival (D), stratification of RFS for early TNM stage (I+II) (E) and tumour size <5 cm (F).
Validation of MMPM by gene expression of prognostic markers
Of 12 of the 13 prognostic markers represented in the microarray (Hepar-1 had no corresponding gene), 7 of them (i.e., AFP, β-catenin, CD31, CD44, CD117, Ki-67, TP53) were overexpressed in >50% of tumour samples. Significant overexpression in tumour samples was observed in AFP, CD31, CD117, Ki-67 genes.
Discussion
Hepatocellular carcinoma is one of the most common malignant tumours worldwide and has poor prognosis and high recurrence rate, regardless of the treatment. Therefore, it is imperative for clinicians and scientists to find new ways to stratify patients for appropriate treatment. Previous reports have attempted to build a model based on the prognostic value of putative hepatic stem cell biomarkers in HCC (Yang et al, 2010). Traditionally, however, tumour staging system (TNM and BCLC staging), tumour size and serum AFP levels are used to predict the outcome of HCC patients, which sometimes cannot accurately predict the outcome of all HCC patients (Qin and Tang, 2004). Till date, there is neither any molecular marker routinely incorporated to staging systems, nor there is a molecular prognostic model. The present study was undertaken to identify a morpho-molecular prognosticator of HCC patients. The criteria for selection of the molecular markers evaluated in this study were tissue-based markers that are routinely available in Pathology Department with a strong basis for their role in hepatocarcinogenesis. Our results showed that the poor prognostic value of the overexpression of these markers was highest when all the four biomarkers and two histological parameters with individual prognostic significance were taken into consideration together. TP53 is a tumour suppressor gene, with a well-known function in DNA repair and apoptosis (Hu et al, 2003) and has been implicated in both hepatocarcinogenesis and HCC tumour recurrence. CD44 has been identified as a tumour stem cell marker in various epithelial cancers, including HCC. It is also a marker for tumour progression and has been previously reported to predict worse survival in HCC patients (Endo and Terada, 2000). CD31 is involved in angiogenesis and microvessel density previously shown in lung cancer and also in HCC (Giatromanolaki et al, 1996; Frachon et al, 2001). Alpha fetoprotein is an oncofetal marker traditionally used to prognosticate and follow up HCC patients (Kawai et al, 2001). We validated our findings in a separate cohort of 228 HCC patients and observed a significant overexpression in tumour samples in AFP, CD31, CD117 and Ki-67 genes. In other studies, significant overexpression of TP53 gene (subgroup G3) (Boyault et al, 2007) and CD44 gene (Yang et al, 2010) were observed in HCC patients. Therefore, the gene expression analysis in these studies was both confirmatory of the protein expression and, despite the possible transcriptional modifications, the overall relevance of the elements forming our proposed MMPM.
Classifications of HCC based on genetic profiles have been reported previously; however in a routine clinical set-up, high-throughput analyses have problems of reproducibility and affordability. Immunohistochemical analysis can provide cheaper, faster and more reproducible results. Few studies have reported HCC stratification based on immunohistochemical analysis (Yamashita et al, 2008). Based on a standard scoring system derived from Cox Regression analysis, we stratified the study cohort into HCClow and HCChigh groups, with considerable differences in OS and RFS between them. When stratified by TNM stage and tumour size, MMPM stood as a good predictor of OS and RFS, regardless of the tumour stage and size (P<0.05). Of interest, the MMPM was valuable in predicting the outcome in early-stage HCC and small size tumour, which are usually difficult to predict by conventional indices (Qin and Tang, 2004). Although the MMPM was validated in a smaller second cohort, a larger independent cohort is required to validate this scoring system.
Hepatocellular carcinoma has a 5-year recurrence rate of approximately 80–90% and currently, size of hepatic nodules, vascular invasion and serum AFP level are used for risk estimation of tumour recurrence. Sorefanib and combination of ribavarin and interferon are few treatment options available for such HCC patients. It is also known that, the HCC recurrence is most frequently observed in the first 1–2 years after curative treatment. Our model shows that HCC patients with score more than 3.240 have shorter RFS period (43 vs 78 months, P<0.0001). Therefore MMPM could be useful in stratifying HCC patients for early recurrence so that a timely intervention could be made.
Immunohistochemical studies have been sometimes criticised by their subjectivity due to their qualitative interpretation. Because of this, and also because of the relatively small amount of tissue analysed per case in the TMA format, we chose two forms of validation. The remarkable concordance between TMA cores and large sections, already reported in other cancer types (Zhang et al, 2003) is highly reassuring of the technical robustness of this approach.
In conclusion, our study identifies p53, CD44, CD31 and AFP as powerful predictors of OS and RFS in HCC patients, and, as such, our proposed MMPM represents a powerful discriminator of prognosis and has implications in future in patient management.
Acknowledgments
We thank Madam Cheong Sok Lian for providing the clinicopathological information and Madam Choo Shoonian for help in immunohistochemistry. This study is funded by Singapore Cancer Syndicate, Agency for Science, Technology and Research, Singapore, and the Grants numbers are MN005, MN005R and MN077.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Boyault S, Rickman DS, de Reynies A (2007) Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 45: 42–52 [DOI] [PubMed] [Google Scholar]
- Burchard J, Zhang C, Liu AM, Poon RT, Lee NP, Wong KF, Sham PC, Lam BY, Ferguson MD, Tokiwa G, Smith R, Leeson B, Beard R, Lamb JR, Lim L, Mao M, Dai H, Luk JM (2010) MicroRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol 6: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XP, Qiu FZ, Wu ZD, Zhang ZW, Huang ZY, Chen YF (2006) Long term outcome of resection of large hepatocellular carcinoma. Br J Surg 93(5): 600–606 [DOI] [PubMed] [Google Scholar]
- Das K, Mohd Omar MF, Ong CW, Bin Abdul Rashid S, Peh BK, Putti TC, Tan PH, Chia KS, Teh M, Shah N, Soong R, Salto-Tellez M (2008) TRARESA: tissue microarray-based hospital system for biomarker validation and discovery. Pathology 40: 441–449 [DOI] [PubMed] [Google Scholar]
- Edmondson HA, Steiner PE (1954) Primary carcinoma of the liver: A study of 100 cases among 48,900 necropsies. Cancer 7: 462–503 [DOI] [PubMed] [Google Scholar]
- Endo K, Terada T (2000) Protein expression of CD44 (standard and variant isoforms) in hepatocellular carcinoma: relationships with tumor grade, clinicopathologic parameters, p53 expression and patient survival. J Hepatol 32: 78–84 [DOI] [PubMed] [Google Scholar]
- Frachon S, Gouysse G, Dumortier J, Couvelard A, Nejjari M, Mion F, Berger F, Paliard P, Boillot O, Scoazec JY (2001) Endothelial cell marker expression in dysplastic lesions of the liver: an immunohistochemical study. J Hepatol 34: 850–857 [DOI] [PubMed] [Google Scholar]
- Giatromanolaki A, Koukourakis M, O’Byrne K, Fox S, Whitehouse R, Talbot DC, Harris AL, Gatter KC (1996) Prognostic value of angiogenesis in operable non-small cell lung cancer. J Pathol 79: 80–88 [DOI] [PubMed] [Google Scholar]
- Heagerty PJ, Lumley T, Pepe MS (2000) Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 56: 337–344 [DOI] [PubMed] [Google Scholar]
- Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M, Watanabe G, Gabriel S, Friedman SL, Kumada H, Llovet JM, Golub TR (2009) Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res 69: 7385–7392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TH, Huang CC, Lin PR, Chang HW, Ger LP, Lin YW, Changchien CS, Lee CM, Tai MH (2003) Expression and prognostic role of tumor suppressor gene PTEN/MMAC1/TEP1 in hepatocellular carcinoma. Cancer 97(8): 1929–1940 [DOI] [PubMed] [Google Scholar]
- Kawai HF, Kaneko S, Honda M, Shirota Y, Kobayashi K (2001) α-fetoprotein-producing hepatoma cell lines share common expression profiles of genes in various categories demonstrated by cDNA microarray analysis. Hepatology 33: 676–691 [DOI] [PubMed] [Google Scholar]
- Lai CL, Ratziu V, Yuen MF, Poynard T (2003) Viral hepatitis B. Lancet 362: 2089–2094 [DOI] [PubMed] [Google Scholar]
- Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, Durnez A, Demetris AJ, Thorgeirsson SS (2004) Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology 40: 667–676 [DOI] [PubMed] [Google Scholar]
- Llovet JM, Bru C, Bruix J (1999) Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis 19: 329–338 [DOI] [PubMed] [Google Scholar]
- Luk JM, Lam CT, Siu AF, Lam BY, Ng IO, Hu MY, Che CM, Fan ST (2006) Proteomic profiling of hepatocellular carcinoma in Chinese cohort reveals heat-shock proteins (Hsp 27, Hsp 70, GRP 78) up-regulation and their associated prognostic values. Proteomics 6(3): 1049–1057 [DOI] [PubMed] [Google Scholar]
- Parkin DM (2000) Global cancer statistics in the year 2000. Lancet Oncol 2: 533–543 [DOI] [PubMed] [Google Scholar]
- Qin LX, Tang ZY (2004) Recent progress in predictive biomarkers for metastatic recurrence of human hepatocellular carcinoma: A review of the literature. J Cancer Res Clin Oncol 130: 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobin LH, Wittekind C (2002) UICC (International Union against Cancer). TNM Classification of Malignant Tumors, 6th edn, pp 1–264. John Wiley & Sons Inc: New York [Google Scholar]
- Vauthey JN, Lauwers GY, Esnaola NF, Do KA, Belghiti J, Mirza N, Curley SA, Ellis LM, Regimbeau JM, Rashid A, Cleary KR, Nagorney DM. (2002) Simplified staging for hepatocellular carcinoma. J Clin Oncol 20: 1527–1536 [DOI] [PubMed] [Google Scholar]
- World Health Organization. Mortality Database. WHO Statistical Information System. Available from URL http://www.who.int/whois
- Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A, Zanetti KA, Chen Y, Qin LX, Tang ZY, Wang XW (2008) EpCAM and α-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res 68(5): 1451–1461 [DOI] [PubMed] [Google Scholar]
- Yang XR, Xu Y, Yu B, Zhou J, Qiu SJ, Shi GM, Zhang BH, Wu WZ, Shi YH, Wu B, Yang GH, Ji Y, Fan J (2010) High expression levels of putative hepatic stem/progenitor cell biomarkers related to tumour angiogenesis and poor prognosis of hepatocellular carcinoma. Gut 59: 953–962 [DOI] [PubMed] [Google Scholar]
- Zhang D, Salto-Tellez M, Putti TC, Do E, Koay ES (2003) Reliability of tissue microarrays in detecting protein expression and gene amplification in breast cancer. Mod Pathol 16(1): 79–85 [DOI] [PubMed] [Google Scholar]
- Zlobec I, Steele R, Terracciano L, Jass JR, Lugli A (2007) Selecting immunohistochemical cut-off scores for novel biomarkers of progression and survival in colorectal cancer. J Clin Pathol 60: 1112–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.