Abstract
Background:
KRAS mutation testing is required to select patients with metastatic colorectal cancer (CRC) to receive anti-epidermal growth factor receptor antibodies, but the optimal KRAS mutation test method is uncertain.
Methods:
We conducted a two-site comparison of two commercial KRAS mutation kits – the cobas KRAS Mutation Test and the Qiagen therascreen KRAS Kit – and Sanger sequencing. A panel of 120 CRC specimens was tested with all three methods. The agreement between the cobas test and each of the other methods was assessed. Specimens with discordant results were subjected to quantitative massively parallel pyrosequencing (MPP). DNA blends were tested to determine detection rates at 5% mutant alleles.
Results:
Reproducibility of the cobas test between sites was 98%. Six mutations were detected by cobas that were not detected by Sanger, and five were confirmed by MPP. The cobas test detected eight mutations which were not detected by the therascreen test, and seven were confirmed by MPP. Detection rates with 5% mutant DNA blends were 100% for the cobas and therascreen tests and 19% for Sanger.
Conclusion:
The cobas test was reproducible between sites, and detected several mutations that were not detected by the therascreen test or Sanger. Sanger sequencing had poor sensitivity for low levels of mutation.
Keywords: KRAS mutation testing, molecular diagnostics, colorectal cancer
Retrospective analyses of pivotal clinical trials for the anti-epidermal growth factor receptor (EGFR) monoclonal antibodies, cetuximab and panitumumab, have revealed that patients with colorectal cancer (CRC) tumours containing activating mutations in the downstream KRAS gene do not receive benefit from therapy (Amado et al, 2008; Karapetis et al, 2008). The findings that KRAS mutation status is a key determinant of response to anti-EGFR therapy are now reflected in the labels for both these agents, with the European Medicines Agency labelling stating that these targeted agents are only indicated in patients with wild-type KRAS tumours, while the US Food and Drug Administration states that such treatment is not recommended in tumours with mutations in codons 12 and 13 of KRAS.
In line with the drug labels, KRAS mutation testing is now recommended by major oncology organisations. Both the American Society for Clinical Oncology and National Comprehensive Cancer Network (NCCN) specify that before treatment, tumours should be tested for the presence of mutations in codons 12 and 13 of the KRAS gene (Allegra et al, 2009; Engstrom et al, 2009), while the European Society for Medical Oncology recommends establishing that tumours are KRAS wild type, without naming specific mutations (Van Cutsem et al, 2010). To date, neither the product labels, nor the oncology guidelines delineate specific codon 12 and 13 mutations.
The recommendation to test for mutations in codons 12 and 13 of the KRAS gene reflects the high frequency of these activating mutations in colorectal tumours: 24–43% of colorectal tumours contain KRAS mutations, of which ∼82% are in codon 12% and ∼17% are in codon 13 (Samowitz et al, 2000; Andreyev et al, 2001). Following the initial retrospective analysis showing that patients with activating KRAS mutations did not respond to treatment, the majority of subsequent clinical trials of anti-EGFR antibodies in CRC have excluded patients with tumours harbouring codon 12 and 13 mutations (Amado et al, 2008; Douillard et al, 2010; Peeters et al, 2010; Bokemeyer et al, 2011; Van Cutsem et al, 2011).
An emerging body of data suggests that KRAS mutations in codon 61 may also be predictors of non-responsiveness to anti-EGFR therapies. Mutations in codon 61 resulted in constitutive activation and increased the transforming activity of cells (Der et al, 1986). Limited clinical data indicate that, similarly to codon 12 and 13 mutations, codon 61 mutations also predict resistance to anti-EGFR monoclonal antibodies (Loupakis et al, 2009; De Roock et al, 2010a; Molinari et al, 2011). Furthermore, the frequency of codon 61 mutations may be greater than initially reported (Richman et al, 2009; Vaughn et al, 2011). In a recent study of CRC samples, 39% were found to be KRAS mutated and 8% of mutations were located in codon 61 (Sundstrom et al, 2010).
The association between defined mutations and response to therapy provides a clear opportunity to increase response rates and reduce the likelihood of treating patients who are unlikely to respond to certain drugs. However, there is a critical need to set standards for this type of mutation testing (van Krieken and Tol, 2009). A number of sequencing and PCR-based methods for detecting KRAS mutations are currently in clinical use. A recent methods comparison study suggested that a variety of techniques might be suitable for KRAS mutation testing. However, it is not clear which technique offers the best performance in terms of sensitivity, specificity, reproducibility, and success rates (Whitehall et al, 2009).
We performed a two-centre study to compare the analytic performance and workflow characteristics of the cobas KRAS Mutation Test to two other methods commonly used in the clinical setting: 2 × bidirectional Sanger sequencing (Sanger) and the therascreen KRAS Mutation Kit (‘therascreen’, Qiagen, Hilden, Germany). We used a blinded panel of formalin-fixed paraffin-embedded tissue (FFPET) CRC specimens and DNA blends.
Materials and methods
Mutation testing methods
The cobas KRAS Mutation Test kit
(Roche Molecular Systems, Inc., Branchburg, NJ, USA) is a CE-IVD marked TaqMelt PCR assay designed to detect the presence of 19 KRAS mutations in codons 12, 13, and 61 in FFPET CRC specimens (Supplementary Figure 1). The test requires 100 ng total DNA input which can typically be obtained by using one 5 μm section. One patient sample is tested in two wells per 96-well plate for a total throughput of 45 specimens per plate. The workflow and testing process have been described previously (Lee et al, 2011).
The therascreen KRAS PCR Kit
(Qiagen, Manchester, UK) is a CE-IVD marked real-time PCR assay that combines an amplification refractory mutation system and a Scorpion fluorescent primer/probe system (ARMS) to detect seven mutations in codons 12 and 13 of the KRAS gene (Supplementary Figure 1). One patient sample is tested by one control assay and seven mutation assays for a total throughput of 10 samples per 96-well plate. All reaction assays include an exogenous internal control for the presence of inhibitors. Reactions were run in the ABI 7500 instrument (Life Technologies, Warrington, UK) and analysed using software v2.0.5 (Life Technologies). The therascreen kit requires ⩾20 ng of amplifiable genomic DNA from FFPET specimens. According to the lab-validated clinical protocol 100–200 ng of total DNA, as measured by spectrophotometry, is used per PCR to account for the partial degradation of FFPET DNA without resulting in oversaturation of the reaction. The test requires a total DNA input of 800–1600 ng. The DNA volume used for each PCR is 5 μl and all samples are diluted to 20–25 ng μl−1. Therascreen kit has been reported to detect mutations at 1% sensitivity in plasmids and cell lines provided the Ct value of the exogenous control is <29.
Sanger sequencing:
Mutation screening for exons 2 and 3 of the KRAS gene was carried out using PCR conditions and 2 × bidirectional direct sequencing following previously described protocols (Fernandez et al, 2004; Conde et al, 2006). Exon 3 primers were designed specifically for this study. Tumour DNA for exon 3 was amplified using the following primers: forward primer 5′-CACTGTAATAATCCAGACTGTG-3′ and reverse: 5′-CCCACCTATAATGGTGAATATC-3′. Sequencing reactions were performed in both direct and reverse directions, and electropherograms were reviewed manually to detect any genetic alteration. All variants were confirmed by resequencing-independent PCR products. Theoretically, 150 ng of DNA is used per PCR performed for a total of 300 ng total DNA input for KRAS exon 2 and exon 3.
454 sequencing
(454 Life Sciences, Branford, CT, USA) is a quantitative massively parallel pyrosequencing (MPP) method, which involves clonal amplification by emulsion PCR of target sequences followed by MPP (Margulies et al, 2005). Each specimen was amplified using primers with 454 tags. The amplicon was quantified, diluted, and pooled with 19 other specimens. Each pool underwent emulsion PCR and four pools were sequenced on a single picotitre plate.
Materials:
FFPET specimens from CRC tumours were purchased from US commercial vendors: Discovery Life Sciences, Inc. (Los Osos, CA, USA); BioServe (Beltsville, MD, USA); ProteoGenex (Culver City, CA, USA); CureLine, Inc. (San Francisco, CA, USA); and Indivumed, Inc. (Kensington, MD, USA).
Study design
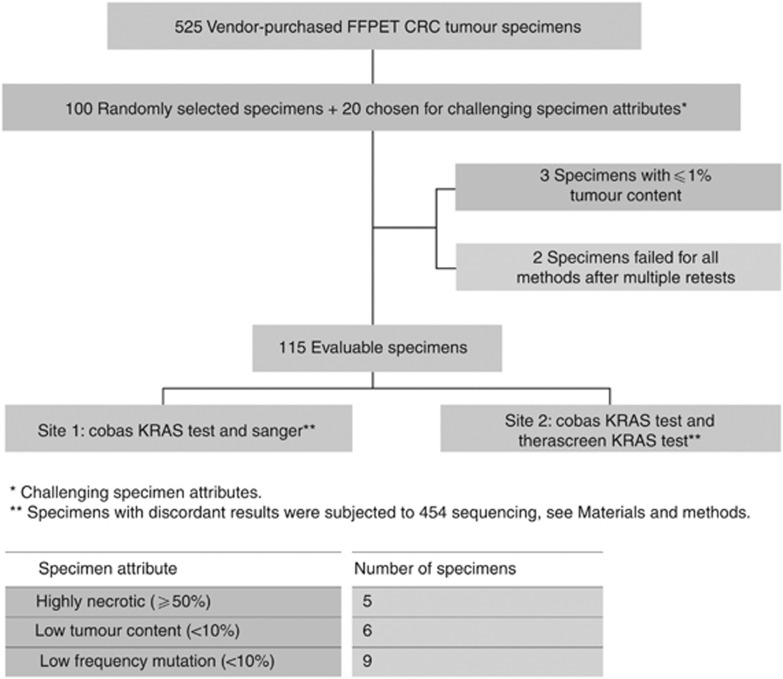
The study was conducted using a blinded panel of FFPET tumour specimens of CRC as well as artificial DNA blends containing low percent mutant alleles as determined by 454 sequencing. The design of the study is depicted in Figure 1. From a panel of 621 vendor-purchased FFPET colorectal tumour specimens, 100 specimens were selected at random; an additional 20 specimens were chosen for challenging specimen attributes such as (a) low tumour content; (b) high levels of necrosis; and (c) presence of less frequent KRAS mutations.
Figure 1.
Study design and specimen selection.
The 120 tumour specimens were each sectioned into five 5 μm curls per panel member and blinded. One section was mounted on a slide and stained with haematoxylin and eosin, coded and reviewed by two pathologists (FL-R and AS-G) to assess the tumour content, lymphocyte infiltration, extent of necrosis and mucin content. Following this assessment, a panel of evaluable specimens was identified. Two 5 μm curls per panel member were sent to clinical site 1 (Hospital Universitario Madrid Sanchinarro, Madrid, Spain) for analysis on the cobas KRAS test and Sanger sequencing, and two 5 μm curls per panel member were sent to clinical site 2 (The Royal Marsden NHS Hospital, London, UK) for analysis on cobas KRAS test and therascreen KRAS PCR Kit test.
DNA for the cobas test was isolated from a single 5 μm section per tumour panel at each site using the cobas DNA Sample Preparation Kit (Roche Molecular Systems). The DNA eluate was subsequently tested using the cobas KRAS test according to the package insert.
DNA for Sanger and the therascreen KRAS PCR Kit was isolated from separate single 5 μm sections per tumour panel member using the QIAamp DNA DNA FFPE tissue kit and automated QIAcube robot (Qiagen, Hilden, Germany). The DNA eluate was then tested with Sanger according to a standard laboratory protocol or the therascreen KRAS PCR Kit test according to the vendor-provided protocol.
Specimens with invalid test results or with discordant results between any of the methods were retested according to manufacturer/procedure instructions. Criteria for retesting were as follows:
cobas KRAS test: <10% tumour content; insufficient DNA concentration;
Sanger: no PCR amplification; or difficult sequence interpretation;
therascreen: positive controls have not amplified specific product, mixed standard delta Ct is not within specified range, no template control has Ct <38, any sample with control gene Ct >34.
Quantitative MPP (454 GS Titanium, 454 Life Sciences, Inc.) was performed on all discordant and invalid specimens.
Of the 120 FFPET specimens, 3 were excluded from analysis because pathologic review revealed ⩽1% tumour content. Two additional specimens failed to generate a valid result using any of the three testing methods at both sites or 454 and were also excluded from the data analysis. This resulted in a panel of 115 FFPET specimens for the methods comparison analysis. The clinical and pathological features of these 115 specimens are described in Table 1.
Table 1. Clinical and pathological characteristics of evaluable specimens.
| Characteristic | n |
|---|---|
| Evaluable patients | 115 |
| Median age (range) | 67 (36–90) |
| Gender, n (%) | |
| Male | 61 (53%) |
| Female | 52 (45.3%) |
| Unknown | 2 (1.7%) |
| Tumour contenta, n (%) | |
| Low | 32 (27.8%) |
| High | 83 (72.2%) |
| Lymphocyte infiltrationa, n (%) | |
| Low | 57 (49.6%) |
| High | 58 (50.4%) |
| Necrosisb, n (%) | |
| Low | 99 (86.1%) |
| High | 16 (13.9%) |
| Mucin contentb, n (%) | |
| Low | 104 (90.4%) |
| High | 11 (9.6%) |
Abbreviation: n=number.
High ⩾10% low <10%.
High ⩾50% low <50%.
Study objectives
Specific objectives of the study were to (1) compare three methods of KRAS mutation detection, by calculating positive percent agreement (PPA) and negative percent agreement (NPA) of the cobas KRAS test with the therascreen KRAS and Sanger sequencing using a panel of human FFPET CRC specimens; (2) assess the reproducibility of the cobas KRAS test at two independent laboratories; (3) assess the frequency of invalid test results for the cobas KRAS test, Sanger sequencing, and therascreen KRAS test methods; (4) assess the effects of tumour content, lymphocyte infiltration, necrosis, and mucin content on the analytical performance of the cobas KRAS test; (5) evaluate the correct call rate for each method at low percentage mutant alleles using plasmid and cell line blends of mutant and wild-type DNA from plasmids; and (6) compare turnaround times between all methods.
Invalid test rate and workflow measures
The number of invalid test results from the evaluable tumour panel was recorded and compared across the three testing methods. Assay turnaround time from DNA isolation to results reporting was compared for all methods, assuming one 8-h shift per day.
Methods correlation
The PPA and NPA of the cobas KRAS test was compared with the two other testing methods (Sanger and therascreen) in sections of evaluable FFPET colorectal tumours. Discrepant analysis by 454 was performed on all specimens for which the cobas KRAS test and the comparison method gave discordant results and/or for which one of the two testing methods gave an invalid result.
Reproducibility
The reproducibility of the cobas KRAS test was evaluated by comparing the results from testing the evaluable FFPET tumour panel at the two independent clinical laboratory sites. Discrepant analysis using 454 was performed on all specimens for which the cobas KRAS test gave discordant results and/or when an invalid result was obtained.
Impact of specimen attributes on analytic performance
The following pathological characteristics – tumour content, lymphocyte infiltration, necrosis, and mucin content – were assessed and graded according to the following criteria:
Tumour content was graded as high (⩾10%) or low (<10%);
Lymphocyte infiltration was graded as high (⩾10%) or low (<10%);
Tumour necrosis was graded as high (⩾50%) or low (<50%);
Mucin content was graded as high (⩾50%) or low (<50%).
Correct call rate at low percentage mutant alleles
Two DNA blends with 5% mutant alleles (as determined by 454) were prepared. One blend was prepared using a composite KRAS mutant plasmid containing both a codon 12 (G12D) and codon 61 (Q61H) mutation and WT genomic DNA. The other blend was prepared from genomic DNA derived from the CCL221 cell line containing a codon 13 (G13D) mutation and WT genomic DNA. In all, 48 replicates (21 replicates of each of the two 5% mutant allele blends and 6 wild-type specimens) were tested by all three methods to assess correct call rate at low percentage mutant alleles.
Results
Invalid test rate
Of the 230 specimens that were evaluated by the cobas KRAS test (115 at each site), 2.2% (5 out of 230) gave an initially invalid test result, and 4 (1.7%) were invalid upon retesting. Of the 115 specimens evaluated by Sanger sequencing, 8 specimens (7.0%) were initially invalid; upon retest, 0.9% (1 out of 115) remained invalid. Of the 115 specimens evaluated by therascreen test, 1.7% (2 out of 115) gave initially invalid test results and remained invalid upon retesting.
Methods correlation with Sanger sequencing
Of the 115 specimens tested at Site 1 using the cobas KRAS test and Sanger sequencing, two specimens were invalid by either cobas or Sanger, leaving 113 evaluable specimens for comparison. The initial agreement analysis showed a PPA of 98.2%, an NPA of 89.7%, and an overall percent agreement (OPA) of 93.8% (Table 2A).
Table 2. Methods correlation between (A) cobas KRAS test and Sanger sequencing at Site 1 and (B) after 454 sequencing of discrepant specimens.
|
(A)
| |||
|---|---|---|---|
|
2 × Bi-Directional Sanger Sequencing |
|||
| N =113 | MD | MND | Totals |
| cobas KRAS | |||
| MD | 54 | 6 | 60 |
| MND | 1 | 52 | 53 |
| Totals | 55 | 58 | 113 |
| Positive agreement: 98.2% (95% CI: 90.4–99.7) | |||
| Negative agreement: 89.7% (95% CI: 79.2–95.2) | |||
| Overall agreement: 93.8% (95% CI: 87.8–97.0) | |||
|
(B)
| |||
|---|---|---|---|
|
Sanger Sequencing post 454 testing
|
|||
| N =113 | MD | MND | Totals |
| cobas KRAS | |||
| MD | 59 | 1 | 60 |
| MND | 0 | 53 | 53 |
| Totals | 59 | 54 | 113 |
| Positive agreement: 100% (95% CI: 93.9–100) | |||
| Negative agreement: 98.1% (95% CI: 90.2–99.7) | |||
| Overall agreement: 99.1% (95% CI: 95.2–99.8) | |||
Abbreviations: CI=confidence interval; MD=mutation detected in codons 12/13 or codon 61; MND=wild-type or non-codons 12/13 or 61 mutant.
The seven specimens with discordant test results were subsequently subjected to 454 sequencing. One specimen reported as ‘mutation not detected’ by the cobas KRAS test and ‘mutation detected’ (12C) by Sanger was reported as ‘mutation not detected’ by 454; this same specimen gave ‘mutation not detected’ results by the cobas and therascreen tests at the other clinical site. Six specimens were reported as ‘mutation detected’ by the cobas KRAS test and ‘mutation not detected’ by Sanger. Sequencing by 454 reported five of these specimens as ‘mutation detected’ (three codon 12, two codon 13, four of these specimens had <20% mutant alleles by 454) and one as ‘mutation not detected’. Of these seven specimens, one was manually selected for less frequent codon 12 mutation. All seven specimens had ⩾10% tumour content and were not macrodissected before testing. Following discrepant resolution with 454 sequencing, where the 454 result was used as the arbiter, the PPA was 100%, the NPA was 98.1%, and the OPA was 99.1% (Table 2B).
Methods correlation with the therascreen KRAS PCR Kit
Of the 115 specimens tested at Site 2, using the cobas KRAS test and the therascreen KRAS PCR Kit, three specimens were invalid by either cobas test or therascreen, six specimens were mutations that the therascreen kit was not designed to detect, including codon 61 mutations, and were therefore removed from the analysis (Table 3A), leaving 106 evaluable specimens for comparison. Of these six specimens, two were G13R, one was G13V, and three were codon 61 mutations. The initial agreement analysis showed a PPA of 100%, an NPA of 96.3%, and an OPA of 98.1% (Table 3B).
Table 3. Methods correlation based on reportable results (A) between (B) cobas KRAS test and therascreen KRAS test at Site 2 and (C) after 454 sequencing of discrepant specimens.
|
(A)
| ||
|---|---|---|
| Mutation | Base change | Cosmic ID |
| Gly12Ala | GGT>GCT | 522 |
| Gly12Asp | GGT>GAT | 521 |
| Gly12Arg | GGT>CGT | 518 |
| Gly12Cys | GGT>TGT | 516 |
| Gly12Ser | GGT>AGT | 517 |
| Gly12Val | GGT>GTT | 520 |
| Gly13Asp | GGC>GAC | 532 |
|
(B)
| |||
|---|---|---|---|
|
therascreen KRAS
|
|||
| N =106 | MD | MND | Totals |
| cobas KRAS | |||
| MD | 52 | 2 | 54 |
| MND | 0 | 52 | 52 |
| Totals | 52 | 54 | 106 |
| Positive agreement: 100% (95% CI: 93.1–100) | |||
| Negative agreement: 96.3% (95% CI: 87.5–99.0) | |||
| Overall agreement: 98.1% (95% CI: 93.4–99.5) | |||
|
(C)
| |||
|---|---|---|---|
|
therascreen KRAS post 454 testing
|
|||
| N =106 | MD | MND | Totals |
| cobas KRAS | |||
| MD | 53 | 1 | 54 |
| MND | 0 | 52 | 52 |
| Totals | 53 | 53 | 106 |
| Positive agreement: 100% (95% CI: 93.2–100) | |||
| Negative agreement: 98.1% (95% CI: 90.1–99.7) | |||
| Overall agreement: 99.1% (95% CI: 94.8–99.8) | |||
Abbreviations: CI=confidence interval; MD=mutation detected in codons 12/13; MND: wild-type or non-codons 12/13.
The two specimens with discordant test results – reporting ‘mutation detected’ with the cobas KRAS test and ‘mutation not detected’ with the therascreen KRAS PCR Kit– were subsequently subjected to 454 sequencing. One of these was subsequently reported as ‘mutation detected’ (G13D mutation) and one was reported ‘mutation not detected’. Following discrepant resolution with 454 sequencing, where the 454 result was used as the arbiter, the PPA was 100%, the NPA was 98.1%, and the OPA was 99.1% (Table 3C).
Reproducibility
In total, 112 specimens were evaluable at each site (224 tests in total) using the cobas KRAS test. Of these, 110 (98.2%) produced concordant results.
Impact of specimen attributes on analytic performance
Pathological assessment of the 115 FFPET specimens revealed specimens with varying degrees of tumour content, lymphocyte infiltration, necrosis, and mucin content (Table 1). None of these pathologic characteristics had a significant effect on agreement analysis, reproducibility, and invalid rate of the cobas KRAS test (data not shown). The mutation results for the 20 specimens selected for challenging specimen attributes are depicted in Supplementary Table 1.
Workflow
Total turnaround time from DNA isolation to result reporting was determined for each method based on one 8-h work shift per day. Turnaround time per run for the cobas KRAS test was ∼1 day for 24 samples (1 kit); for Sanger it was ∼5 days for 24 samples; and for the therascreen KRAS PCR Kit it was ∼1 day for 10 samples (1/2 kit).
Correct call rate
Correct call rate at low (5%) mutant alleles was assessed using 48 replicates (21 replicates of each of the two 5% mutant allele blends and 6 wild-type) by each method. Correct call rates were as follows: cobas KRAS test (100%); Sanger (18.8%); and therascreen KRAS PCR Kit (100%). Cobas and therascreen rates of correct calls were equal, while the difference between the rates (proportions) of correct calls between cobas or therascreen and Sanger is 48/48–9/48=39/48. Such difference is statistically significant with exact two-sided P-value (calculated using StatXact) of 0.000001 (<0.05).
Discussion
The recognition that activating mutations in the KRAS gene render patients non-responsive to therapy with anti-EGFR monoclonal antibodies has resulted in the need for tumour mutation status to be ascertained before treatment. Treating patients who are unlikely to respond to a therapy is both costly and exposes them unnecessarily to potential adverse effects. In the case of the anti-EGFR monoclonal antibodies, panitumumab and cetuximab, treating patients with activating KRAS mutations may result in detrimental effects on progression-free survival (Bokemeyer et al, 2009; Douillard et al, 2010), further emphasising the need to maximise the detection of KRAS activating mutations. The detection of KRAS activating mutations in CRC has largely only focused on mutations in codons 12 and 13; however, mutations in codon 61 are known to result in activation and emerging data suggest that these patients are also unlikely to benefit from anti-EGFR therapy (Der et al, 1986; Loupakis et al, 2009; De Roock et al, 2010a; Molinari et al, 2011), although additional clinical data are needed to confirm these observations.
At present, there are numerous ways of testing for KRAS mutations, and there have been comparative studies and analyses of the sensitivity of these assays in the clinical setting (Whitehall et al, 2009; Angulo et al, 2010; Franklin et al, 2010; Tol et al, 2010). There remains an unmet need to set standards in the field of mutation testing. In this current study, the analytical performance of the cobas KRAS Mutation Test was compared with Sanger sequencing and the ARMS-Scorpion therascreen test using a blinded panel of FFPET tumour specimens of CRC as well as artificial DNA blends. The study was conducted at two independent clinical laboratories, one site comparing the cobas KRAS test with Sanger sequencing and the second site comparing the cobas KRAS test with the therascreen KRAS test.
The cobas KRAS test was highly reproducible (>98%) between sites. Reproducibility of mutation test results between different clinical laboratories sites is a critical clinical diagnostic issue. The current use of a variety of testing methodologies to detect specific mutations, both commercially available and in-house designed, increases the potential variability of results in different laboratories with different levels of expertise, especially when labour-intensive methods that require subjective analysis and interpretation are used. A recent analysis of KRAS testing at 59 laboratories from 8 European countries found that only 70% of laboratories correctly identified the mutational status in all samples (Bellon et al, 2011); similar findings were observed in a study of KRAS and EGFR mutation testing in non-small cell lung cancer (Beau-Faller et al, 2011).
Both the cobas KRAS test and therascreen tests had short turnaround times (1 day) and were more sensitive for detecting KRAS mutations at 5% mutant alleles compared with Sanger sequencing.
The cobas KRAS test requires 100 ng of total DNA input and detects 19 mutations in codon 12/13 and codon 61. The therascreen test required >160 ng of amplifiable DNA (equivalent to 800 ng of total DNA input in our experience) and detects seven mutations in codon 12/13. With the Sanger sequencing method, two PCRs were performed (one reaction for exon 2 and one reaction for exon 3) using, when possible, ∼150 ng DNA per PCR to theoretically detect any mutation in these two exons.
Analysis of discordant results suggests that the cobas KRAS test is more sensitive than both Sanger and therascreen for detecting KRAS mutations. In some instances, the increased rate of detection by the cobas KRAS test reflected the ability of the cobas KRAS test to detect more mutations (19) in codons 12, 13, and 61, than the therascreen assay, which is designed to detect seven mutations. In addition, macrodissection of the tissue sample before DNA extraction can increase the sensitivity of Sanger sequencing, although this also introduces a higher chance of human error.
Current guidelines for KRAS mutation testing in CRC do not delineate specific mutations within codons 12 and 13. However, one group has suggested, based on retrospective studies of small cohorts, that patients with tumours harbouring G13D mutations may benefit from therapy with anti-EGFR antibody therapy, suggesting that all codon 12 and 13 mutations may not be equal in terms of their clinical impact (De Roock et al, 2010b). These findings have been challenged by a recent retrospective analysis of patients treated with panitumumab in three large randomised controlled trials (Peeters et al, 2011). In this study, there was no evidence that patients with G13D mutations benefitted from anti-EGFR antibody treatment. Thus, the treatment guidelines for selecting patients for anti-EGFR antibodies are unchanged and recommend that patients with any mutations of codons 12 and 13 should not be treated with these therapies.
In summary, we have presented a methods comparison study of three assays for the detection of KRAS mutation in FFPET specimens of CRC. The three methods had comparable invalid rates. The cobas and therascreen assays had greater sensitivity for low levels of mutation than Sanger sequencing. The cobas test detected a number of KRAS mutations that the therascreen test was not designed to detect. The cobas KRAS test was highly reproducible, had the most rapid turnaround time, and was the only test that offered automated results reporting.
Acknowledgments
We wish to acknowledge Jeffrey Vaks for the statistical analysis; and Julie Tsai, Minet Negash, and Grantland Hillman for their technical assistance. This study was sponsored by Roche Molecular Systems, Inc.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
FS, MV, VHB, and HJL are employees of Roche Molecular Systems, Inc. The remaining authors declare no conflict of interest.
Supplementary Material
References
- Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL (2009) American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 27(12): 2091–2096 [DOI] [PubMed] [Google Scholar]
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD (2008) Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 26(10): 1626–1634 [DOI] [PubMed] [Google Scholar]
- Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, Beranek M, Jandik P, Benamouzig R, Jullian E, Laurent-Puig P, Olschwang S, Muller O, Hoffmann I, Rabes HM, Zietz C, Troungos C, Valavanis C, Yuen ST, Ho JW, Croke CT, O'Donoghue DP, Giaretti W, Rapallo A, Russo A, Bazan V, Tanaka M, Omura K, Azuma T, Ohkusa T, Fujimori T, Ono Y, Pauly M, Faber C, Glaesener R, de Goeij AF, Arends JW, Andersen SN, Lovig T, Breivik J, Gaudernack G, Clausen OP, De Angelis PD, Meling GI, Rognum TO, Smith R, Goh HS, Font A, Rosell R, Sun XF, Zhang H, Benhattar J, Losi L, Lee JQ, Wang ST, Clarke PA, Bell S, Quirke P, Bubb VJ, Piris J, Cruickshank NR, Morton D, Fox JC, Al-Mulla F, Lees N, Hall CN, Snary D, Wilkinson K, Dillon D, Costa J, Pricolo VE, Finkelstein SD, Thebo JS, Senagore AJ, Halter SA, Wadler S, Malik S, Krtolica K, Urosevic N (2001) Kirsten ras mutations in patients with colorectal cancer: the 'RASCAL II' study. Br J Cancer 85(5): 692–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angulo B, Garcia-Garcia E, Martinez R, Suarez-Gauthier A, Conde E, Hidalgo M, Lopez-Rios F (2010) A commercial real-time PCR kit provides greater sensitivity than direct sequencing to detect KRAS mutations: a morphology-based approach in colorectal carcinoma. J Mol Diagn 12(3): 292–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beau-Faller M, Degeorges A, Rolland E, Mounawar M, Antoine M, Poulot V, Mauguen A, Barbu V, Coulet F, Pretet JL, Bieche I, Blons H, Boyer JC, Buisine MP, de Fraipont F, Lizard S, Olschwang S, Saulnier P, Prunier-Mirebeau D, Richard N, Danel C, Brambilla E, Chouaid C, Zalcman G, Hainaut P, Michiels S, Cadranel J (2011) Cross-validation study for epidermal growth factor receptor and KRAS mutation detection in 74 blinded non-small cell figure lung carcinoma samples: a total of 5550 exons sequenced by 15 molecular French laboratories (evaluation of the EGFR mutation status for the administration of EGFR-TKIs in non-small cell lung carcinoma [ERMETIC] project--part 1). J Thorac Oncol 6(6): 1006–1015 [DOI] [PubMed] [Google Scholar]
- Bellon E, Ligtenberg MJ, Tejpar S, Cox K, de Hertogh G, de Stricker K, Edsjo A, Gorgoulis V, Hofler G, Jung A, Kotsinas A, Laurent-Puig P, Lopez-Rios F, Hansen TP, Rouleau E, Vandenberghe P, van Krieken JJ, Dequeker E (2011) External quality assessment for KRAS testing is needed: setup of a European program and report of the first joined regional quality assessment rounds. Oncologist 16(4): 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokemeyer C, Bondarenko I, Hartmann JT, de Braud F, Schuch G, Zubel A, Celik I, Schlichting M, Koralewski P (2011) Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol 22(7): 1535–1546 [DOI] [PubMed] [Google Scholar]
- Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, Donea S, Ludwig H, Schuch G, Stroh C, Loos AH, Zubel A, Koralewski P (2009) Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol 27(5): 663–671 [DOI] [PubMed] [Google Scholar]
- Conde E, Angulo B, Tang M, Morente M, Torres-Lanzas J, Lopez-Encuentra A, Lopez-Rios F, Sanchez-Cespedes M (2006) Molecular context of the EGFR mutations: evidence for the activation of mTOR/S6K signaling. Clin Cancer Res 12(3 Pt 1): 710–717 [DOI] [PubMed] [Google Scholar]
- De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Van Cutsem E, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S (2010a) Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 11(8): 753–762 [DOI] [PubMed] [Google Scholar]
- De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M, Piessevaux H, Van Cutsem E, O'Callaghan CJ, Khambata-Ford S, Zalcberg JR, Simes J, Karapetis CS, Bardelli A, Tejpar S (2010b) Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 304(16): 1812–1820 [DOI] [PubMed] [Google Scholar]
- Der CJ, Finkel T, Cooper GM (1986) Biological and biochemical properties of human rasH genes mutated at codon 61. Cell 44(1): 167–176 [DOI] [PubMed] [Google Scholar]
- Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocakova I, Ruff P, Blasinska-Morawiec M, Smakal M, Canon JL, Rother M, Oliner KS, Wolf M, Gansert J (2010) Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol 28(31): 4697–4705 [DOI] [PubMed] [Google Scholar]
- Engstrom PF, Arnoletti JP, Benson AB, Chen YJ, Choti MA, Cooper HS, Covey A, Dilawari RA, Early DS, Enzinger PC, Fakih MG, Fleshman J, Fuchs C, Grem JL, Kiel K, Knol JA, Leong LA, Lin E, Mulcahy MF, Rao S, Ryan DP, Saltz L, Shibata D, Skibber JM, Sofocleous C, Thomas J, Venook AP, Willett C (2009) NCCN Clinical Practice Guidelines in Oncology: colon cancer. J Natl Compr Canc Netw 7(8): 778–831 [DOI] [PubMed] [Google Scholar]
- Fernandez P, Carretero J, Medina PP, Jimenez AI, Rodriguez-Perales S, Paz MF, Cigudosa JC, Esteller M, Lombardia L, Morente M, Sanchez-Verde L, Sotelo T, Sanchez-Cespedes M (2004) Distinctive gene expression of human lung adenocarcinomas carrying LKB1 mutations. Oncogene 23(29): 5084–5091 [DOI] [PubMed] [Google Scholar]
- Franklin WA, Haney J, Sugita M, Bemis L, Jimeno A, Messersmith WA (2010) KRAS mutation: comparison of testing methods and tissue sampling techniques in colon cancer. J Mol Diagn 12(1): 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR (2008) K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359(17): 1757–1765 [DOI] [PubMed] [Google Scholar]
- Lee S, Brophy VH, Cao J, Velez M, Hoeppner C, Soviero S, Lawrence HJ (2011) Analytical performance of a PCR assay for the detection of KRAS mutations (codons 12/13 and 61) in formalin-fixed paraffin-embedded tissue samples of colorectal carcinoma. Virchows Arch 460(2): 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, Masi G, Stasi I, Canestrari E, Rulli E, Floriani I, Bencardino K, Galluccio N, Catalano V, Tonini G, Magnani M, Fontanini G, Basolo F, Falcone A, Graziano F (2009) KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer 101(4): 715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437(7057): 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari F, Felicioni L, Buscarino M, De Dosso S, Buttitta F, Malatesta S, Movilia A, Luoni M, Boldorini R, Alabiso O, Girlando S, Soini B, Spitale A, Di Nicolantonio F, Saletti P, Crippa S, Mazzucchelli L, Marchetti A, Bardelli A, Frattini M (2011) Increased detection sensitivity for KRAS mutations enhances the prediction of anti-EGFR monoclonal antibody resistance in metastatic colorectal cancer. Clin Cancer Res 17(14): 4901–4914 [DOI] [PubMed] [Google Scholar]
- Peeters M, Douillard JY, Van Cutsem E, Siena S, Zhang K, WIlliams R, Wiezorek J (2011) Evaluation of individual codon 12 and 13 mutant KRAS alleles as prognostic and predictive biomarkers of response to panitumumab in patients with metastatic colorectal cancer. European Multidisciplinary Cancer Congress. abstract 33LBA [DOI] [PubMed]
- Peeters M, Price TJ, Cervantes A, Sobrero AF, Ducreux M, Hotko Y, Andre T, Chan E, Lordick F, Punt CJ, Strickland AH, Wilson G, Ciuleanu TE, Roman L, Van Cutsem E, Tzekova V, Collins S, Oliner KS, Rong A, Gansert J (2010) Randomized phase III study of panitumumab with fluorouracil, leucovorin, and irinotecan (FOLFIRI) compared with FOLFIRI alone as second-line treatment in patients with metastatic colorectal cancer. J Clin Oncol 28(31): 4706–4713 [DOI] [PubMed] [Google Scholar]
- Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH, Quirke P (2009) KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol 27(35): 5931–5937 [DOI] [PubMed] [Google Scholar]
- Samowitz WS, Curtin K, Schaffer D, Robertson M, Leppert M, Slattery ML (2000) Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: a population-based study. Cancer Epidemiol Biomarkers Prev 9(11): 1193–1197 [PubMed] [Google Scholar]
- Sundstrom M, Edlund K, Lindell M, Glimelius B, Birgisson H, Micke P, Botling J (2010) KRAS analysis in colorectal carcinoma: analytical aspects of Pyrosequencing and allele-specific PCR in clinical practice. BMC Cancer 10: 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tol J, Dijkstra JR, Vink-Borger ME, Nagtegaal ID, Punt CJ, Van Krieken JH, Ligtenberg MJ (2010) High sensitivity of both sequencing and real-time PCR analysis of KRAS mutations in colorectal cancer tissue. J Cell Mol Med 14(8): 2122–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cutsem E, Kohne CH, Lang I, Folprecht G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D, Tejpar S, Schlichting M, Zubel A, Celik I, Rougier P, Ciardiello F (2011) Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol 29(15): 2011–2019 [DOI] [PubMed] [Google Scholar]
- Van Cutsem E, Nordlinger B, Cervantes A (2010) Advanced colorectal cancer: ESMO Clinical Practice Guidelines for treatment. Ann Oncol 21(Suppl 5): v93–v97 [DOI] [PubMed] [Google Scholar]
- van Krieken H, Tol J (2009) Setting future standards for KRAS testing in colorectal cancer. Pharmacogenomics 10(1): 1–3 [DOI] [PubMed] [Google Scholar]
- Vaughn CP, Zobell SD, Furtado LV, Baker CL, Samowitz WS (2011) Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosomes Cancer 50(5): 307–312 [DOI] [PubMed] [Google Scholar]
- Whitehall V, Tran K, Umapathy A, Grieu F, Hewitt C, Evans TJ, Ismail T, Li WQ, Collins P, Ravetto P, Leggett B, Salto-Tellez M, Soong R, Fox S, Scott RJ, Dobrovic A, Iacopetta B (2009) A multicenter blinded study to evaluate KRAS mutation testing methodologies in the clinical setting. J Mol Diagn 11(6): 543–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.