The simple gas nitric oxide (NO) controls a variety of complex biological processes, including blood pressure homeostasis, platelet aggregation, and transmission of signals by the nervous system. NO is also important for immune system function, playing key roles in the activation of macrophages and cellular defenses against microbial pathogens. Recent studies have revealed that some responses to NO are similar to those among very distantly related organisms; these findings suggest that many of the biological functions of NO have an evolutionarily ancient origin.
This session addressed NO function in animals, microorganisms, and plants. Some effects of NO in animals are mediated by interactions with hemoglobin (Hb) that facilitate the delivery of oxygen to tissues with low oxygen tension. Interactions between NO and Hb have also been observed in bacteria, where they serve an alternative function in protecting bacteria from nitrosative stresses in their environment. Similarities between the mechanisms that control responses to pathogen attack in plants and innate immunity in animals led to a search for a role of NO in plant defense. This search has now borne fruit.
Hb in NO Delivery and Nitrosative Stress.
Hb and NO have been inextricably linked from the earliest studies of globin function (1) to the recent identification of NO with biological activity (2). The standard model of Hb interaction with NO is based on two reactions, addition (or nitrosylation) and oxidation, expressed in Eqs. 1 and 2, respectively, below.
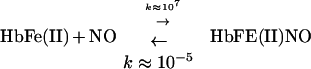 |
1 |
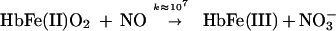 |
2 |
Both of these reactions were thought to be effectively irreversible. It was recently shown, however, that nitrosylHb is not stable at physiological ratios of NO to Hb—i.e., when NO:Hb ≪ 1. Rather, it is redox active—liberating NO− in the deoxy or T structure to form N2O (Eq. 3), and transferring NO+ to cysteine β93 within the R structure to form S-nitrosoHb (Eq. 4) (see ref. 3).
 |
3 |
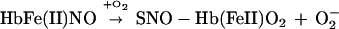 |
4 |
Furthermore, the NO oxidation reaction, purportedly the dominant reaction in NO biology and the major route of NO elimination from the body, is actually of little physiological significance. Rather, NO binds to oxyHb in a cooperative manner to form S-nitrosoHb and nitrosylHb (4). Thus, under physiologically relevant conditions, NO binding to hemes and thiols in oxygen-ligated Hb effectively competes with the oxidation reaction (Eq. 5).
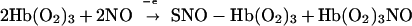 |
5 |
These new observations have led to a revision of our understanding of the respiratory cycle to include a third gas, NO (3). Specifically, deoxygenated erythrocytes, transiting the capillaries, carry NO ligated to hemes (and CO2). On entering the lung, Hb undergoes an oxygen-driven allosteric transition (from T to R) that is coupled with NO group transfer from the hemes to cysteine β93. The molecule S-nitroso-oxyHb (where NO is bound to thiol and O2 to heme) thus enters the arterial circuit. There, it is exposed to low O2 tension in resistance vessels that induces a transition back to the T state, releasing the NO group, which dilates blood vessels and thereby facilitates O2 delivery (5).
Hbs are not restricted to cells of erythroid origin; they are expressed in other mammalian cells (6), plants, and microorganisms. In bacteria, Hbs are two-domain proteins that share significant sequence homology to Vitreoscilla globin in their N termini and to flavoprotein cytochrome P450 reductases in their C termini. These flavohemoglobins (HMPs) function to protect against nitrosative stress (NO-related toxicity), as demonstrated by the finding that Escherichia coli and Salmonella that harbor deletions in the HMP gene also display increased sensitivity to NO and S-nitrosothiols (7, 8), antimicrobial compounds that are produced by the infected host. Moreover, the adaptive response to sublethal doses of NO and S-nitrosothiols involves induction of HMP.
HMP catalytically transforms NO into NO3− or N2O (8). The nitrate-forming reaction consumes an equivalent of oxygen and half an equivalent of NADH. During steady-state turnover, the heme is in the Fe(II)O2 state. In the absence of oxygen, HMP(FeII) reduces NO to nitrous oxide (N2O). Both reactions involve an Fe(III) intermediate, which is reduced by NAD(P)H (8). That is, HMP uses P450-reductase activity to support the classical NO oxidation (Eq. 2) and reduction (Eq. 3) reactions. Thus, in contrast with mammalian Hb, which functions to secure and deliver NO/S-nitrosothiols, HMP is designed to consume NO.
These results raise important questions with respect to Hb evolution. In particular, what are the factors that determine NO consumption or delivery by Hb, and when in evolution did this change in function take place? Future studies of Hbs that are placed in phylogeny at the evolutionary divide between plants and animals (9) may provide answers to these fundamental questions. Such Hbs have lost the P450-reductase domain characteristic of ancient microbes and some have evolved cysteines in close proximity to the ligand-binding site analogous to mammalian Hb.
In summary, the ancient Hb is an enzyme that uses redox chemistry to consume NO and resist a nitrosative threat: it uses the NO-oxidation reaction aerobically and reduces NO anaerobically, generating mainly nitrate and N2O, respectively. The evolution of Hb has led to the loss of the reductase domain and the incorporation of thiols for NO-related functions, as exemplified in Ascaris and human Hb.
The Role of NO in Plant Defense Responses.
Plants react to pathogen attack by activating an elaborate defense mechanism that acts both locally and systemically. In many cases, local resistance is manifested as a hypersensitive response, characterized by the development of lesions (programmed cell death) which restrict pathogen growth and/or spread. The hypersensitive response is associated with the induction of defense-related genes that play important roles in containing pathogen growth, either indirectly, by helping to reinforce plant cell walls, or directly, by providing antimicrobial enzymes and toxic secondary metabolites, such as phytoalexins, which kill pathogens.
Reactive oxygen species (ROS) appear to play key roles in early and later stages of the plant response against pathogens. During the hypersensitive response, a massive production of ROS can be observed. Although ROS seem to act as both cellular signals and direct weapons, their precise modes of action are still not known. The participation of ROS in the induction of host cell death and pathogen killing seems to be necessary, but not sufficient (10). In animals, ROS (generated by NADPH oxidase), collaborate with NO and related species, generated mainly by inducible NO synthase (NOS) to regulate apoptosis and kill invading pathogens (11). The discovery of plant homologs of the NADPH oxidase (13) prompted several groups to examine whether NOS also plays a role during plant–pathogen interactions.
The presence of NO in plants well documented. Previously, NO was shown to induce leaf expansion, root growth, and the production of phytoalexins. Initial evidence for the presence of a mammalian-type NOS in plants was reported in 1996 (14). More recently, it was shown that NO and possibly NOS play a prominent role in plant defense against microbial pathogens. Infection of tobacco with a tobacco-specific virus resulted in enhanced NOS-like activity (15). Corresponding results were obtained with soybean cells and the model plant Arabidopsis by measuring both the activity of NOS and the subsequent release of NO in response to either a bacterial pathogen or an elicitor (a signaling molecule that indicates the presence of a pathogen). Strikingly, NOS inhibitors were able to compromise the resistance response in Arabidopsis and the induction of programmed cell death in pathogen-treated soybean suspension cells (16). These experiments clearly suggest that NO plays an essential role in the early events of plant resistance responses.
NO is not only a weapon, but also an important signaling molecule. In mammalian systems, one of its most important targets is guanylate cyclase. cGMP, produced upon NO binding to heme in the cyclase, regulates many cellular functions (17). In plants, cGMP can stimulate the induction of stress-associated genes and can initiate biosynthesis of secondary metabolites involved in defense responses (18). Strikingly, the tobacco defense genes induced by NO were also induced by cGMP and cyclic ADP-ribose. These two molecules serve as second messengers for NO signaling in mammals. The NO-induced increases in tobacco cGMP levels were of similar magnitude to those detected during NO-induced smooth muscle relaxation in animals (15). Thus, plants and animals appear to use common mechanisms to transduce NO signals.
Interestingly, NO signaling in plants is closely intertwined with another important signaling molecule, salicylic acid (SA). SA is a crucial signal during plant pathogen interactions, when it might serve as a general redox signal. NO activity is at least partially SA-dependent (15). In addition, NO acts synergistically with SA as well as with ROS to potentiate defense responses (16). Although the relations among NO, SA, and ROS in the activation of defense genes and/or induction of host cell death are currently unresolved, these data suggest a self-amplifying process, during which redox signaling through NO and ROS is enhanced by SA.
Although these findings are fascinating, many questions remain. Macrophages execute pathogens through a deadly mixture of NO and reactive oxygen radicals. Currently, it is unclear whether NO and/or its derivatives, such as peroxynitrite (ONOO−), are directly toxic to microbial plant pathogens. Furthermore, we do not know whether NOS-like activity in plants originates from a true mammalian-type NOS. A plant NOS gene has yet to be cloned.
In summary, NO appears to play important roles during plant–pathogen interactions, including the regulation of programmed host cell death, a mechanism that restricts growth and spreading of the pathogen, and activation of genes that encode defense proteins. Furthermore, components of animal NO signal-transduction pathways are also operative in plant and microbial defense responses. Deciphering its mechanisms of action and rigorously establishing its involvement in plant disease resistance and in microbial resistance to environmental stresses could provide important new insights into signal transduction and disease resistance.
Acknowledgments
This work was supported in part by National Institutes of Health Grants HL52529 and HL59130 to J.S.S.
Abbreviations
- HMG
flavohemoglobin
- NOS
NO synthase
- ROS
reactive oxygen species
- SA
salicylic acid
Footnotes
This paper is a summary of a session presented at the fifth annual German-American Frontiers of Science symposium, held June 10–13, 1999, at the Alexander von Humboldt Foundation in Potsdam, Germany.
References
- 1.Keilin D, Hartree E F. Nature (London) 1937;139:548. [Google Scholar]
- 2.Ignarro L J, Buga G M, Wood K S, Byrns R E, Chaudhuri G. Proc Natl Acad Sci USA. 1987;84:9265–9269. doi: 10.1073/pnas.84.24.9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gow A J, Stamler J S. Nature (London) 1998;391:169–173. doi: 10.1038/34402. [DOI] [PubMed] [Google Scholar]
- 4.Gow A J, Luchsinger B P, Pawloski J P, Singel D J, Stamler J S. Proc Natl Acad Sci USA. 1999;96:9027–9032. doi: 10.1073/pnas.96.16.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stamler J S, Jia L, Eu J P, McMahon T J, Demchenko I T, Bonaventura J, Gernert K, Piantadosi C A. Science. 1997;276:2034–2037. doi: 10.1126/science.276.5321.2034. [DOI] [PubMed] [Google Scholar]
- 6.Liu L, Zeng M, Stamler J S. Proc Natl Acad Sci USA. 1999;96:6643–6647. doi: 10.1073/pnas.96.12.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford M J, Goldberg D E. J Biol Chem. 1998;273:12543–12547. doi: 10.1074/jbc.273.20.12543. [DOI] [PubMed] [Google Scholar]
- 8.Hausladen A, Gow A J, Stamler J S. Proc Natl Acad Sci USA. 1998;95:14100–14105. doi: 10.1073/pnas.95.24.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardison R. J Exp Biol. 1998;201:1099–1117. doi: 10.1242/jeb.201.8.1099. [DOI] [PubMed] [Google Scholar]
- 10.Dangl J. Nature (London) 1998;394:525–526. doi: 10.1038/28958. [DOI] [PubMed] [Google Scholar]
- 11.Brüne B, von Knethen A, Sandau K B. Eur J Pharmacol. 1998;351:261–272. doi: 10.1016/s0014-2999(98)00274-x. [DOI] [PubMed] [Google Scholar]
- 12.Mayer B, Hemmens B. Trends Biochem Sci. 1997;22:477–481. doi: 10.1016/s0968-0004(97)01147-x. [DOI] [PubMed] [Google Scholar]
- 13.Scheel D. Curr Opin Plant Biol. 1998;1:305–310. doi: 10.1016/1369-5266(88)80051-7. [DOI] [PubMed] [Google Scholar]
- 14.Durner J, Klessig D F. Curr Opin Plant Biol. 1999;2:369–374. doi: 10.1016/s1369-5266(99)00007-2. [DOI] [PubMed] [Google Scholar]
- 15.Durner J, Wendehenne D, Klessig D F. Proc Natl Acad Sci USA. 1998;95:10328–10333. doi: 10.1073/pnas.95.17.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delledonne M, Xia Y, Dixon R A, Lamb C. Nature (London) 1998;394:585–588. doi: 10.1038/29087. [DOI] [PubMed] [Google Scholar]
- 17.McDonald L J, Murad F. Adv Pharmacol Chemother. 1995;34:263–276. doi: 10.1016/s1054-3589(08)61091-1. [DOI] [PubMed] [Google Scholar]
- 18.Bowler C, Neuhaus G, Yamagata H, Chua N-H. Cell. 1994;77:73–81. doi: 10.1016/0092-8674(94)90236-4. [DOI] [PubMed] [Google Scholar]


