Abstract
Retroviral integrase (IN) is responsible for two consecutive reactions, which lead to insertion of a viral DNA copy into a host cell chromosome. Initially, the enzyme removes di- or trinucleotides from viral DNA ends to expose 3′-hydroxyls attached to the invariant CA dinucleotides (3′-processing reaction). Second, it inserts the processed 3′-viral DNA ends into host chromosomal DNA (strand transfer). Herein, we report a crystal structure of prototype foamy virus IN bound to viral DNA prior to 3′-processing. Furthermore, taking advantage of its dependence on divalent metal ion cofactors, we were able to freeze trap the viral enzyme in its ground states containing all the components necessary for 3′-processing or strand transfer. Our results shed light on the mechanics of retroviral DNA integration and explain why HIV IN strand transfer inhibitors are ineffective against the 3′-processing step of integration. The ground state structures moreover highlight a striking substrate mimicry utilized by the inhibitors in their binding to the IN active site and suggest ways to improve upon this clinically relevant class of small molecules.
Keywords: inhibitor, integrase, mechanism, retrovirus, structure
Introduction
The retroviral lifecycle depends on insertion of viral DNA into a host cell chromosome, and integrase (IN) is the enzyme that orchestrates the key catalytic events involved in this process (reviewed in Craigie, 2002; Lewinski and Bushman, 2005). IN is responsible first for 3′-processing, the reaction in which two or three nucleotides are removed from one or both 3′-viral DNA ends, leaving 3′-hydroxyl groups. IN subsequently catalyses strand transfer, wherein it uses the 3′-hydroxyls to attack a pair of phosphodiester bonds in host cell DNA.
Retroviral INs contain three canonical domains: a zinc-binding N-terminal domain, a catalytic core domain harbouring the active site and a C-terminal domain (Engelman and Craigie, 1992; Kulkosky et al, 1992; Hare et al, 2010a). The IN active site contains a triad of invariant acidic residues, referred to as the D,DX35E motif (Engelman and Craigie, 1992; Kulkosky et al, 1992; Dyda et al, 1994). The carboxylates coordinate a pair of divalent metal cations, essential for both enzymatic activities. Abundant in vivo, Mg2+ is considered to be the natural cofactor, although Mn2+ fully supports IN function in vitro (Bushman and Craigie, 1991; Engelman and Craigie, 1995; Andrake et al, 2009; Hare et al, 2010a). Both reactions catalysed by the IN active site proceed via bimolecular nucleophilic substitution (SN2), shared by metal-dependent nucleotidyl transferases and some nucleases, including transposases and RNase H enzymes (Engelman et al, 1991; Mizuuchi and Adzuma, 1991; Davies et al, 2000; Kennedy et al, 2000; Nowotny and Yang, 2006). The metal ion cofactors are thought to play dual roles during catalysis. Owing to the preferred octahedral geometry of the Mg2+ and Mn2+ coordination spheres (Harding, 2006), the ions initially help to select and position the reacting groups. Second, they help to destabilize the scissile phosphodiester and promote formation of the phosphorane intermediate (Nowotny and Yang, 2006; Yang et al, 2006).
Retroviral integration shares a common set of intermediates with many DNA transposition systems (Figure 1A) (Li et al, 2006; Li and Craigie, 2009). Initially, a tetramer of IN assembles on the viral DNA ends, forming the intasome. Following 3′-processing, the intasome binds target DNA, in a transient target capture complex (TCC). Following strand transfer, the post-catalytic strand transfer complex (STC) likely requires energy-dependent disassembly prior to 5′-end joining by the host DNA repair machinery. In the context of this work it is helpful to discriminate between two forms of the intasome: its initial state containing unprocessed (blunt) viral DNA ends and its post-3′-processing state. Herein, we refer to these complexes as uncleaved and cleaved intasomes (UI and CI), respectively. The intasome in its post-3′-processing state (CI), TCC and STC were recently structurally characterized using the IN from prototype foamy virus (PFV) (reviewed in Cherepanov et al, 2011). The CI comprises a dimer-of-dimers of IN, wherein the central pair of IN subunits form a network of protein–protein and protein–DNA interactions and engage the processed 3′-viral DNA ends within their active sites (Hare et al, 2010a). All three canonical domains of the inner IN subunits participate in protein–protein and protein–DNA interactions. The outer IN chains interact with the inner chains via the catalytic core domains. In the TCC, target DNA binds in a groove formed between the inner IN subunits in a severely bent conformation, forced to yield its target phosphodiesters to the IN active sites (Maertens et al, 2010).
Figure 1.
Retroviral integration intermediates and detection of 3′-processing and strand transfer in crystallo. (A) Schematic of IN–DNA complexes observed in in vitro systems; a tetramer of IN (cyan oval) assembles on viral DNA ends, forming the intasome; this initial intasomal complex is referred to as the UI. In the presence of Mg2+ or Mn2+ cations, IN catalyses 3′-processing, resulting in the CI. The CI binds target DNA to form the target capture complex (TCC). The STC represents the final post-catalytic state. The reactive and nonreactive DNA strands at each viral DNA end and target DNA are shown as gold, orange and purple lines, respectively; arrowheads represent 3′-ends. Strand transfer inhibitors bind to the CI active sites and prevent formation of the TCC. Letter codes indicate DNA species observed in crystals: N, 19-mer nonreactive viral DNA strand; r and R, 19-mer unprocessed and 17-mer processed reactive strand, respectively; T, self-complementary 30-mer target DNA strands; R+T and t, 34-mer and 13-mer strand transfer products, respectively. (B) Denaturing PAGE analysis of DNA species isolated from UI crystals prior to (lane 3) or following soaking in the presence of Mn2+ (lanes 4–9) or Mg2+ (lane 10). Reaction times are indicated above the gel image; based on relative intensities of the R and r bands, the reaction was ∼10, 20 and 50% complete after 20, 45 and 120 min, respectively. 5′-Labelled N:r and N:R duplexes were loaded in lanes 1 and 2, respectively. Sizes of substrate and product oligonucleotides are indicated to the right of gel. The 5′-end of the nonreactive strand was blocked with a non-radioactive phosphate group to improve detection of the reactive strand during labelling, without perturbing the active site (Supplementary Figure S2). (C) DNA species isolated from TCC crystals prior to (lane 7) or following incubation in Mn2+ (lanes 8–13) or Mg2+ (lane 14) for indicated periods of time; based on relative intensities of the T and t bands, the reaction was ∼30, 70, 90% complete after 30, 120 and 300 s, respectively. Lanes 1–6 contained t, S, T, N, R and annealed N:R DNA samples, respectively. Note that although intasome does not bind target DNA in a sequence-specific fashion, only the symmetric complex crystallizes under the conditions employed (Maertens et al, 2010).
HIV-1 IN is a validated drug target with raltegravir currently being used for treatment of AIDS, and several related inhibitors are in clinical trials (Summa et al, 2008) (reviewed in Marchand et al, 2009; McColl and Chen, 2010). These small molecules are classed as IN strand transfer inhibitors, as they specifically target the second catalytic step of the integration process (Espeseth et al, 2000). Although inhibition of 3′-processing can be observed in the presence of elevated concentrations of these molecules (Metifiot et al, 2010), it is unlikely to contribute to their antiviral activity. Strand transfer inhibitor scaffolds comprise two functional moieties, a heterocyclic core displaying a triad of metal chelating atoms (typically three oxygens) and a halo-benzyl side chain, connected via a short, torsionally flexible linker. A high degree of sequence conservation in the active sites of retroviral INs allowed the use of the PFV intasome as a surrogate for its HIV-1 counterpart (Hare et al, 2010a, 2010b, 2011). In the PFV CI-inhibitor cocrystal structures, the metal chelating triad interacts with the Mg2+ cations in the IN active site, while the halo-benzyl side chain displaces the base of the reactive deoxyadenosine at the processed 3′-viral DNA end (Hare et al, 2010a, 2010b, 2011).
Our prior work focused on post-catalytic (CI and STC) or inactivated (TCC lacking 3′-hydroxyls or catalytic metal ions) IN–DNA complexes (Hare et al, 2010a; Maertens et al, 2010). Herein, we present crystal structures of the functional UI and TCC in their ground states committed for 3′-processing and strand transfer, respectively. The structures provide unprecedented insight into the positions of the metal ions and chemical reacting groups in the IN active site, highlight a substrate mimicry utilized by strand transfer inhibitors in their mode of binding to IN and explain why these small molecules are ineffective against the 3′-processing reaction and why inhibitors of 3′-processing have been more difficult to develop.
Results
Detection of 3′-processing and strand transfer in crystallo
We were able to assemble and crystallize the PFV intasome containing a blunt-ended 19-bp DNA mimicking the unprocessed U5 end of the viral DNA (the complex referred to as UI). We also prepared crystals of the PFV TCC, by cocrystallization of the CI and a 30-bp target DNA molecule (Maertens et al, 2010). Both types of crystals were grown in the presence of EDTA to chelate traces of divalent metal ions and prevent catalysis during crystallization.
To evaluate the functionality of the IN active site in the crystallized forms, we soaked the UI and TCC crystals in solutions containing Mn2+ or Mg2+ salts. The crystals were dissolved, and the DNA material, 5′-labelled by incubation with a polynucleotide kinase and γ-32P-ATP, was fractionated through denaturing polyacrylamide gels. The two 19-mer strands of the non-processed viral DNA substrate duplex migrate as a doublet, with clear separation of the 17-mer reaction product (Figure 1B, lanes 1 and 2). Neither 3′-processing nor strand transfer products were detected prior to exposure of the UI and TCC crystals to Mg2+ or Mn2+ salts (Figure 1B, lane 3; Figure 1C, lane 7). The products of 3′-processing became detectable after soaking UI crystals in MnCl2 for 5 min, and the reaction was 50% complete after 120 min (based on relative intensities of R and r bands; Figure 1B). Under optimized in vitro conditions, strand transfer reactions using PFV IN and blunt viral DNA mimics are delayed by ∼30 min compared with using pre-processed viral DNA (Valkov et al, 2009). Thus, the 3′-processing reaction time scale in crystallo was not drastically different to that in solution. Strand transfer ensued faster than 3′-processing, with products appearing after 30 s of soaking TCC crystals in the presence of Mn2+ (Figure 1C, lane 8), and the reaction was over 50% complete after 2 min (lane 10). Importantly, both Mn2+ and Mg2+ could act as cofactors for both IN-catalysed reactions in crystallo (Figure 1B lanes 9 and 10; Figure 1C, lanes 13 and 14). These results confirmed that the intasome and TCC complexes are functional in their crystallized forms.
IN binding to blunt-ended viral DNA and the mechanism of 3′-processing
We collected X-ray diffraction data and refined structures of the UI in the absence of metal cofactors (UIApo) and after soaking the crystals in solutions containing Mn2+ for various periods of time (Supplementary Table S1; Supplementary Figure S1). As a more electron dense element than Mg, Mn allows for more precise determination of metal positions in crystal structures at the given medium–low resolution range (2.5–3 Å). A 5-min soak allowed us to freeze trap the Michaelis complex with full occupancy for both metal ions, and the structure was refined to 2.5 Å resolution (UIMn; Figure 2A). Prolonged soaks (24–48 h) revealed a structure (CIMn) that was indistinguishable from that of the intasome assembled on pre-processed viral DNA ends (Hare et al, 2010a, 2010b); the cleaved dinucleotide was not seen in the electron density, indicating that it had dissociated from the active site (Supplementary Figure S1). We also determined the structure of the UI with 5′-phosphorylated nonreactive viral DNA strands (UIMn-PO3; Supplementary Figure S2A). Addition of the phosphate group had no effect on the configuration of the IN active site (Supplementary Figure S2B). This was expected, since the 5′-end of the nonreactive viral DNA strand is threaded outside of the active site (Figure 2B) (Hare et al, 2010a).
Figure 2.
Overall architecture of the retroviral intasome prior to 3′-processing. (A) The UIMn structure viewed in two orientations with IN chains shown in a space-fill mode (top) or as cartoons (bottom); the inner subunits are coloured dark cyan and green and outer chains grey. The reactive and nonreactive viral DNA strands are depicted as yellow and orange cartoons, respectively. Locations of the active sites (asterisks) and the scissile dinucleotides are indicated. Note that the crystallized construct is symmetrized by the presence of two identical viral DNA termini derived from the U5 viral DNA end. The native retroviral nucleoprotein complexes are less symmetric due to the sequence differences of the left (U3) and right (U5) viral DNA termini. In the native PFV UI, only the U5 DNA end is expected to have a scissile dinucleotide, while the U3 end, naturally terminating on a CA sequence, does not undergo 3′-processing (Juretzek et al, 2004). (B) Fraying of the viral DNA end prior to 3′-processing. IN is shown as cartoons (green) with secondary structure elements indicated; selected side chains are shown in sticks. The reactive and nonreactive viral DNA strands are shown as yellow and orange cartoons, respectively, with six terminal nucleotides indicated; grey spheres are Mn atoms.
3′-Processing does not grossly affect the overall structure of the intasome, and the r.m.s. deviation between IN main chain atom positions in UIMn (Figure 2A) and CIMn (Hare et al, 2010a, 2010b) structures is only ∼0.35 Å. While sharing all of the major architectural features described for the cleaved complex (Hare et al, 2010a), the UI structures reveal the IN active site committed for 3′-processing and the configuration of the reaction substrate. Three bases are unpaired at the viral DNA end due to the insertion of the IN 310 helix η2 and η2/β5 loop between the reactive and nonreactive viral DNA strands (Figure 2B). The fraying of viral DNA ends prior to 3′-processing is fully consistent with biochemical observations (Scottoline et al, 1997; Katz et al, 2011). The scissile pApTOH dinucleotide passes over Pro214 and between Tyr212 and Gln186 and projects into the groove between the halves of the symmetric intasome structure (Figures 2A and 3A). The interactions between the bases of the scissile dinucleotide and IN are limited to Van der Waals contacts with IN residues 212–214 (Figure 3A), and its 3′-terminal thymidine is only partially ordered in the electron density. The bonding interactions of the scissile dinucleotide within the IN active site are made exclusively via the DNA phosphodiester backbone, which is well defined in electron density (Supplementary Figure S1). The internal phosphodiester of the dinucleotide is hydrogen bonded to the backbone amides of Tyr212 and Gln186, and makes Van der Waals interactions with the side chain of Tyr212 (Figure 3A). Two Mn2+ ions bind at the active site, inducing relocation of the scissile phosphodiester towards the active site (1.6-Å shift in the position of the P atom; Figure 3B). In the absence of metal ions the phosphodiester is less constrained, leading to higher B factors for the scissile dinucleotide and the disorder of the 3′-thymidine (Supplementary Figure S1).
Figure 3.
The IN active site engaged with the non-processed 3′-viral DNA end. (A) Stereo view (wall-eye) on the 3′-end of viral DNA in UIApo. The protein is shown as a semitransparent blue surface and the DNA as a stick representation with the scissile phosphodiester in grey. Hydrogen bonds between the internal phosphodiester of the scissile dinucleotide and protein backbone amides are shown as dashes. (B) Stereo view of the active site bound to non-processed 3′-end of viral DNA end with and without Mn2+. Carbon atoms belonging to the UIApo structure are coloured blue, and those belonging to UIMn are green, while other atoms follow standard coloration: blue for nitrogen, red for oxygen and orange for phosphorus. Spheres represent the manganese ions; black arrow indicates the relocation of the scissile phosphodiester upon metal binding. (C) Stereo view of the active site in the UIMn structure. Metal ions are shown in purple and associated water molecules in red. The large red sphere indicates the water molecule poised to act as a nucleophile in 3′-processing, with potential path indicated with a red dash. (D) The inter metal distances in the different structures, with metals shown as purple spheres, carbons of protein in green, viral DNA in yellow and target DNA in magenta.
The octahedral coordination sphere of metal A in the UIMn is nearly perfect, comprising an oxygen from each of the Asp128 and Asp185 carboxylates, the pro-S p oxygen atom of the scissile phosphodiester plus three water molecules, one of which (shown as a large sphere in Figure 3C) is poised for in-line nucleophilic attack on the scissile phosphodiester. Metal B shares the non-bridging pro-S p oxygen with metal A and also contacts the bridging 3′-oxygen atom of the scissile phosphodiester, both oxygen atoms from the Glu221 carboxylate and one from the Asp128 side chain, plus one water molecule. Due to the bidentate coordination to Glu221 and the scissile phosphodiester, the coordination sphere of metal B is far from the optimal octahedral geometry (Harding, 2006). Thus, similar to the case of RNase H–substrate complex (Nowotny and Yang, 2006), the non-ideal ligand environment of metal B may promote destabilization of the scissile bond.
The distance between metal ions in the UIMn complex is 3.9 Å (Figure 3D), which is very close to that seen in structures harbouring the RNase H–substrate state (Nowotny et al, 2005). Dissociation of the cleaved dinucleotide allows the metals to move closer to each other, separated by 3.1 Å in CIMn. Nowotny and Yang (2006) observed variable metal positions in the RNase H active site, depending on the stage in the catalytic process. Their suggestion that movement of metal ions during catalysis may allow the attacking water molecule to approach the scissile phosphodiester was supported by molecular dynamics simulations (De Vivo et al, 2008; Rosta et al, 2011). In our ground state UIMn structure, the water nucleophile is separated from the target phosphorus atom by 3.3 Å, a distance identical to that observed in RNase H–substrate complexes (Nowotny et al, 2005). Of note, the earlier work had to rely on inactivating mutations in the RNase H active site in order to stabilize the substrate-bound state. Our observation of the water in the identical position within the functional IN active site strongly argues for its functional significance.
The configuration of the UIMn active site explains observations made by Brown and colleagues over a decade ago that the HIV-1 IN 3′-processing reaction is strongly inhibited when the pro-S p oxygen of the scissile phosphodiester is replaced with sulphur, while no such effect was present at the pro-R p position (Gerton et al, 1999). In the UIMn structure, the pro-S p oxygen is shared between metal ions (Figure 3C). As a more electronegative and smaller atom, oxygen would be preferred at this position (Brautigam and Steitz, 1998), while substitutions of the pro-R p oxygen, which is pointing away from the metal cofactors (Figure 3C), are less likely to affect the catalysis.
Nucleophilic substitution requires two proton transfer events: initially from the water nucleophile (generating a reactive hydroxide anion) and then onto the 3′-alkoxide (generating the leaving 3′-hydroxyl). It was proposed that in RNase H a pro-R p oxygen atom of the phosphodiester group located 3′ of the cleavage site serves as a general base to abstract a proton from the attacking molecule; and that a protonated carboxylate coordinated to metal B donates a proton to the 3′-alkoxide completing the catalytic cycle (Rosta et al, 2011). In the UIMn structure, the internal phosphodiester of the scissile dinucleotide is unlikely to act as a general base in the initial proton transfer, since its non-bridging oxygen atoms point away from the attacking water molecule and are hydrogen bonded to IN main chain amides. Second, unlike the situation in RNase H, the coordination sphere of metal B in the UIMn includes a water molecule, which may provide a proton for the leaving 3′-alkoxide. Thus, despite close similarities, the RNase H and IN systems likely differ in some details of their catalytic mechanisms. Quantum molecular mechanics simulations may help to explain the mechanism of proton transfer during 3′-processing.
Target DNA binding and strand transfer
Because strand transfer in the TCC crystals is relatively fast (Figure 1C), considerable effort was expended to optimize metal-soaking and snap-freezing conditions, with over 200 TCC crystals utilized. Very short soaks (<1 min) resulted in predominant binding of a single Mn2+ ion at position B, leaving position A only partially occupied (TCCMn*, the asterisk indicates an early intermediate), while incubations for 2.5 min or longer resulted in complete strand transfer. Soaking TCC crystals with Mn2+ for intermediate periods often had deleterious effects on the diffraction limit, presumably due to induced crystal heterogeneity. Following a large number of trials we were able to acquire a 3.0-Å resolution data set from a 1.5-min soak, which revealed a metal-occupied TCC active site poised for strand transfer (TCCMn). Two independent structures of the post-strand transfer states, STCMn* (early intermediate) and STCMn (late), were refined using diffraction data collected from TCC crystals incubated in the presence of Mn2+ for 2.5 min and 2 h, respectively. In addition, the structure of the metal-free TCC (TCCApo) was refined to 3.15 Å resolution (Supplementary Table S1; see also Supplementary Figure S3 for the final and simulated annealing omit electron density maps of the refined models). Of note, kinetic analysis of strand transfer through dissolving soaked TCC crystals and radiolabelling substrate and product DNAs (Figure 1C) suggested a faster reaction to that observed by X-ray crystallography; the apparent discrepancy is explained by the different methods used to stop the reactions—back-soaking in EDTA versus instant freezing.
The phosphodiester backbone of the target DNA in the TCC overlaps with that of the scissile dinucleotide in the UI (Figure 4A). However, the target DNA in the TCC runs in the opposite direction to that of the viral DNA strand in the UI. The bases and ribose moieties of the targeted strand, being additionally base-paired as part of the duplex, are not in the same position as those of the scissile viral dinucleotide. In addition, the target phosphodiester group (marked 0 in the figure) has an inverted orientation compared with that of the viral scissile phosphodiester (Figure 4A).
Figure 4.
Target DNA binding and the strand transfer reaction. (A) Stereo view of target DNA (magenta and purple) overlaid with the position of reactive viral DNA strand before processing (yellow) in TCCMn and UIMn, respectively. The targeted, the upstream and downstream phosphodiesters of the target DNA are labelled in black with ‘0’, ‘−1’ and ‘+1’, respectively; the bases of the reactive viral DNA strand labelled in gold. (B) Relocation of the viral–target DNA phosphodiester bond following strand transfer (black arrow).
Upon target DNA loading, the metal cofactors move further apart from each other, with inter-metal distance increasing from 3.1 Å in CIMn to 3.8 Å in TCCMn due to the interaction with the target phosphodiester (Figure 3D). This inter-metal distance is very close to that observed in the ground state UIMn and RNase H–substrate complexes. The phosphodiester located 5′ of the target site (marked −1 in Figure 4A) interacts with the main chain amides of residues Tyr212 and Gln186, akin to the similarly positioned viral DNA phosphodiester in the UI (Figure 3A). As in the case of the UIMn active site, the pro-S p oxygen atom of the target phosphodiester is shared between the metal ions, while the pro-R p atom is pointing away from the metals (Figure 4B). This configuration explains the negative effects of thiosubstitutions at pro-S p positions in target DNA on IN strand transfer activity (Gerton et al, 1999). Metal A additionally interacts with the 3′-bridging oxygen atom from the target phosphodiester and one oxygen atom from each of the Asp128 and Asp185 carboxylates. Metal B is bound to both oxygen atoms from the Glu221 side chain, the second carboxylate atom of Asp128, and the 3′-hydroxyl group from the viral DNA. Water molecules not fully discernable at the resolution of the diffraction data presumably complete the coordination spheres of both metals. The 3′-hydroxyl of viral DNA is poised for in-line nucleophilic attack on the target phosphodiester, separated from its phosphorus atom by ∼2.8 Å (path marked with red dash in Figure 4B).
As predicted by Nowotny et al (2005), the roles of metal ion cofactors switch between 3′-processing and strand transfer, and it is metal B that activates the attacking 3′-hydroxyl group in the TCC. Following transesterification, the metal ions move closer again (3.2 Å in STCMn*; Figure 3D), accompanied by ejection of the phosphodiester linking the viral and host DNA strands from the active site (Figure 4B). Comparing structures of individually crystallized PFV TCC and STC, we observed a similar relocation of the newly made phosphodiester bond from the active site and postulated that this change serves to make the strand transfer reaction irreversible (Maertens et al, 2010). However, because the original TCC and STC crystals were grown under different conditions, it was not clear whether the conformation change took place during the reaction, or was somehow enforced during crystallization of the STC. The fact that the phosphodiester ejection is observed upon strand transfer in crystallo strongly supports its functional relevance.
The STCMn* structure, determined from TCC crystals soaked for 2.5 min in the presence of MnCl2, contains two Mn2+ ions, refined at full occupancy. Longer soaks (>1 h) yielded structures with a Mn2+ ion bound at position A, while position B was at most partially occupied, such as in the case of the STCMn (Supplementary Figure S3), suggesting a slow dissociation of metal B after strand transfer. Similarly, STC crystals grown in the presence of Mg2+ had a single metal in the active site (Maertens et al, 2010). We speculate that the apparent loss of metal B binding affinity following strand transfer is caused by the ejection of the newly formed phosphodiester form the active site.
Discussion
The relatively slow kinetics and metal dependence of IN-catalysed reactions allowed us to freeze trap the relevant Michaelis complexes formed prior to 3′-processing and strand transfer without resorting to the use of inactivating mutations or detuned reaction conditions. The resulting structures not only shed light on the IN active site mechanics, but also provide many insights relevant to HIV IN inhibitor development. Strand transfer inhibitor binding to the retroviral intasome that has undergone 3′-processing requires displacement of the 3′-deoxyadenosine from the IN active site (Hare et al, 2010a, 2010b). The UIMn structure provides an obvious explanation for the pronounced selectivity of this class of small molecules towards inhibiting strand transfer (Espeseth et al, 2000). Indeed, strand transfer inhibitor binding to the intasome prior to 3′-processing would require that the deoxyadenosine would leave the active site along with the uncleaved 3′-dinucleotide (Figure 5), at the cost of disruption of additional interactions (phosphate-metal and phosphate-amide; Figure 3A and C). Yet, the active site configuration in the UIMn complex does not necessarily preclude development of specific 3′-processing inhibitors, which would not require ejection of the trinucleotide. A small molecule binding at the viral DNA–IN interface and replacing the attacking water molecule with an inert metal chelating atom (such as an sp2 oxygen) would hinder the reaction. Formation of 5′-ester dinucleotide derivatives during 3′-processing by retroviral INs in the presence of glycerol and other alcohols (Vink et al, 1991; Dotan et al, 1995) indicates that a range of nucleophiles can access the catalytic centre. In principle, with a cleverly designed ligand it may be possible to replace all three water molecules bound to metal A in the pre-3′-processing state. However, the apparent mobility of the scissile dinucleotide can pose a problem for 3′-processing inhibitor design. The 3′-nucleotide is only partially ordered in our UIApo structure and displays higher than average B factors even in UIMn. It is not surprising therefore that its 3′-hydroxyl can sometimes replace the water nucleophile, explaining formation of the cyclic dinucleotide adduct, detectable amounts of which have been observed during 3′-processing by various retroviral INs in vitro (Engelman et al, 1991; Dotan et al, 1995).
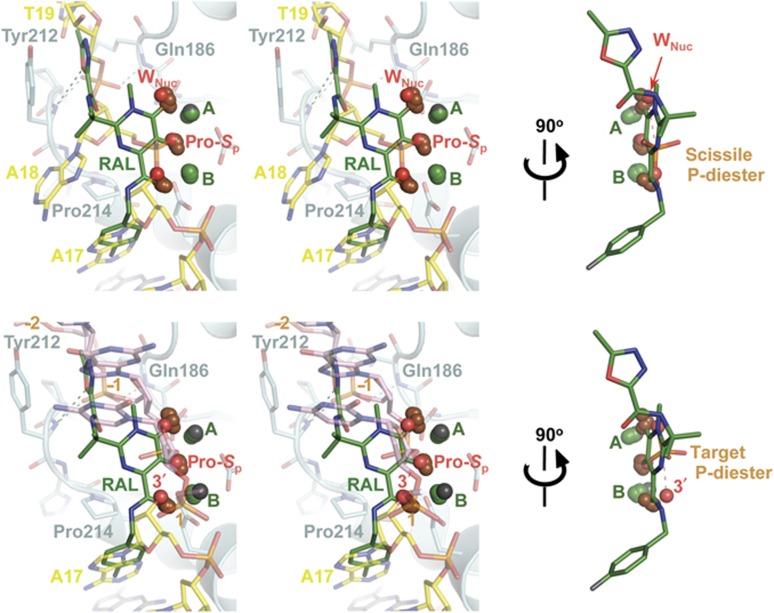
Figure 5.
Substrate mimicry by the HIV IN strand transfer inhibitors. Active sites of inhibitor-bound CI structures (PDB codes 3OYA, 3OYB, 3OYH, 3OYG, 3S3M) were superposed onto the UIMn (top) or TCCMn (bottom) based on Cα atoms of the active site residues. In the stereo views, raltegravir (3OYA) is shown as sticks with carbon atoms coloured green and fluorine in grey; only the metal chelating oxygen atoms of all other strand transfer inhibitors are shown (brown spheres). Protein is shown as semitransparent cartoons; viral and target DNA and selected IN residues as semitransparent sticks. Metal ions from the UIMn and TCCMn structures are shown as grey and those from the inhibitor-bound structures as green spheres. The pro-S p and the bridging 3′-oxygen atoms of the scissile or target phosphodiester (sticks), water (WNuc) and 3′-hydroxyl nucleophiles are shown as red spheres. The images to the right are related to the stereo views by a 90° rotation, as indicated. Hydrogen bonds discussed in the text are shown as dashed lines. The direction of the nucleophilic attack during 3′-processing or strand transfer is indicated with a pink dashed line. The chemical structure of raltegravir is shown in Supplementary Figure S4.
Comparison of the ground state structures presented here with the strand transfer inhibitor-bound forms of the CI (Hare et al, 2010a, 2010b, 2011) highlights an intriguing case of substrate mimicry utilized by the drugs. Superposition of the UIMn, TCCMn and raltegravir-bound CI (PDB ID 3OYA) places the middle oxygen atom of the metal chelating triad at the position normally occupied by the pro-S p oxygen atoms of the target or viral DNA scissile phosphodiester (Figure 5; Supplementary Figure S4). The oxygen atom distal from the inhibitor halo-benzyl side chain simultaneously mimics the attacking water molecule during 3′-processing and a bridging oxygen atom of the target phosphodiester during strand transfer. The third oxygen of the triad moreover mimics the attacking 3′-hydroxyl group during strand transfer and the bridging oxygen of the scissile phosphodiester during 3′-processing (Figure 5). The ground state structures presented here may allow detailed molecular dynamics simulations of both IN-catalysed reactions. Calculation of the associated transition states of the scissile/target phosphodiesters will likely highlight more optimal positions for metal chelating atoms and the design of tighter binding strand transfer inhibitors. The structural incompatibility of target DNA binding to the intasome prior to 3′-processing (Figure 4A) suggests that the scissile dinucleotide may in fact serve as a natural strand transfer inhibitor. The necessity to complete 3′-processing may restrict the ability of incompletely assembled viral nucleoprotein complexes to engage target DNA and perhaps reduce the chance of suicidal auto-integration.
Interactions between IN backbone atoms and reactive phosphodiesters observed in the UI and STC structures (Figures 3C and 4A) likely help to align the substrate DNA backbone and the enzyme active site. The PFV IN main chain amides involved in these contacts belong to Tyr212 and Gln186 (corresponding to HIV-1 IN Tyr143 and Asn117, respectively) and directly abut the drug-binding pocket. Of the strand transfer inhibitors characterized structurally (Hare et al, 2010a, 2010b, 2011), only raltegravir forms a hydrogen bond with the Tyr amide (Figure 5). A small molecule ligand that optimally reproduces both of these interactions could be predicted to bind tighter to the IN active site, regardless of local sequence variations. The IN surface involved in Van der Waals interactions with the first base of the scissile dinucleotide may represent another important contact point (Figure 3A). Notably, the second-generation strand transfer inhibitor MK2048 and its analogues make contacts in this area (Vacca et al, 2007; Hare et al, 2010b). Recent biochemical data indicated a correlation between inhibitor activity against raltegravir-resistant HIV-1 variants and tighter binding to wild-type IN (slow off-rate) (Grobler et al, 2009; Hightower et al, 2011). Our results highlight functionally important contact points that have not been fully explored in the current HIV IN inhibitors, raising hopes that next-generation compounds can be developed based on the now available structural information.
Materials and methods
Crystal preparation
The UI was prepared and crystallized as described previously for the cleaved complex (Hare et al, 2010b) using oligonucleotides 5′-TGCGAAATTCCATGACAAT-3′ and 5′-ATTGTCATGGAATTTCGCA-3′, mimicking the transferred and nonreactive strands, respectively. For some experiments, the nonreactive DNA strand was synthetized with 5′-phosphate, to suppress its detection on radioactive gels. The TCC was prepared as described previously (Maertens et al, 2010) in the presence of 2.5 mM EDTA and was crystallized at 18°C by vapour diffusion against a reservoir solution of 2.5 mM EDTA, 180 mM Li2SO4, 34% PEG-400 and 0.1 M Tris–HCl pH 7.4, in drops of 1 μl IN–DNA complex plus 1 μl reservoir solution.
Reactions in crystallo
Crystals were transferred to drops containing reservoir solution plus 25 mM MnCl2 or MgCl2 and incubated at 18–20°C. To detect reaction products, the crystals were washed through three 2-μl drops of reservoir solution with no metal ions plus 5 mM EDTA, prior to transfer into 10 μl solubilization buffer containing 5 mM EDTA, 1% (w/v) SDS, 0.1 M Tris–HCl, pH 9.0. The samples, supplemented with 10 μg proteinase K, were incubated at 37°C for 1 h. The protease and SDS were removed by extraction with two changes of phenol–chloroform–isoamyl alcohol (25:24:1, pH 8.0). Following two additional extractions with chloroform, the samples were desiccated to dryness and re-dissolved in 10 μl of 10 mM MgCl2, 50 mM Tris–HCl, pH 7.4. The DNA products were 5′-end labelled using γ-32P-ATP and thermostable polynucleotide kinase Clp1 at 80°C (Jain and Shuman, 2009). Labelled DNA species, separated on denaturing urea 17% polyacrylamide gels, were detected by phosphorimaging. RNA-specific 5′-OH kinases such as Clp1 or T4 polynucleotide kinase act on unpaired 5′-ends (Wang et al, 2002). Although both kinases were biased towards phosphorylating the nonreactive strand of the viral DNA duplex (strand N; Figure 1A), which displays a protruding 5′-end following 3′-processing, the thermostable enzyme performed better at labelling other DNA species present in crystals. For X-ray diffraction studies, after soaking in the presence of 25 mM MnCl2 for set incubations times, crystals were flash-frozen in liquid nitrogen and stored until data collection at 100 K.
Diffraction data collection and structure refinement
X-ray diffraction data were collected on the beamlines I02, I03 and I04-1 of the Diamond Light Source (Oxfordshire, UK). Data, integrated using Mosflm (Leslie, 1992) or XDS (Kabsch, 2010), were merged and scaled in Scala (Evans, 2006). The structures were solved by isomorphous replacement using PDB ID 3OY9 (for UI structures), 3OS0 (for STC) or 3OS1 (for TCC) as starting models. Coot (Emsley and Cowtan, 2004) was used for manual model building, and Refmac (Murshudov et al, 1997) and Phenix (Zwart et al, 2008) for structure refinement. All final models had good geometry (Supplementary Table S1), as assessed by Molprobity (Davis et al, 2007). Composite simulated annealing omit maps (Supplementary Figure S3) were generated in Phenix. All structural illustrations were created using PyMol (www.pymol.org).
Accession codes
The coordinates and structure factors for UIApo, UIMn, TCCApo, TCCMn and STCMn* were deposited with the protein data bank under accession codes 4E7H, 4E7I, 4E7J, 4E7K and 4E7L, respectively. Structure factors for UIApo-PO3, CIMn (analogous to 3OY9) and STCMn (similar to 3OS0) will be available on request.
Supplementary Material
Acknowledgments
We thank Dr A Engelman, Dr F Dyda and Dr M Nowotny for critical reading of the manuscript and Dr S Schuman (Sloan Kettering Institute) for a generous gift of a PhoClp1 expression construct (Jain and Shuman, 2009). This work was funded by the UK Medical Research Council Grant G1000917 and National Institutes of Health grant AI070042.
Author contributions: SH and GNM grew the crystals, collected and processed X-ray diffraction data. SH analysed in crystallo reactions by denaturing PAGE. SH, GNM and PC refined and analysed the structures and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andrake MD, Ramcharan J, Merkel G, Zhao XZ, Burke TR Jr., Skalka AM (2009) Comparison of metal-dependent catalysis by HIV-1 and ASV integrase proteins using a new and rapid, moderate throughput assay for joining activity in solution. AIDS Res Ther 6: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam CA, Steitz TA (1998) Structural principles for the inhibition of the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates. J Mol Biol 277: 363–377 [DOI] [PubMed] [Google Scholar]
- Bushman FD, Craigie R (1991) Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci USA 88: 1339–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Maertens GN, Hare S (2011) Structural insights into the retroviral DNA integration apparatus. Curr Opin Struct Biol 21: 249–256 [DOI] [PubMed] [Google Scholar]
- Craigie R (2002) Retroviral DNA Integration. InMobile DNA II Craig NL, Craigie R, Gellert M, Lambowitz AM (eds), pp613–630. Washington DC: ASM Press [Google Scholar]
- Davies DR, Goryshin IY, Reznikoff WS, Rayment I (2000) Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289: 77–85 [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB 3rd, Snoeyink J, Richardson JS, Richardson DC (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35: W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vivo M, Dal Peraro M, Klein ML (2008) Phosphodiester cleavage in ribonuclease H occurs via an associative two-metal-aided catalytic mechanism. J Am Chem Soc 130: 10955–10962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotan I, Scottoline BP, Heuer TS, Brown PO (1995) Characterization of recombinant murine leukemia virus integrase. J Virol 69: 456–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR (1994) Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266: 1981–1986 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Engelman A, Craigie R (1992) Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol 66: 6361–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, Craigie R (1995) Efficient magnesium-dependent human immunodeficiency virus type 1 integrase activity. J Virol 69: 5908–5911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, Mizuuchi K, Craigie R (1991) HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 67: 1211–1221 [DOI] [PubMed] [Google Scholar]
- Espeseth AS, Felock P, Wolfe A, Witmer M, Grobler J, Anthony N, Egbertson M, Melamed JY, Young S, Hamill T, Cole JL, Hazuda DJ (2000) HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc Natl Acad Sci USA 97: 11244–11249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P (2006) Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62: 72–82 [DOI] [PubMed] [Google Scholar]
- Gerton JL, Herschlag D, Brown PO (1999) Stereospecificity of reactions catalyzed by HIV-1 integrase. J Biol Chem 274: 33480–33487 [DOI] [PubMed] [Google Scholar]
- Grobler JA, McKenna PM, Ly S, Stillmock KA, Bahnck CM, Danovich RM, Dornadula G, Hazuda DJ, Miller MD (2009) HIV integrase inhibitor dissociation rates correlate with efficacy in vitro. Antivir Ther 14: A27 [Google Scholar]
- Harding MM (2006) Small revisions to predicted distances around metal sites in proteins. Acta Crystallogr D Biol Crystallogr 62: 678–682 [DOI] [PubMed] [Google Scholar]
- Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P (2010a) Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464: 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Smith SJ, Metifiot M, Jaxa-Chamiec A, Pommier Y, Hughes SH, Cherepanov P (2011) Structural and functional analyses of the second-generation integrase strand transfer inhibitor dolutegravir (S/GSK1349572). Mol Pharmacol 80: 565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Vos AM, Clayton RF, Thuring JW, Cummings MD, Cherepanov P (2010b) Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc Natl Acad Sci USA 107: 20057–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hightower KE, Wang R, Deanda F, Johns BA, Weaver K, Shen Y, Tomberlin GH, Carter HL 3rd, Broderick T, Sigethy S, Seki T, Kobayashi M, Underwood MR (2011) Dolutegravir (S/GSK1349572) exhibits significantly slower dissociation than raltegravir and elvitegravir from wild-type and integrase inhibitor-resistant HIV-1 integrase-DNA complexes. Antimicrob Agents Chemother 55: 4552–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Shuman S (2009) Characterization of a thermostable archaeal polynucleotide kinase homologous to human Clp1. RNA 15: 923–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretzek T, Holm T, Gartner K, Kanzler S, Lindemann D, Herchenroder O, Picard-Maureau M, Rammling M, Heinkelein M, Rethwilm A (2004) Foamy virus integration. J Virol 78: 2472–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010) Xds. Acta Crystallogr D Biol Crystallogr 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz RA, Merkel G, Andrake MD, Roder H, Skalka AM (2011) Retroviral integrases promote fraying of viral DNA ends. J Biol Chem 286: 25710–25718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy AK, Haniford DB, Mizuuchi K (2000) Single active site catalysis of the successive phosphoryl transfer steps by DNA transposases: insights from phosphorothioate stereoselectivity. Cell 101: 295–305 [DOI] [PubMed] [Google Scholar]
- Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM (1992) Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol 12: 2331–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW (1992) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4+ESF-EAMCB Newsletter on Protein Crystallography [Google Scholar]
- Lewinski MK, Bushman FD (2005) Retroviral DNA integration--mechanism and consequences. Adv Genet 55: 147–181 [DOI] [PubMed] [Google Scholar]
- Li M, Craigie R (2009) Nucleoprotein complex intermediates in HIV-1 integration. Methods 47: 237–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Mizuuchi M, Burke TR Jr, Craigie R (2006) Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J 25: 1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens GN, Hare S, Cherepanov P (2010) The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468: 326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand C, Maddali K, Métifiot M, Pommier Y (2009) HIV-1 IN inhibitors: 2010 update and perspectives. Curr Top Med Chem 9: 1016–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl DJ, Chen X (2010) Strand transfer inhibitors of HIV-1 integrase: bringing IN a new era of antiretroviral therapy. Antiviral Res 85: 101–118 [DOI] [PubMed] [Google Scholar]
- Metifiot M, Maddali K, Naumova A, Zhang X, Marchand C, Pommier Y (2010) Biochemical and pharmacological analyses of HIV-1 integrase flexible loop mutants resistant to raltegravir. Biochemistry 49: 3715–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K, Adzuma K (1991) Inversion of the phosphate chirality at the target site of Mu DNA strand transfer: evidence for a one-step transesterification mechanism. Cell 66: 129–140 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nowotny M, Gaidamakov SA, Crouch RJ, Yang W (2005) Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Nowotny M, Yang W (2006) Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J 25: 1924–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosta E, Nowotny M, Yang W, Hummer G (2011) Catalytic mechanism of RNA backbone cleavage by ribonuclease H from quantum mechanics/molecular mechanics simulations. J Am Chem Soc 133: 8934–8941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scottoline BP, Chow S, Ellison V, Brown PO (1997) Disruption of the terminal base pairs of retroviral DNA during integration. Genes Dev 11: 371–382 [DOI] [PubMed] [Google Scholar]
- Summa V, Petrocchi A, Bonelli F, Crescenzi B, Donghi M, Ferrara M, Fiore F, Gardelli C, Gonzalez Paz O, Hazuda DJ, Jones P, Kinzel O, Laufer R, Monteagudo E, Muraglia E, Nizi E, Orvieto F, Pace P, Pescatore G, Scarpelli R, Stillmock K, Witmer MV, Rowley M (2008) Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J Med Chem 51: 5843–5855 [DOI] [PubMed] [Google Scholar]
- Vacca J, Wai J, Fisher T, Embrey M, Hazuda D, Miller M, Felock P, Witmer M, Gabryelski L, Lyle T (2007) Discovery of MK-2048—subtle changes confer unique resistance properties to a series of tricyclic hydroxypyrrole integrase strand transfer inhibitors. In4th International AIDS Socienty Conference Sydney, Australia [Google Scholar]
- Valkov E, Gupta SS, Hare S, Helander A, Roversi P, McClure M, Cherepanov P (2009) Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res 37: 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C, Yeheskiely E, van der Marel GA, van Boom JH, Plasterk RH (1991) Site-specific hydrolysis and alcoholysis of human immunodeficiency virus DNA termini mediated by the viral integrase protein. Nucleic Acids Res 19: 6691–6698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LK, Lima CD, Shuman S (2002) Structure and mechanism of T4 polynucleotide kinase: an RNA repair enzyme. EMBO J 21: 3873–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Lee JY, Nowotny M (2006) Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol Cell 22: 5–13 [DOI] [PubMed] [Google Scholar]
- Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD (2008) Automated structure solution with the PHENIX suite. Methods Mol Biol 426: 419–435 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.