Abstract
Phosphatidylinositol-4,5-bisphosphate, PtdIns(4,5)P2, is an essential signalling lipid that regulates key processes such as endocytosis, exocytosis, actin cytoskeletal organization and calcium signalling. Maintaining proper levels of PtdIns(4,5)P2 at the plasma membrane (PM) is crucial for cell survival and growth. We show that the conserved PtdIns(4)P 5-kinase, Mss4, forms dynamic, oligomeric structures at the PM that we term PIK patches. The dynamic assembly and disassembly of Mss4 PIK patches may provide a mechanism to precisely modulate Mss4 kinase activity, as needed, for localized regulation of PtdIns(4,5)P2 synthesis. Furthermore, we identify a tandem PH domain-containing protein, Opy1, as a novel Mss4-interacting protein that partially colocalizes with PIK patches. Based upon genetic, cell biological, and biochemical data, we propose that Opy1 functions as a coincidence detector of the Mss4 PtdIns(4)P 5-kinase and PtdIns(4,5)P2 and serves as a negative regulator of PtdIns(4,5)P2 synthesis at the PM. Our results also suggest that additional conserved tandem PH domain-containing proteins may play important roles in regulating phosphoinositide signalling.
Keywords: phosphatidylinositol-4,5-bisphosphate; phosphoinositide kinase complex; tandem PH domain-containing proteins
Introduction
Phosphorylated derivatives of phosphatidylinositol, collectively known as phosphoinositide (PIP) lipids, regulate cell polarity, growth and development, and membrane trafficking (Martin, 1998; Behnia and Munro, 2005; Di Paolo and De Camilli, 2006; Vicinanza et al, 2008). A wide range of protein effectors are directly regulated by PIP lipids through conserved PIP-binding modules that specify effector recruitment and activation at distinct organelle membranes (Lemmon, 2008). However, how levels of distinct PIP isoforms are accurately maintained by the opposing actions of PIP kinases and PIP phosphatases is not fully understood. One of the major PIP speies, PtdIns(4,5)P2, is enriched in the inner leaflet of the plasma membrane (PM). PtdIns(4,5)P2 was initially shown to function as a precursor of the second messengers diacylglycerol (DAG) and 1,4,5-inositol trisphosphate (IP3) generated by phospholipase C (Hokin, 1985). More recent studies have highlighted direct roles for PtdIns(4,5)P2 in the regulation of the endocytic machinery (Haucke, 2005), exocytosis (Liu et al, 2007; James et al, 2008), the actin cytoskeleton (Yin and Janmey, 2003) and septin assembly during cell division (Logan and Mandato, 2006; Bertin et al, 2010).
In the yeast Saccharomyces cerevisiae, PtdIns(4,5)P2 is synthesized by the conserved PtdIns(4)P 5-kinase Mss4. Mss4 localizes to the PM and generates essential pools of PtdIns(4,5)P2 (Desrivieres et al, 1998; Homma et al, 1998; Audhya and Emr, 2002). Mss4 also undergoes nuclear-cytoplasmic shuttling and nuclear sequestration may control Mss4 function (Audhya and Emr, 2003). However, little is known about how regulation of Mss4 at the PM is achieved. We have found that Mss4 assembles into oligomeric protein complexes at the PM that we term PIK (PIP kinase) patches. Mss4 PIK patches are dynamic and are distinct from other previously described cortical structures, such as actin patches (Pruyne and Bretscher, 2000a, 2000b) and eisosomes (Walther et al, 2006). Assembly of Mss4 PIK patches requires the C-terminus of Mss4 and its substrate PtdIns(4)P. In addition, by genetic and quantitative proteomic approaches, we identify the dual PH domain-containing protein Opy1 as a novel regulator of Mss4. We propose that assembly of Mss4 PIK patches permits coordinate regulation of multiple Mss4 kinase molecules by accessory factors, including the conserved Opy1 protein.
Results
Mss4 forms dynamic protein complexes, PIK patches, at the PM
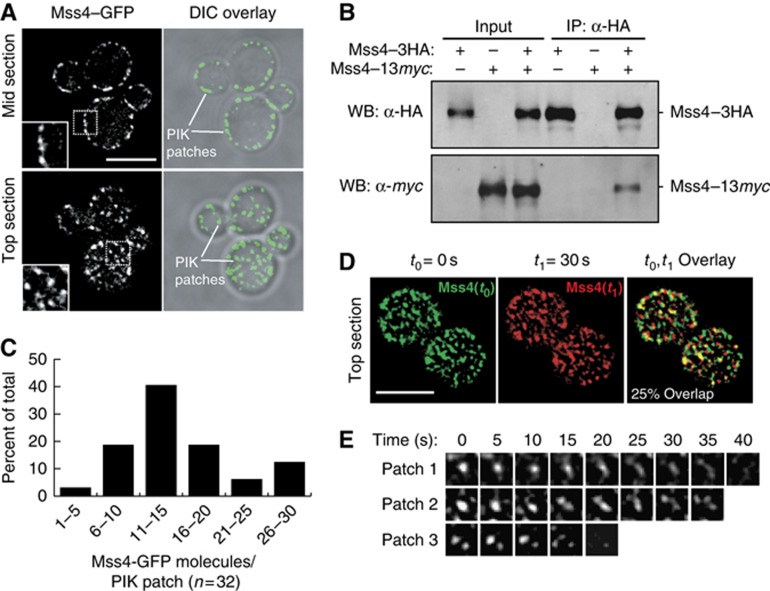
Mss4 is a cytoplasmic protein that associates with the inner face of the PM (Audhya and Emr, 2003). To understand how Mss4 is regulated, we employed cells solely expressing a functional Mss4–GFP fusion. Mss4–GFP formed cortical, punctate structures at the PM: PIK (phosphoinositide kinase) patches (Figure 1A, observed at both mid and top sections of cells; Supplementary Movie S1) similar to previously reported patterns for Mss4 localization (Audhya and Emr, 2002; Sun et al, 2007; Smaczynska-de et al, 2008). Reconstructions (2D projections of Z series) of cells expressing Mss4–GFP indicated that there are ∼30–50 Mss4 PIK patches in each individual yeast cell. However, the oligomeric status or the dynamics of Mss4 PIK patches at the PM has not been addressed. To determine whether multiple copies of Mss4 assemble at PIK patches, we tested if an Mss4–13xmyc fusion isolated with an Mss4–3xHA fusion in coimmunoprecipitation experiments. Mss4–13xmyc was present in anti-HA immunoprecipitates from cell lysates coexpressing Mss4–3xHA, but not from control cell lysates lacking Mss4–3xHA (Figure 1B), suggesting that Mss4 oligomerizes. To estimate the copy numbers of Mss4 molecules present in PIK patches, we expressed Mss4–GFP in cells coexpressing a Cse4–3xGFP fusion (both are integrated; Supplementary Figure S1A). Cse4–3xGFP forms a complex containing 96 GFP molecules in the nucleus of yeast cells (Markus et al, 2009). By comparing the GFP signal intensities of Mss4 PIK patches and Cse4–3xGFP in the nucleus, we found a distribution of ∼5–30 copies of Mss4–GFP in each PIK patch (Figure 1C). However, most PIK patches contained ∼10–20 Mss4–GFP molecules (Figure 1C). Thus, multiple copies of the Mss4 lipid kinase assemble at PIK patches.
Figure 1.
Mss4 forms oligomeric, dynamic cortical structures at the PM. (A) Mss4–GFP localization in mid and top sections of yeast cells. Cells shown are representative of over 100 cells observed. Lines indicate individual Mss4 PIK patches at the PM. The inset (boxed area) shows a region magnified two-fold. Levels of the colour images overlayed on DIC images were adjusted with Adobe Photoshop. Scale bar, 5 μm. (B) Mss4–13myc was coimmunoprecipitated with Mss4–3HA. Lysates from cells expressing Mss4–3HA, Mss4–13myc or both were incubated with crosslinker and immunoprecipitated with anti-HA beads, and analysed by immunoblotting to detect Mss4–Mss4 interaction. (C) Quantification of numbers of Mss4 molecules in PIK patches (n=32). Cells coexpressing integrated Mss4–GFP and Cse4–3xGFP were analysed by fluorescence microscopy. Numbers of Mss4 were calculated based on fluorescence signal intensity of Mss4 PIK patches and Cse4–3xGFP in the nucleus as an internal standard (corresponding to 96 GFP molecules; see Supplementary Figure S1A; Supplementary data set 1). Using this approach, the average number of Mss4 molecules per PIK patch was 15±6 (s.d.). (D) Mss4 PIK patches are dynamic structures. Cells expressing Mss4–GFP were examined by time-lapse fluorescence microscopy. Images of Mss4–GFP at the cell surface were captured every 5 s. At time t 0=0 s, Mss4–GFP is shown in green, at t 1=30 s Mss4–GFP is shown in red. Scale bar, 5 μm. (E) Three representative examples of Mss4 PIK patch lifetimes at the cell surface, images were captured every 5 s. Figure source data can be found with the Supplementary data.
To address the dynamics of Mss4 PIK patches, we performed time-lapse imaging experiments following Mss4–GFP at the cell surface by focusing on the top of cells. Strikingly, Mss4 PIK patches were highly dynamic and short-lived structures (Supplementary Movie S2). More than 75% of Mss4 PIK patches appear to change localization within 30 s (Figure 1D), and the lifetime of Mss4 PIK patches range from 10 to 40 s (Figure 1E; Supplementary Figure S3A). This dramatic rearrangement in distribution likely occurs by the dynamic assembly and disassembly of Mss4 PIK patches as well as lateral movements along the surface of the PM, as Mss4 PIK patches did not move into the interior of the cell by following Mss4–GFP in mid sections of cells (Supplementary Movie S3). Mss4 PIK patches were distinct from cortical actin patches (Pruyne and Bretscher, 2000a, 2000b) and did not require actin polymerization for assembly (Supplementary Figure S1B). Likewise, Mss4 PIK patches were distinct from eisosomes and independent of the eisosome component Pil1 (Supplementary Figure S1C and D). These results suggested that Mss4 assembles into unique dynamic structures at the PM. We thus sought to further understand how Mss4 organization and function are regulated.
The Mss4 kinase domain and PtdIns(4)P are required for PIK patch assembly
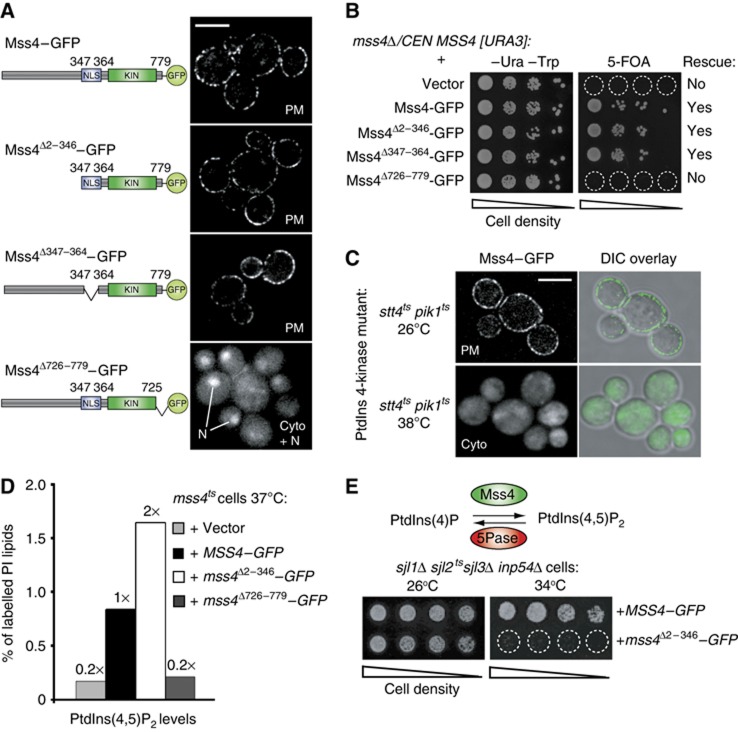
Mss4 consists of an uncharacterized N-terminal domain, a central nuclear localization signal (NLS), and a conserved C-terminal PIP 5-kinase domain (Figure 2A). To map regions in Mss4 necessary for PIK patch assembly, we generated a series of truncated Mss4 mutants tagged with GFP. The large N-terminal region of Mss4 (residues 2–346) and the NLS in Mss4 (residues 347–364) were dispensable for PM localization (Figure 2A). In contrast, a mutant form lacking the last 54 amino acids of Mss4 (residues 726–779), did not localize to the PM and instead accumulated in the cytoplasm and nucleus (Figure 2A). We then tested if the truncated Mss4–GFP fusions were functional using a plasmid shuffle growth assay. For this, we transformed an mss4Δ strain carrying an URA3-marked wild-type MSS4 plasmid or with plasmids encoding truncated forms of Mss4–GFP. As expected, cells expressing Mss4Δ726–779–GFP alone failed to grow on 5-FOA media (due to loss of the URA3-marked wild-type MSS4 plasmid). However, neither the N-terminal region nor the NLS were required for Mss4 function, as cells expressing mutant forms of Mss4 lacking these regions were able to grow on 5-FOA plates (Figure 2B).
Figure 2.
The Mss4 kinase domain and PtdIns(4)P are required for PIK patch assembly. (A) Localization of wild-type and truncated Mss4–GFP proteins. From top to bottom: full-length Mss4–GFP, N-terminally truncated Mss4–GFP (lacking residues 2–346), NLS-deleted Mss4–GFP (lacking residues 347–364), and C-terminally truncated Mss4–GFP (lacking residues 726–779). Lines indicate the nucleus of cells. Scale bar, 5 μm. Cyto, cytoplasm, N, nucleus. (B) Complementation assays of the truncated Mss4 mutants. The mss4Δ cells carrying a centromeric URA3-marked MSS4 plasmid was cotransformed with plasmids expressing various mutant Mss4–GFP forms as indicated. Cells were spotted onto –Ura –Trp plates to retain both plasmids or plates containing 5-FOA to select for loss of the URA3-marked MSS4 plasmid; only cells harbouring functional MSS4–GFP plasmids grew on 5-FOA plates. (C) Mss4–GFP localization in stt4tspik1ts double-mutant cells at 26 and 38°C. Cells were grown at 26°C to mid-log phase and shifted to 38°C for 60 min prior to observation by fluorescence microscopy. Scale bar, 5 μm. (D) Cellular PtdIns(4,5)P2 levels measured by 3H-inositiol labelling and HPLC analysis of mss4ts cells expressing empty vector, full-length Mss4–GFP, N-terminally truncated Mss4–GFP (lacking residues 2–346), or C-terminally truncated Mss4–GFP (lacking residues 726–779). The lipid labelling was performed at 37°C, a non-permissive temperature for the mss4ts cells. Results from a representative experiment are shown. Additional data are provided in Supplementary Table S1. (E) Expression of N-terminally truncated Mss4 impairs the growth of sjl1Δ sjl2tssjl3Δ inp54Δ cells deficient in PIP 5-phosphatase activity. PtdIns(4,5)P2 metabolism is regulated by the Mss4 PIP 5-kinase and a set of conserved 5-phosphatases, the synaptojanin-like (Sjl) proteins and Inp54. Full-length Mss4–GFP or N-terminally truncated Mss4Δ2–346–GFP was expressed in sjl1Δ sjl2tssjl3Δ inp54Δ cells as indicated. Serial dilutions of yeast cells were grown on –Ura plates to retain the MSS4–GFP plasmids at either 26 or 34°C for 4 days.
The truncation resulting in Mss4 mislocalization occurs in a conserved region of the C-terminus, termed the activation loop (Supplementary Figure S2A; Kunz et al, 2000). The activation loops of mammalian PtdIns(4)P 5-kinases control substrate recognition and subcellular targeting (Kunz et al, 2000). Substitution of two highly conserved lysine residues to negatively charged aspartate residues in the activation loop resulted in Mss4 mislocalization (Supplementary Figure S2B). In contrast, substitutions with arginine, retaining the positive charge, did not affect Mss4 PM localization (Supplementary Figure S2B). Thus, binding to the anionic substrate PtdIns(4)P may be important for Mss4 localization. Stt4 and Pik1 are the two major PtdIns 4-kinases responsible for PtdIns(4)P synthesis in yeast (Audhya et al, 2000). To deplete PtdIns(4)P, we used a temperature conditional stt4tspik1ts double-mutant strain. Mss4–GFP localized to the cytoplasm in stt4tspik1ts double-mutant cells at the restrictive temperature (Figure 2C). However, Mss4–GFP localized to the PM in stt4ts and pik1ts single-mutant cells at the non-permissive temperature (Supplementary Figure S2C), suggesting that both PtdIns 4-kinases contribute to Mss4 PM targeting. Mss4 localization was not dependent on PtdIns(4,5)P2 levels, as a kinase inactive form of Mss4–GFP localized to the PM when PtdIns(4,5)P2 is depleted in mss4ts cells (Supplementary Figure S2D; Stefan et al, 2002). Consistent with a role for PtdIns(4)P in targeting Mss4 to the PM, the purified Mss4 kinase domain, His6–SUMO–Mss4454–779, bound to PtdIns(4)P, as well as PtdIns(3)P and weakly to PtdIns(4,5)P2, in lipid overlay experiments (Supplementary Figure S2E). Thus, the C-terminus of Mss4 and PtdIns(4)P target Mss4 to the PM.
The N-terminal region of Mss4 functions as a negative regulatory domain
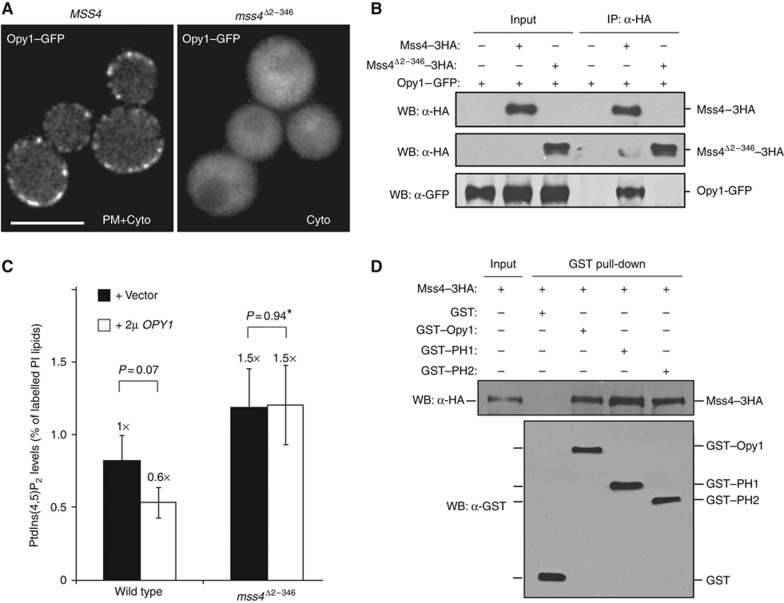
Our initial results implicated the conserved C-terminus of Mss4 in PIK patch assembly. However, the role of the Mss4 N-terminal region remained unclear. We further examined Mss4 regulation by performing 3H-inositol labelling experiments to measure cellular PtdIns(4,5)P2 levels in mss4ts cells coexpressing various forms of Mss4–GFP. As expected, the C-terminal truncated Mss4Δ726–779–GFP was defective in PtdIns(4,5)P2 synthesis (Figure 2D; Supplementary Table S1). However, deletion of the N-terminus increased Mss4 activity, as PtdIns(4,5)P2 levels were ∼2-fold higher in mss4ts cells expressing Mss4Δ2–346–GFP compared with cells expressing full-length Mss4–GFP (Figure 2D; Supplementary Table S1). Both Mss4Δ726–779–GFP and Mss4Δ2–346–GFP were expressed at levels similar to full-length Mss4–GFP, indicating that the altered activities of the mutant proteins were not due to changes in protein stability (Supplementary Figure S2F). In addition, PIK patches containing Mss4–GFP and Mss4Δ2–346–GFP displayed similar dynamics and GFP fluorescence intensity distributions (Supplementary Figure S3A and B; Supplementary Movie S4). Thus, the increase in PtdIns(4,5)P2 levels in cells expressing Mss4Δ2–346–GFP was not due to increases in Mss4 assembly or PIK patch lifetime. Elevated PtdIns(4,5)P2 levels are toxic in cells with impaired PtdIns(4,5)P2 phosphatase activity (Stefan et al, 2002). Expression of the N-terminal truncated form but not full-length Mss4 impaired the growth of cells deficient in PIP 5-phosphatase function (sjl1Δ sjl2tssjl3Δ inp54Δ cells) at a semi-permissive temperature (Figure 2E), further suggesting that deletion of the N-terminal region results in increased Mss4 PIP kinase activity.
The PH domain-containing protein Opy1 binds Mss4
The mechanisms of Mss4 localization and regulation at the PM are poorly characterized. We reasoned that the localization and regulation of Mss4 are mediated in part by associated factors. To identify candidate proteins that interact with Mss4, we undertook a quantitative proteomics approach (Figure 3A). Cells were grown in media containing either light (for the control strain) or heavy (for cells expressing Mss4–3xFlag) isotope amino acids. We then performed crosslinking immunopurification (IP) experiments and processed the protein samples for quantitative mass spectrometry analysis. This approach detects the enrichment of proteins that specifically interact with Mss4, and allows for the identification of weak/transient interactions. One protein, Opy1, was highly enriched with purified Mss4–3xFlag (Figure 3B). To confirm the Mss4–Opy1 interaction, we repeated the crosslinking coIP experiment and monitored the results by immunoblotting. As expected, Opy1–3xHA was present in immunoprecipitates from cells expressing Mss4–3xFlag, but not control cells lacking Mss4–3xFlag (Figure 3C).
Figure 3.
Identification of Mss4-interacting proteins by chemical crosslinking experiments and quantitative mass spectrometry. (A) Outline of the quantitative SILAC–MS approach. See the Results and Materials and methods for additional details. (B) Expression ratio (Xpress ratio=Mss4–3xFlag IP/ control IP) for proteins identified by the SILAC–MS experiments. An Xpress ratio of >10 was used as a set point to define specific Mss4-interacting proteins. Opy1 was enriched >50-fold in the Mss4–3xFlag IP compared with control IP. The inset shows the number of Mss4 and Opy1 peptides identified in the heavy (containing Mss4–3xFlag) and light samples. (C) Opy1–3HA crosslinks and coimmunoprecipitates with Mss4–Flag. Lysates from cells expressing Opy1–3HA or Mss4–Flag and Opy1–3HA were incubated with crosslinker and incubated with anti-Flag beads. Immunoprecipitates were analysed by immunoblotting to detect Mss4–Opy1 interactions. (D) Opy1 localizes to cortical structures and the cytoplasm. Wild-type cells expressing Opy1–GFP were grown to mid-log and examined by fluorescence microscopy. Cells shown are representative of over 100 cells observed. Scale bar, 4 μm. Diagram of the Opy1 protein is shown under the fluorescence images. PH, pleckstrin homology domain. Figure source data can be found with the Supplementary data.
Opy1 consists of two PH domains and the C-terminal PH domain has been proposed to bind PtdIns(4,5)P2 (Figure 3D; Szentpetery et al, 2009). By examining full-length Opy1–GFP in vivo, we observed Opy1 in the cytoplasm and at cortical punctate structures (Figure 3D). Interestingly, the cortical Opy1 structures partially colocalized with Mss4 PIK patches at the PM (Supplementary Figure S3D), Taken together, these results suggested that Opy1 interacts with Mss4 at PIK patches.
Opy1 is a novel regulator of PtdIns(4,5)P2 synthesis
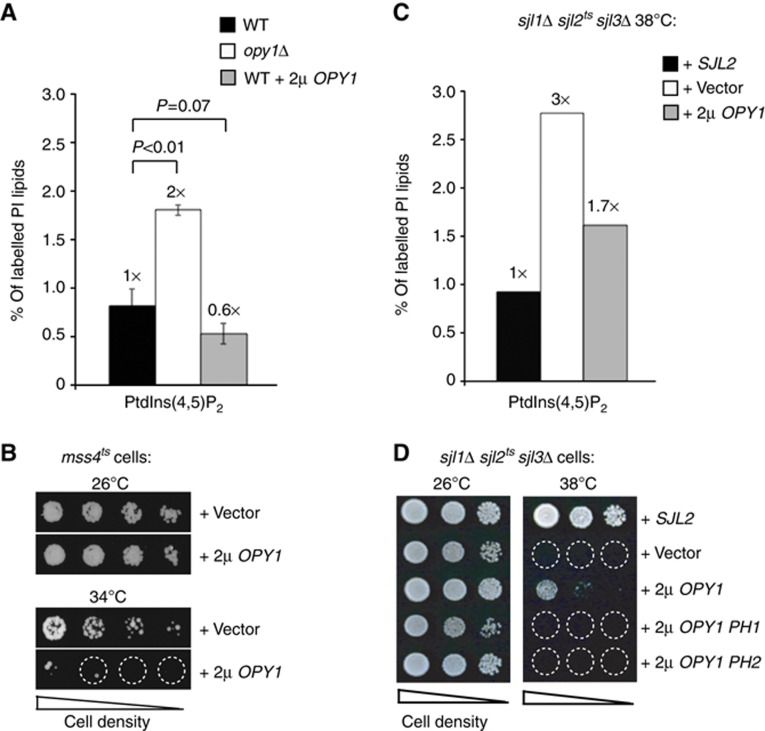
To study the function of Opy1 in vivo, we first deleted the OPY1 gene. Mss4 assembled into PIK patches at the PM in cells lacking OPY1 with wild-type dynamics and Mss4–GFP fluorescence intensities, and thus Opy1 was not essential for Mss4 PIK patch formation (Supplementary Figure S3A and C; Supplementary Movie S5). To test if Opy1 regulates PtdIns(4,5)P2 metabolism, we performed 3H-inositol labelling experiments to measure cellular PIP levels. We detected a two-fold increase in PtdIns(4,5)P2 levels in opy1Δ cells compared with wild-type cells (Figure 4A; Supplementary Table S1). In addition, overexpression of Opy1 in wild-type cells resulted in a 40% decrease in PtdIns(4,5)P2 levels (Figure 4A; Supplementary Table S1). This was not due to decreases in Mss4 PIK patch lifetime (Supplementary Figure S3A; Supplementary Movie S6) or assembly (as measured by relative Mss4–GFP fluorescence intensity; Supplementary Figure S3C). Moreover, overexpression of Opy1 impaired the growth of mss4ts cells at a semi-permissive temperature (34°C; Figure 4B), further suggesting that Opy1 regulates PtdIns(4,5)P2 metabolism, either by inhibiting its synthesis or by promoting its turnover.
Figure 4.
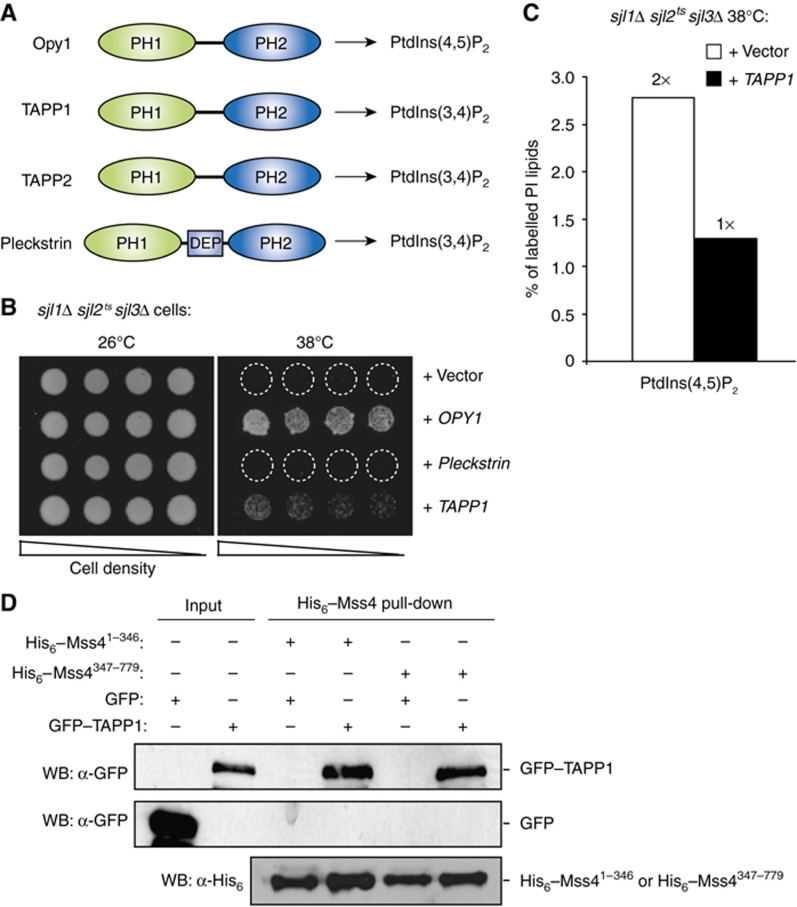
Opy1 inhibits PtdIns(4,5)P2 synthesis at the PM. (A) Cellular PtdIns(4,5)P2 levels were determined in wild-type, opy1Δ cells, or wild-type cells overexpressing OPY1 by 3H-inositiol labelling and HPLC analysis. Three independent labelling experiments were performed at 26°C. Data represent the mean of three independent experiments (±s.d.). (B) Overexpression of Opy1 impairs the growth of mss4ts cells with impaired PIP 5-kinase activity at a semi-permissive temperature (34oC). Serial dilutions of mss4ts cells were grown on –Ura plates to retain the high-copy OPY1 plasmid at either 26 or 34°C for 3 days. (C) Cellular PtdIns(4,5)P2 levels as measured by 3H-inositiol labelling and HPLC analysis in sjl1Δ sjl2tssjl3Δ cells carrying empty vector, SJL2, or OPY1 plasmids. Two independent labelling experiments were performed at 38°C. Results from a representative experiment are shown. Additional data are provided in Supplementary Table S1. (D) Overexpression of full-length Opy1, but not the PH1 or PH2 domains from Opy1 alone, rescues the growth defect of sjl1Δ sjl2tssjl3Δ cells at 38°C. Serial dilutions of sjl1Δ sjl2tssjl3Δ cells carrying empty vector, SJL2, full-length OPY1 or truncated opy1 (PH1 domain or PH2 domain) plasmids as indicated were spotted onto –Ura plates to retain the plasmids and grown at either 26 or 38°C for 4 days.
In an independent genetic screen to identify regulators of PtdIns(4,5)P2 signalling, we isolated OPY1 as a high copy suppressor of synaptojanin mutant cells deficient in PIP 5-phosphatase activity (sjl1Δ sjl2tssjl3Δ cells; Figure 4D). Consistent with this, OPY1 overexpression resulted in decreased PtdIns(4,5)P2 levels in sjl1Δ sjl2tssjl3Δ cells at the restrictive temperature (Figure 4C; Supplementary Table S1). Thus, Opy1 modulates PtdIns(4,5)P2 metabolism independently of the synaptojanin-like PIP phosphatases. Consistent with this, loss of Opy1 did not alter the cortical localization of Sjl2 (Supplementary Figure S4A). Furthermore, triple deletion of SJL1, SJL2, and OPY1 resulted in an additive growth defect, as sjl1Δ sjl2Δ opy1Δ cells failed to grow upon loss of a URA3-marked SJL2 plasmid on 5-FOA media (Supplementary Figure S4B), suggesting that Opy1 and the synaptojanins regulate PtdIns(4,5)P2 by distinct mechanisms. In control experiments, OPY1 overexpression did not rescue the growth defects of other PIP phosphatase mutant cells that accumulate toxic levels of PtdIns(4)P and PtdIns(3)P, sac1tssjl2Δ sjl3Δ cells and ymr1tssjl2Δ sjl3Δ cells, respectively (Supplementary Figure S4C; Foti et al, 2001; Parrish et al, 2004). We observed that PtdIns(4)P levels were slightly elevated (1.5-fold) in cells lacking Opy1 (opy1Δ cells; Supplementary Table S1). It was therefore possible that Opy1 could activate a PIP 4-phosphatase such as the Sac1 enzyme (Foti et al, 2001; Stefan et al, 2011), and thus loss of Opy1 could lead to elevated levels of both PtdIns(4)P and PtdIns(4,5)P2. However, PtdIns(4,5)P2 levels were increased (1.8-fold) in sac1Δ opy1Δ double-mutant cells, as compared with sac1Δ single-mutant cells (Supplementary Figure S4D; Supplementary Table S1). Thus, coupled with our co-IP and colocalization results, we propose that Opy1 may inhibit Mss4 activity at PIK patches rather than activate a PIP phosphatase, such as the synaptojanins or Sac1.
Opy1 is a coincidence detector of PtdIns(4,5)P2 and Mss4
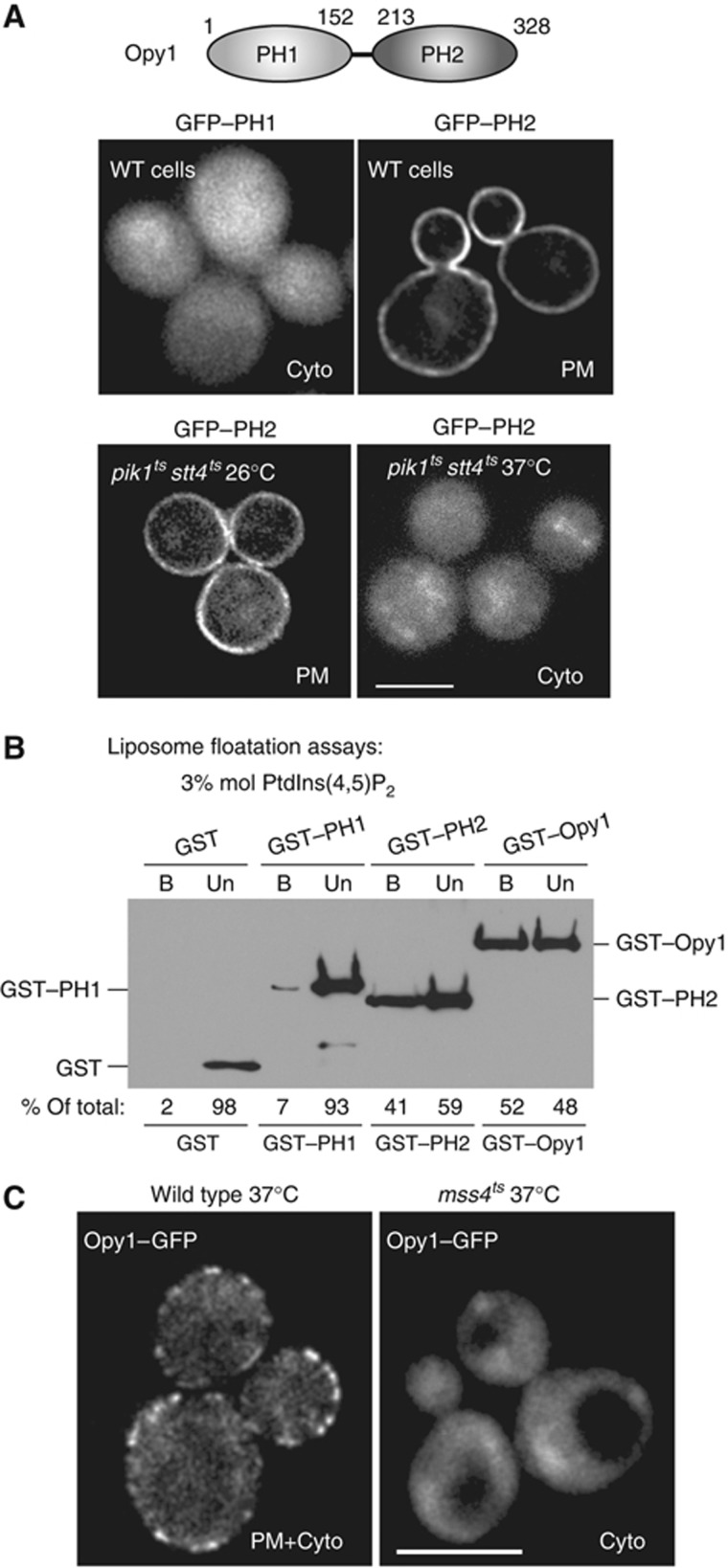
As Opy1 consists of two PH domains, we next addressed how Opy1 localizes to PIK patches at the PM. The N-terminal PH domain of Opy1 fused to GFP (GFP–PH1) did not localize to the PM in vivo (Figure 5A; Yu et al, 2004). In contrast, the C-terminal PH domain fused to GFP (GFP–PH2) was sufficient for PM targeting (Figure 5A; Yu et al, 2004). Similar to a previous study, the GFP–PH2 fusion still localized to the PM in mss4ts cells at the non-permissive temperature (Yu et al, 2004; our unpublished observations). However, we found that GFP–PH2 became mis-localized from the PM to the cytoplasm in pik1tsstt4ts double-mutant cells upon an extended shift to 37°C (Figure 5A). A previous study reported that GFP–PH2 localized to the PM in pik1tsstt4ts mutant cells (Yu et al, 2004). However, GFP–PH2 was overexpressed from a high copy plasmid in this study. In addition, Yu et al found that overexpression of the PH2 domain caused growth defects in pik1tsstt4ts cells, suggesting that the PH2 domain may compete for some factor that becomes limiting in these cells. As pik1tsstt4ts double-mutant cells have significantly reduced levels of PtdIns(4)P, PtdIns(4,5)P2, and Mss4 PIK patches at the PM (Audhya et al, 2000; Figure 2C in this study), we further addressed the lipid and protein binding activities for full-length Opy1 and the Opy1 PH domains.
Figure 5.
The Opy1 PH2 domain is sufficient for PM targeting in vivo and PtIns(4,5)P2-binding in vitro. (A) Top panels: Localization of GFP-tagged Opy1 PH1 or PH2 domains in wild-type cells. Wild-type cells expressing GFP–PH1 or GFP–PH2 were grown to mid-log and examined by fluorescence microscopy. Bottom panels: Localization of GFP-tagged Opy1 PH2 domain in cells with impaired PtdIns 4-kinase activity. Double-mutant pik1tsstt4ts cells expressing GFP–PH2 were grown to mid-log at the permissive temperature (26°C) and examined by fluorescence microscopy at 26°C and following a 1-h incubation at the restrictive temperature (37°C). Scale bar, 3 μm. Cyto, cytoplasm. (B) Full-length Opy1 and the Opy1 PH2 domain bind PtdIns(4,5)P2-containing liposomes in vitro. Recombinant GST, GST–PH1, GST–PH2, and GST–Opy1 fusion proteins were expressed, purified from bacteria, and incubated with PC:PtdIns(4,5)P2-containing liposomes (0.3 mM total lipid, 3% mol PtdIns(4,5)P2). Liposomes were separated from unbound protein by floatation on Nycodenz equilibrium gradients (see Materials and methods). GST fusion proteins in unbound (Un) and liposome-bound (B) fractions were detected by immunoblotting with GST antisera. (C) Opy1 PM targeting is dependent on PtdIns(4,5)P2 synthesis. Opy1–GFP localization in wild-type and mss4ts cells incubated at 37°C. Cells were grown at 26°C to mid-log phase, shifted to 37°C for 60 min, and examined by fluorescence microscopy. Scale bar, 5 μm. Cyto, cytoplasm. Figure source data can be found with the Supplementary data.
First, we examined whether Opy1 bound PtdIns(4,5)P2-containing liposomes in vitro. For these experiments, we used GST, GST–PH1, GST–PH2, and GST–Opy1 fusion proteins purified from bacteria. GST–Opy1 (52% of the total protein) efficiently bound and floated with PC:PtdIns(4,5)P2-containing liposomes (3% mol PtdIns4,5P2) on an equilibrium density gradient (Figure 5B). Likewise, GST–PH2 (41% of the total protein) was present with liposomes following equilibrium density fractionation, suggesting that the PH2 domain was sufficient for lipid binding (Figure 5B). In control experiments employing liposome sedimentation assays, neither GST–Opy1 nor GST–PH2 bound liposomes lacking PtdIns(4,5)P2 (e.g., PC:PtdIns-containing liposomes; Supplementary Figure S5A). GST alone or the GST–PH1 fusion did not efficiently bind liposomes (Figure 5B; Supplementary Figure S5).
Next, we examined the PIP-binding specificities of Opy1 and the PH2 domain in liposome sedimentation experiments. GST–Opy1 (32% of the total protein) specifically bound to liposomes containing 3% PtdIns(4,5)P2, but not to liposomes containing equal concentrations of other PIP isoforms (PtdIns(3)P, PtdIns(4)P, or PtdIns(3,5)P2; Supplementary Figure S5B). The GST–PH2 fusion sedimented with PtdIns(4,5)P2- and PtdIns(4)P-containing liposomes (30% and 18% of the total protein, respectively; Supplementary Figure S5B), but not with PtdIns(3)P- or PtdIns(3,5)P2-containing liposomes (2 and 1% of the total protein, respectively; Supplementary Figure S5B). As expected, GST alone or the GST–PH1 fusion did not efficiently sediment with PIP-containing liposomes (Supplementary Figure S5). PtdIns(4)P binding by the PH2 domain may have resulted in the Pik1- and Stt4-dependent PM localization of the GFP–PH2 fusion in vivo (Figure 5A). However, unlike the PH2 domain alone, full-length Opy1 specifically bound PtdIns(4,5)P2in vitro. To confirm whether full-length Opy1 localizes to the PM in a PtdIns(4,5)P2-dependent fashion, we examined GFP–Opy1 localization in mss4ts cells that have reduced levels of PtdIns(4,5)P2 at the restrictive temperature (Stefan et al, 2002). The PM localization of full-length Opy1–GFP was reduced in mss4ts cells shifted to the restrictive temperature but not in wild-type cells (Figure 5C), suggesting that the recruitment of Opy1 to Mss4 PIK patches was dependent on PtdIns(4,5)P2 synthesis.
Since the Mss4 N-terminal domain attenuates PtdIns(4,5)P2 synthesis (Figure 2D), we addressed whether it regulates Opy1 function. Strikingly, Opy1–GFP localized diffusely in the cytoplasm in cells expressing only the N-terminal truncated form of Mss4 (mss4Δ2–346 cells; Figure 6A). To test if the Mss4 N-terminal region was necessary for the Mss4–Opy1 interaction, we performed crosslinking co-IP experiments using either full-length or N-terminally truncated Mss4. As in our previous results (Figure 3C), Opy1 copurified with full-length Mss4 (Figure 6B). However, Opy1 did not interact with the mutant form of Mss4 lacking its N-terminal region (Figure 6B), even though this truncated Mss4 localized to the PM and was functional (Figure 2A and B). Therefore, the Mss4 N-terminal region may serve as a scaffold for Opy1 recruitment to PIK patches. Consistent with this, overexpression of OPY1 in mss4Δ2–346 cells did not result in a reduction in PtdIns(4,5)P2 levels (Figure 6C; Supplementary Table S1). Together, these results suggest that the tandem PH domain protein Opy1 serves dual roles as a coincidence detector of PtdIns(4,5)P2 and Mss4 as well as a regulator of PtdIns(4,5)P2 synthesis at the PM (Supplementary Figure S6C).
Figure 6.
Opy1 PM targeting and function is dependent on the Mss4 N-terminus. (A) Opy1–GFP localization in mss4Δ cells expressing either full-length Mss4 or the N-terminally truncated Mss4. Scale bar, 5 μm. Cyto, cytoplasm. (B) Opy1–GFP crosslinks and coimmunoprecipitates with Mss4–3HA but not with N-terminally truncated Mss4–3HA (lacking residues 2–346). Lysates from cells expressing Opy1–GFP, Opy1–GFP and Mss4–3HA, or Opy1–GFP and Mss4Δ2–346–3HA were incubated with crosslinker and incubated with anti-HA beads. Immunoprecipitates were analysed by immunoblotting to detect Mss4–Opy1 interactions. (C) Cellular PtdIns(4,5)P2 levels measured by 3H-inositiol labelling and HPLC analysis for wild-type cells and mss4Δ2–346 cells expressing only the N-terminally truncated form Mss4 carrying a high-copy OPY1 plasmid or vector alone. Three independent labelling experiments were performed at 26°C. Data represent the mean (±s.d.). *P=1 indicates that there is no statistical difference between sample averages. (D) GST–Opy1, GST–PH1, and GST–PH2 bind to Mss4–3HA from yeast cell lysates. GST alone does not bind to Mss4–3HA. Figure source data can be found with the Supplementary data.
To further dissect the functions of the two PH domains in Opy1, we overexpressed either the PH1 domain or the PH2 domain in sjl1Δ sjl2tssjl3Δ cells. Neither of the PH domains was sufficient to complement the growth defect of sjl1Δ sjl2tssjl3Δ cells at the non-permissive temperature (Figure 4D), suggesting they may function together to regulate PtdIns(4,5)P2 synthesis. However, to test whether the PH1 domain was functional when artificially targeted to the PM, we fused a palmitoylation motif (from Psr1) to the N-terminus of the PH1 domain, which resulted in its efficient targeting to the PM (Supplementary Figure S6A). Interestingly, this fusion partially rescued the growth defect of sjl1Δ sjl2tssjl3Δ cells (Supplementary Figure S6B). These results predicted that both PH1 and PH2 might interact independently with Mss4 and have distinct roles in PIK patch targeting and Mss4 PIP 5-kinase regulation. To test this idea, cell lysates containing Mss4–3xHA were incubated with GST, GST–PH1, GST–PH2, or GST–Opy1 fusion proteins bound to glutathione sepharose beads. As expected, full-length GST–Opy1 pulled down Mss4–3xHA from cell lysates but GST alone did not (Figure 6D) Interestingly, Mss4–3xHA was isolated with both the GST–PH1 and GST–PH2 fusion proteins in pull-down experiments (Figure 6D). Thus, the PH2 domain may target Opy1 to the PM by binding PtdIns(4,5)P2 and Mss4, and the PH1 domain may serve as a negative regulator of PtdIns(4,5)P2 synthesis.
Mammalian TAPP1 rescues the growth defect of yeast synaptojanin mutant cells
Mammalian cells have several putative Opy1 homologues with tandem PH domains. Three members of this protein family, pleckstrin, TAPP1, and TAPP2, bind PtdIns(3,4)P2 through their PH2 domains (Figure 7A; Haslam et al, 1993; Dowler et al, 2000; Allam and Marshall, 2005; Edlich et al, 2005). To test whether these proteins have conserved functions in PIP signalling, we overexpressed pleckstrin or TAPP1 in sjl1Δ sjl2tssjl3Δ cells that accumulate toxic levels of PtdIns(4,5)P2. Intriguingly, TAPP1, but not pleckstrin, weakly rescued the growth defect of sjl1Δ sjl2tssjl3Δ cells at the non-permissive temperature (Figure 7B). And consistently, PtdIns(4,5)P2 levels were restored to normal in sjl1Δ sjl2tssjl3Δ cells upon overexpression of TAPP1 (Figure 7C). To address whether inhibition of PtdIns(4,5)P2 synthesis by TAPP1 occurred through Mss4–TAPP1 interactions, cell lysates containing GFP–TAPP1 were incubated with His6–SUMO–Mss41–346 or His6–SUMO–Mss4347–779 fusion proteins bound to nickel-charged agarose beads. Interestingly, both His6–SUMO–Mss41–346 and His6–SUMO–Mss4347–779 fusion proteins pulled down GFP–TAPP1 from cell lysates, but not GFP alone (Figure 7D). TAPP1 and TAPP2 downregulate insulin and PtdIns(3,4,5)P3 signalling upon generation of PtdIns(3,4)P2 by the PIP 5-phosphatase SHIP2 (Wullschleger et al, 2011). Our results suggests TAPP1 may inhibit PIP 5-kinase activity thus reducing PtdIns(4,5)P2 levels as substrate for the PIP 3-kinase (PI3K) isoforms that synthesize PtdIns(3,4,5)P3. Alternatively, TAPP1 could inhibit PI3K itself given the similarity between PIP kinases (Supplementary Figure S6D). Importantly, our results have identified conserved roles for this family of tandem PH domain proteins as monitors and regulators of PIP metabolism at the PM.
Figure 7.
TAPP1 rescues the growth defects of yeast synaptojanin mutant cells. (A) Schematic diagrams of the Opy1, TAPP1, TAPP2, and pleckstrin tandem PH domain proteins are shown. PH, pleckstrin homology domain; DEP, dishevelled/EGL-10/Pleckstrin homology domain. (B) Overexpression of full-length Opy1 or TAPP1, but not pleckstrin, rescues the growth defect of sjl1Δ sjl2tssjl3Δ cells at 38°C. Serial dilutions of sjl1Δ sjl2tssjl3Δ cells carrying empty vector or plasmids encoding Opy1, TAPP1, or pleckstrin as indicated were spotted onto –Ura plates to retain the plasmids and grown at either 26 or 38°C for 4 days. (C) Cellular PtdIns(4,5)P2 levels as measured by 3H-inositiol labelling and HPLC analysis in sjl1Δ sjl2tssjl3Δ cells carrying empty vector or TAPP1 plasmids. Two independent labelling experiments were performed at 38°C. Results from a representative experiment are shown. Additional data are provided in Supplementary Table S1. (D) His6–SUMO–Mss41–346 and His6–SUMO–Mss4347–779 bind to GFP–TAPP1 from yeast cell lysates. GFP alone does not bind to His6–SUMO–Mss41–346 and His6–SUMO–Mss4347–779. Inputs for His6–SUMO–Mss41–346 and His6–SUMO–Mss4347–779 for each binding reaction are shown (bottom).Figure source data can be found with the Supplementary data.
Discussion
PtdIns(4,5)P2 is essential for a variety of cellular processes and its distribution and abundance must be tightly controlled (Stefan et al, 2002). We demonstrate that Mss4 assembles into dynamic, oligomeric structures: PIK patches. The dynamic assembly and disassembly of Mss4 PIK patches may serve as a mechanism for localized synthesis of PtdIns(4,5)P2 at the PM. Furthermore, oligomerization of Mss4 provides a mechanism to precisely modulate multiple copies of PIP kinase activity, as needed, for localized increases in PtdIns(4,5)P2 synthesis through the recruitment of specific activators such as small GTPases and clathrin adaptors (Nakano-Kobayashi et al, 2007; Smaczynska-de et al, 2008). In addition, we found that the tandem PH domain protein, Opy1, associates with Mss4 at PIK patches and inhibits PtdIns(4,5)P2 synthesis. Thus, Mss4 PIP 5-kinase activity is attenuated by negative regulatory factors, such as Opy1, prior to the disassembly of PIK patches.
Mss4 PIK patches are distinct from other known cortical structures such as actin patches and eisosomes. The PtdIns 4-kinase Stt4 also oligomerizes at the PM to form PIK patches (Baird et al, 2008). However, Stt4 and Mss4 PIK patches are distinct structures and show unique characteristics (Audhya and Emr, 2002; Baird et al, 2008). Stt4 PIK patches are relatively static at the PM, with a lifetime on the order of 3 min (Baird et al, 2008). Unexpectedly, Mss4 PIK patches are highly dynamic structures undergoing rapid assembly/disassembly and lateral movements. The biological function of PIK patches is not completely clear. However, the assembly of Mss4 into PIK patches may provide spatial and temporal control of PtdIns(4,5)P2 synthesis necessary to coordinate the numerous processes regulated by PtdIns(4,5)P2 (e.g., polarized secretion, endocytosis, MAPK activation). Accordingly, individual PIK patches may perform specialized functions and have different compositions (distinct nucleation and regulatory factors). Consistent with this idea, mammalian PtdIns(4)P 5-kinase isoforms are differentially recruited to focal adhesion sites and clathrin-coated pits by talin and the μ2 AP-2 adaptor subunit, respectively (Ling et al, 2002; Krauss et al, 2006; Nakano-Kobayashi et al, 2007).
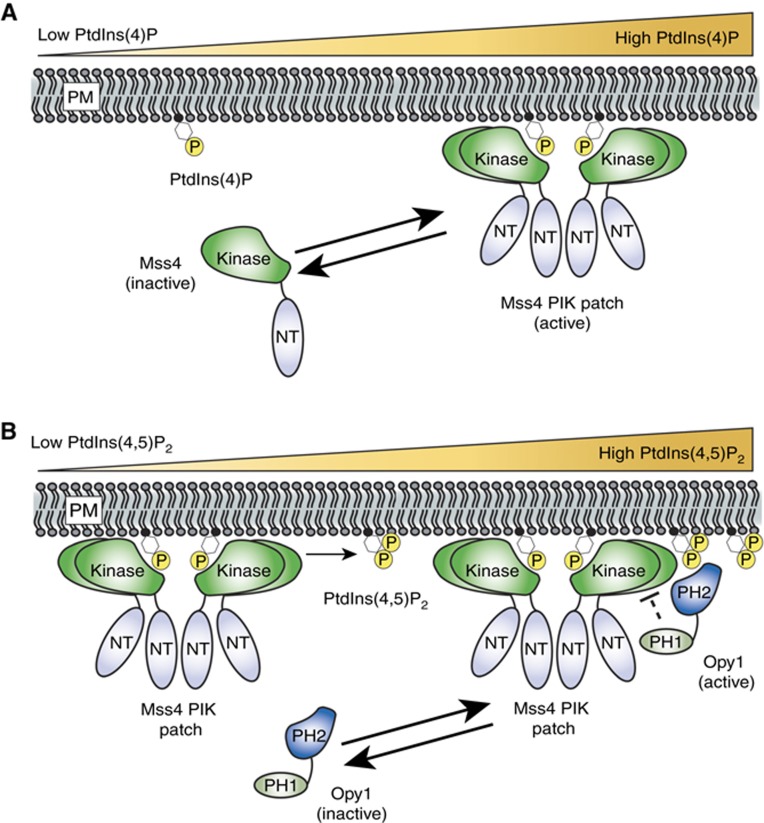
A striking feature of Mss4 PIK patches is their rapid assembly and disassembly. The assembly of Mss4 PIK patches involves the activation loop in the Mss4 lipid kinase domain and its substrate PtdIns(4)P at the PM (Figure 8). Although the kinase–substrate interaction is necessary for PIK patch formation, PtdIns(4)P is not sufficient to recruit Mss4 to membranes as Mss4 was not observed at Golgi compartments that are enriched in PtdIns(4)P (Audhya et al, 2000; Stefan et al, 2002). In addition, elevation of PtdIns(4)P levels (e.g., in mutant cells deficient in Sac1 PtdIns(4)P phosphatase activity) does not result in increased PtdIns(4,5)P2 levels (Foti et al, 2001; Stefan et al, 2011). Thus, there must be factors in addition to PtdIns(4)P that specify the PM localization and activity of Mss4. We speculate that the conversion of PtdIns(4)P to PtdIns(4,5)P2 by Mss4 may lead to PIK patch disassembly as PtdIns(4)P becomes locally depleted (Figure 8). This elegant mechanism would provide an intrinsic switch for the localized control of Mss4 assembly and disassembly reactions at the PM. An electrostatic switch mechanism has been proposed for the membrane association of a mammalian PIP 5-kinase during phagocytosis (Fairn et al, 2009). However, this process involves PIP turnover (by lipases and phosphatases) rather than inherent PIP kinase activity.
Figure 8.
A speculative model for Mss4 PIK patch dynamics and Opy1 function. (A) Above a threshold PtdIns(4)P concentration, Mss4 is recruited to its substrate at the PM where it oligomerizes and assembles into PIK patches. Localized depletion of PtdIns(4)P levels by Mss4 PIP kinase activity may promote the disassembly of Mss4 PIK patches. (B) Opy1 is in an inactive form when PtdIns(4,5)P2 levels are low. Upon binding PtdIns(4,5)P2, Opy1 is activated and recruited to Mss4 PIK patches in a negative regulatory feedback loop to shut down Mss4 PIP kinase activity. Opy1 may inhibit Mss4 PIP 5-kinase directly or indirectly through an unknown effector (indicated with the dashed line).
Using independent proteomic and genetic approaches, we identified the tandem PH domain protein Opy1 as a negative regulator of Mss4 PIP kinase activity. Our results show that Opy1 has dual functions as a PtdIns(4,5)P2 sensor and regulator. First, localization of Opy1 to cortical patches is dependent on PtdIns(4,5)P2 as well as the N-terminus of Mss4. In addition, overexpression or deletion of Opy1 results in mis-regulation of PtdIns(4,5)P2 synthesis at the PM. Full-length Opy1 protein is mostly cytoplasmic under conditions when PtdIns(4,5)P2 levels at the PM are low (e.g., mss4ts cells). Upon activation of PtdIns(4,5)P2 signalling in response to extracellular or intracellular cues, Opy1 may be initially recruited to the PM (Figure 8). In support of this, Opy1 directly binds PtdIns(4,5)P2-containing liposomes in vitro. We propose that Opy1 is recruited to PIK patches through its interaction with the Mss4 N-terminus to subsequently downregulate Mss4 PIP kinase activity. Consistent with this model, the N-terminal truncated form of Mss4 displays increased activity compared with full-length Mss4. Thus, together with PIP 5-phosphatases, Opy1 may be part of a rapid negative regulatory feedback system that is activated as PtdIns(4,5)P2 levels rise (Supplementary Figure S6C). Following attenuation of PtdIns(4,5)P2 synthesis and disassembly of Mss4 PIK patches, Opy1 may then disassociate from the PM.
Mammalian pleckstrin, TAPP1, and TAPP2 are tandem PH domain proteins that specifically bind PtdIns(3,4)P2in vitro (Haslam et al, 1993; Dowler et al, 2000; Allam and Marshall, 2005; Edlich et al, 2005). Intriguingly, TAPP1, TAPP2, and pleckstrin inhibit PI3K signalling (Abrams et al, 1996; Wullschleger et al, 2011). The inhibition of PI3Kγ activity by pleckstrin has been proposed to occur by direct interaction between PI3Kγ and pleckstrin, or alternatively through binding of pleckstrin to PI3Kγ activators, the βγ subunits of heterotrimeric GTP-binding proteins (Abrams et al, 1996). TAPP1 and TAPP2 may regulate PtdIns(3,4,5)P3 levels in cells by analogous mechanisms. Our results suggest yet another possible model for the control of PI3K signalling by dual PH domain proteins–inhibition of PIP 5-kinases and PtdIns(4,5)P2 synthesis that would impinge on PI3K activity (Supplementary Figure S6D). Though the PIP-binding specificities of pleckstrin, TAPP1, TAPP2, and Opy1 have diverged, they appear to share similar roles in attenuating PIP signalling. Diversity in PIP-binding specificities is consistent with a widespread role for this protein family in the regulation of multiple PIP kinases (e.g., PIP 3-kinase, 4-kinase, and 5-kinase activities). In support of this idea, we found that the PH2 domain of Opy1 bound PtdIns(4)P-containing liposomes in vitro. In addition, PtdIns(4)P levels were modestly increased in cells lacking Opy1. Thus, future studies may reveal new roles for Opy1 and other tandem PH domain proteins in the regulation of additional PIP kinase activities.
We propose that the C-terminal PH2 domains of these tandem PH domain proteins serve as PIP sensors, while the N-terminal PH1 domains regulate PIP kinases. Consistent with this, PH domains are involved in protein–protein interactions in addition to PIP binding (Wang et al, 1994; Yao et al, 1994). It remains unclear whether these tandem PH domain proteins directly inhibit PIP kinases or indirectly act through additional factors. Opy1 was originally identified as a regulator of GPCR and MAPK signalling in yeast (Edwards et al, 1997) and MAPK activation in yeast requires localized PtdIns(4,5)P2 signalling (Garrenton et al, 2010). Our study links the regulation of GPCR, MAPK, and PIP signalling through Opy1 function. It is therefore interesting that pleckstrin and TAPP1, TAPP2 control PI3K activity in response to GPCR and insulin receptor signalling. Thus, future studies on this family of dual PH domain proteins will likely provide new insight into our understanding of PIP signalling pathways.
Materials and methods
Additional details of the Materials and methods are provided in the Supplementary data.
Yeast strains and plasmid construction
A list of S. cerevisiae strains and plasmids used in this study and their genotypes can be found in Supplementary Tables S2 and S3. Homologous recombination was used to tag or delete genes in yeast (Longtine et al, 1998). All integrations and deletions were verified by PCR analysis and expression of fusion proteins was confirmed by western blot analysis. The yeast shuttle vectors used in this study have been previously described (Sikorski and Hieter, 1989).
Fluorescence microscopy and quantification of fluorescence intensity
Microscopy was performed using a fluorescence microscope (DeltaVision RT; Applied Precision) equipped with FITC and rhodamine filters. Images were captured with a digital camera (Cool Snap HQ; Photometrics) and deconvolved using softWoRx 3.5.0 software (Applied Precision). To quantify numbers of Mss4–GFP molecules in PIK patches in Figure 1C, cells coexpressing Mss4–GFP and Cse4–3xGFP were grown to mid-log phase. Cells were examined by fluorescence microscopy and undeconvolved data were used for quantification of intensities. Regions of interest (with identical areas) that contained a single Mss4 PIK patch or Cse4–3xGFP structure were selected and quantified using softWoRx 3.5.0 software (Applied Precision). Background signal was subtracted from measured fluorescence intensities of PIK patches and Cse4–GFP. Additional methods for comparing PIK patch intensities are described in the Supplementary data.
In-vivo analysis of PIPs
PIPs levels were analysed as previously described (Stefan et al, 2002; Baird et al, 2008). Briefly, 5 OD600 units of cells were labelled with 50 μCi of myo-[2-3H]-inositol (Perkin-Elmer) in media lacking inositol. After precipitation in 4.5% perchloric acid for 5 min, phospholipids were deacylated by incubation in methylamine reagent at 53°C. Samples were dried in a vacuum chamber, washed, dried again, and resuspended in 300 μl sterile water. Extraction reagent (1-butanol/ethyl-ether/formic acid ethyl ester at a ratio of 20:4:1) was added and [3H]glycero-PIPs were separated into the aqueous phase by vortexing and centrifugation at 14 000 g. The extraction was repeated twice and the final aqueous phase was collected and dried. Dried pellets were resuspended in water and separated on a Partisphere SAX column (Whatman, Florham Park, NJ) attached to a Shimadzu HPLC system (Shimadzu, Kyoto, Japan) and a 610TR on-line radiomatic detector (Perkin-Elmer, Waltham, MA) using Ultima Flo scintillation fluid (Perkin-Elmer). The HPLC and on-line detector were controlled with EZStart 7.2.1 and ProFSA 3.3 software, respectively, and data were analysed using ProFSA 3.3 software.
Analysis of cellular protein expression levels
In all, 5 OD600 equivalents of mid-log cells were harvested by precipitation in 10% trichloroacidic acid (TCA). Precipitates were washed in acetone, aspirated, resuspended in lysis buffer (150 mM NaCl, 50 mM Tris pH 7.5, 1 mM EDTA, 1% SDS), and mechanically lysed with glass beads. Protein sample buffer (150 mM Tris pH 6.8, 6 M Urea, 6% SDS, 10% β-mercaptoethanol, 20% Glycerol) was added and extracts were analysed by SDS–PAGE and immunoblotting with anti-G6PDH (Sigma) and anti-GFP (Santa Cruz Biotechnology) antibodies.
In-vivo crosslinking and coimmunoprecipitation
Yeast cells expressing epitope-tagged proteins were grown to an OD600 of 0.5–0.8. Cells were harvested and spheroplasted with Zymolyase 100T (Seikagaku Biobusiness). Spheroplasts were washed and crosslinked by 1 mM DSP (Pierce) in crosslinking buffer (20 mM HEPES, 0.7 M Sorbitol, 100 mM KOAc pH=7.4) for 30 min. In all, 100 mM Tris–HCl (pH=7.5) was added to the crosslinking buffer to stop the reaction. Spheroplasts were lysed by vortexing with glass beads in Urea cracking buffer (6 M Urea, 50 mM Tris–HCl, 1 mM EDTA, 1% SDS, pH=7.5). Cell lysates were diluted in Tris IP buffer (50 mM Tris–HCl, 150 mM NaCl, 5 mM EDTA, 0.5% Tween-20, pH=7.5 with protease inhibitors added) and cleared by centrifugation at 13 000 g. Immunoprecipitations were performed by adding either HA or Flag affinity gel (Sigma-Aldrich) to the supernatant at 4°C. The affinity matrix was washed with IP buffer four times. Immunoprecipitated proteins were eluted and reduced with elution buffer (100 mM Tris–HCl pH=8.0, 1% SDS, 10 mM DTT) at 65°C. Elutes were mixed with sample buffer (6 M Urea, 150 mM Tris–HCl pH=6.8, 6% SDS, Bromophenol Blue) and analysed by SDS–PAGE and immunoblotting.
SILAC and quantitative mass spectrometry analysis
For quantitative mass spectrometry analysis using SILAC (Stable isotope labelled amino acids in cell culture), yeast strains auxotrophic for lysine and arginine were grown to mid-log phase in either heavy or light lysine and arginine isotopes. Following affinity purification, light and heavy elutes were mixed, reduced with 10 mM DTT and alkylated with 20 mM iodoacetimide (Sigma-Aldrich). Proteins were precipitated by adding (3:1) 50% acetone, 49.9% ethanol, and 0.1% acetic acid, resuspended in 8 M urea, 50 mM Tris–HCl pH=8.0, diluted 3:1 with water and digested with 1 μg of trypsin overnight. Tryptic peptides were fractionated by hydrophilic interaction chromatography (HILIC), dried, and reconstituted in 0.1% trifluoroacetic acid. Each fraction was analysed by LC–MS/MS using an Orbitrap XL mass spectrometer (Thermo). Peptide analysis and SILAC quantitation was performed using Sorcerer software.
Recombinant protein expression and purification
The bacterial expression vectors pGEX6P-1 (GE Healthcare) and ppSUMO (Sondermann Lab) were used to generate recombinant fusion proteins. C41 (DE3) cells (Lucigen) transformed with protein expression plasmids were grown at 37°C to an OD600 of 0.4. The bacteria were then shifted to 25°C and protein expression was induced by the addition of 0.1 mM IPTG for 16 h. GST and His6 fusion proteins were purified from C41 (DE3) cells with glutathione sepharose 4B (GE Healthcare) and Ni-NTA agarose (Qiagen) according to the manufacture's instructions. Protein concentrations were determined by the Bradford assay (Bio-Rad). Purified proteins were stored at −80°C in phosphate-buffered saline (PBS) or in storage buffer (PBS, 50% glycerol) until further use.
Lipid-binding assays
For liposome flotation assays, 97%PC: 3%PtdIns(4,5)P2 (Avanti Polar Lipids) liposomes were generated by dehydration in a chloroform:methanol mix using a speed vacuum centrifuge, and then rehydrated in PBS with 1 mM DTT at a final concentration of 1.2 mM total lipid. To create unilamellar liposomes, the liposomes were sonciated for 1 min at room temperature in a water bath. Liposomes (1.2 mM) were then mixed with an equal volume (50 μl) of GST fusion proteins (resulting in 1.7 μM final protein concentration and 0.6 mM final lipid concentration) and allowed to incubate for 30 min at room temperature. Samples were then resuspended in 200 μl buffer (PBS, 1 mM DTT) containing 30% Optiprep density gradient medium (Sigma) and pipetted into thick walled 2.2 ml centrifuge tubes. In all, 150 μl of 15% Optiprep buffer was overlaid over the samples, followed by 50 μl of buffer (PBS, 1 mM DTT). Samples were centrifuged for 1 h at 55 000 r.p.m. at 10°C in a TLS-55 ultracentrifuge rotor in desktop ultracentrifuge (Beckman Coulter). The top 200 μl of the gradient containing liposomes and bound proteins was then collected as the ‘bound’ fraction. The bottom 200 μl ‘unbound’ fraction was collected separately. Samples were resuspended in protein sample buffer, run on 10% SDS–PAGE gels, and GST fusion proteins were detected by immunoblotting using GST antisera. The relative amounts of fusion proteins in bound and unbound fractions were determined using NIH ImageJ software.
For liposome sedimentation assays, 100%PC, 97%PC: 3%PtdIns and 97%PC:3%PIP-containing liposomes were prepared as described above except that the final lipid concentration was 0.3 mM, incubated with 1.7 μM recombinant GST, GST–Opy1, GST–PH1, or GST–PH2 fusion proteins for 30 min at room temperature, and centrifuged at 13 000 g for 20 min at 4°C. The resulting supernatant and pellet fractions were prepared for SDS–PAGE analysis and Coomassie stained to detect recombinant proteins. The gels were scanned and relative amounts of fusion proteins in pellet (bound) and in supernatant (unbound) fractions were determined using LI-COR software.
Methods for lipid overlay assays are described in the Supplementary data.
Protein-binding assays
For Mss4–3xHA and GST–Opy1 binding, GST, GST–Opy1, GST–PH1, or GST–PH2 fusion proteins immobilized on glutathione sepharose 4B were incubated with Mss4–3xHA lysates solubilized in Tris IP buffer (50 mM Tris–HCl, 150 mM NaCl, 5 mM EDTA, 0.5% Tween-20, pH=7.5, with protease inhibitors added) for 1 h at 4°C. Beads were washed with IP buffer three times and elutes were analysed by SDS–PAGE and immunoblotting using anti-HA antibodies. Similarly, For Mss4 and TAPP1 binding, His6–SUMO–Mss41–346 and His6–SUMO–Mss4347–779 proteins immobilized on Ni-NTA beads were incubated with cell lysates expressing GFP–TAPP1 or GFP alone in Tris IP buffer for 1 h at 4°C. Beads were washed with IP buffer three times and elutes were analysed by SDS–PAGE and immunoblotting using GFP antisera.
Supplementary Material
Acknowledgments
We thank Haiyuan Yu for providing the TAPP1 and pleckstrin plasmids. We thank Qi Wang and Holger Sondermann for providing the His6–SUMO vector. We thank the members of the SDE laboratory, David Holowka, Yuxin Mao, Marcus Smolka, and John York (Duke University) for helpful discussions and comments on the manuscript. We are grateful to Marcus Smolka for assistance with the quantitative mass spectrometry experiments. This work was supported by funds from the Weill Institute for Cell and Molecular Biology and a Cornell University Research Grant (to SDE) and by a Sam and Nancy Fleming Research Fellowship (to JAM).
Author contributions: YL, CJS, and SDE designed the experiments; YL and CJS performed the experiments; JAM performed the quantitative mass spectrometry analysis; AA isolated OPY1 in a genetic screen; YL, CJS, and SDE analysed the data; YL, CJS, and SDE wrote the manuscript. All authors discussed the results and contributed to the final manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abrams CS, Zhang J, Downes CP, Tang X, Zhao W, Rittenhouse SE (1996) Phosphopleckstrin inhibits gbetagamma-activable platelet phosphatidylinositol-4,5-bisphosphate 3-kinase. J Biol Chem 271: 25192–25197 [DOI] [PubMed] [Google Scholar]
- Allam A, Marshall AJ (2005) Role of the adaptor proteins Bam32, TAPP1 and TAPP2 in lymphocyte activation. Immunol Lett 97: 7–17 [DOI] [PubMed] [Google Scholar]
- Audhya A, Emr SD (2002) Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev Cell 2: 593–605 [DOI] [PubMed] [Google Scholar]
- Audhya A, Emr SD (2003) Regulation of PI4,5P2 synthesis by nuclear-cytoplasmic shuttling of the Mss4 lipid kinase. EMBO J 22: 4223–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Foti M, Emr SD (2000) Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell 11: 2673–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird D, Stefan C, Audhya A, Weys S, Emr SD (2008) Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J Cell Biol 183: 1061–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R, Munro S (2005) Organelle identity and the signposts for membrane traffic. Nature 438: 597–604 [DOI] [PubMed] [Google Scholar]
- Bertin A, McMurray MA, Thai L, Garcia G 3rd, Votin V, Grob P, Allyn T, Thorner J, Nogales E (2010) Phosphatidylinositol-4,5-bisphosphate promotes budding yeast septin filament assembly and organization. J Mol Biol 404: 711–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrivieres S, Cooke FT, Parker PJ, Hall MN (1998) MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J Biol Chem 273: 15787–15793 [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657 [DOI] [PubMed] [Google Scholar]
- Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR (2000) Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem J 351(Part 1): 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlich C, Stier G, Simon B, Sattler M, Muhle-Goll C (2005) Structure and phosphatidylinositol-(3,4)-bisphosphate binding of the C-terminal PH domain of human pleckstrin. Structure 13: 277–286 [DOI] [PubMed] [Google Scholar]
- Edwards MC, Liegeois N, Horecka J, DePinho RA, Sprague GF Jr, Tyers M, Elledge SJ (1997) Human CPR (cell cycle progression restoration) genes impart a Far- phenotype on yeast cells. Genetics 147: 1063–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairn GD, Ogata K, Botelho RJ, Stahl PD, Anderson RA, De Camilli P, Meyer T, Wodak S, Grinstein S (2009) An electrostatic switch displaces phosphatidylinositol phosphate kinases from the membrane during phagocytosis. J Cell Biol 187: 701–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti M, Audhya A, Emr SD (2001) Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell 12: 2396–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrenton LS, Stefan CJ, McMurray MA, Emr SD, Thorner J (2010) Pheromone-induced anisotropy in yeast plasma membrane phosphatidylinositol-4,5-bisphosphate distribution is required for MAPK signaling. Proc Natl Acad Sci USA 107: 11805–11810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam RJ, Koide HB, Hemmings BA (1993) Pleckstrin domain homology. Nature 363: 309–310 [DOI] [PubMed] [Google Scholar]
- Haucke V (2005) Phosphoinositide regulation of clathrin-mediated endocytosis. Biochem Soc Trans 33(Part 6): 1285–1289 [DOI] [PubMed] [Google Scholar]
- Hokin LE (1985) Receptors and phosphoinositide-generated second messengers. Annu Rev Biochem 54: 205–235 [DOI] [PubMed] [Google Scholar]
- Homma K, Terui S, Minemura M, Qadota H, Anraku Y, Kanaho Y, Ohya Y (1998) Phosphatidylinositol-4-phosphate 5-kinase localized on the plasma membrane is essential for yeast cell morphogenesis. J Biol Chem 273: 15779–15786 [DOI] [PubMed] [Google Scholar]
- James DJ, Khodthong C, Kowalchyk JA, Martin TF (2008) Phosphatidylinositol 4,5-bisphosphate regulates SNARE-dependent membrane fusion. J Cell Biol 182: 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss M, Kukhtina V, Pechstein A, Haucke V (2006) Stimulation of phosphatidylinositol kinase type I-mediated phosphatidylinositol (4,5)-bisphosphate synthesis by AP-2mu-cargo complexes. Proc Natl Acad Sci USA 103: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J, Wilson MP, Kisseleva M, Hurley JH, Majerus PW, Anderson RA (2000) The activation loop of phosphatidylinositol phosphate kinases determines signaling specificity. Mol Cell 5: 1–11 [DOI] [PubMed] [Google Scholar]
- Lemmon MA (2008) Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol 9: 99–111 [DOI] [PubMed] [Google Scholar]
- Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA (2002) Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature 420: 89–93 [DOI] [PubMed] [Google Scholar]
- Liu J, Zuo X, Yue P, Guo W (2007) Phosphatidylinositol 4,5-bisphosphate mediates the targeting of the exocyst to the plasma membrane for exocytosis in mammalian cells. Mol Biol Cell 18: 4483–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan MR, Mandato CA (2006) Regulation of the actin cytoskeleton by PIP2 in cytokinesis. Biol Cell 98: 377–388 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Markus SM, Punch JJ, Lee WL (2009) Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr Biol 19: 196–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TF (1998) Phosphoinositide lipids as signaling molecules: common themes for signal transduction, cytoskeletal regulation, and membrane trafficking. Annu Rev Cell Dev Biol 14: 231–264 [DOI] [PubMed] [Google Scholar]
- Nakano-Kobayashi A, Yamazaki M, Unoki T, Hongu T, Murata C, Taguchi R, Katada T, Frohman MA, Yokozeki T, Kanaho Y (2007) Role of activation of PIP5Kgamma661 by AP-2 complex in synaptic vesicle endocytosis. EMBO J 26: 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish WR, Stefan CJ, Emr SD (2004) Essential role for the myotubularin-related phosphatase Ymr1p and the synaptojanin-like phosphatases Sjl2p and Sjl3p in regulation of phosphatidylinositol 3-phosphate in yeast. Mol Biol Cell 15: 3567–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A (2000a) Polarization of cell growth in yeast. J Cell Sci 113(Part 4): 571–585 [DOI] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A (2000b) Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J Cell Sci 113(Part 3): 365–375 [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczynska-de R II, Costa R, Ayscough KR (2008) Yeast Arf3p modulates plasma membrane PtdIns(4,5)P2 levels to facilitate endocytosis. Traffic 9: 559–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ, Audhya A, Emr SD (2002) The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol Biol Cell 13: 542–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD (2011) Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 144: 389–401 [DOI] [PubMed] [Google Scholar]
- Sun Y, Carroll S, Kaksonen M, Toshima JY, Drubin DG (2007) PtdIns(4,5)P2 turnover is required for multiple stages during clathrin- and actin-dependent endocytic internalization. J Cell Biol 177: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentpetery Z, Balla A, Kim YJ, Lemmon MA, Balla T (2009) Live cell imaging with protein domains capable of recognizing phosphatidylinositol 4,5-bisphosphate; a comparative study. BMC Cell Biol 10: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicinanza M, D'Angelo G, Di Campli A, De Matteis MA (2008) Function and dysfunction of the PI system in membrane trafficking. EMBO J 27: 2457–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Brickner JH, Aguilar PS, Bernales S, Pantoja C, Walter P (2006) Eisosomes mark static sites of endocytosis. Nature 439: 998–1003 [DOI] [PubMed] [Google Scholar]
- Wang DS, Shaw R, Winkelmann JC, Shaw G (1994) Binding of PH domains of beta-adrenergic receptor kinase and beta-spectrin to WD40/beta-transducin repeat containing regions of the beta-subunit of trimeric G-proteins. Biochem Biophys Res Commun 203: 29–35 [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Wasserman DH, Gray A, Sakamoto K, Alessi DR (2011) Role of TAPP1 and TAPP2 adaptor binding to PtdIns(3,4)P2 in regulating insulin sensitivity defined by knock-in analysis. Biochem J 434: 265–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Kawakami Y, Kawakami T (1994) The pleckstrin homology domain of Bruton tyrosine kinase interacts with protein kinase C. Proc Natl Acad Sci USA 91: 9175–9179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin HL, Janmey PA (2003) Phosphoinositide regulation of the actin cytoskeleton. Annu Rev Physiol 65: 761–789 [DOI] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA (2004) Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell 13: 677–688 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.