Abstract
The small GTPase RhoG plays a central role in actin remodelling during diverse biological processes such as neurite outgrowth, cell migration, phagocytosis of apoptotic cells, and the invasion of pathogenic bacteria. Although it is known that RhoG stimulates neurite outgrowth in the rat pheochromocytoma PC12 cell line, neither the physiological function nor the regulation of this GTPase in neuronal differentiation is clear. Here, we identify RhoG as an inhibitor of neuronal process complexity, which is regulated by the microRNA miR-124. We find that RhoG inhibits dendritic branching in hippocampal neurons in vitro and in vivo. RhoG also inhibits axonal branching, acting via an ELMO/Dock180/Rac1 signalling pathway. However, RhoG inhibits dendritic branching dependent on the small GTPase Cdc42. Finally, we show that the expression of RhoG in neurons is suppressed by the CNS-specific microRNA miR-124 and connect the regulation of RhoG expression by miR-124 to the stimulation of neuronal process complexity. Thus, RhoG emerges as a cellular conductor of Rac1 and Cdc42 activity, in turn regulated by miR-124 to control axonal and dendritic branching.
Keywords: Cdc42, ELMO–Dock180–Rac1 signalling, miR-124, neuronal process complexity, RhoG
Introduction
MicroRNAs, a class of small noncoding RNAs that posttranscriptionally regulate gene expression, are likely to have key roles in neuronal development and plasticity (Schratt et al, 2006; Fineberg et al, 2009; Gao, 2010). In particular, the nervous system-specific microRNA miR-124 has been pointed out to promote neuronal differentiation at the transcriptional as well as the posttranscriptional level. miR-124 contributes to the regulation of chromatin assembly and accessibility by facilitating a switch of BAF53 subunits in ATP-dependent chromatin-remodelling complexes (Yoo et al, 2009). This switching of subunits relieves the repression of BAF53b, which is critical for activity-dependent dendritic outgrowth.
Additionally, at the level of transcription, miR-124 inhibits the NRSF/REST complex, a global repressor of nervous system-specific transcription, by targeting the expression of the SCP1 phosphatase, which in turn is implicated in the function of the NRSF/REST complex (Visvanathan et al, 2007). Posttranscriptionally, miR-124 stimulates neuronal differentiation by downregulating the expression of the RNA-binding protein PTBP1, inducing a switch towards nervous system-specific alternative splicing (Makeyev et al, 2007). Consequently, miR-124 induces neurite outgrowth in the CNS catecholaminergic CAD cell line. Furthermore, miR-124 reduces the expression of several transcription factors including Lhx2 and CREB1 either to prevent apoptosis in the developing retina and regulate proper axonal development of dentate gyrus granule neurons (Sanuki et al, 2011), or to constrain serotonin-induced synaptic facilitation (Rajasethupathy et al, 2009), respectively. In addition, miR-124 promotes adult neurogenesis in the subventricular zone stem cell niche by repressing the expression of the transcription factor Sox9 (Cheng et al, 2009).
By modulating chromatin-remodelling complexes, global transcriptional and alternative splicing regulators as well as transcription factors, miR-124 indirectly affects the expression of several target genes involved in neuronal differentiation (Gao, 2010). So far, little is known about miR-124 target interactions that directly control signalling pathways to define morphological characteristics of differentiated neurons such as axonal shape or dendritic tree complexity. We were interested in identifying novel target interactions of miR-124 that directly connect gene expression to a discernable phenotype related to neuronal process differentiation. Using the algorithms PicTar (Krek et al, 2005), miRanda (John et al, 2004), TargetScan (Lewis et al, 2005), and MiRtarget2 (Wang and El Naqa, 2008) for microRNA target prediction, we found the small GTPase RhoG as a strong candidate for miR-124-regulated gene expression. RhoG has been demonstrated to be critical for proper regulation of the actin cytoskeleton in several organisms and cell types: this GTPase mediates phagocytosis of apoptotic cells (deBakker et al, 2004), ICAM1-dependent endothelial cup assembly necessary for trans-endothelial migration of leucocytes (van Buul et al, 2007), and clathrin-independent T-cell receptor internalization and trogocytosis (Martínez-Martín et al, 2011). Furthermore, RhoG is involved in the invasion of pathogenic bacteria (Patel and Galán, 2006; Roppenser et al, 2009), and the migration of cancer cells (Hiramoto-Yamaki et al, 2010). In neural development, RhoG was demonstrated to promote progenitor cell proliferation in the cerebral cortex (Fujimoto et al, 2009). However, most of the previous research on the role of RhoG in neuronal differentiation has been based on experimental investigations using the rat pheochromocytoma PC12 cell line (Katoh et al, 2000; Estrach et al, 2002; Katoh and Negishi, 2003). In this cell line, RhoG stimulates neurite outgrowth depending on an active ELMO/Dock180 signalling module (Katoh and Negishi, 2003).
Here, we set out to explore the functions of RhoG in neuronal process differentiation in primary hippocampal neurons in culture and in the native brain in vivo. Our results demonstrate that RhoG inhibits axonal branching via the ELMO/Dock180/Rac1 signalling pathway. In addition, RhoG reduces dendritic tree complexity through the small GTPase Cdc42. We validate the predicted regulation of RhoG expression by the microRNA miR-124 in hippocampal neurons, and connect this regulation to the stimulation of neuronal process complexity.
Results
RhoG reduces dendritic tree complexity
Up to now it was found that RhoG stimulates neurite outgrowth in the rat pheochromocytoma PC12 cell line (Katoh et al, 2000; Estrach et al, 2002; Katoh and Negishi, 2003). This GTPase is expressed in brain tissue (Ishikawa et al, 2002) and in hippocampal neurons in culture (Supplementary Figure S1). To elucidate the function of RhoG in neuronal process elaboration in primary neurons, we performed loss-of-function and gain-of-function experiments in hippocampal neurons in cell culture and in the native mouse brain in vivo.
For loss-of-function experiments, we used the independent shRNA constructs RhoG-kd1 and RhoG-kd4 (Meller et al, 2008) to reduce the amount of RhoG endogenously expressed in hippocampal neurons. To assess the efficiency and specificity of these RhoG knockdown constructs, we performed three different experiments: (1) HA-tagged RhoG was co-expressed with the knockdown constructs RhoG-kd1 and RhoG-kd4, respectively, in HEK293 cells. Cell extracts were subjected to immunoblot analysis with an antibody directed to the HA tag. The quantification of the immunopositive bands confirmed that RhoG-kd4 very efficiently reduced the amount of overexpressed RhoG compared with the control (kdcontrol). Similarly, RhoG-kd1 significantly decreased RhoG expression, although to a lesser extent (Figure 1B). (2) Hippocampal neurons were transfected with eitherRhoG-kd1, RhoG-kd4, or the corresponding control construct. Two days after transfection, the neurons were stained with a monoclonal antibody directed to RhoG (Supplementary Figure S1). The quantification of the RhoG-specific immunopositive signals in neuronal cell bodies confirmed that both knockdown constructs, RhoG-kd1 and RhoG-kd4, significantly reduced the amount of endogenously expressed neuronal RhoG (Supplementary Figure S2A and B). (3) HA-tagged RhoG was co-expressed with the knockdown constructs RhoG-kd1 and RhoG-kd4, respectively, in hippocampal neurons. These neurons were stained with an antibody to the HA tag (Supplementary Figure S2C and E) and the immunopositive signals in the primary dendrites were quantified. RhoG-kd4 almost entirely prevented the expression of HA-tagged RhoG in hippocampal neurons (Supplementary Figure S2D). RhoG-kd1 significantly reduced the amount of overexpressed RhoG in neurons, but less efficiently than RhoG-kd4 (Supplementary Figure S2F). Finally, we co-expressed RhoG-kd4 together with the RhoG overexpression construct RhoGresist(kd4) harbouring two silent point mutations in the binding site for the RhoG-kd4-derived siRNA (see Materials and methods). When co-expressing RhoG-kd4 together with RhoGresist(kd4) in hippocampal neurons, RhoG-kd4 was not able to reduce the expression of RhoGresist(kd4) (Supplementary Figure S2C and D).
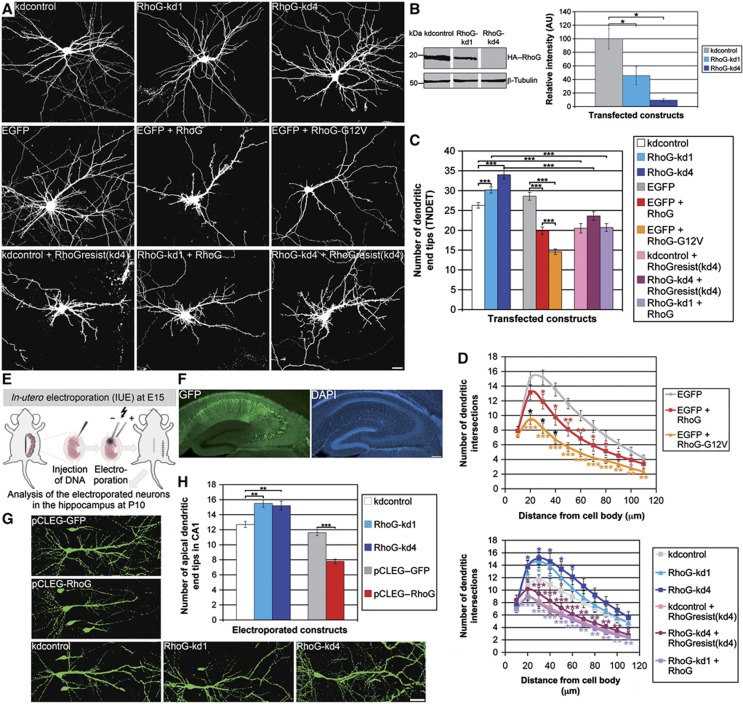
Figure 1.
RhoG reduces dendritic tree complexity. (A) The dendritic phenotype of hippocampal neurons, transfected with the indicated constructs at days in vitro 7 (DIV7), was analysed after staining for GFP at DIV7+3. Scale bar, 15 μm. (B) Immunoblotting for HA–RhoG expression showed that RhoG-kd1 and RhoG-kd4 reduced the amount of overexpressed HA–RhoG in HEK293 cells with RhoG-kd4 being more effective than RhoG-kd1. Immunoblotting for β-tubulin served as a loading control. The knockdown efficiency of both knockdown constructs was confirmed by quantifying the HA–RhoG-specific bands. Mean values (n=4 experiments)±s.e.m. (*P<0.05). (C) The shRNA constructs RhoG-kd1 and RhoG-kd4 increased, while RhoG and RhoG-G12V decreased the total number of dendritic end tips (TNDET). RhoGresist(kd4), like RhoG, reduced TNDET, and led to a rescue of TNDET when co-expressed with the stronger knockdown construct RhoG-kd4. Overexpression of RhoG was sufficient to rescue TNDET when co-expressed with the weaker knockdown construct RhoG-kd1. Mean values (n=80 neurons for all shRNA constructs, n=40 neurons for all overexpression constructs)±s.e.m. (***P<0.0005; *P<0.05). (D) Neurons transfected as described above were analysed by Sholl analysis for all experimental conditions. For each point of the Sholl graph, the mean values (n≥24)±s.e.m. are shown (***P<0.0005; **P<0.005; *P<0.05). Red stars, EGFP+RhoG compared with EGFP; orange stars, EGFP+RhoG-G12V compared with EGFP; black stars, EGFP+RhoG-G12V compared with EGFP+RhoG; dark blue stars, RhoG-kd4 compared with kdcontrol; light blue stars, RhoG-kd1 compared with kdcontrol; red brown stars, RhoG-kd4+RhoGresist(kd4) compared with RhoG-kd4; purple stars, RhoG-kd1+RhoG compared with RhoG-kd1. (E) Scheme of the experimental design for the in vivo analysis by IUE. (F) Example of a hippocampal slice with electroporated pyramidal neurons stained for GFP and DAPI. (G) Dendritic morphology of electroporated neurons was analysed at P10 after staining for GFP. The pCLEG vector (see Materials and methods) was used for gain-of-function experiments. Scale bar, 40 μm. (H) RhoG decreased, while RhoG-kd1 and RhoG-kd4 increased the number of apical dendritic end tips in CA1 pyramidal neurons. Mean values (n=40 neurons)±s.e.m. (***P<0.0005; **P<0.005 ).
We then employed the validated shRNA constructs RhoG-kd1 and RhoG-kd4 to investigate the function of RhoG in neuronal process elaboration. We found that knockdown of endogenous RhoG expression by RNA interference using the shRNA constructs RhoG-kd1 and RhoG-kd4 promoted dendritic tree complexity by increasing the total number of dendritic end tips (TNDET) of days in vitro 7 (DIV7) cultured primary hippocampal neurons (Figure 1A and C). Consistent with this finding, overexpression of either RhoG or the dominant-positive mutant RhoG-G12V reduced TNDET (Figure 1A and C). Next, a rescue experiment was performed using RhoGresist(kd4) to prove that the RhoG-kd4-mediated dendritic phenotype was caused specifically by a reduction of endogenously expressed RhoG. In fact, RhoGresist(kd4) decreased TNDET similar to RhoG even in the presence of co-expressed RhoG-kd4, which led to a depletion of endogenously expressed RhoG (Figure 1A and C). In the presence of the weaker knockdown construct RhoG-kd1 (Supplementary Figure S2D and F), RhoG function could be rescued simply by overexpression of RhoG (Figure 1A and C).
The relevance of RhoG for dendritic tree differentiation was further investigated by conducting Sholl analysis, which quantifies the number of times dendrites from a neuron cross concentric circles of increasing diameters around the neuronal cell body (Brandt et al, 2007). The Sholl analysis confirmed that overexpression of RhoG decreased dendritic tree complexity by reducing dendritic branching, while knockdown of endogenous RhoG increased dendritic branching (Figure 1D). Additionally, the results of the rescue experiments were corroborated. The Sholl analysis further indicated that the inhibitory effect of RhoG on dendritic tree complexity became manifest predominantly in the proximal to middle part of the dendritic tree.
To demonstrate the in vivo relevance of these results, we employed the in-utero electroporation (IUE) technique (Brandt et al, 2007) to elicit in mouse brains acute manipulations of the amount of functional RhoG in cornu ammonis 1 (CA1) pyramidal neurons (Figure 1E and F). Knockdown of endogenous RhoG expression by the respective shRNA constructs resulted in an increase in the number of apical dendritic end tips of CA1 pyramidal neurons due to enhanced dendritic branching. In line with this outcome, the number of apical dendritic end tips of CA1 neurons overexpressing RhoG was significantly reduced (Figure 1G and H). Together, we demonstrate that RhoG reduces dendritic tree complexity in primary neurons in culture and in the native mouse brain in vivo.
RhoG inhibits axonal branching
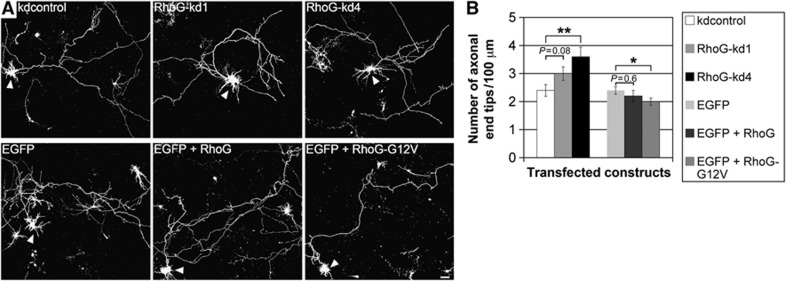
The expression of RhoG not only during dendritogenesis but additionally during axonogenesis (Supplementary Figure S1) suggests that this protein may also be involved in regulating axonal morphology. We verified this assumption by employing the same constructs as described above either to reduce endogenous RhoG expression or to overexpress wild-type RhoG as well as dominant-positive RhoG in DIV2 hippocampal neurons. In contrast to the weaker knockdown construct RhoG-kd1, the stronger knockdown construct RhoG-kd4 (Figure 1B; Supplementary Figure S2) significantly increased the number of axonal end tips/100 μm without having an effect on axonal length (Figure 2A and B; Supplementary Figure S3). Furthermore, the dominant-positive construct RhoG-G12V, but not wild-type RhoG led to a reduction in the number of axonal end tips/100 μm. As it was seen for the RhoG knockdown constructs, neither RhoG nor RhoG-G12V affected the axonal length compared with the EGFP control (Figure 2A and B; Supplementary Figure S3). The fact that only dominant-positive RhoG-G12V but not RhoG elicit a functional effect was also observed in another context. Namely it was found that RhoG-G12V, but not RhoG stimulates neural progenitor cell proliferation (Fujimoto et al, 2009). One possible explanation for this difference could be that RhoG-activating guanine nucleotide exchange factors (GEFs) are limiting during axonogenesis. To support this hypothesis experimentally, we expressed the deletion construct SGEFΔN, which is derived from the RhoG-activating GEF SGEF (SH3-containing GEF) in hippocampal neurons. This construct comprises the DH/PH module of SGEF harbouring the GEF activity for RhoG and a more C-terminal located SH3 domain conveying an intracellular targeting function (Ellerbroek et al, 2004). We observed that SGEFΔN significantly reduced axonal branching (Supplementary Figure S4). To ascertain whether SGEFΔN reduces axonal branching via activation of RhoG, SGEFΔN was co-expressed together with RhoG-kd4 in hippocampal neurons, and axonal branching was analysed. Knockdown of endogenous RhoG expression by RhoG-kd4 was able to rescue axonal branching under the condition SGEFΔN+RhoG-kd4 when compared with the corresponding control condition SGEFΔN+kdcontrol (Supplementary Figure S4). This finding suggests that a substantial amount of neuronal RhoG is inactive (particularly during RhoG overexpression) but could be activated by a GEF (e.g., SGEFΔN) to reduce axonal branching. Collectively, these results reveal that RhoG inhibits axonal branching.
Figure 2.
RhoG inhibits axonal branching. (A) The axonal morphology of hippocampal neurons, transfected with the indicated constructs at DIV2, was analysed after staining for GFP at DIV2+2 (arrowheads). Scale bar, 15 μm. (B) RhoG-kd4 increased, while RhoG-G12V decreased the number of axonal end tips. Mean values (n=80 neurons for EGFP and EGFP+RhoG-G12V, n=40 neurons for all other conditions)±s.e.m. (**P<0.005; *P<0.05).
RhoG inhibits axonal branching via ELMO/Dock180/Rac1 signalling
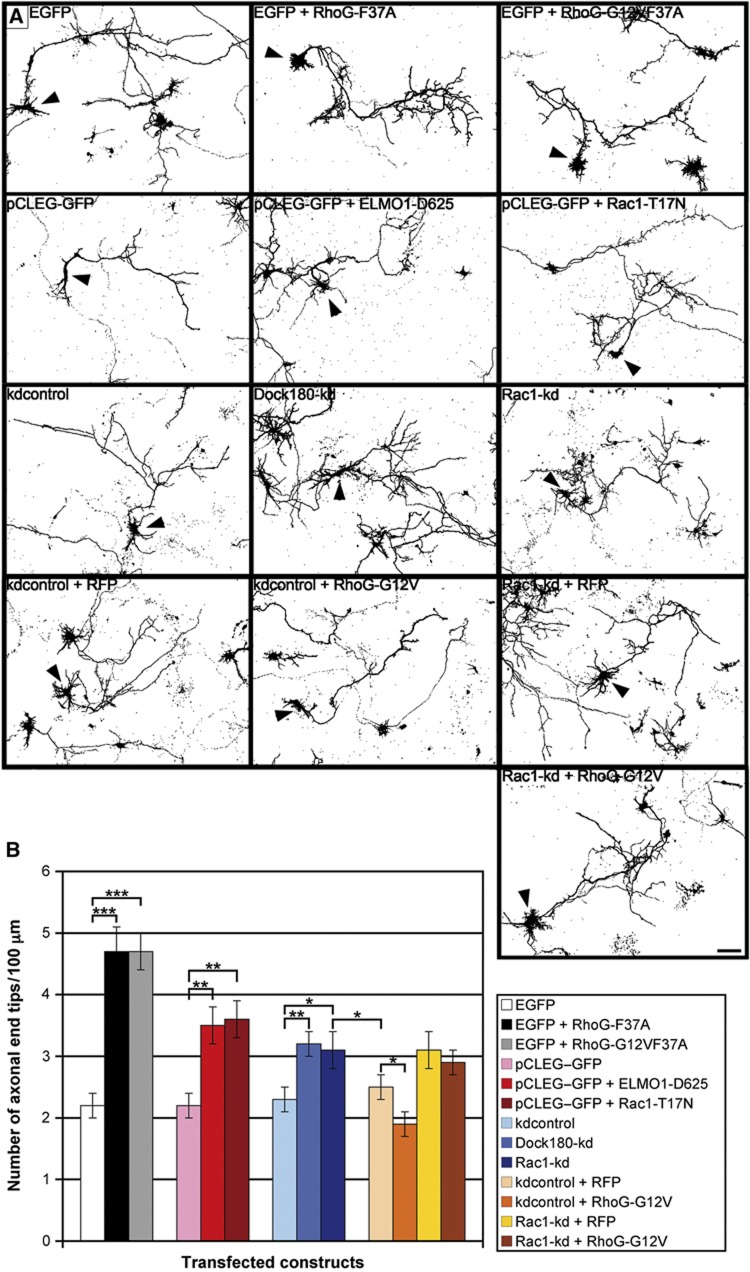
A functional ELMO/Dock180 signalling module leading to Rac1 activation has been presented to be essential for RhoG stimulating neurite outgrowth in PC12 cells (Katoh and Negishi, 2003). The functional significance of the ELMO/Dock180 module was also ascertained for phagocytosis as well as engulfment of apoptotic cells (Gumienny et al, 2001; deBakker et al, 2004; Elliott et al, 2010; Lu et al, 2011), and the invasion of pathogenic bacteria (Handa et al, 2007; Roppenser et al, 2009). Based on these published data, the question arose whether this signalling pathway is also fundamental for RhoG-inhibited axonal branching. Overexpression of the dominant-negative RhoG-F37A and RhoG-G12VF37A mutants, unable to bind to ELMO (Katoh and Negishi, 2003), in DIV2 hippocampal neurons resulted in a considerable increase in axonal branching (Figure 3A and B). These results suggest that the ELMO/Dock180/Rac1 signalling pathway is important for RhoG-mediated inhibition of axonal branching. We further corroborated these data by examining the effects of the dominant-negative ELMO deletion construct ELMO-D625, which cannot bind to Dock180 (deBakker et al, 2004; Meller et al, 2008), and the dominant-negative Rac1 construct Rac1-T17N on axonal branching. We found that both constructs significantly increased the number of axonal end tips/100 μm (Figure 3A and B). Moreover, the knockdown of either Dock180 or Rac1 expression by the shRNA constructs Dock180-kd and Rac1-kd (Meller et al, 2008; Supplementary Figure S5A and B), respectively, resulted in increased axonal branching (Figure 3A and B). In addition, RhoG knockdown led to a reduction in the amount of active, GTP-bound Rac1 in axons (Supplementary Figure S6). To further demonstrate the importance of the ELMO/Dock180/Rac1 signalling module for RhoG-mediated inhibition of axonal branching, overexpression of RhoG-G12V was combined with knockdown of Rac1. We found that dominant-positive RhoG did not inhibit axonal branching if Rac1 expression was reduced by RNA interference with Rac1-kd (Figure 3A and B). Therefore, these findings demonstrate that RhoG inhibits axonal branching via the ELMO/Dock180/Rac1 signalling pathway.
Figure 3.
RhoG inhibits axonal branching via ELMO/Dock180/Rac1 signalling. (A) The axonal morphology of hippocampal neurons, transfected with the indicated constructs at DIV2, was analysed after staining for GFP at DIV2+2 (arrowheads). Scale bar, 20 μm. (B) RhoG-F37A and RhoG-G12VF37A, RhoG mutants that cannot bind to ELMO, strongly increased the number of axonal end tips. The same effect was determined for ELMO-D625, Rac1-T17N, Dock180-kd, and Rac1-kd. Under the condition of Rac1 knockdown, RhoG-G12V did not decrease the number of axonal end tips. Mean values (n=80 neurons for Dock180-kd and kdcontrol, n=40 neurons for all other conditions)±s.e.m. (***P<0.0005; **P<0.005; *P<0.05).
RhoG inhibits dendritic branching dependent on the small GTPase Cdc42
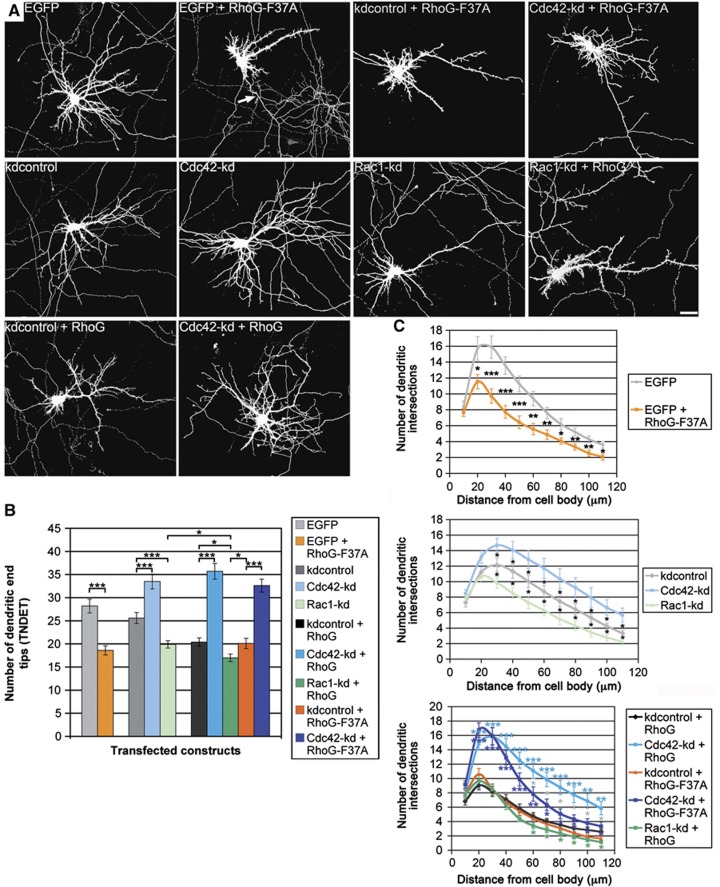
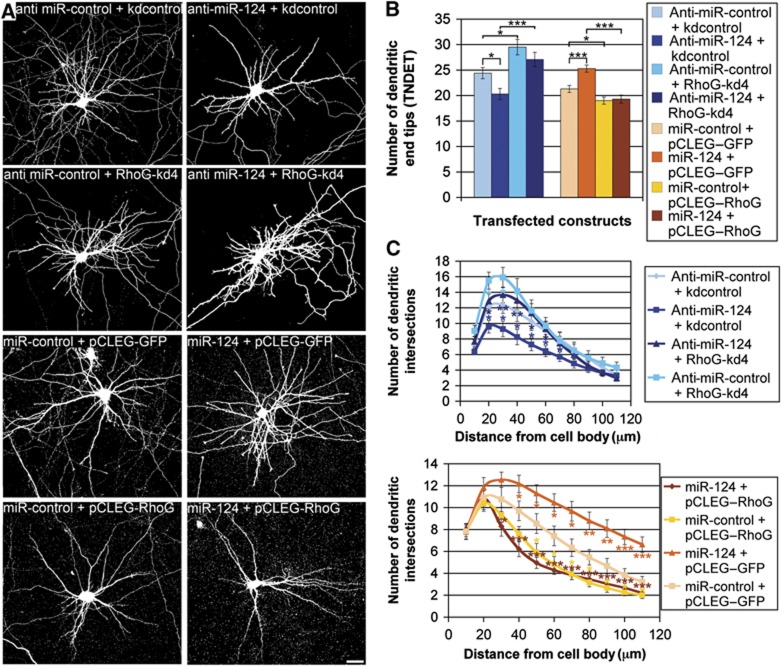
Surprisingly, however, RhoG-F37A, which is unable to bind to ELMO and to stimulate Rac1 activation via Dock180, did not increase but rather decreased the total number of dendritic end tips in DIV7 hippocampal neurons (Figure 4A and B). In addition, knockdown of Rac1 also decreased TNDET, and overexpression of RhoG combined with knockdown of Rac1 synergistically affects TNDET (Figure 4A and B). These results imply that RhoG does not reduce dendritic tree complexity via the ELMO/Dock180/Rac1 signalling pathway. Instead, we found that the small GTPase Cdc42 is imperative for RhoG-mediated inhibition of dendritic branching. Knockdown of Cdc42 with the shRNA construct Cdc42-kd (Leemhuis et al, 2010; Supplementary Figure S8C) led to an increase in TNDET. Additionally, the decrease in TNDET mediated by RhoG was completely reversed in the presence of Cdc42-kd. Actually, even though Cdc42 knockdown was performed concurrent to RhoG overexpression, we could not determine any reduction of TNDET when compared with the respective Cdc42 knockdown alone (Figure 4A and B). A similar outcome could be observed when combining overexpression of RhoG-F37A with knockdown of Cdc42 (Figure 4A and B). Sholl analysis of all experimental conditions confirmed the results presented above (Figure 4C). Only a subtle difference could be noticed when comparing the effects of RhoG overexpression with RhoG-F37A overexpression in the distal part of the Sholl graph. RhoG-F37A decreased the number of dendritic intersections in the distal part of the dendritic tree to a higher extent than RhoG. Taken together, these data reveal that RhoG reduces dendritic branching via Cdc42 signalling.
Figure 4.
RhoG inhibits dendritic branching dependent on Cdc42. (A) The dendritic phenotype of hippocampal neurons, transfected with the indicated constructs at DIV7, was analysed after staining for GFP at DIV7+3. Note that RhoG-F37A led to a considerable reduction in dendritic tree complexity, but still increased axonal branching (arrow). Scale bar, 15 μm. (B) RhoG-F37A decreased TNDET to a similar degree as RhoG did. Knockdown of Cdc42 increased, whereas knockdown of Rac1 decreased TNDET. Combining Rac1 knockdown with RhoG overexpression synergistically decreased TNDET. Under the condition of Cdc42 knockdown performed by expression of Cdc42-kd, RhoG no longer decreased TNDET. Under these conditions, TNDET equals that of Cdc42-kd expression alone. Similarly, knockdown of Cdc42 led to a rescue of TNDET when co-expressed with RhoG-F37A. Mean values (n=40 neurons)±s.e.m. (***P<0.0005; *P<0.05). (C) Neurons transfected as described above were analysed by Sholl analysis for all experimental conditions. For each point of the Sholl graph, the mean values (n≥24 neurons)±s.e.m. are shown (***P<0.0005; **P<0.005; *P<0.05). Light blue stars, Cdc42-kd+RhoG compared with kdcontrol+RhoG; dark blue stars, Cdc42-kd+RhoG-F37A compared with kdcontrol+RhoG-F37A; green stars, Rac1-kd+RhoG compared with kdcontrol+RhoG; orange stars, kdcontrol+RhoG-F37A compared with kdcontrol+RhoG; grey stars, Cdc42-kd+RhoG compared with Cdc42-kd+RhoG-F37A.
The microRNA miR-124 regulates the expression of RhoG
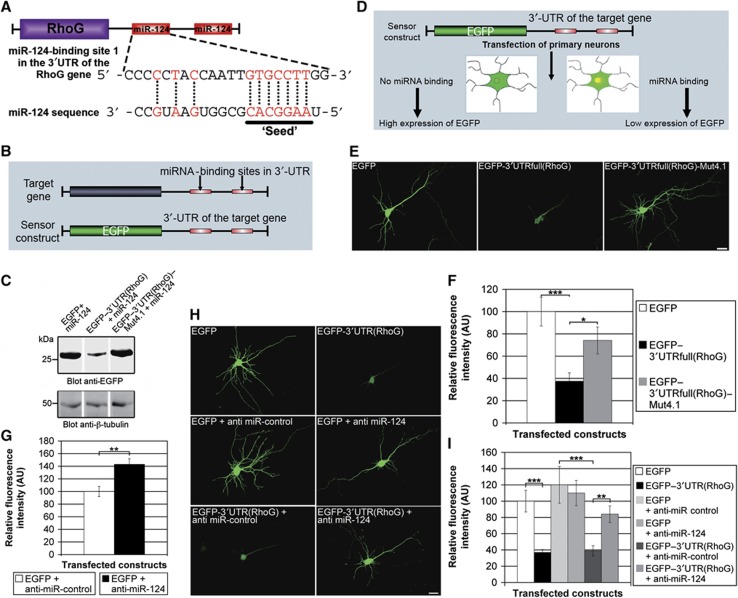
According to the algorithms PicTar (Krek et al, 2005), miRanda (John et al, 2004), TargetScan (Lewis et al, 2005), and MiRtarget2 (Wang and El Naqa, 2008) for microRNA target prediction, the 3′UTR of the RhoG gene comprises two miR-124-binding sites (Figure 5A; Supplementary Figure S7). These sites are highly conserved in mice, rats, and humans, while the 3′UTRs derived from the genes coding for Mtl and Mig-2 (the functional equivalents of RhoG in Drosophila and Caenorhabditis elegans, respectively) do not contain miR-124-binding sites. To experimentally verify the miR-124-regulated gene expression of RhoG, we used a sensor construct comprising large parts of the 3′UTR of the RhoG target gene with the two miR-124-binding sites downstream of an EGFP expression cassette (Figure 5B). This sensor construct, EGFP–3′UTR(RhoG), was transfected into HEK293 cells together with vector-based miR-124 and the expression of EGFP was analysed by immunoblotting. It was conspicuously reduced in comparison to the expression of the EGFP control construct, which contains the EGFP expression cassette but lacks the 3′UTR of the RhoG target gene (Figure 5C). In contrast, the construct RhoG–3′UTR(RhoG)–Mut4.1 harbouring two point mutations in the ‘seeds’ of both miR-124-binding sites (see Materials and methods) was expressed as high as the unregulated EGFP control construct (Figure 5C). To examine whether endogenously expressed miR-124 (Krichevsky et al, 2003; Deo et al, 2006; Sanuki et al, 2011) was able to reduce the sensor construct expression in primary hippocampal neurons, we measured the fluorescence intensity of EGFP 4 h after transfection of these neurons (Figure 5D; Supplementary Figure S8A). A clear reduction of EGFP expression was observed when using the sensor construct EGFP–3′UTR(RhoG) (Supplementary Figure S8A). A rescue of EGFP expression was observed in neurons expressing EGFP–3′UTR(RhoG)–Mut4.1. This indicated an expression regulation depending on the miR-124-binding sites in the 3′UTR of the sensor. To further substantiate this finding, we analysed a sensor construct comprising the full-length RhoG 3′UTR including the endogenous polyadenylation signal [EGFP–3′UTRfull(RhoG)]. Nearly identical results were observed (Figure 5E and F). If miR-124 regulated RhoG expression, an increased expression of endogenous RhoG would be expected when inhibiting miR-124. Therefore, anti-miR-124, which should capture the endogenously expressed miR-124, was co-transfected together with EGFP into hippocampal neurons. Then a quantification of the RhoG-specific immunopositive signals in neuronal cell bodies of transfected neurons was performed (Supplementary Figure S8B). We detected a significant increase in RhoG expression when inhibiting endogenous miR-124 by the transfected anti-miR-124 (Figure 5G). Finally, we wanted to verify that miR-124 regulates RhoG expression by an interaction with the miR-124-binding sites in the 3′UTR of the RhoG-encoding message. To this end, anti-miR-124 was co-transfected with the sensor construct EGFP–3′UTR(RhoG) or the corresponding EGFP control construct, respectively, in hippocampal neurons. Anti-miR-control was used as a control to exclude unspecific effects (Figure 5H). Indeed, we found that the anti-miR-124 completely abolished the expression regulation of the sensor (Figure 5I). Together, these results demonstrate that the expression of RhoG in hippocampal neurons is regulated by miR-124.
Figure 5.
RhoG expression is regulated by miR-124-binding sites in the 3′UTR of the RhoG mRNA. (A) Scheme of the RhoG message including the miR-124-binding sites in the 3′UTR. (B) Diagram of the sensor construct. (C) Immunoblotting for EGFP expression showed the importance of the miR-124-binding sites for sensor expression. Immunoblotting for β-tubulin served as a loading control. (D) Scheme for experimental design (upper panel). (E) The EGFP expression in hippocampal neurons for the indicated constructs is shown. (F) The EGFP expression of the sensor construct EGFP–3′UTRfull(RhoG) was downregulated by endogenous miR-124. Point mutations in the miR-124-binding sites (see Materials and methods) reduced the degree of regulation. Mean values (n=22 neurons)±s.e.m. (**P<0.005; *P<0.05). Scale bar, 20 μm. (G) Quantification of relative fluorescence intensities of cell bodies of hippocampal neurons transfected with the indicated constructs at DIV8 and stained for endogenous RhoG at DIV8+2. Mean values (n=21 neurons for EGFP+anti-miR-control, and n=18 neurons for EGFP+anti-miR-124)±s.e.m. (**P<0.005). (H, I) Anti-miR-124 inhibited the expression regulation of the sensor construct. Mean values (n=76 neurons)±s.e.m. (***P<0.001). Scale bar, 20 μm.
miR-124 increases dendritic and axonal complexity by inhibiting RhoG expression
We showed that RhoG expression is endogenously regulated by the microRNA miR-124 (Figure 5). Therefore, the question arose whether miR-124 could have an impact on dendrite elaboration by inhibiting RhoG expression. To tackle this question, we first determined the effects of either enhanced miR-124 expression or capture of endogenous miR-124 by anti-miR-124 on dendritic tree morphology. We found that miR-124 increased, while anti-miR-124 decreased TNDET (Figure 6A and B). Obviously, RhoG is not the only target for expression regulation by miR-124 (Gao, 2010; Sanuki et al, 2011). To substantiate that miR-124 increases dendritic tree complexity through inhibition of RhoG expression, we co-expressed miR-124 together with RhoG lacking the miR-124-binding sites in the 3′UTR. This overexpressed RhoG, which is not susceptible for miR-124, abolished the increase of TNDET induced by miR-124. In addition, TNDET could be rescued when reducing endogenous RhoG expression with the shRNA construct RhoG-kd4 in the presence of anti-miR-124 (Figure 6A and B). All these results were confirmed by performing Sholl analysis (Figure 6C).
Figure 6.
miR-124 increases dendritic complexity by inhibiting RhoG expression. (A) The dendritic phenotype of hippocampal neurons, transfected with the indicated constructs at DIV7, was analysed after staining for GFP at DIV7+3. Scale bar, 15 μm. (B) Anti-miR-124 decreased TNDET. RhoG-kd4, co-expressed with anti-miR-124, rescued TNDET. miR-124 increased TNDET. Co-expression of RhoG with miR-124 abolished the increase of TNDET induced by miR-124. Mean values (n=40 neurons for all experiments with anti-miR, n=80 neurons for all experiments with miR precursors)±s.e.m. (***P<0.0005; *P<0.05). (C) Neurons transfected as described above were analysed by Sholl analysis for all experimental conditions. For each data point of the Sholl graph, the mean values (n≥24 neurons)±s.e.m. are shown (***P<0.0005; **P<0.005; *P<0.05). Blue stars, anti-miR-124+kdcontrol compared with anti-miR-control+kdcontrol; light blue stars, anti-miR-control+RhoG-kd4 compared with anti-miR-control+kdcontrol; dark blue stars, anti-miR-124+RhoG-kd4 compared with anti-miR-124+kdcontrol; orange stars, miR-124+pCLEG–GFP compared with miR-control+pCLEG–GFP; yellow stars, miR-control+pCLEG–RhoG compared with miR-control+pCLEG–GFP; brown stars, miR-124+pCLEG–RhoG compared with miR-124+pCLEG–GFP.
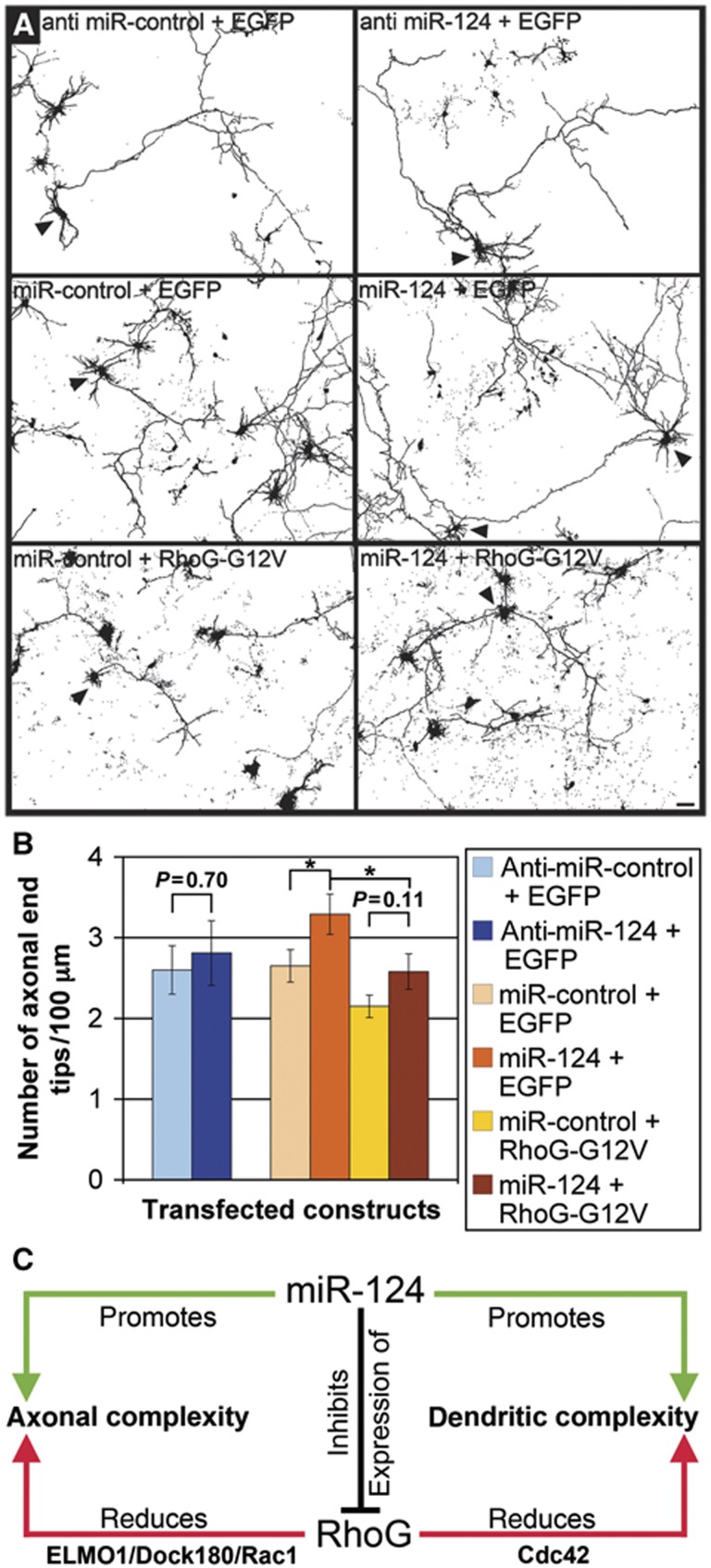
Overexpression of wild-type RhoG was sufficient to reduce dendritic tree complexity but not to inhibit axonal branching (Figures 1C, D, H and 2B). Thus, we assumed that increasing endogenous RhoG expression by means of anti-miR-124 would not reduce axonal branching. In fact, we found that miR-124, leading to a cellular knockdown of RhoG, but not anti-miR-124, resulting in an upregulation of RhoG expression, affected axonal branching by increasing the number of axonal end tips/100 μm (Figure 7A and B). This effect of miR-124 on axonal branching was likely to be mediated specifically by RhoG, because RhoG-G12V was shown to abrogate the increase in axonal branching, which was induced by miR-124 (Figure 7A and B). Taken together, these findings indicate that miR-124 regulates axonal and dendritic tree complexity by inhibition of RhoG expression (Figure 7C).
Figure 7.
miR-124 increases axonal branching by inhibiting RhoG expression. (A) The axonal morphology of hippocampal neurons, transfected with the indicated constructs at DIV2, was analysed after staining for GFP at DIV2+2 (arrowheads). Scale bar, 20 μm. (B) Anti-miR-124 did not affect the number of axonal end tips, while miR-124 increased the number of axonal end tips. This effect of miR-124 on axonal branching is likely to be mediated specifically by RhoG because RhoG-G12V abrogated the increase in axonal branching, which was induced by miR-124. Mean values (n=40 neurons)±s.e.m. (*P<0.05). (C) Schematic of miR-124 and RhoG-regulated neuronal process complexity.
Discussion
We unravelled RhoG as a novel target for miR-124-dependent expression regulation, and assigned a precise developmental function to this regulation: miR-124 stimulates axonal and dendritic tree complexity by inhibiting RhoG expression (Figure 7C). RhoG, in fact, was uncovered to reduce axonal and dendritic tree complexity in primary neuronal cell culture and in the mouse brain in vivo. These results are surprising as it was shown that RhoG overexpression promotes neurite outgrowth both in the PC12 cell line (Katoh et al, 2000; Estrach et al, 2002; Katoh and Negishi, 2003) and in rat superior cervical ganglion (SCG) neurons in culture (May et al, 2002). The apparent discrepancies in the outcome of RhoG-regulated process differentiation in PC12 cells and SCG neurons in comparison to hippocampal neurons could be attributable to the fact that most of the previous studies were based on RhoG overexpression, or did employ dominant-negative as well as constitutively active mutants of RhoG, which can exhibit nonspecific or nonphysiological effects resulting from perturbation of several Rho GTPases (Wang and Zheng, 2007). In our present study, we combine overexpression of wild-type RhoG, constitutively active, or dominant-negative RhoG mutants with different knockdown approaches employing RNA interference, and get concordant results. More likely, however, the observed difference in the functional outcome of RhoG-driven neuronal process morphogenesis in PC12 cells and SCG neurons compared with hippocampal neurons may stem from differences in the cellular systems analysed. It is tempting to speculate that RhoG could function in a lineage-specific manner. In fact, SCG neurons belong to the sympathetic nervous system, and PC12 cells are derived from a tumour of the sympathoadrenal lineage (Anderson, 1993; Unsicker, 1993), thus differing from hippocampal neurons in origin.
Although RhoG reduces neuronal process complexity in both dendrites and axons, we found subtle differences. Whereas the dominant-positive RhoG mutant RhoG-G12V reduced dendritic and axonal branching, wild-type RhoG only inhibited dendritic but not axonal branching. Furthermore, while both RhoG knockdown constructs increased dendritic tree complexity, only the stronger knockdown construct RhoG-kd4 stimulated axonal branching. In line with these results is our finding that anti-miR-124, leading to an increase in RhoG expression, reduced dendritic but not axonal complexity. All of these observations may be explained by a limited availability of RhoG-activating GEFs during axon, but not during dendrite differentiation. Several GEFs being able to activate RhoG have been described as Vav family members (Schuebel et al, 1998; Samson et al, 2010), Trio (Bellanger et al, 2000; Blangy et al, 2000; Estrach et al, 2002), Kalirin (May et al, 2002), and Ephexin4 (Hiramoto-Yamaki et al, 2010). Hence, it is also conceivable that different GEFs activate RhoG for reducing either axonal or dendritic tree complexity, respectively.
Different aspects support our hypothesis that overexpressed RhoG may be functionally inactive due to limited RhoG-stimulating endogenous GEF activity under certain experimental conditions: (1) Several previous studies on RhoG function in different experimental settings only used dominant-positive RhoG-G12V or RhoG-Q61L constructs but not wild-type RhoG to demonstrate RhoG-specific biological effects (Katoh and Negishi, 2003; van Buul et al, 2007; Yamaki et al, 2007; Meller et al, 2008). (2) A recent study showed that dominant-positive RhoG-G12V but not wild-type RhoG promotes the proliferation of neural progenitor cells whereas reduction of endogenous RhoG expression via RNA interference decreased this proliferation (Fujimoto et al, 2009). (3) In this study, we demonstrate that overexpression of the SGEFΔN construct, which comprises the DH/PH module harbouring the GEF activity for RhoG, reduced axonal branching in hippocampal neurons in a RhoG-dependent manner (Supplementary Figure S4). The SGEFΔN is derived from SGEF, a GEF that is active on RhoG but not on Rac1 (Ellerbroek et al, 2004).
The striking upregulation of miR-124 during the time of axonal and dendritic development suggests that this microRNA and its target genes may play specific roles during these processes. Actually, several recent studies reported on a relevance of miR-124 in neuronal process formation. In the CAD cell line, derived from CNS catecholaminergic neurons, miR-124 induces the outgrowth of long neurites, which branch and form complex networks (Makeyev et al, 2007). In addition, in mouse P19 embryonal carcinoma cells differentiating to neurons, miR-124 promotes neurite outgrowth (Yu et al, 2008). Interestingly, this outgrowth can be blocked by co-expression of constitutive-active Cdc42, but only to a lesser extent by constitutive-active Rac1, suggesting that inhibition of Cdc42 is necessary for miR-124 function. In a well-characterized model system to study neuronal process differentiation (Dotti et al, 1988), we specified the function of miR-124 and established this microRNA to contribute to dendritic tree complexity by repressing the expression of RhoG, which reduces dendritic tree complexity through Cdc42. On the first view contrary to our results, Edbauer et al (2010) did not observe any effects of miR-124 expression on dendritic growth or arborization in hippocampal neurons in culture. However, they used DIV14+3 hippocampal neurons, while our experiments were performed with DIV7+3 neurons. When using DIV14+3 neurons in our experimental setting, in agreement with Edbauer et al (2010), we neither found a significant increase in TNDET elicited by miR-124 nor a significant shift of the Sholl graph caused by miR-124 (Supplementary Figure S9). However, miR-124 still led to a small (although not significant) shift of the Sholl curve to more dendritic tree complexity at DIV14+3 (Supplementary Figure S9B). Dendritic growth in hippocampal neurons in culture starts around DIV4, and the dendritic tree of most neurons is well elaborated around DIV14. It is therefore likely that miR-124 has no longer a significant impact on dendritic growth and arborization during the time when dendritogenesis is largely completed but spinogenesis proceeds. In line with our data are also the results obtained by Yoo et al (2009), who showed that miR-124 stimulates activity-dependent dendritic growth by switching BAF53a to BAF53b expression. Together, a model arises, in which miR-124 promotes the elaboration of dendritic trees.
Finally, we delineated different signalling pathways involved: RhoG inhibits axonal branching via ELMO/Dock180/Rac1, while reducing dendritic tree complexity dependent on Cdc42 (Figure 7C). Previously, it has been shown that RhoG is able to drive two different signalling pathways: the ELMO/Dock180-stimulated activation of Rac1 (Katoh and Negishi, 2003) and the phosphatidylinositide 3-kinase (PI3K)-mediated phosphorylation of Akt (Murga et al, 2002; Harada et al, 2011). The latter pathway was published to be important for RhoG-dependent regulation of cell survival and anoikis as well as RhoG-promoted neural progenitor cell proliferation in the cerebral cortex (Fujimoto et al, 2009; Harada et al, 2011). The former pathway was identified as critical for phagocytosis and engulfment of apoptotic cells (Gumienny et al, 2001; deBakker et al, 2004; Elliott et al, 2010; Lu et al, 2011), the invasion of pathogenic bacteria (Handa et al, 2007, Roppenser et al, 2009), cell migration (Katoh et al, 2006), and neurite outgrowth of PC12 cells (Katoh and Negishi, 2003). However, the relevance of RhoG-induced, and Rac1-dependent effects on the actin cytoskeleton has been discussed controversially (Meller et al, 2008). Furthermore, it has been suggested that RhoG may independently activate Rac1 and Cdc42 (Gauthier-Rouvière et al, 1998).
Our results support the hypothesis that RhoG can independently activate Rac1 and Cdc42. The first indication was the finding of the divergent effects of the mutant RhoG-F37A on axonal versus dendritic branching. RhoG-F37A contains a mutation in the effector region of RhoG that prevents RhoG-F37A from binding to ELMO (Katoh and Negishi, 2003). However, based on studies of a F37A mutant derived from Rac1, which is structurally similar to RhoG, it is likely that RhoGF37A still can bind GEFs (Lamarche et al, 1996; Schwartz et al, 1998). This way, RhoG-F37A would function in a dominant-negative manner for the RhoG/ELMO/Dock180/Rac1 signalling pathway by inhibiting the activation of endogenous RhoG through these GEFs. Consequently, RhoG-F37A increases axonal branching. On the other hand, it was shown biochemically and functionally that the F37A mutant of Rac1 can still activate several signalling molecules (Lamarche et al, 1996; Schwartz et al, 1998). Based on these results, it is likely that RhoG-F37A can still activate other signalling molecules independent from ELMO1. Our data support the view that RhoG-F37A decreased dendritic branching by means of two different effects: (1) RhoG-F37A, like RhoG, stimulates the activity of Cdc42, which is inhibitory for dendritic branching, probably through a so far unknown signalling intermediate; (2) RhoG-F37A cannot increase Rac1 activity, which is stimulatory for dendritic branching (Figure 4B and C). This interpretation could also give an explanation for the subtle difference, which could be observed when comparing the effects of RhoG and RhoG-F37A on the distal part of the dendritic tree (Figure 4C). Together, the results of our present study strengthen the view that RhoG is capable of activating either of these two key molecules (Rac1 and Cdc42) dependent on the cellular and developmental context.
A relevance of Rac1 signalling for the regulation of axonal branching in the Drosophila giant fibre system was formerly proposed (Allen et al, 2000). Another study highlighted the dependence of axon growth, guidance, and branching regulation of Drosophila mushroom body neurons on differential activation of Rac1, Rac2, and Mtl (the functional equivalent of RhoG in Drosophila) signalling (Ng et al, 2002). The same study showed that even Mtl single mutants represent an axon branching phenotype. Similarly, redundant functions of Ced-10, Rac2, and Mig-2 (the functional equivalent of RhoG in C. elegans) in axon branching and pathfinding of several neuron classes in C. elegans were demonstrated (Lundquist et al, 2001; Struckhoff and Lundquist, 2003).
Finally, a potential significance of Cdc42 for inhibition of dendritic branching in Drosophila neurons was suggested (Gao et al, 1999). Furthermore, Cdc42 knockout as well as dominant-negative Cdc42 led to increased branching of the endfeet of radial glial cells, which function as neural stem cells (Yokota et al, 2010). Thus, our results strengthen an unappreciated facet of Rac1 and Cdc42 functions in neuronal process development, and point to the view that RhoG is a cellular conductor of Rac1 and Cdc42 activity, in turn regulated by the microRNA miR-124.
Materials and methods
DNA constructs
Most of the 3′UTR sequence of the RhoG mRNA (shown in Supplementary Figure S7), which comprised the miR-124-binding sites, was amplified from rat brain cDNA with the following primer pair: 5′-CACGAATTCCCCTGGCACTCAGCT-TGGAGGCTATCCCTA-3′ and 3′-CACGGATCCGGCTCCTCCCTTGGGAGAGGGAGAGA-ACAT-5′. The amplified sequence was inserted into the 3′UTR of the EGFP gene carried on a modified pEGFP-C1 vector (Clontech) with a stop codon inserted at the 5′end of the MCS to give the sensor construct EGFP–3′UTR(RhoG). The construct EGFP–3′UTR(RhoG)–Mut4.1 carries the following mutations in the ‘seeds’ of the conserved miR-124-binding sites: ‘GTGCCTT’ of site 1 is mutated to ‘GTGCCCG’ and ‘GTGCCTTA’ of site 2 is mutated to ‘GTGCCTGC’. The sensor construct EGFP–3′UTRfull(RhoG) comprising the full-length 3′UTR sequence of the RhoG message including the endogenous polyadenylation site was derived from an PCR amplification product from rat brain cDNA with the following primer pair: 5′-CACGAATTCCCCTGGCACTCAGCTTGGAGGCTATCCCTA-3′ and 5′-CACGGATCCACTGGAGACAGGAGGTTTTATTGATGCTGATGG-3′, and was cloned into the modified pEGFP-C1 vector as described above. The construct EGFP–3′UTRfull(RhoG)–Mut4.1 was cloned analogous to EGFP–3′UTR(RhoG)–Mut4.1.
The construct RhoG was cloned into the vectors pCMV–HA or pCMV–myc (Clontech). The primer pair used for amplification from rat brain cDNA was as follows: 5′-CACGAATTCCAATGCAGAGCATCAAGTGTGTGGTGGTGGGTGATGGGGCCGTAG-3′and 5′-CACAGATCTTCACAAGAGGATGCAGGACCGCCCACGCTTTATCGGTGTTGGG-3′. The construct RhoGresist(kd4) was derived from the construct RhoG by introducing two point mutations into the binding site for RhoG-kd4 (‘CGTCTTCGTCATATGCTTC’) using the QuickChange Site-directed Mutagenesis kit (Stratagene). For the IUE experiments, RhoG was subcloned into the pCLEG vector (Brandt et al, 2007). This vector, derived from the pCLE vector and used for gain-of-function experiments, contains a CMV/MLV promoter, an EF1α enhancer, and an internal ribosomal entry site (IRES) for the translation of GFP as a reporter (Chen et al, 2005).
Point mutations of RhoG were introduced using the QuickChange Site-directed Mutagenesis kit (Stratagene) and the following primer pairs were used to generate the constructs RhoG-G12V and RhoG-F37A: 5′-CACGAATTCCAATGCAGAG-CATCAAGTGTGTGGTGGTGGGTGATGTGGCCGTAG-3′ and 5′-CACAGATCTTCACAAGAGGATGCAGGACCGCCCACGCTTTATCGGTGTTGGG-3′ (RhoG-G12V), and 5′-ATCCCCACTGTGGCCGACAATTACAGCGCC-3′ and 5′-GGCGCTGTAATTGTCGGCCACAGTGGGGAT-3′ (RhoG-F37A). The shRNA construct RhoG-kd1 was generated using the pCGLH vector and the following primer pair: 5′-GATCTCGCCACTGTGTTTGACAATTATTCAAGAGATAATTGTCAAACACAGTGGTTTTTTGGAAC-3′ and 5′-TCGAGTTCCAAAAAACCACTGTGTTTGACAATTATCTCTTGAATAATTGTCAAACACAGTGGCGA-3′. The primer pair to generate the control constructs RhoG–kdcontrol and Luciferase-kd were as follows: 5′-GATCTCGGTCTCCTATGAAACGTATTTTCAAGAGAAATACGTTTCATAGGAGACTTTTTTGGAAC-3′ and 5′-TCGAGTTCCAAAAAAGTCTCCTATGAAACGTATTTCTCTTGAAAATACGTTTCATAGGAGACCGA-3′ (RhoG–kdcontrol), and 5′-GATCTCGCGTACGCGGAATACTTCGATTCAAGAGATCGAAGTATTCCGCGTACGTTTTTTGGAAC-3′ and 5′-TCGAGTT-CCAAAAAACGTACGCGGAATACTTCGATCTCTTGAATCGAAGTATTCCGCGTACGCGA-3′ (Luciferase-kd). The constructs RhoG-kd4, Dock180-kd, and Rac1-kd match the shRNA constructs for RhoG, Dock180, and Rac1, which were already published (Meller et al, 2008). The shRNA construct Cdc42-kd was derived from the siRNA sequence 5′-GACTACGACCGCTAAGTTA-3′ (Leemhuis et al, 2010). The deletion constructs ELMO-T625 and ELMOs-T625 were cloned into the vectors pCMV–HA and pCMV–myc (Clontech) after amplification with the following primer pair: 5′-CACGAATTCGGATGCCGCCGCCTTCGGACATCGTGAA-3′ and 5′-CACCTCGAGCATATGCGGGCAGTCCTTTCCCGTC-3′. For generating the construct SGEFΔN, the PCR amplification product from rat brain cDNA with the primers 5′-CACCTCGAGTTACACGTTGGTTTCCAGTCCTAGCAAGCGTCC-3′ and 5′-CACGAATTCGGGGGCTATCTCAGACAGTGAGCCAGGAGGAGAG-3′ was cloned into the vectors pCMV–myc and pCMV–HA.
MicroRNA target prediction
The following algorithms were used for microRNA target prediction: PicTar (http://pictar.mdc-berlin.de; Krek et al, 2005), miRanda (http://www.microrna.org; John et al, 2004), TargetScan (http://www.targetscan.org; Lewis et al, 2005), and MiRtarget2 (http://mirdb.org; Wang and El Naqa, 2008).
Biochemical procedures
Hippocampal or HEK293 cell extracts were prepared with Tris-buffered saline, pH 7.4 containing 1.2% Triton X-100 and a protease inhibitor cocktail containing leupeptin, pepstatin, aprotinin, and PMSF (all from FLUKA). Protein concentrations were determined with the BCA micro protein assay (Pierce). After performing an SDS–PAGE under reducing conditions, a subsequent western blot was analysed either with a polyclonal antibody to GFP (Abcam) or with antibodies to the HA tag (Sigma) or to β-tubulin (Santa Cruz).
Cell culture and transfection
Primary hippocampal neurons were prepared from embryonic days 18–19 Wistar rat pups as described (Brandt et al, 2007). The cells were cultured on poly-L-lysine-coated glass coverslips in neurobasal A medium supplemented with 2% B27 (Invitrogen), 0.5 mM glutamine, and the antibiotics penicillin and streptomycin at a density of 150 000 cells/well.
Hippocampal neurons were transfected with Effectene (Qiagen) according to the manufacturer’s instructions. HEK293 cells were transfected with FuGene6 according to the manufacturer’s instructions. Anti-miR-124, anti-miR-control, miR-124 precursor, and miR-control precursor were purchased (Ambion). The final concentration in the cell culture experiments was 40 nM for anti-miR-124 and anti-miR-control, and 30 nM for miR-124 precursor and miR-control precursor.
Immunohistochemistry and immunocytochemistry
P10 mice were deeply anaesthetised and sacrificed by transcardial perfusion with 4% paraformaldehyde. Vibratom sections (50 μm thick) were prepared and, after permeabilization in PBS supplemented with 5% fetal calf serum and 0.2% Triton X-100, incubated with a polyclonal antibody to GFP (ab6556, Abcam). Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (Molecular Probes) was used. Hippocampal cells in culture were indirectly stained after permeabilization with a polyclonal antibody to GFP (ab6556, Abcam), and co-stained with a monoclonal antibody to either the HA (clone HA-7, Sigma) or myc (clone 9E10, Roche) tag. Stainings for endogenously expressed RhoG, Dock180, and Rac1 were performed with a monoclonal anti-RhoG antibody (clone 1F3B3E5, Millipore #04-486), a polyclonal anti-Dock180 antibody (sc-6167, Santa Cruz Biotechnology), and a monoclonal anti-Rac1 antibody (clone 23A8, Millipore). Active, GTP–Rac1 was detected with a monoclonal anti-active Rac antibody (NewEast, #26903). T7-Cdc42 was visualized by an indirect immunostaining with a monoclonal antibody to the T7 tag (#69522-3, Novagen). The secondary antibodies Alexa Fluor 488-conjugated goat anti-rabbit, Alexa Fluor 568-conjugated goat anti-mouse (Molecular Probes), and Cy3-conjugated goat anti-mouse (Dianova) were used according to the manufacturer’s instructions. Cells were imaged with a fluorescence microscope (Olympus, BX 50) equipped with a Cool SNAP ES digital camera (Roper Scientific). For fluorescence imaging, the filters U-MWIG, U-MNIBA, and U-MWU2 (Olympus) were used. For higher magnification pictures, an oil immersion objective (PLAN APO × 60, 1.4 NA) was used.
In-utero electroporations
The IUE experiments were carried out with C57BL/6 mice as described in accordance with a protocol approved by the local animal welfare committee (Brandt et al, 2007). The morning of a detectable vaginal plug and the first neonatal day were considered to be embryonic day 0.5 (E0.5) and postnatal day 0 (P0), respectively. Plasmids were prepared using EndoFree Plasmid Kit (Qiagen, Hilden, Germany). Pregnant mice were anaesthetised at E15. The uterine horns were exposed and two of the pubs were randomly chosen for injecting plasmids. In all, 1–2 μl of DNA solution (4 μg/μl, supplemented with Fast Green) was injected through the uterine wall into the lateral ventricle of the embryo using a pulled glass capillary (World Precision Instruments). Electric pulses were delivered to embryos through the uterine wall with a CUY21 EDIT (Nepagene) or a ECM830 (BTX) square wave electroporator and a pair of platinum electrodes (CUY650P5) (five 50-ms pulses of 40 V with 950-ms intervals). The uterine horns were repositioned in the abdominal cavity, and the abdominal wall and skin were sewed up with surgical sutures. In all, five to eight animals were electroporated for each condition and the electroporated animals were sacrificed at P10 for analysis.
Image acquisition and data analysis
HEK293 cells and hippocampal neurons were cultured and processed as described (Brandt et al, 2008). The analysis of expression regulation by the miR-124-binding sites of the 3′UTR of RhoG message in HEK293 cells was carried out 48 h after transfection. To assay sensor construct expression in hippocampal neurons, neurons were transfected at days in vitro 8 (DIV8) and fixed 4 h later for analysis. The quantification of fluorescence intensities (GFP, HA, T7, RhoG, Dock180, Rac1) was performed after staining with the corresponding antibodies as described (Brandt et al, 2007). For the quantification of the intensity of immunofluorescence intensities the same acquisition settings for each set of control and experimental samples were used. All the imaging was done in the same session. Data acquisition was performed with wide-field epifluorescence using the equipment described above and Metamorph image analysis software (Universal Imaging Corporation).
All other images were captured on a Leica TCS SP2 or SL scanning confocal microscope with × 40 HCX PL APO, 1.25 NA, × 63 HCX PL APO, 1.4 NA or × 100 PLAN APO, 1.4 NA oil immersion objectives. For dendritic tree analysis, z-stacks were collected at 0.7-μm intervals. Dendritic branching after transfection of DIV7 old hippocampal neurons was explored 3 days later. For quantification of dendritic end tips in pyramidal neurons from slices of animals treated by IUE, 15-μm thick z-stacks of confocal images of dendritic trees of electroporated neurons collected at 1-μm intervals were analysed. All morphometric measurements were done with Metamorph image analysis software. Dendritic trees of all healthy transfected neurons with more than five dendritic end tips were analysed. All apical end tips of dendrites longer than 8 μm were counted. We used EGFP stainings to clearly visualize all dendritic branches, because in some cases autofluorescence of GFP is too weak to detect all dendritic branches.
Axonal morphology was examined 2 days after transfection of DIV2 hippocampal neurons. Following EGFP staining, all axons longer than 150 μm were analysed. In case of elaborating more than one axon, the longest axon of that neuron was investigated. Axonal identity could be determined by staining for sodium channels, which are concentrated in the axon initial segment, with a pan-antibody to sodium channels (K58/35, Sigma). All axonal branches longer than 8 μm were counted.
For performing Sholl analysis, the number of dendrites crossing a series of concentric circles centred upon the cell body (dendritic intersection) of each neuron was counted manually.
Acquisition and data analysis for all experiments were performed by investigators blind to the experimental conditions. Comparison of data and calculation of P-values were performed by two-tailed Student’s t-test.
Supplementary Material
Acknowledgments
We thank Monika Dulinski, Elke Hoffmann, and Anne Lehner for excellent technical assistance, and Jens Baron for cloning the constructs EGFP–3′UTR(RhoG), EGFP–3′UTR(RhoG)–Mut4.1, RhoG, and RhoG-V12. Heiko Fuchs endowed us with vector-based miR-124. Dr H Bock and Elisabeth Bouché generously provided the Cdc42-kd construct. This study was supported by the DFG (Grants SFB 665-A2 to SS and RN, and SCHU 1406/3-1 to SS).
Author contributions: KF and SS performed and analysed most of the experiments in this study. WO contributed to the sensor experiments and documented the RhoG knockdown in the immunoblot. SJ performed the expression analysis of RhoG. JB performed the in-utero electroporations. RN was involved in the design of this study and participated in writing of the manuscript. SS designed the experiments, supervised the study, and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allen MJ, Shan X, Murphey RK (2000) A role for Drosophila Drac1 in neurite outgrowth and synaptogenesis in the giant fiber system. Mol Cell Neurosci 16: 754–765 [DOI] [PubMed] [Google Scholar]
- Anderson DJ (1993) Molecular control of cell fate in the neural crest: the sympathoadrenal lineage. Ann Rev Neurosci 16: 129–158 [DOI] [PubMed] [Google Scholar]
- Bellanger JM, Astier C, Sardet C, Ohta Y, Stossel TP, Debant A (2000) The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat Cell Biol 113: 729–739 [DOI] [PubMed] [Google Scholar]
- Blangy A, Vignal E, Schmidt S, Debant A, Gauthier-Rouvière C, Fort P (2000) TrioGEF1 controls Rac- and Cdc42-dependent cell structures through the direct activation of RhoG. J Cell Sci 113: 729–739 [DOI] [PubMed] [Google Scholar]
- Brandt N, Franke K, Johannes S, Buck F, Harder S, Hassel B, Nitsch R, Schumacher S (2008) B56beta, a regulatory subunit of protein phosphatase 2A, interacts with CALEB/NGC and inhibits CALEB/NGC-mediated dendritic branching. FASEB J 22: 2521–2533 [DOI] [PubMed] [Google Scholar]
- Brandt N, Franke K, Rasin MR, Baumgart J, Vogt J, Khrulev S, Hassel B, Pohl EE, Sestan N, Nitsch R, Schumacher S (2007) The neural EGF family member CALEB/NGC mediates dendritic tree and spine complexity. EMBO J 26: 2371–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Rasin MR, Kwan KY, Sestan N (2005) Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Si USA 102: 17792–17797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F (2009) miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci 12: 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- deBakker CD, Haney LB, Kinchen JM, Grimsley C, Lu M, Klingele D, Hsu PK, Chou BK, Cheng LC, Blangy A, Sondek J, Hengartner MO, Wu YC, Ravichandran KS (2004) Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and armadillo repeats of CED-12/ELMO. Curr Biol 14: 2208–2216 [DOI] [PubMed] [Google Scholar]
- Deo M, Yu JY, Chung KH, Tippens M, Turner D (2006) Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn 235: 2538–2548 [DOI] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, Banker GA (1988) The establishment of polarity by hippocampal neurons in culture. J Neurosci 8: 1454–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M (2010) Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellerbroek SM, Wennerberg K, Arthur WT, Dunty JM, Bowman DR, DeMali KA, Der C, Burridge K (2004) SGEF, a RhoG guanine nucleotide exchange factor that stimulates macropinocytosis. Mol Biol Cell 15: 3309–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott MR, Zheng S, Park D, Woodson RI, Reardon MA, Juncadella IJ, Kinchen JM, Zhang J, Lysiak JJ, Ravichandran KS (2010) Unexpected requirement for ELMO1 in clearance of apoptotic germ cells in vivo. Nature 467: 333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrach S, Schmidt S, Diriong S, Penna A, Blangy A, Fort P, Debant A (2002) The human Rho-GEF Trio and its target GTPase RhoG are involved in the NGF pathway, leading to neurite outgrowth. Curr Biol 12: 307–312 [DOI] [PubMed] [Google Scholar]
- Fineberg SK, Kosik KS, Davidson BL (2009) MicroRNAs potentiate neural development. Neuron 64: 303–309 [DOI] [PubMed] [Google Scholar]
- Fujimoto S, Negishi M, Katoh H (2009) RhoG promotes neural progenitor cell proliferation in mouse cerebral cortex. Mol Biol Cell 20: 4941–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB (2010) Context-dependent functions of specific microRNAs in neuronal development. Neural Dev 5: 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Brenman JE, Jan LY, Jan YN (1999) Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev 13: 2549–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier-Rouvière C, Vignal E, Mériane M, Roux P, Montcourier P, Fort P (1998) RhoG GTPase controls a pathway that independently activates Rac1 and Cdc42Hs. Mol Biol Cell 9: 1379–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, Walk SF, Nemergut ME, Macara IG, Francis R, Schedl T, Qin Y, Van Aelst L, Hengartner MO, Ravichandran KS (2001) CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107: 27–41 [DOI] [PubMed] [Google Scholar]
- Handa Y, Suzuki M, Ohya K, Iwai H, Ishijima N, Koleske AJ, Fukui Y, Sasakawa C (2007) Shigella IpgB1 promotes bacterial entry through the ELMO/Dock180 machinery. Nat Cell Biol 9: 121–128 [DOI] [PubMed] [Google Scholar]
- Harada K, Hiramoto-Yamaki N, Negishi M, Katoh H (2011) Ephexin4 and EphA2 mediate resistance to anoikis through RhoG and phosphatidylinositol 3-kinase. Exp Cell Res 317: 1701–1713 [DOI] [PubMed] [Google Scholar]
- Hiramoto-Yamaki N, Takeuchi S, Ueda S, Harada K, Fujimoto S, Negishi M, Katoh H (2010) Ephexin4 and EphA2 mediate cell migration through a RhoG-dependent mechanism. J Cell Biol 190: 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa Y, Katoh H, Nakamura K, Mori K, Negishi M (2002) Developmental changes in expression of small GTPase RhoG mRNA in the rat brain. Brain Res Mol Brain Res 106: 145–150 [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS (2004) Human MicroRNA targets. PloS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H, Hiramoto K, Negishi M (2006) Activation of Rac1 by RhoG regulates cell migration. J Cell Sci 119: 56–65 [DOI] [PubMed] [Google Scholar]
- Katoh H, Negishi M (2003) RhoG activates Rac1 by direct interaction with the Dock180-binding protein ELMO. Nature 424: 461–464 [DOI] [PubMed] [Google Scholar]
- Katoh H, Yasui H, Yamaguchi Y, Aoki J, Fujita H, Mori K, Negishi M (2000) Small GTPase RhoG is a key regulator for neurite outgrowth in PC12 cells. Mol Cell Biol 20: 7378–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N (2005) Combinatorial microRNA target predictions. Nat Genet 37: 495–500 [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS (2003) A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9: 1274–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarche N, Tapon N, Stowers L, Burbelo PD, Aspenström P, Bridges T, Chant J, Hall A (1996) Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell 87: 519–529 [DOI] [PubMed] [Google Scholar]
- Leemhuis J, Bouché E, Frotscher M, Henle F, Hein L, Herz J, Meyer DK, Pichler M, Roth G, Schwan C, Bock HH (2010) Reelin signals through apolipoprotein E receptor 2 and Cdc42 to increase growth cone motility and filopodia formation. J Neurosci 30: 14759–14772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Lu Z, Elliott MR, Chen Y, Walsh JT, Klibanov AL, Ravichandran KS, Kipnis J (2011) Phagocytic activity of neuronal progenitors regulates adult neurogenesis. Nat Cell Biol 13: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, Bargmann CI (2001) Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development 128: 4475–4488 [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T (2007) The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 27: 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Martín N, Fernández-Arenas E, Cemerski S, Delgado P, Turner M, Heuser J, Irvine DJ, Huang B, Bustelo XR, Shaw A, Alarcón B (2011) T Cell Receptor Internalization from the Immunological Synapse Is Mediated by TC21 and RhoG GTPase-Dependent Phagocytosis. Immunity 35: 208–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May V, Schiller MR, Eipper BA, Mains RE (2002) Kalirin Dbl-homology guanine nucleotide exchange factor 1 domain initiates new axon outgrowths via RhoG-mediated mechanisms. J Neurosci 22: 6980–6990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller J, Vidali L, Schwartz MA (2008) Endogenous RhoG is dispensable for integrin-mediated cell spreading but contributes to Rac-independent migration. J Cell Sci 121: 1981–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga C, Zohar M, Teramoto H, Gutkind JS (2002) Rac1 and RhoG promote cell survival by the activation of PI3K and Akt, independently of their ability to stimulate JNK and NF-kappaB. Oncogene 21: 53–64 [DOI] [PubMed] [Google Scholar]
- Ng J, Nardine T, Harms M, Tzu J, Goldstein A, Sun Y, Dietzl G, Dickson BJ, Luo L (2002) Rac GTPases control axon growth, guidance and branching. Nature 416: 442–447 [DOI] [PubMed] [Google Scholar]
- Patel JC, Galán JE (2006) Differential activation and function of Rho GTPases during Salmonella-host cell interactions. J Cell Biol 175: 453–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, Sander C, Tuschl T, Kandel E (2009) Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron 63: 803–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roppenser B, Röder A, Hentschke M, Ruckdeschel K, Aepfelbacher M (2009) Yersinia enterocolitica differentially modulates RhoG activity in host cells. J Cell Sci 122: 696–705 [DOI] [PubMed] [Google Scholar]
- Samson T, Welch C, Monaghan-Benson E, Hahn KM, Burridge K (2010) Endogenous RhoG is rapidly activated after epidermal growth factor stimulation through multiple guanine-nucleotide exchange factors. Mol Biol Cell 21: 1629–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanuki R, Onishi A, Koike C, Muramatsu R, Watanabe S, Muranishi Y, Irie S, Uneo S, Koyasu T, Matsui R, Chérasse Y, Urade Y, Watanabe D, Kondo M, Yamashita T, Furukawa T (2011) miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci 14: 1125–1134 [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME (2006) A brain-specific microRNA regulates dendritic spine development. Nature 439: 283–289 [DOI] [PubMed] [Google Scholar]
- Schuebel KE, Movilla N, Rosa JL, Bustelo XR (1998) Phosphorylation-dependent and constitutive activation of Rho proteins by wild-type and oncogenic Vav-2. EMBO J 17: 6608–6621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MA, Meredith JE, Kiosses WB (1998) An activated Rac mutant functions as dominant negative for membrane ruffling. Oncogene 17: 625–629 [DOI] [PubMed] [Google Scholar]
- Struckhoff EC, Lundquist EA (2003) The actin-binding protein UNC-115 is an effector of Rac signaling during axon pathfinding in C. elegans. Development 130: 693–704 [DOI] [PubMed] [Google Scholar]
- Unsicker K (1993) The chromaffin cell: paradigm in cell, development and growth factor biology. J Anat 183: 207–221 [PMC free article] [PubMed] [Google Scholar]
- van Buul JD, Allingham MJ, Samson T, Meller J, Boulter E, García-Mata R, Burridge K (2007) RhoG regulates endothelial apical cup assembly downstream from ICAM1 engagement and is involved in leukocyte trans-endothelial migration. J Cell Biol 178: 1279–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK (2007) The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev 21: 744–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki N, Negishi M, Katoh H (2007) RhoG regulates anoikis through a phosphatidylinositol 3-kinase-dependent mechanism. Exp Cell Res 313: 2821–2832 [DOI] [PubMed] [Google Scholar]
- Yokota Y, Eom TY, Stanco A, Kim WY, Rao S, Snider WD, Anton ES (2010) Cdc42 and Gsk3 modulate the dynamics of radial glial growth, inter-radial glial interactions and polarity in the developing cerebral cortex. Development 137: 4101–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR (2009) MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460: 642–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, Chung KH, Deo M, Thompson RC, Turner DL (2008) MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp Cell Res 314: 2618–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, El Naqa IM (2008) Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics 24: 325–332 [DOI] [PubMed] [Google Scholar]
- Wang L, Zheng Y (2007) Cell type-specific functions of Rho GTPases revealed by gene targeting in mice. Trends Cell Biol 17: 58–64 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.