Abstract
The stability of epigenetic patterns is critical for genome integrity and gene expression. This highly coordinated process involves interrelated positive and negative regulators that impact distinct epigenetic marks, including DNA methylation and dimethylation at histone H3 lysine 9 (H3K9me2). In Arabidopsis, mutations in the DNA methyltransferase MET1, which maintains CG methylation, result in aberrant patterns of other epigenetic marks, including ectopic non-CG methylation and the relocation of H3K9me2 from heterochromatin into gene-rich chromosome regions. Here, we show that the expression of the H3K9 demethylase IBM1 (increase in BONSAI methylation 1) requires DNA methylation. Surprisingly, the regulatory methylated region is contained in an unusually large intron that is conserved in IBM1 orthologues. The re-establishment of IBM1 expression in met1 mutants restored the wild-type H3K9me2 nuclear patterns, non-CG DNA methylation and transcriptional patterns at selected loci, which included DNA demethylase genes. These results provide a mechanistic explanation for long-standing puzzling observations in met1 mutants and reveal yet another layer of control in the interplay between DNA methylation and histone modification, which stabilizes DNA methylation patterns at genes.
Keywords: DNA methylation, epigenetic stability, gene-body methylation, H3K9 demethylase, intron methylation
Introduction
Silent chromatin is typically associated with specific patterns of epigenetic modifications, which in plants include high levels of DNA methylation in all three cytosine contexts (CG, CHG and CHH, where H=A, T or C) and dense dimethylation at lysine 9 of histone H3 (H3K9me2) (Bender, 2004; Chan et al, 2005; Grewal and Jia, 2007). H3K9 methylation is conserved from plants to mammals and relies on the activities of histone lysine methyltransferases in the Su(var)3-9 family (Rea et al, 2000). The Arabidopsis genome encodes nine homologues of the Drosophila Su(var)3-9 protein, which are referred to as SUVH proteins (for Su(var)3-9 homologues) (Baumbusch et al, 2001). Although SUVH4 (also known as KRYPTONITE or KYP), SUVH5 and SUVH6 seem to act redundantly in the maintenance of H3K9me2 at transposable elements (TEs) and repeats, only KYP appears to function at genes (Jackson et al, 2004; Ebbs and Bender, 2006; Inagaki et al, 2010). Conversely, enzymes of the JHDM2 family contain a jumonji C (jmjC) domain and can remove H3K9 methylation (Klose et al, 2006; Tsukada et al, 2006; Yamane et al, 2006). In Arabidopsis, experimental evidence supports the hypothesis that the jmjC domain-containing protein increase in BONSAI methylation 1 (IBM1) is a histone demethylase that is specific for H3K9me2 and H3K9 monomethylation (Saze et al, 2008; Inagaki et al, 2010). Mutants for IBM1 display ectopic H3K9me2 accumulation in the transcribed regions of a large number of genes, whereas TEs are unaffected (Inagaki et al, 2010).
In Arabidopsis, CG methylation is propagated during DNA replication by the maintenance DNA methyltransferase METHYLTRANSFERASE 1 (MET1), which robustly copies methylation patterns on newly synthesized DNA strands. The maintenance of asymmetrical CHH methylation is mostly ensured by DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) in a process known as RNA-directed DNA methylation (RdDM), which involves the polymerases IV and V (Law and Jacobsen, 2010). The perpetuation of CHG methylation patterns is largely ensured by the plant-specific chromomethylase CMT3, and genetic analyses suggest that targeting of CMT3 to chromatin relies on H3K9me2, which indicates that H3K9me2 acts upstream of CHG methylation (Lindroth et al, 2004; Feng and Jacobsen, 2011). These two repressive marks are intimately associated, and at the genome level, ∼90% of CHG methylation coincides with H3K9me2-enriched regions (Bernatavichute et al, 2008). Additionally, the loss-of-function kyp and cmt3 alleles show a similar loss of cytosine methylation at CHG sites and induce transcriptional reactivation of a common set of silent targets (Jackson et al, 2002; Lippman et al, 2003; Lindroth et al, 2004; Ebbs et al, 2005; Tran et al, 2005; Ebbs and Bender, 2006). The SET and RING-associated (SRA) domain of KYP and SUVH6 binds to DNA that is methylated at CHGs in vitro, which suggests that CHG methylation also feeds back onto H3K9me2 (Johnson et al, 2007).
In heterochromatin, methylation at CG sites and H3K9me2 are also linked. In wild-type (WT) plant nuclei, H3K9me2 is largely confined to heterochromatic chromosomal regions that are densely CG methylated. In the nuclei of the loss-of-function met1-3 mutant, CG methylation is lost, and H3K9me2 is redistributed away from the chromocentres (Soppe et al, 2002; Tariq et al, 2003; Mathieu et al, 2007). Therefore, although the molecular mechanism remains elusive, CG methylation appears to direct H3K9me2 in heterochromatin. Importantly, CG methylation is not restricted to heterochromatin, and genome-wide methylation profiling studies have highlighted that approximately one-third of Arabidopsis genes are CG methylated (Tran et al, 2005; Zhang et al, 2006; Zilberman et al, 2007; Cokus et al, 2008; Lister et al, 2008). Noticeably, gene-body methylation is almost exclusively restricted to CG sites, and it is not associated with H3K9me2 (Bernatavichute et al, 2008). Therefore, CG methylation and H3K9me2 show distinct interactions at different chromosomal locations.
In addition, met1 mutants exhibit ectopic non-CG methylation (Soppe et al, 2002; Tariq et al, 2003; Zhang et al, 2006; Mathieu et al, 2007; Cokus et al, 2008; Lister et al, 2008). At heterochromatic sequences, this has been shown to result primarily from the misdirection of the RdDM pathway, while aberrant methylation at a few hundred genes likely originates from the transcriptional downregulation of the DNA demethylases (which notably include REPRESSOR OF SILENCING 1 (ROS1)), in the absence of CG methylation (Huettel et al, 2006; Mathieu et al, 2007). Importantly, several thousand genes specifically display CHG hypermethylation in their body sequence in the met1 background, and the majority of these genes contain CG methylation in the WT background (Lister et al, 2008; Reinders et al, 2008). This suggests that CG methylation (and/or MET1 itself) may exclude CHG methylation from genes; however, the molecular mechanism that links CG and CHG methylation at genes remains to be elucidated. The recent analyses of the IBM1 loss-of-function mutant have revealed widespread ectopic CHG DNA methylation and H3K9me2 at genes (Saze et al, 2008; Miura et al, 2009; Inagaki et al, 2010). These new epigenetic patterns are dependent on the function of CMT3 and KYP and interestingly, genes that contain CHG hypermethylation in the met1 and ibm1 mutant backgrounds largely overlap (Miura et al, 2009).
These similarities between the met1 and ibm1 mutants have led us to hypothesize that CG methylation and/or MET1 may protect genes from ectopic CHG methylation and H3K9me2 because they are required for proper IBM1 expression. IBM1 encodes two mRNA variants; the longer variant (IBM1-L) specifically encodes the functional IBM1 protein that contains the jmjC domain. We found that the proper accumulation of IBM1-L mRNA is controlled by DNA methylation and depends on the simultaneous presence of CG and CHG methylation in an unusually large intron of the IBM1 gene. The re-establishment of IBM1-L expression in met1 mutants largely suppressed the abnormal H3K9me2, DNA methylation and transcriptional patterns that were induced by the mutation at selected target genes. Interestingly, the expression of the ROS1 DNA demethylase was recovered when IBM1-L accumulation was restored in the met1 background, which thereby also contributed to the removal of aberrant DNA methylation patterns that occur in this mutant background. Therefore, by controlling the proper expression of H3K9 and DNA demethylases, CG methylation insures the maintenance of proper genic DNA methylation and histone modification patterns through a self-regulatory loop. These results highlight the importance of CG methylation as a central coordinator of epigenetic stability at genes and provide mechanistic explanations for long-standing enigmatic observations in met1 mutants.
Results
DNA methylation is required for proper IBM1 expression
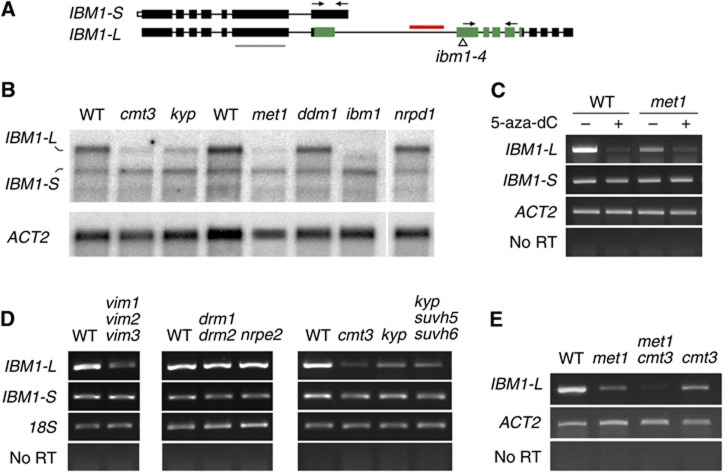
To understand how CHG gene-body methylation occurs in met1 mutant plants, we examined the transcription of the IBM1 gene with respect to DNA methylation. IBM1 is predicted to produce two different transcripts; only the longer transcript (hereafter referred to as IBM1-L) is predicted to encode the jmjC domain and thus the functional IBM1 protein (Figure 1A). Northern blotting and reverse transcription (RT)–PCR analyses confirmed the presence of the two predicted IBM1 transcripts (Figure 1B and C). In WT plants that were treated with the DNA methylation inhibitor 5-aza-2′-deoxycytidine (5-aza-dC) and in plants that were homozygous for the met1-3 null MET1 allele (Saze et al, 2003), the accumulation of IBM1-L mRNA was specifically downregulated, whereas the accumulation of the short IBM1 RNA transcript (IBM1-S) was not significantly affected (Figure 1B and C). To further investigate the impact of DNA methylation on IBM1 expression, we assayed transcript accumulation in mutants of additional regulators of genomic DNA methylation. Triple mutants for the VARIATION IN METHYLATION (VIM) 1, 2 and 3 genes, which encode co-factors that are required for CG methylation maintenance (Woo et al, 2007, 2008; Kraft et al, 2008), also showed specific downregulation of the IBM1-L mRNA transcript (Figure 1D). Together with the results from the met1 mutants, this suggests that CG methylation is required for the proper accumulation of IBM1 transcripts that contain the putative H3K9 demethylase domain jmjC.
Figure 1.
DNA methylation is required for IBM1-L transcript accumulation. (A) Schematic representation of the two IBM1 mRNA variants, IBM1-S and IBM1-L. The boxes represent exons; coding regions are indicated in black and untranslated regions are indicated in white. The region encoding the jmjC domain is shown in green (Saze et al, 2008). The intronic DNA-methylated zone (horizontal red bar), the positions of the probe that was used for the northern blot analysis (horizontal grey bar) and the primers that were used for the RT–PCR analysis of the IBM1-L and IBM1-S variants (arrows) are shown. The position of the T-DNA insertion in ibm1-4 is indicated with a triangle. (B) Northern blot analysis of the IBM1 transcripts using poly(A)+ RNAs for the indicated mutant genotypes. Hybridization with a probe corresponding to ACTIN2 (ACT2) is shown as a loading control. (C) RT–PCR analysis of IBM1-S and IBM1-L transcripts in WT and met1-3 plants that were grown on medium containing (+) or lacking (−) 5-aza-dC. (D) RT–PCR analysis of the IBM1-S and IBM1-L transcripts in the indicated DNA methylation-deficient mutant backgrounds. (E) RT–PCR analysis of IBM1-L expression in met1 cmt3 compared with the WT and single mutants. The amplification of ACT2 or 18S rRNA (18S) was used to normalize the RNA template levels. The negative controls (no RT) lacked reverse transcriptase.
Noticeably, the downregulation of IBM1-L that was observed in the met1 plants was further enhanced when the met1 plants were grown on 5-aza-dC, which suggests that non-CG methylation may also participate in the control of IBM1 expression (Figure 1C). In the nrpd1 mutant of polymerase IV, as well as in the nrpe2 (nrpd2a) mutant of the common subunit of polymerases IV and V, IBM1 transcription was similar to the WT, and the drm1 drm2 double mutant also exhibited no detectable IBM1 transcriptional variation, which indicates that the RdDM pathway/CHH methylation does not control IBM1 expression (Figure 1B and D). H3K9me2 and DNA methylation at CHG sites are intimately coupled, and a self-reinforcing feedback loop between KYP and CMT3 maintains these two marks in heterochromatin. We analysed the cmt3 and kyp mutants to determine whether CHG methylation influences IBM1 expression. RNA-gel blot and RT–PCR analyses showed that accumulation of the IBM1-L transcript was drastically downregulated in both cmt3 and kyp (Figure 1B and D). Triple mutants of the histone H3K9 methyltransferases KYP(SUVH4)/SUVH5/SUVH6 mimic the kyp single mutant, indicating that SUVH5 and SUVH6 do not significantly contribute IBM1 transcriptional control (Figure 1D). Together, these observations indicate that CG and CHG methylation are required for the proper expression of IBM1. This reveals another layer of interdependence and control between DNA methylation and H3K9 methylation, in which, somehow paradoxically with their role in the maintenance of DNA methylation, MET1, CMT3 and KYP are also involved in the exclusion of CHG methylation/H3K9me2 from genes. The mutation of DECREASE IN DNA METHYLATION 1 (DDM1) did not alter IBM1 transcript accumulation (Figure 1B).
Intronic DNA methylation controls IBM1 expression
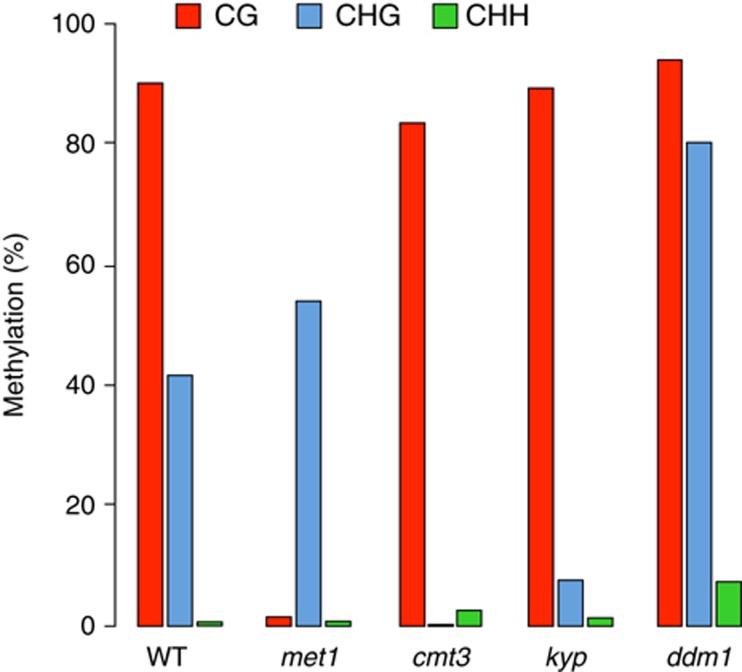
To understand how DNA methylation controls IBM1 expression, we examined DNA methylation profiles of the IBM1 gene by bisulphite sequencing. It has been reported that gene-body methylation is preferentially targeted to nucleosomes on exons (Chodavarapu et al, 2010), and accordingly, IBM1 carries body methylation at CG sites in several exons in WT plants (http://neomorph.salk.edu/epigenome/epigenome.html). In Arabidopsis, the average intron size is ∼180 bp (The Arabidopsis Genome Initiative, 2000). The IBM1 gene contains an unusually large intron that is >2 kb; this intron must be spliced out to generate the IBM1-L mRNA that encodes the jmjC histone demethylase domain. Interestingly, the large intron contains a zone that is densely methylated at both CG and CHG positions in the WT (http://neomorph.salk.edu/epigenome/epigenome.html; Figure 2). In the met1-3 null mutant, exonic CG methylation was lost in the IBM1 gene (http://neomorph.salk.edu/epigenome/epigenome.html). In the large intron, bisulphite sequencing confirmed that CG methylation was also erased; however, non-CG methylation, which was mostly represented by CHG methylation, was essentially maintained (Figure 2).
Figure 2.
Bisulphite sequencing analysis of the DNA methylation pattern in the large intron of IBM1. The position of the methylated zone is indicated in Figure 1. The proportions of methylated cytosines at the CG (red), CHG (blue) and CHH (green) sites are given as percentages.
In the kyp mutant, CG methylation remained largely intact. However, methylation at CHG positions drastically decreased from ∼40 to 10% (Figure 2). This indicates that CHG methylation in the large IBM1 intron strongly depends on KYP activity. The remaining low level of CHG methylation in the mutant probably results from the redundant activity of other SUVH proteins, such as SUVH5 and/or SUVH6, which was previously shown for other loci (Ebbs et al, 2005; Ebbs and Bender, 2006); although the contribution of SUVH5 and/or SUVH6 was not visualized at the transcriptional level (Figure 1D). DRM2 and CMT3 have been reported to be redundantly required for the maintenance of CHG methylation at particular genomic loci (Cao and Jacobsen, 2002; Cao et al, 2003). At the large intron of IBM1, CHG methylation was completely lost in cmt3, which indicates that CMT3 is solely responsible for CHG methylation at this locus; this is in agreement with the absence of detectable difference in IBM1-L accumulation in the drm1 drm2 background (Figures 1D and 2).
Based on the impact of the met1, cmt3 and kyp mutations on IBM1-L transcript accumulation, these data suggest that the simultaneous presence of CG and CHG methylation in the large IBM1 intron is required for the proper production of IBM1-L transcripts. Accordingly, the met1 cmt3 double mutant showed a stronger reduction in IBM1-L accumulation relative to each single mutant (Figure 1E). As mentioned above, the accumulation of IBM1 transcripts was not altered by the ddm1-2 mutation (Figure 1B), and consistent with our previous conclusion, the large IBM1 intron was not hypomethylated but rather hypermethylated at all of the cytosine sequence contexts in this mutant (Figure 2). Regardless of the mechanism responsible for IBM1 hypermethylation in the ddm1 mutant background, this result supports the conclusion that DNA methylation in the large IBM1 intron positively correlates with IBM1-L accumulation.
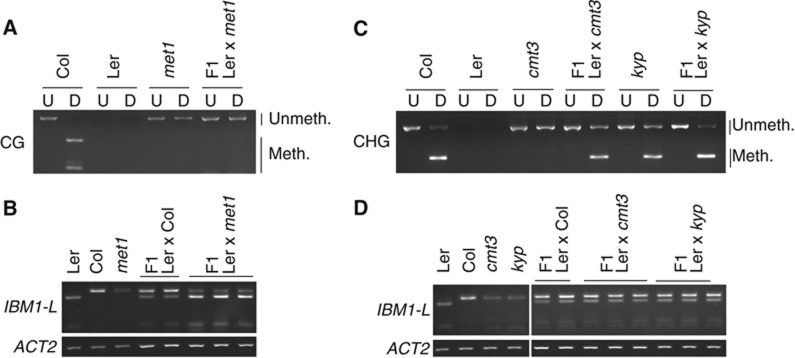
To further confirm that DNA methylation in the large IBM1 intron controls the accumulation of RNA that encodes the protein that contains the H3K9me2 demethylase jmjC domain, we outcrossed the met1, cmt3 and kyp mutants (all in the Col-0 background) with WT Ler-0 plants and assayed for DNA methylation and the transcription of the mutant-derived IBM1 allele in the resultant F1 plants. Once they are altered in met1 mutants, the CG methylation patterns cannot be re-established upon the reintroduction of MET1 activity (Soppe et al, 2000; Kankel et al, 2003; Saze et al, 2003; Mathieu et al, 2007). Consistently, PCR from bisulphite-treated DNA followed by digestion with a restriction enzyme showed that the met1-derived IBM1 allele was still unmethylated at CG sites in the Ler × met1 F1 hybrids (Figure 3A). This absence of remethylation correlated with a low accumulation of IBM1-L mRNA that originated from the met1-derived IBM1 allele, which was similar to the IBM1 transcription in the mutant parent (Figure 3B). In contrast to MET1, the reintroduction of CMT3 largely restores the developmental phenotypes induced by the loss of silencing that is associated with non-CG methylation in the drm1/2 cmt3 mutants (Chan et al, 2006). Methylation at CHG sites, which was completely lost in the cmt3 mutants, reappeared to a certain level in the large intron of the mutant-derived IBM1 allele in the Ler × cmt3 F1 individuals (Figure 3C). RT–PCR analyses revealed that the accumulation of the IBM1-L transcript from the cmt3-derived allele was restored to WT levels, such that the IBM1 transcriptional patterns in the Ler × cmt3 F1s and the Ler × Col control F1s were indistinguishable (Figure 3D). Likewise, the accumulation of IBM1-L mRNA from the kyp-derived IBM1 allele was re-established to WT levels in the F1 progeny of the Ler × kyp cross (Figure 3D). Restoration of CHG methylation at the mutant-derived IBM1 allele appeared to be more efficient in the Ler × kyp F1s when compared with the Ler × cmt3 F1 plants (Figure 3C), and this was confirmed with bisulphite sequencing (Supplementary Figure S1). Because CHG methylation was completely lost in cmt3 but not in kyp, CHG remethylation is likely more efficient or occurs faster in the presence of residual CHG methylation. Together, these observations indicate that DNA methylation in the large intron of IBM1 is required for the proper accumulation of the IBM1-L transcript that encodes the jmjC domain.
Figure 3.
Inheritance of IBM1 intronic methylation and IBM1-L transcript accumulation. (A, C) DNA methylation of the IBM1 large intron was assayed in the indicated genotypes by Col-0-specific PCR from bisulphite-treated DNA that was followed by digestion with TaqI (A) or HpyCH4V (C). CG and CHG methylation protect the TaqI and HpyCH4V sites, respectively, from bisulphite conversion, which facilitates restriction digestion after bisulphite treatment and PCR. Undigested (U) and digested (D) samples are shown. (B, D) Allele-specific RT–PCR analysis of IBM1-L transcript accumulation in Ler-0 × met1 (B) Ler-0 × cmt3 and Ler-0 × kyp (D) F1 individuals. The Ler × Col-0 individuals are shown as controls. The ACTIN 2 (ACT2) gene was used as a control.
RT–PCR analyses with primer sets that were designed along the length of the large IBM1 intron revealed that the reduction of the IBM1-L transcript accumulation in the met1 background occurred inside the methylated region of the large intron (Supplementary Figure S2). When primer sets located upstream of the intron methylation region were used, no clear difference in IBM1 pre-mRNA level was detected between the WT and met1. Furthermore, northern blot analyses using poly(A)+ RNA samples revealed no stable alternative polyadenylated IBM1 transcript variants in met1, cmt3 or kyp relative to the WT, indicating no differential transcript polyadenylation in these mutants (Figure 1B). Collectively, these results favour the hypothesis that intronic DNA methylation at CG and CHG sites is required for proper IBM1-L transcript elongation.
Enhanced IBM1-L mRNA accumulation complements the ibm1 mutation and largely restores H3K9me2 patterns in met1
The loss of CG methylation in met1 mutants results in aberrant patterns of other epigenetic marks, including non-CG methylation and H3K9me2. In met1-3 mutant nuclei, H3K9me2 is relocated away from heterochromatic chromocentres into euchromatic chromosomal regions (Tariq et al, 2003; Mathieu et al, 2007). To date, the underlying molecular mechanism that is responsible for this relocation has not been elucidated. Because IBM1 targets genes for H3K9 demethylation, we hypothesized that the lower accumulation of IBM1-L may account for the ectopic accumulation of H3K9me2 at genes in met1.
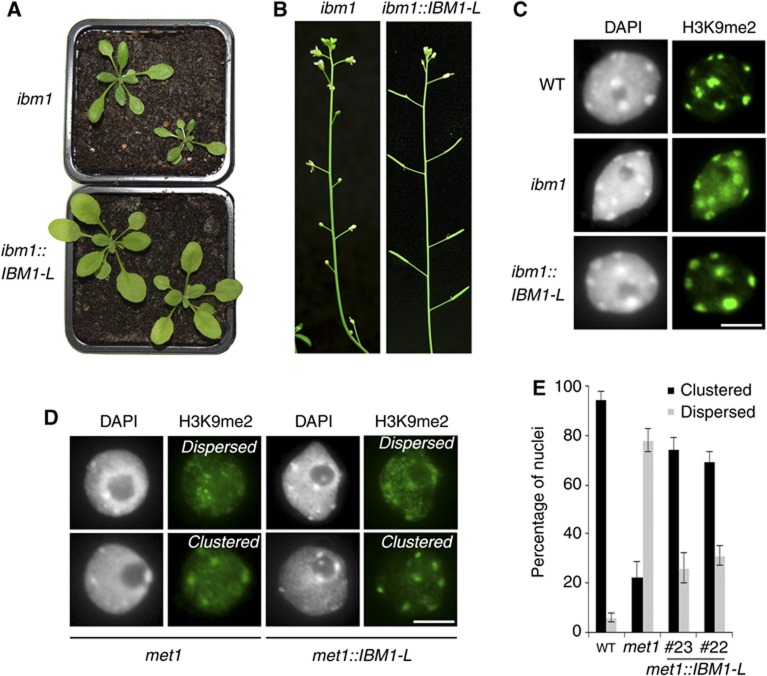
To test this hypothesis, we cloned the IBM1-L cDNA under the endogenous IBM1 promoter and used the resulting construct (pIBM1:IBM1-L) to transform ibm1-4/IBM1 and met1-3/MET1 heterozygous plants. Unlike untransformed ibm1 mutant segregants, the ibm1 individuals expressing IBM1-L (ibm1::IBM1-L) exhibited WT-like leaf size and morphology, as well as restored fertility (Figure 4A and B). Additionally, ibm1::IBM1-L plants showed reduced ectopic DNA methylation at the BONSAI gene (Supplementary Figure S3). These observations indicate that the pIBM1:IBM1-L construct efficiently complements the ibm1-4 mutation. In WT nuclei, H3K9me2 is essentially clustered at heterochromatic chromocentres (Figure 4C and E). The H3K9me2 signal in euchromatic regions was notably enhanced in nuclei of the ibm1-4 mutant compared with the WT (Figure 4C), which is consistent with the fact that IBM1 targets a large number of genes for H3K9 demethylation (Inagaki et al, 2010). The nuclei of complemented ibm1::IBM1-L lines displayed H3K9me2 patterns that were indistinguishable from the WT (Figure 4C). In nuclei from first-generation met1-3 mutants, H3K9me2 was frequently associated with euchromatic nuclear regions (Figure 4D and E; Tariq et al, 2003; Mathieu et al, 2007). The restored accumulation of IBM1-L in the met1-3 background in met1::IBM1-L plants (see Figure 5C and Supplementary Figure S5) drastically increased the proportion of nuclei that exhibited WT-like H3K9me2 patterns; this mark was preferentially associated with heterochromatin and less-intense euchromatin staining (Figure 4D and E). These results indicate that the downregulation of IBM1-L largely accounts for the relocation of H3K9me2 at gene-rich euchromatic regions that occurs in met1 mutants.
Figure 4.
IBM1-L expression complements the ibm1-4 mutation and restores H3K9me2 patterns in met1::IBM1-L plants. (A) Representative images of 3-week-old siblings segregating from self-pollination of an ibm1-4/IBM1::IBM1-L/+ parent are shown. (B) Inflorescences of ibm1 and ibm1::IBM1-L plants. H3K9me2 patterns in WT, ibm1-4, ibm1::IBM1-L (C), met1 and met1::IBM1-L plants (D) were analysed by immunocytology with a specific antibody against this mark. Representative images from one experiment are shown; two independent ibm1::IBM1-L and met1::IBM1-L (#22 and #23) lines were analysed, and they provided similar results. Two different H3K9me2 patterns were commonly observed in the met1-3 and met1::IBM1-L plants, and the signals were either dispersed from or clustered at the chromocentres. (E) Proportion of both types of nuclei was monitored and is represented as a histogram (±s.d.). Between 60 and 80 nuclei from three independent experiments were scored. Scale bar=5 μm.
Figure 5.
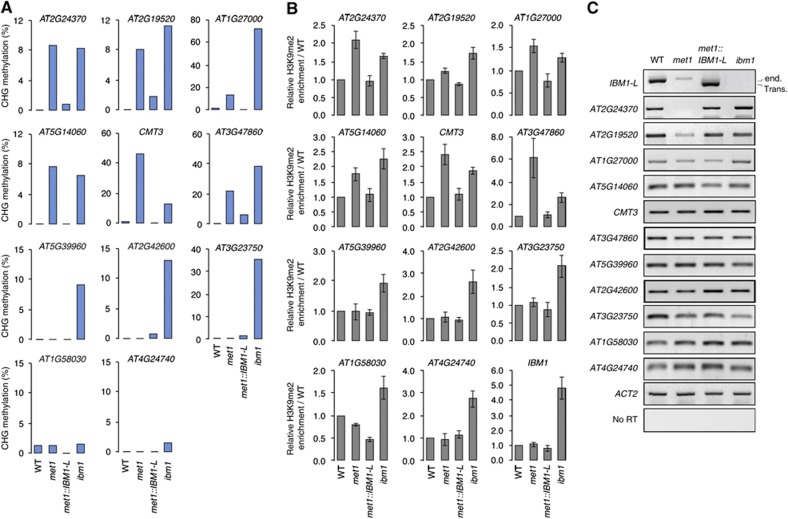
Restoration of IBM1-L transcript accumulation in met1 plants suppresses ectopic CHG hypermethylation and H3K9me2 enrichment at genes. (A) CHG methylation was determined in the WT, met1-3, met1::IBM1-L (#23) and ibm1-4 at the body of the indicated genes using bisulphite sequencing. Each region analysed contains 11–23 CHG sites. The full DNA methylation profiles are provided in Supplementary Figure S4. (B) Association of the indicated genes with H3K9me2 was determined by ChIP with a specific antibody against this mark and is represented as a relative enrichment over the WT. Quantifications are from three independent experiments (±standard error of the mean). (C) RT–PCR analysis of transcript accumulation at the indicated genes. ‘end.’ endogenous IBM1 transcript, ‘trans.’ transcript from the IBM1-L transgene. Amplification of ACTIN2 (ACT2) was used to normalize the RNA template levels. The negative controls (no RT) lacked reverse transcriptase.
Downregulation of IBM1-L is responsible for ectopic CHG methylation at genes in met1 mutants
Because the maintenance of H3K9me2 and CHG methylation are mechanistically linked, we sought to determine whether the relocation of H3K9me2 in heterochromatin in met1::IBM1-L was accompanied by the suppression of ectopic CHG methylation at genes. Using bisulphite sequencing, we analysed DNA methylation at 11 genes that are body methylated at CG positions in the WT (http://neomorph.salk.edu/epigenome/epigenome.html). Methylation at CHG sites was virtually absent in the WT at all these genes (Figure 5A). In agreement with the fact that IBM1 targets a large number of genes for H3K9me2 demethylation, all genes but one (AT1G58030) showed ectopic CHG methylation in the ibm1 mutant background compared with the WT. Among these, six also exhibited ectopic CHG methylation (∼10–40%) in the met1 background (Figure 5A; Supplementary Figure S4). In met1::IBM1-L, CHG methylation levels were strongly decreased in the body of these six genes, indicating that the enhanced expression of IBM1-L in met1 largely suppresses ectopic body methylation at CHG sites (Figure 5A; Supplementary Figure S4). Interestingly, four genes (AT5G39960, AT2G42600, AT3G23750, AT4G24740) were CHG hypermethylated in ibm1 but not in met1 (Figure 5A), suggesting that some genes are protected from ectopic CHG methylation in met1, although they are targeted for H3K9me2 demethylation by IBM1. Chromatin immunoprecipitation (ChIP) assays confirmed that the gene-body CHG hypermethylation in both met1 and ibm1 mutants was associated with enrichment in H3K9me2 at the same regions analysed for DNA methylation relative to the WT (Figure 5B). Consistent with the fact that IBM1-L encodes the functional form of the H3K9 demethylase, nearly WT H3K9me2 levels were restored in met1::IBM1-L at genes, showing ectopic H3K9me2 in the met1 mutant background (Figure 5B). Interestingly, the intronic DNA-methylated region of the IBM1 gene also showed H3K9me2 enrichment in ibm1, indicating that IBM1 targets its large intron for H3K9me2 demethylation (Figure 5B). However, the persistence of high CHG methylation levels at this genomic region in WT plants (see Figure 2), suggests that KYP-mediated H3K9me2 deposition surpasses IBM1-mediated demethylation activity.
We next determined whether these changes in DNA methylation and H3K9me2 patterns were associated with changes in transcriptional patterns. All 11 genes were transcribed in the WT and their transcription was not significantly altered in ibm1, although most of these genes showed ectopic CHG methylation and H3K9me2 in this mutant background and intact CG methylation (Figure 5A–C; Supplementary Figure S4). This suggests that aberrant CHG body methylation does not apparently impede transcription in the presence of WT CG gene-body methylation. Previous studies have revealed a modest impact of met1 mutation on gene expression compared with the massive transcriptional activation of pseudogenes and TEs (Zhang et al, 2006; Lister et al, 2008). Accordingly, of the six genes analysed showing ectopic CHG methylation in met1, only two (AT2G24370 and AT2G19520) were transcriptionally downregulated in met1 (Figure 5C; Supplementary Figure S5), which indicates that ectopic CHG body methylation does not necessarily lead to transcriptional silencing, even in the absence of CG methylation. Transcription from the genes that did not show ectopic CHG methylation in met1 was also not detectably altered in this mutant background (Figure 5C).
Importantly, suppression of both ectopic CHG methylation and CG methylation at AT2G24370 and AT2G19520 in met1::IBM1-L (Figure 5A; Supplementary Figure S4) restored WT transcript accumulation from these two genes (Figure 5C; Supplementary Figure S5). Therefore, CG body methylation appears to act as a protective mark at certain genes, safeguarding their transcription from the deleterious consequences of aberrant non-CG methylation. Why only a subset of genes do need such protection remains to be elucidated.
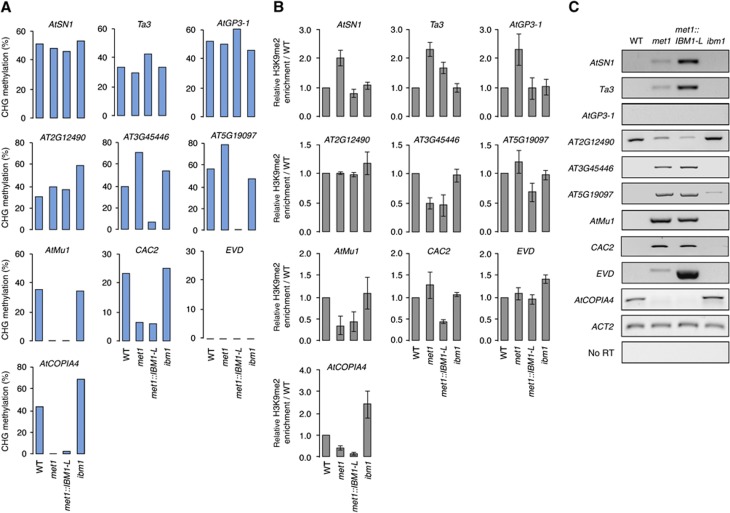
Unlike a large number of genes, it has been reported that TEs are not targeted by IBM1 (Inagaki et al, 2010). Accordingly, of the 10 elements that we analysed, only AtCOPIA4 was enriched in H3K9me2 and CHG hypermethylated in the ibm1 mutant (Figure 6A and B). Methylation at CG sites was shown to direct H3K9me2 at heterochromatic sequences, and H3K9me2 depletion was observed at selected TEs in met1 (Soppe et al, 2002; Lippman et al, 2003; Tariq et al, 2003). Although AtMu1, AtCOPIA4, AT3G45446 showed markedly decreased H3K9me2 levels in met1, no/subtle changes (AT5G19097, AT2G12490, CAC2, EVD) or clear enrichment in H3K9me2 (AtSN1, Ta3, AtGP3-1) were observed at the other elements (Figure 6B), indicating that the dependence of H3K9me2 over CG methylation is not conserved between distinct TEs. Noticeably, H3K9me2 levels in met1 and in met1::IBM1-L were highly similar, except at TEs showing increased H3K9me2 levels in met1 (e.g., AtGP3-1; Figure 6B). At these elements, enhanced expression of IBM1-L in met1::IBM1-L resulted in decreased H3K9me2 compared with met1 (Figure 6B). This suggests that these elements can be targeted by IBM1 for H3K9me2 demethylation in the absence of MET1/CG methylation. In contrast to genes, the changes in H3K9me2 levels at TEs were not necessarily mirrored by changes in CHG methylation. For instance, although AtSN1, Ta3 and AtGP3-1 were enriched in H3K9me2 in met1, CHG methylation remained nearly unchanged (Figure 6A).
Figure 6.
The mechanisms governing H3K9me2 and CHG methylation largely differ at genes and TEs. CHG methylation (A), H3K9me2 enrichment (B) and transcript accumulation (C) was determined at the indicated TEs in the WT, met1-3, met1::IBM1-L (#23) and ibm1-4 as described in the legend of Figure 5. Quantifications shown in (B) are from three independent experiments (±standard error of the mean).
RT–PCR analyses revealed that most of the elements were silent in the WT and transcriptionally activated in met1 mutants except the AtGP3-1 retrotransposon, which was still transcriptionally repressed in this mutant background as previously reported (Tsukahara et al, 2009; Figure 6C). Interestingly, silencing of AtGP3-1 in met1 suggests that IBM1 targeting at this locus occurs independent of transcription, although we cannot rule out transcriptional activity from this TE at specific developmental stages or in particular tissues of met1 plants. Conversely, the active transcription from AT2G12490 and AtCOPIA4 that was detected in the WT was downregulated in the met1 mutant background (Figure 6C), indicating that CG methylation differently impacts the transcription of distinct elements. These TEs were also transcribed in ibm1, suggesting that its transcriptional activity in the WT does not rely on IBM1 H3K9me2 demethylase activity. When compared with met1, the transcription of TEs was not significantly affected in met1::IBM1-L, except that of AtSN1, Ta3 and EVD, which was upregulated in met1::IBM1-L (Figure 6C). However, since this transcriptional upregulation was not necessarily associated with H3K9me2 depletion or decreased CHG methylation at these elements, it probably results from indirect effects of IBM1 on loci restraining the transcription of these elements in the met1 background. Together, these results confirm that IBM1 does not generally target TEs and highlight that the mechanisms governing H3K9me2 and CHG methylation largely differ at genes and TEs.
IBM1-L downregulation influences the expression of DNA demethylases
The Arabidopsis genome contains four genes that encode DNA demethylases, which are named DEMETER (DME), ROS1/DEMETER-LIKE1 (ROS1/DML1), DML2 and DML3. We previously showed that the expression of ROS1 and DME is repressed at the transcriptional level upon the loss of CG methylation in the met1-3 background (Mathieu et al, 2007). Because these demethylases preferentially target genic regions for demethylation (Penterman et al, 2007), their downregulation likely contributes to the aberrant DNA methylation patterns that are generated in met1. The effect of DME downregulation was confirmed by the sporadic appearance of abnormal flower phenotypes in met1 lines; these phenotypes were highly reminiscent of the developmental abnormalities that were observed in some met1 dme homozygotes (Supplementary Figure S6; Xiao et al, 2003).
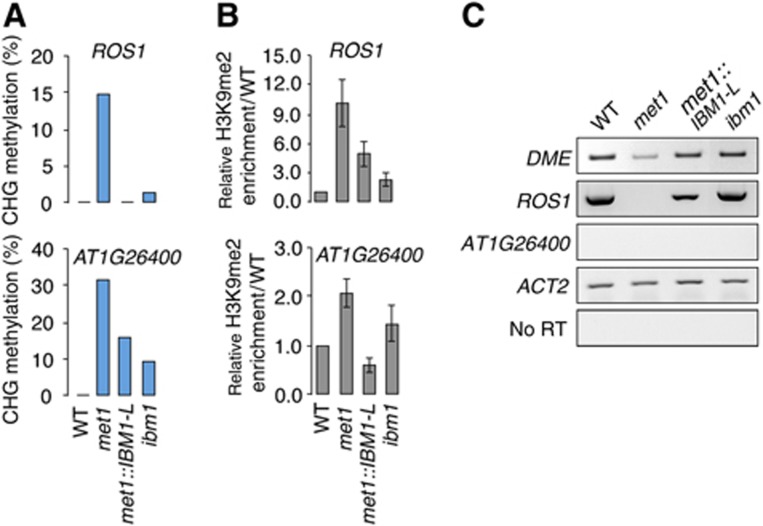
DME primarily functions in the central cell of the female gametophyte, whereas ROS1 additionally acts in the sporophyte. Interestingly, similar to the other genes that were analysed above (Figure 5A), bisulphite sequencing revealed that the coding region of ROS1 was ectopically hypermethylated at non-CG sites in met1 (Figure 7A; Supplementary Figure S4). Moreover, this methylation correlated with H3K9me2 enrichment in met1 and was directly associated with the downregulation of IBM1-L because it was suppressed in the met1::IBM1-L background (Figure 7B and C). The DME gene is similarly hypermethylated at CHG sites in met1 plants (http://neomorph.salk.edu/epigenome/epigenome.html). Importantly, the restored expression of IBM1-L in met1 also restored DME transcription and resumed ROS1 transcription, which was completely absent in the met1-3 background (Figure 7C; Supplementary Figure S5). This result indicates that the downregulation of IBM1-L in met1 mutants contributes to the creation of aberrant DNA methylation patterns at genes; this occurs because of its lack of direct activity at genes and also through the negative control of DNA demethylases expression. To further support this conclusion, we analysed DNA methylation at a target gene of ROS1 (AT1G26400), which is hypermethylated in the ros1-3 mutant (Penterman et al, 2007). As anticipated, this locus was also enriched in H3K9me2 and hypermethylated in met1, primarily at non-CG sites, but it was much less DNA hypermethylated and showed reduced H3K9me2 levels in met1::IBM1-L (Figure 7A; Supplementary Figure S4). This locus was enriched in H3K9me2 and CHG hypermethylated in the ibm1 mutant (Figure 7A and B), indicating that it is also targeted for H3K9me2 demethylation by IBM1.
Figure 7.
IBM1 targets DNA demethylase genes in the met1 background. (A) CHG methylation in the gene body of ROS1 and a ROS1-target gene (AT1G26400) was determined in the WT, met1-3 and a met1::IBM1-L line (#23) using bisulphite sequencing. The full DNA methylation profiles are provided in Supplementary Figure S4. (B) Association of ROS1 and AT1G26400 with H3K9me2 was determined by ChIP and is represented as a relative enrichment over the WT. Quantifications are from three independent experiments (±standard error of the mean). (C) RT–PCR analysis of the ROS1, DME and AT1G26400 transcript accumulation in the indicated genotypes. The amplification of ACTIN 2 (ACT2) was used to normalize the RNA template levels. The negative controls (no RT) lacked reverse transcriptase.
Discussion
Controlling gene expression through intron DNA methylation
DNA methylation has been widely associated with the repression of gene expression, for which two mechanisms have been proposed (Bird and Wolffe, 1999; Bird, 2002). The methylation of promoter sequences can directly inhibit transcription by blocking transcription factor binding; alternatively, DNA methylation can indirectly prevent transcription by recruiting various transcriptional repressors to form an inactive chromatin conformation. Here, we reveal an unorthodox role for DNA methylation, in which its presence in an intron is required for the proper expression of a gene. The large intron of IBM1 contains a region that is densely methylated at both CG and CHG positions, and a reduction in the level of either type of methylation results in the decreased accumulation of the IBM1-L transcript. In Arabidopsis, the average intron size is 180 bp (The Arabidopsis Genome Initiative, 2000). The IBM1 gene contains an unusually large intron of over 2 kb that must be spliced out to generate the IBM1-L mRNA, which encodes the jmjC histone demethylase domain. Less than 1% of Arabidopsis introns are >1 kb (Wang and Brendel, 2006). Because large introns may require more energy for transcription and splicing, they are expected to be selected against during evolution unless they provide regulatory features (Castillo-Davis et al, 2002). IBM1 orthologues in distantly related plant species and honeybee also contain introns that are much larger than the mean intron size of their respective genomes, which supports the evolutionary ancient origin of the large intron size in the regulation of IBM1 expression (Supplementary Figure S7).
At least three hypotheses can be formulated to explain how intronic methylation may promote IBM1-L accumulation. First, DNA methylation may prevent premature transcription termination by obscuring transcription termination sites. Indeed, DNA methylation tends to be distributed away from the 3′ ends of genes in Arabidopsis, suggesting that methylation is deleterious for transcription termination (Tran et al, 2005; Zhang et al, 2006; Zilberman et al, 2007; Lister et al, 2008). Second, DNA methylation may be required to maintain chromatin in an open state that is permissive for transcription elongation over the large intron. Specific methyl-binding proteins may bind to the methylated intronic region and recruit proteins that can modulate the chromatin compaction state, such as histone acetyltransferases. For example, the Arabidopsis METHYL-CpG BINDING DOMAIN 9 (AtMBD9) protein has been shown to bind to the FLOWERING LOCUS C (FLC) locus, where it triggers local histone acetylation, and enhances FLC expression (Peng et al, 2006; Yaish et al, 2009). A third possibility is that the presence of DNA methylation may directly or indirectly prevent the recruitment of a transcriptional repressor. In support of the latter hypothesis, DNA methylation was shown in mammals to interfere with the binding of the CCCTC-binding factor (CTCF), which induces the transcriptional repression of its target genes (Lobanenkov et al, 1990; Bell and Felsenfeld, 2000; Hark et al, 2000; Lai et al, 2010; Rodriguez et al, 2010). Moreover, CTCF binding influences polymerase II elongation dynamics and promotes RNA polymerase II pausing (Shukla et al, 2011). Although no direct CTCF homologues have been described in Arabidopsis (Heger et al, 2009), the loss of DNA methylation in the large IBM1 intron may recruit proteins with similar repressor activities, which could induce polymerase II pausing or reduce its elongation rate in the large intron; this would thereby lead to the premature dissociation of the polymerase, which has already been demonstrated in yeast (Uptain et al, 1997; Mason and Struhl, 2005; Gromak et al, 2006), and would therefore result in low IBM1-L transcript accumulation. Although our results tend to suggest that intronic DNA methylation may indeed be required for proper IBM1-L transcript elongation, the identification of additional mutations that affect IBM1 transcription will help to discriminate between these alternatives. Our results set the stage for future work to determine the molecular mechanism by which intronic DNA methylation affects Arabidopsis gene expression. Interestingly, this mechanism may be conserved in other eukaryotes because the expression of the EGR2 gene in mammals also appears to be positively regulated by intronic methylation (Unoki and Nakamura, 2003).
CG methylation and epigenome stability
From observations in Neurospora crassa and Arabidopsis, it has been long understood that DNA methylation and H3K9 methylation interact in heterochromatin. In N. crassa, DNA methylation by the DIM-2 DNA methyltransferase depends on the methylation of H3K9, which is deposited by DIM-5 (Rountree and Selker, 2010). In Arabidopsis, the efficient maintenance of CHG methylation involves a self-reinforcing feedback loop between CMT3, which is structurally similar to DIM-2, and KYP (Johnson et al, 2007). Dense CG methylation, which is characteristic of heterochromatin, also directs H3K9me2 and the loss of CG methylation in met1 null mutants leads to the depletion of H3K9me2 at heterochromatic sequences and the enrichment of H3K9me2 at euchromatic regions (Soppe et al, 2002; Tariq et al, 2003; Mathieu et al, 2007). Moreover, we and others previously showed that CG methylation directly prevents the accumulation of H3K4me2 and H3K27me3 in heterochromatin (Tariq et al, 2003; Mathieu et al, 2005; Zhang et al, 2009). In addition to the post-translational modification of histones, CG methylation also influences the deposition of histone variants, and there is a strong anti-correlative relationship between DNA methylation and H2A.Z distribution (Zilberman et al, 2008). Finally, CG methylation is required to stabilize proper patterns of non-CG methylation. In a manner that resembles CpG island methylation that occurs in mammalian tumourigenesis, the loss of CG methylation induces the CHH hypermethylation of several heterochromatic sequences and ectopic CHG methylation in the body of a large number of genes (Mathieu et al, 2007; Cokus et al, 2008; Lister et al, 2008; Reinders et al, 2008). The hypermethylation of heterochromatin was linked to the misdirection of the RdDM pathway, and the hypermethylation of several genes was attributed to inefficient DNA demethylation (Mathieu et al, 2007). Somehow paradoxically, the efficient transcription of genes that encode DNA demethylases, including ROS1 and DME, requires CG methylation, such that ROS1 transcription is totally impaired in met1 null mutants (Mathieu et al, 2007).
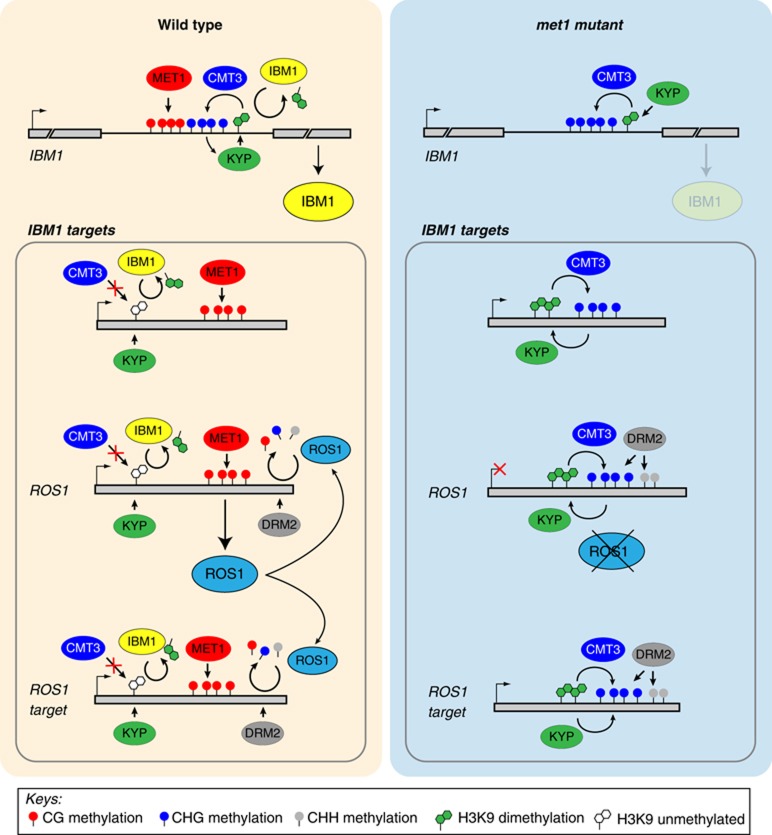
Mechanistically, it is unclear how CG methylation affects all of these epigenetic factors and epigenetic marks. Here, we demonstrated that the proper expression of a gene that encodes another demethylase, the IBM1 H3K9 demethylase, surprisingly also requires DNA methylation. Through this effect, CG methylation directly contributes to the stability of DNA methylation patterns at genes. In the absence of MET1, the loss of CG methylation in the large intron of IBM1 results in the downregulation of the functional form IBM1-L, which demethylates a large number of genes. By restoring IBM1-L accumulation in the met1-3 background, we showed that IBM1-L downregulation is responsible for the relocation of H3K9me2 in euchromatin and accounts for ectopic DNA methylation and H3K9me2 at genes in met1 mutants. Ectopic genic DNA methylation results from at least two distinct causes at different loci: (i) decreased IBM1-mediated H3K9 demethylation and (ii) the loss of ROS1 DNA demethylase activity. Remarkably, we demonstrated that these two mechanisms are also interconnected because the restored IBM1-L expression in met1 resumes ROS1 transcription (Figure 8). Moreover, we showed that the functions of KYP and CMT3 are also required to promote proper IBM1-L accumulation. These interconnections illustrate the sophisticated interplay between the methylation and demethylation pathways at the DNA and the histone levels.
Figure 8.
Model for the appearance of ectopic non-CG methylation and H3K9me2 at genes in met1 mutants. In WT plants, the simultaneous presence of MET1-mediated CG methylation and CMT3-mediated CHG methylation at the large intron of the IBM1 gene is required for proper accumulation of the transcript encoding the functional IBM1 activity. IBM1 targets a large number of CG body-methylated genes for H3K9me2 demethylation, including the gene encoding the ROS1 DNA demethylase as well as ROS1-target genes. Through removal of H3K9me2, the IBM1 activity prevents CHG methylation by protecting genes from CMT3 recruitment. Additionally, ROS1-mediated demethylation contributes to the stabilization of DNA methylation patterns by counteracting small RNA-mediated DNA methylation involving DRM2 at selected loci, including ROS1. Loss of CG methylation at the large intron of IBM1 in met1 mutants results in lower IBM1 accumulation, and thus to H3K9me2 enrichment at IBM1 target genes. In parallel, ROS1 transcription is totally turned off in met1. These two outcomes of CG methylation erasure are responsible for the appearance of alternative epigenetic patterns at genes: ectopic enrichment in H3K9me2 leads to ectopic CHG body methylation through CMT3 recruitment, while the absence of ROS1 permits DRM2-mediated methylation at CHG and CHH sequence contexts at ROS1-target loci.
Transcribed genes tend to be more CG methylated than poorly transcribed genes, and in met1, which displays reduced IBM1 activity, CG gene-body methylation is largely replaced by CHG methylation (Zhang et al, 2006; Zilberman et al, 2007; Lister et al, 2008). A comparison of CG body-methylated genes in the WT (Lister et al, 2008) with genes that show ectopic DNA hypermethylation in ibm1 mutants (Miura et al, 2009) revealed that the majority of genes most hypermethylated in ibm1 (about 73 and 57% of Class I and Class II genes, respectively) are strongly enriched in methylated CG positions (>25 methylated CGs) compared with the proportion of all genes with such CG methylation density (about 23%; Supplementary Figure S8). Therefore, we propose that CG methylation is involved in the recruitment of IBM1 to genes in WT plants, and CHG methylation can substitute for this signal in the met1 background. In support of this hypothesis, the CG and CHG methylation profiles are very similar in WT and met1 plants, respectively (Lister et al, 2008). If DNA methylation is the signal, then why is IBM1 not recruited to silent, heavily DNA-methylated heterochromatin? Some proteins require a combinatorial code of epigenetic marks to facilitate their recruitment to chromatin. For instance, CMT3 has been shown to bind in vitro to histone H3 peptides that are methylated at both H3K9 and H3K27 positions (Lindroth et al, 2004), and in mammals, the Origin recognition complex 2 (Orc2) protein is more efficiently recruited to nucleosomes that exhibit CG methylation in addition to H3K9me3 or H3K27me3 (Bartke et al, 2010). Therefore, it is plausible that the efficient recruitment of IBM1 to euchromatin may require DNA methylation and another epigenetic mark. Because we determined that transcriptionally silent genes can potentially be targeted by IBM1 in a met1 background (Figure 5), this aforementioned additional mark would need to be maintained in euchromatin regions independent of transcription and DNA methylation. However, no epigenetic mark that fulfils these criteria has been described in Arabidopsis. Alternatively, IBM1 recruitment may be prevented by the combinatorial presence of DNA methylation and a heterochromatin-specific mark, such that decreasing this mark would result in IBM1 binding and H3K9 demethylation. In support of this scenario, a reduction of the heterochromatin-associated histone modification H4K20me1 in suvh2 mutants is accompanied by decreased H3K9me2 at heterochromatic chromocentres (Naumann et al, 2005). Forward genetics approaches that are designed to identify additional mutants that display ectopic CHG methylation at genes will certainly provide more insights into IBM1 targeting.
The stability of epigenetic patterns along the entire genome is a highly orchestrated process that involves several molecular players that contribute to various epigenetic marks. These factors do not act independently, and in this epigenetic ‘symphony’, CG methylation may be observed as ‘the conductor’ because it plays a pivotal role in influencing the homeostasis of many other epigenetic marks. An understanding of the mechanisms that direct genomic CG methylation patterns remains an important challenge for future studies.
Materials and methods
Plant material
The met1-3 (Saze et al, 2003), cmt3-11 (Chan et al, 2006), kyp-7 (Mathieu et al, 2005), nrpd2a-2 (Onodera et al, 2005), drm1-2 drm2-2 (Chan et al, 2006), ddm1-2 (Vongs et al, 1993), vim1 vim2 vim3 (Woo et al, 2008), nrpd1a-4 (SALK_083051) and ibm1-4 (Saze et al, 2008) mutants were previously described. The met1-3 cmt3-11 double mutant, and the kyp-4 suvh5-2 suvh6-1 triple mutant were a kind gift from S Jacobsen and J Bender, respectively. All of the mutants are in the Columbia (Col-0) genetic background. The plants were grown in soil under long-day conditions (16 h light/8 h dark) at 22°C. For the 5-aza-dC treatment, seeds were germinated and grown for 9 days on 1/2 Murashige and Skoog medium that contained 4 μM 5-aza-dC (Sigma) as previously described (Mathieu et al, 2007).
RNA analyses
Total RNA was extracted from immature flower buds (or from leaves for Figure 1C and E) using the TRI Reagent (Sigma). For the RNA gel blot analyses, poly(A)+ RNA was purified from 18 μg of total RNA using the Oligotex Kit (Qiagen), and the entirety of the resultant samples was loaded onto the gel. The IBM1 probe was amplified from Col-0 genomic DNA using IBM1-F10 and IBM1-R12 primers (Supplementary Table 1) and was labelled with [α-32P]dCTP using random hexamer priming (Megaprime DNA labeling system, GE Healthcare). The RT–PCR reactions were performed using the OneStep RT–PCR Kit (Qiagen); the amplification of ACTIN2 or 18S rRNA was used as an internal control for the RNA amount. A derived-CAPS (dCAPS) assay was used to distinguish allele-specific IBM1-L transcript accumulation in Ler-0 × met1-3, Ler-0 × cmt3-11 and Ler-0 × kyp-7 F1 hybrids. The IBM1_polyR1 primer introduces a Rsa I restriction site specifically on Ler-0-derived alleles. The primers that were used are listed in Supplementary Table 1.
Transgene construction and transformation
The IBM1-L transgene—which consisted of 1124, bp of IBM1 promoter sequence fused to the full-length IBM1-L cDNA and 382 bp of sequence downstream of the IBM1-L stop codon—was cloned into the pHm43GW and pKm43GW binary vectors (Karimi et al, 2005) using the MultiSite Gateway® Three-Fragment Vector Construction technology (Invitrogen). The resultant IBM1-L transgenes (pHm43GW:IBM1-L and pKm43GW:IBM1-L) contain an additional BsrGI restriction site that was used to distinguish them from the endogenous IBM1 gene in the RT–PCR assays. The transgenes were introduced into Agrobacterium strain C58C1 and were used to transform plants that were heterozygous for the met1-3 (pKm43GW:IBM1-L) or ibm1-4 (pHm43GW:IBM1-L) mutations by floral dipping. The T1 generation transformants were genotyped for the respective mutation, and the homozygous mutants were analysed in comparison with their corresponding homozygous siblings that lacked the IBM1-L transgene.
DNA methylation analyses
Bisulphite conversion of genomic DNA was performed as previously described (Mirouze et al, 2009) using the Epitect Kit (Qiagen). The PCR products were cloned using the pGEM-Teasy vector (Promega). For each sample, 8–12 clones were sequenced. The sequencing data were visualized using the KisMeth software program (Gruntman et al, 2008), and the methylation percentage was calculated for each cytosine sequence context (CG, CHG and CHH). The primers that were used are listed in Supplementary Table 1.
For the IBM1 methylation analysis in the F1 hybrids, the intronic IBM1 methylated zone was amplified from bisulphite-treated DNA by nested-PCR with the IBM1-BS-F/IBM1-BS-R and IBM1-BS-Fbis/IBM1-BS-R primer sets (Supplementary Table 1), which facilitated the specific amplification of the Col-0 alleles. The PCR products were digested with TaqI (5′-TCGA-3′; 3 sites) and HpyCH4V (5′-TGCA-3′; 1 site) to diagnose the CG and CHG methylation, respectively. Methylation at the BNS locus was analysed by the double digestion of 30 ng of genomic DNA with BglII and EcoRI in a 30-μl reaction volume. The control ‘undigested’ samples were digested with EcoRI alone. One microlitre of each sample was amplified by PCR with the BNS-F2 and BNS-R3 primers (Supplementary Table 1) (Saze and Kakutani, 2007).
Chromatin immunoprecipitation
Leaves of 4-week-old seedlings (0.1 g) were harvested and chromatin was crosslinked and prepared as previously described (Mathieu et al, 2005). ChIP was performed with a mouse monoclonal antibody specific for dimethyl H3K9 (Abcam Ab1220) or a negative control IgG from rabbit (Diagenode) using the LowCell# ChIP kit (Diagenode) following the manufacturer’s instructions with some modifications. Chromatin samples were pre-cleared with Protein A-coated paramagnetic beads and immunoprecipitated with antibody overnight. Immune complexes were then collected with Protein A-coated paramagnetic beads. ChIP experiments were performed in three independent biological replicates. ChIP samples were amplified in duplicates or triplicates and target amplifications were normalized for amplification of TUBULIN 8 (met1 and met1::IBM1-L samples) or for amplification of the Ta3 element (ibm1 samples), for which H3K9me2 patterns are unaltered in ibm1 (Saze et al, 2008; Inagaki et al, 2010) using previously described procedures (Mathieu et al, 2005). The primers that were used are listed in Supplementary Table 1.
Immunocytology
The immunodetection of H3K9me2 was performed with young rosette leaves as previously described (Mathieu et al, 2005) using a mouse monoclonal antibody specific for dimethyl H3K9 (Abcam Ab1220, dilution 1:100). The nuclei were visualized with a Zeiss Axio Imager Z.1 microscope that was equipped with the Zeiss AxioCam MRm camera system and the Zeiss Axiovision software program.
Supplementary Material
Acknowledgments
We thank Tetsuji Kakutani, Hidetoshi Saze, Steve Jacobsen, Hervé Vaucheret and Judith Bender for providing Arabidopsis seeds; Ryan Lister for providing the list of methylated cytosines in the WT and met1-3 mutant; Huan Shu for technical advice; Marie-Noëlle Pouch Pélissier for assistance; and Charles White and Yoko Ikeda for helpful comments. The work in the Mathieu laboratory was supported by the CNRS, the INSERM, the Université Blaise Pascal, the Université d’Auvergne, and by the European Community’s Seventh Framework Programme (FP7 2007/2013) through a Starting Independent Researcher Grant from the European Research Council to OM (I2ST- Grant agreement #260742).
Author contributions: MR, TP and OM conceived the study. MR, ZK, TP and OM performed the experiments and analysed the data. OM supervised the work and wrote the paper together with MR.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T (2010) Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143: 470–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB (2001) The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res 29: 4319–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485 [DOI] [PubMed] [Google Scholar]
- Bender J (2004) DNA methylation and epigenetics. Annu Rev Plant Biol 55: 41–68 [DOI] [PubMed] [Google Scholar]
- Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE (2008) Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One 3: e3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP (1999) Methylation-induced repression--belts, braces, and chromatin. Cell 99: 451–454 [DOI] [PubMed] [Google Scholar]
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE (2003) Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13: 2212–2217 [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE (2002) Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12: 1138–1144 [DOI] [PubMed] [Google Scholar]
- Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA (2002) Selection for short introns in highly expressed genes. Nat Genet 31: 415–418 [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6: 351–360 [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Zhang X, Shah G, Chien JS, Jacobsen SE (2006) RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in arabidopsis. PLoS Genet 2: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, Casero D, Bernal M, Huijser P, Clark AT, Kramer U, Merchant SS, Zhang X, Jacobsen SE, Pellegrini M (2010) Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs ML, Bartee L, Bender J (2005) H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol Cell Biol 25: 10507–10515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs ML, Bender J (2006) Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18: 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE (2011) Epigenetic modifications in plants: an evolutionary perspective. Curr Opin Plant Biol 14: 179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S (2007) Heterochromatin revisited. Nat Rev Genet 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Gromak N, West S, Proudfoot NJ (2006) Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol 26: 3986–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntman E, Qi Y, Slotkin RK, Roeder T, Martienssen RA, Sachidanandam R (2008) Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489 [DOI] [PubMed] [Google Scholar]
- Heger P, Marin B, Schierenberg E (2009) Loss of the insulator protein CTCF during nematode evolution. BMC Mol Biol 10: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJ, Matzke M (2006) Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J 25: 2828–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki S, Miura-Kamio A, Nakamura Y, Lu F, Cui X, Cao X, Kimura H, Saze H, Kakutani T (2010) Autocatalytic differentiation of epigenetic modifications within the Arabidopsis genome. EMBO J 29: 3496–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Johnson L, Jasencakova Z, Zhang X, PerezBurgos L, Singh PB, Cheng X, Schubert I, Jenuwein T, Jacobsen SE (2004) Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112: 308–315 [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560 [DOI] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE (2007) The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 17: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ (2003) Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y (2006) JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7: 715–727 [DOI] [PubMed] [Google Scholar]
- Kraft E, Bostick M, Jacobsen SE, Callis J (2008) ORTH/VIM proteins that regulate DNA methylation are functional ubiquitin E3 ligases. Plant J 56: 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AY, Fatemi M, Dhasarathy A, Malone C, Sobol SE, Geigerman C, Jaye DL, Mav D, Shah R, Li L, Wade PA (2010) DNA methylation prevents CTCF-mediated silencing of the oncogene BCL6 in B cell lymphomas. J Exp Med 207: 1939–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, Jenuwein T, Khorasanizadeh S, Jacobsen SE (2004) Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J 23: 4286–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, May B, Yordan C, Singer T, Martienssen R (2003) Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol 1: E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH (1990) A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5'-flanking sequence of the chicken c-myc gene. Oncogene 5: 1743–1753 [PubMed] [Google Scholar]
- Mason PB, Struhl K (2005) Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell 17: 831–840 [DOI] [PubMed] [Google Scholar]
- Mathieu O, Probst AV, Paszkowski J (2005) Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J 24: 2783–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J (2007) Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130: 851–862 [DOI] [PubMed] [Google Scholar]
- Mirouze M, Reinders J, Bucher E, Nishimura T, Schneeberger K, Ossowski S, Cao J, Weigel D, Paszkowski J, Mathieu O (2009) Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461: 427–430 [DOI] [PubMed] [Google Scholar]
- Miura A, Nakamura M, Inagaki S, Kobayashi A, Saze H, Kakutani T (2009) An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J 28: 1078–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann K, Fischer A, Hofmann I, Krauss V, Phalke S, Irmler K, Hause G, Aurich AC, Dorn R, Jenuwein T, Reuter G (2005) Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J 24: 1418–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS (2005) Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622 [DOI] [PubMed] [Google Scholar]
- Peng M, Cui Y, Bi YM, Rothstein SJ (2006) AtMBD9: a protein with a methyl-CpG-binding domain regulates flowering time and shoot branching in Arabidopsis. Plant J 46: 282–296 [DOI] [PubMed] [Google Scholar]
- Penterman J, Uzawa R, Fischer RL (2007) Genetic interactions between DNA demethylation and methylation in Arabidopsis. Plant Physiol 145: 1549–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- Reinders J, Delucinge Vivier C, Theiler G, Chollet D, Descombes P, Paszkowski J (2008) Genome-wide, high-resolution DNA methylation profiling using bisulfite-mediated cytosine conversion. Genome Res 18: 469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez C, Borgel J, Court F, Cathala G, Forne T, Piette J (2010) CTCF is a DNA methylation-sensitive positive regulator of the INK/ARF locus. Biochem Biophys Res Commun 392: 129–134 [DOI] [PubMed] [Google Scholar]
- Rountree MR, Selker EU (2010) DNA methylation and the formation of heterochromatin in Neurospora crassa. Heredity (Edinb) 105: 38–44 [DOI] [PubMed] [Google Scholar]
- Saze H, Kakutani T (2007) Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J 26: 3641–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Mittelsten Scheid O, Paszkowski J (2003) Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet 34: 65–69 [DOI] [PubMed] [Google Scholar]
- Saze H, Shiraishi A, Miura A, Kakutani T (2008) Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science 319: 462–465 [DOI] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S (2011) CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479: 74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJ (2000) The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell 6: 791–802 [DOI] [PubMed] [Google Scholar]
- Soppe WJ, Jasencakova Z, Houben A, Kakutani T, Meister A, Huang MS, Jacobsen SE, Schubert I, Fransz PF (2002) DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J 21: 6549–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq M, Saze H, Probst AV, Lichota J, Habu Y, Paszkowski J (2003) Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci USA 100: 8823–8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Tran RK, Henikoff JG, Zilberman D, Ditt RF, Jacobsen SE, Henikoff S (2005) DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr Biol 15: 154–159 [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- Tsukahara S, Kobayashi A, Kawabe A, Mathieu O, Miura A, Kakutani T (2009) Bursts of retrotransposition reproduced in Arabidopsis. Nature 461: 423–426 [DOI] [PubMed] [Google Scholar]
- Unoki M, Nakamura Y (2003) Methylation at CpG islands in intron 1 of EGR2 confers enhancer-like activity. FEBS Lett 554: 67–72 [DOI] [PubMed] [Google Scholar]
- Uptain SM, Kane CM, Chamberlin MJ (1997) Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem 66: 117–172 [DOI] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ (1993) Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928 [DOI] [PubMed] [Google Scholar]
- Wang BB, Brendel V (2006) Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci USA 103: 7175–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Dittmer TA, Richards EJ (2008) Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet 4: e1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Pontes O, Pikaard CS, Richards EJ (2007) VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev 21: 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Gehring M, Choi Y, Margossian L, Pu H, Harada JJ, Goldberg RB, Pennell RI, Fischer RL (2003) Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev Cell 5: 891–901 [DOI] [PubMed] [Google Scholar]
- Yaish MW, Peng M, Rothstein SJ (2009) AtMBD9 modulates Arabidopsis development through the dual epigenetic pathways of DNA methylation and histone acetylation. Plant J 59: 123–135 [DOI] [PubMed] [Google Scholar]
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y (2006) JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125: 483–495 [DOI] [PubMed] [Google Scholar]
- Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE (2009) Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol 10: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S (2008) Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S (2007) Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39: 61–69 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.