Abstract
Peroxisomes undergo rapid, selective autophagic degradation (pexophagy) when the metabolic pathways they contain are no longer required for cellular metabolism. Pex3 is central to the formation of peroxisomes and their segregation because it recruits factors specific for these functions. Here, we describe a novel Saccharomyces cerevisiae protein that interacts with Pex3 at the peroxisomal membrane. We name this protein Atg36 as its absence blocks pexophagy, and its overexpression induces pexophagy. We have isolated pex3 alleles blocked specifically in pexophagy that cannot recruit Atg36 to peroxisomes. Atg36 is recruited to mitochondria if Pex3 is redirected there, where it restores mitophagy in cells lacking the mitophagy receptor Atg32. Furthermore, Atg36 binds Atg8 and the adaptor Atg11 that links receptors for selective types of autophagy to the core autophagy machinery. Atg36 delivers peroxisomes to the preautophagosomal structure before being internalised into the vacuole with peroxisomes. We conclude that Pex3 recruits the pexophagy receptor Atg36. This reinforces the pivotal role played by Pex3 in coordinating the size of the peroxisome pool, and establishes its role in pexophagy in S. cerevisiae.
Keywords: autophagy, mitophagy, peroxin, pexophagy, receptor
Introduction
Saccharomyces cerevisiae depends exclusively on the extracellular medium for its supply of nutrients to sustain growth. It is able to respond rapidly to changing environmental conditions, and some organelles, such as peroxisomes, fluctuate in size and number. Peroxisomes carry out a number of metabolic functions, including a variety of hydrogen peroxide-producing oxidation reactions, and their abundance reflects the requirement of the pathways they contain for cellular metabolism.
Peroxisomes multiply by growth and division and under certain conditions can form directly from the ER. Genetic dissection of peroxisome biogenesis has identified a large number of genes (known as PEX genes) required for peroxisome multiplication and segregation, as well as for import of peroxisomal luminal proteins and peroxisomal membrane biogenesis. Many of the genes required for peroxisome biogenesis are evolutionarily conserved, and mutations in the human orthologues cause Zellweger spectrum disorders (ZSDs). Two of these genes, PEX3 and PEX19, are required for peroxisomal membrane biogenesis. Peroxisomal structures are absent in pex3 and pex19 mutants, and as a consequence most peroxisomal membrane proteins (PMPs) are broken down. The exact role of Pex3 and Pex19 in peroxisomal membrane biogenesis is under intensive study, and several roles have been proposed, including import of PMPs into the ER, exit of PMPs from the ER, and direct import of PMPs into peroxisomal structures (for reviews, see Ma et al, 2011; Rucktaschel et al, 2011). Additionally, Pex3 has been implicated in peroxisome segregation in S. cerevisiae (Munck et al, 2009), and a Pex3-like protein has been implicated in this process in Yarrowia lipolytica (Chang et al, 2009). An additional role for Pex3 comes from work in methylotrophic yeast where it has been implicated in the selective autophagic breakdown of peroxisomes, a process known as pexophagy (Bellu et al, 2002; Farre et al, 2008; van Zutphen et al, 2011).
Macroautophagy (hereafter referred to as autophagy) is a catabolic process that allows recycling of cytoplasmic components, and in yeast functions primarily as an adaptive mechanism for survival in response to changes in the availability of nutrients in the environment (de Duve and Wattiaux, 1966; Mitchener et al, 1976; Takeshige et al, 1992; Levine and Klionsky, 2004; Yorimitsu and Klionsky, 2005b). During autophagy, cytoplasmic contents are enclosed by a de novo-synthesised double membrane to form a structure termed an autophagosome (Baba et al, 1994; Mizushima et al, 2001). The outer layer of the autophagosome then fuses with the vacuolar membrane and a single membrane structure, referred to as an autophagic body, enters the vacuole lumen where it is degraded by the vacuolar hydrolases and recycled into reusable components. Although this process is mainly non-selective, some cargoes are selectively sequestered by preautophagosomal structures (PAS). These selective autophagy pathways include mitophagy, ER-phagy, ribophagy, xenophagy, aggrephagy, pexophagy and the cytoplasm-to-vacuole targeting (Cvt) pathway, a biosynthetic pathway that delivers some vacuolar hydrolases from the cytosol to the vacuole (Harding et al, 1995, 1996; Campbell and Thorsness, 1998; Hutchins et al, 1999; Kim et al, 2002; Ravikumar et al, 2002; Suzuki et al, 2002; Rich et al, 2003; Bernales et al, 2006; Xie and Klionsky, 2007; Kraft et al, 2008; Lynch-Day and Klionsky, 2010). In selective processes, cargo recognition is an important step and requires a specific receptor-like protein that links the protein/organelle to the autophagy machinery at the PAS (Scott et al, 2001; Lynch-Day and Klionsky, 2010; Weidberg et al, 2011).
Genetic approaches in yeasts have identified >30 components required for selective or non-selective autophagy, and these have been designated Atg proteins (Xie and Klionsky, 2007; Suzuki and Ohsumi, 2010). A subset of these Atg proteins is required for autophagosome formation. This subset is referred to as the ‘core’ autophagy machinery, and its role in double-membrane vesicle formation has been well studied (Suzuki et al, 2007; Xie and Klionsky, 2007; Nakatogawa et al, 2009). The core machinery is modulated with additional Atg components to accommodate the non-selective sequestration of bulk cytosol or the selective sequestration of specific cargoes. In S. cerevisiae, cargo-specific receptors for the Cvt pathway (Atg19 and Atg34) (Shintani et al, 2002; Suzuki et al, 2010) and mitophagy (Atg32) (Kanki et al, 2009; Okamoto et al, 2009) have been identified, and in Pichia pastoris, the pexophagy receptor has been identified (PpAtg30) (Farre et al, 2008). These cargo-specific receptors link to the core autophagy machinery via Atg11. The Cvt receptors Atg19 and Atg34 also interact stably with Atg8 via an Atg8-interacting motif (AIM), and this interaction is required for cargo delivery to the vacuole (Noda et al, 2008; Suzuki et al, 2010). Likewise, in mammalian cells, known autophagy receptors bind to Atg8-family orthologues via conserved LIRs (LC3-interacting region), and these interactions are essential for function (Kirkin et al, 2009; Novak et al, 2010; Wild et al, 2011). For one of these receptors, optineurin, a serine residue directly flanking the LIR can be phosphorylated, and this phosphorylation stimulates interaction with LC3 (Wild et al, 2011). This has been proposed to be a general mechanism to regulate selective autophagy (Wild et al, 2011). Surprisingly, although the yeast mitophagy receptor Atg32 contains an AIM that interacts with Atg8, inactivation of this AIM has only a mild effect on function (Okamoto et al, 2009; Kondo-Okamoto et al, 2012). No interaction between the pexophagy receptor Atg30 and Atg8 has been described.
To date, most mechanistic insight into pexophagy has come from dissection in methylotrophic yeasts, namely P. pastoris and Hansenula polymorpha (for reviews, see Dunn et al, 2005; Sakai et al, 2006; Manjithaya et al, 2010b). In P. pastoris, Atg30 marks peroxisomes for breakdown by linking peroxisomes with the autophagy machinery (Farre et al, 2008). Atg30 binds two PMPs, Pex3 and Pex14, and although these proteins have also been implicated in pexophagy in mammalian cells (Hara-Kuge and Fujiki, 2008) and H. polymorpha (van Zutphen et al, 2008), their role in pexophagy is not understood. In mammalian cells, attaching ubiquitin to peroxisomes via Pex3 or PMP34 targets them for autophagic degradation (Kim et al, 2008), whereas removal of Pex3 from peroxisomes was shown to precede pexophagy in H. polymorpha (Bellu et al, 2002; van Zutphen et al, 2011). Studies into the role of Pex3 in pexophagy are complicated by the multiple roles of Pex3 in peroxisome formation and segregation.
In this paper, we report the isolation of pex3 alleles, including pex3-177, affected specifically in pexophagy. We describe the identification and initial characterisation of a new Pex3-interacting protein, which we have named Atg36, as it is required for pexophagy in S. cerevisiae: its overexpression induced pexophagy, and its absence prevented pexophagy. We found that pex3-177 cells are defective in the recruitment of Atg36 to peroxisomes. Furthermore, we show that Atg36 binds Atg8 and Atg11 in vivo, and is required to link peroxisomes to Atg11-positive structures. We analysed eight motifs in Atg36 that weakly resemble the AIM/LIR consensus (Noda et al, 2008; Johansen and Lamark, 2011), and found that although one of these sequences is required for Atg36-driven pexophagy, it is not conserved and does not constitute a functional AIM. We show that Atg36 is present on peroxisomes, and that it is cotransported with peroxisomes into the vacuole during pexophagy where it is broken down. We found that Atg36–GFP targets to mitochondria if Pex3 is mislocalised there, where it can drive mitophagy. Finally, we found that endogenous Atg36 can restore mitophagy in atg32Δ cells expressing a mitochondrial version of Pex3. This shows that Pex3 can act as a transplantable module that links its cargo to the autophagy apparatus via Atg36. We conclude that Atg36 comprises the pexophagy receptor and that Pex14 is not required for pexophagy in S. cerevisiae. We show the role of Pex3 in pexophagy is to recruit Atg36 to peroxisomes, which in turn links peroxisomes to the autophagy apparatus. This finding reinforces the central role that Pex3 plays in coordinating peroxisome formation, segregation and breakdown.
Results
Atg36 is required for autophagic breakdown of peroxisomes
S. cerevisiae Pex3 is a membrane protein with a short luminal N-terminus followed by a single transmembrane segment and a cytosolic domain (residues 40–441) (Hohfeld et al, 1991). We performed a yeast two-hybrid screen with the cytosolic domain of S. cerevisiae Pex3 and identified a novel uncharacterised protein, encoded by YJL185c, as a potential Pex3-interacting protein. We have named this gene ATG36. ATG36 encodes a 34-kDa protein that is not well conserved. Using BLASTP searches we identified a few putative orthologues among Saccharomycetaceae species, including Zygosaccharomyces rouxii (CAQ43500), Ashbya gossypii (NP_984715), Lanchancea thermotolerans (XP-002554387) and Vanderwaltozyma polyspora (XP-001646846).
To investigate the possible function of Atg36 in peroxisome biogenesis, we compared atg36Δ cells with WT cells for their ability to form peroxisomes and segregate them during cell division. GFP appended with a peroxisomal targeting signal type 1 (GFP–PTS1) is imported post-translationally into peroxisomes, and cytosolic labelling is observed in cells that lack peroxisomes or are defective in lumenal protein import. When import is intact, GFP–PTS1 is imported into puncta that represent individual peroxisomes or small clusters of peroxisomes. The number of peroxisomes in a cell is a reflection of the processes of peroxisome formation and the mechanisms that regulate peroxisome number including degradation and segregation. The relative distribution of puncta between mother and bud is a measure for segregation of peroxisomes during cell division.
Peroxisome number is increased in atg36Δ cells under most growth conditions (Supplementary Figure S1A and B). This increase is best illustrated by switching cells from conditions of peroxisome proliferation (oleate medium) to glucose starvation medium (SD-N), a condition that induces pexophagy. In WT cells, peroxisome numbers decreased dramatically whereas peroxisome numbers remained high in atg36Δ cells.
Prolonged growth on glucose medium at log-phase conditions, however, results in normalisation of peroxisome number (Supplementary Figure S1B). This clearly demonstrates that replicative peroxisome multiplication is unaffected. Both mother and daughter cells contain peroxisomes under all growth conditions tested, indicating that segregation is unaffected. Since atg36Δ cells grow normally on oleate (Supplementary Figure S1C), peroxisome function is not severely affected. This indicates the β-oxidation machinery is properly induced, and that peroxisomes proliferate normally in response to oleate. Indeed, Pex11 levels are induced on oleate to a level comparable to that in WT cells. Import of the luminal marker GFP–PTS1 is unaffected.
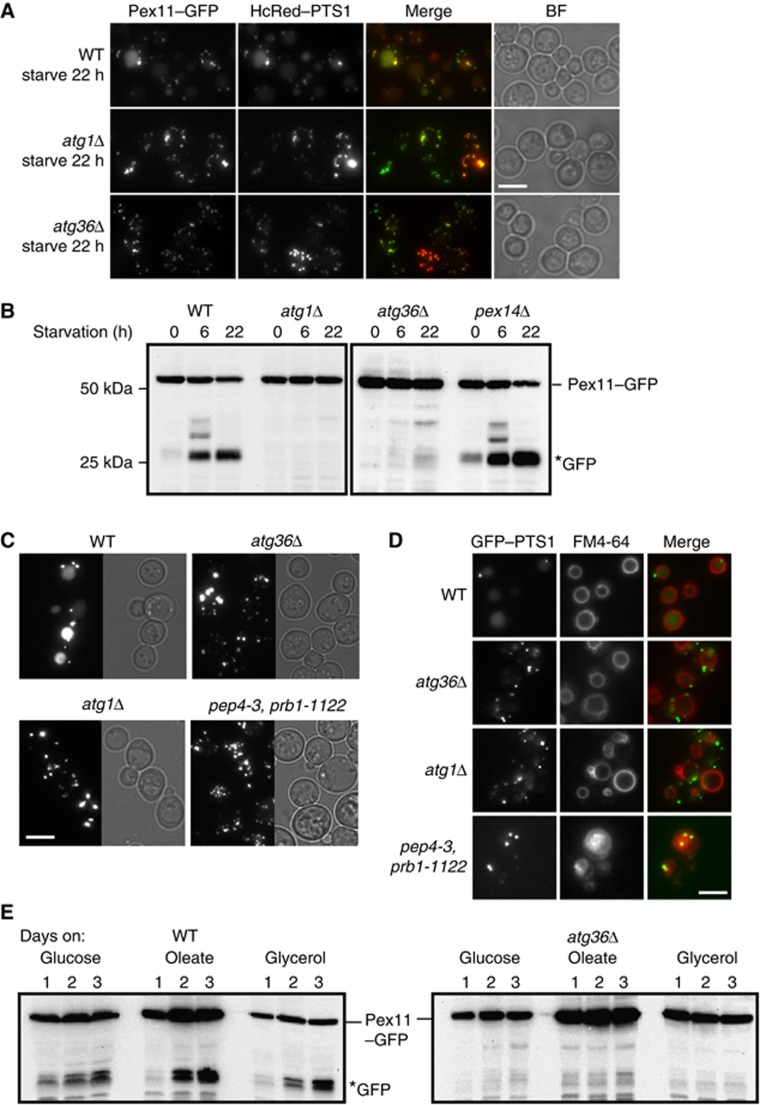
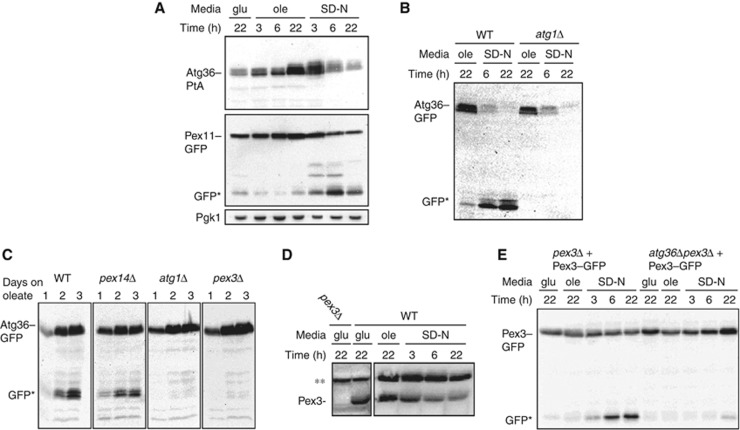
The finding that post-log phase and nitrogen-starved atg36Δ cells have increased peroxisome numbers suggests that Atg36 may be required for peroxisome turnover. We investigated this phenotype further using biochemical and microscopy-based pexophagy assays. The biochemical assay measures degradation of Pex11 expressed from its own promoter and tagged at its C-terminus with GFP. Pex11 is the most abundant PMP and is induced on oleate. Knoblach and Rachubinski (2010) show that a minor fraction of Pex11 locates to the peripheral ER during growth on oleate, and subsequently translocates to peroxisomes when cells are switched to glucose medium. We find Pex11–GFP colocalises with HcRed–PTS1 in cells grown on glucose. In cells grown overnight on oleate, Pex11–GFP-labelled peroxisomes tend to cluster somewhat, although even under this condition, the overlap is almost complete, with just occasional Pex11–GFP-containing membrane extensions not containing HcRed–PTS1 (Supplementary Figure S1E). This clustering also occurs with Pex11-monomeric GFP (Supplementary Figure S1D). Switching the Pex11–GFP-labelled cells from oleate to SD-N medium restores complete overlap of the two peroxisomal markers: 3 h after switching most of the clustered structures have disappeared, punctate structures are present, and some faint labelling of the vacuole became apparent (Supplementary Figure S1E). A further reduction in peroxisome number and a clear vacuolar labelling can be seen in WT cells after 22 h (Figure 1A). This shows that Pex11–GFP can be used as a marker to follow peroxisome degradation. Vacuolar labelling in WT cells corresponds by western blot to the appearance of GFP, the protease-resistant breakdown product of Pex11–GFP (Figure 1B). In WT cells, this breakdown product is evident after 6 h on starvation medium, in contrast to in atg36Δ and atg1Δ cells, where peroxisomes remain intact (Figure 1A) and Pex11–GFP remains stable even after 22 h on starvation medium (Figure 1B).
Figure 1.
Atg36 is required for pexophagy. Validation of Pex11–GFP as a marker for pexophagy in WT, atg1Δ, and atg36Δ cells by (A) microscopy and (B) western blot including pex14Δ cells. The fluorescence assay shows that like HcRed–PTS1, Pex11–GFP accumulates in the vacuole in starvation medium (A), which corresponds to the accumulation of GFP on a western blot (B). (B) Cells were grown 18 h in oleate medium and switched to glucose medium lacking nitrogen (SD-N) for the times indicated. GFP* indicates the relatively protease-resistant degradation product indicative of vacuolar breakdown. (C) Pexophagy assay by fluorescence of GFP–PTS1 expressed from the CTA1 promoter in WT, atg36Δ, atg1Δ, and pep4-3, prb1-1122 strains. Cells were grown as above for Pex11–GFP pexophagy assay and imaged after 22 h in SD-N medium. Multiple fluorescent images were acquired in Z-axis and flattened into a single image. Bright-field image is a single plane. (D) FM4-64 labelling of cells grown as in (C). The three central images of a Z-stack were flattened into a single plane. Bright-field image is a single plane. Bar, 5 μm. (E) Pexophagy as assayed by Pex11–GFP western blot of cells grown for up to 3 days in glucose, oleate or glycerol medium. Samples of WT cells (left panel) and atg36Δ cells (right panel) were taken from the cultures after 24, 48 and 72 h.
We tested a series of mutants for their ability to induce Pex11–GFP breakdown under starvation conditions. Immunoblotting of pex1Δ, pex2Δ, pex4Δ, pex5Δ, pex6Δ, pex7Δ, pex10Δ, pex11Δ, pex12Δ, pex13Δ, pex14Δ, pex15Δ, pex17Δ, pex18Δ, pex22Δ, pex25Δ, pex27Δ, pex28Δpex29Δ, pex30Δpex31Δpex32Δ, pex34Δ, djp1Δ, inp1Δ, inp2Δ, pxa1Δ, pxa2Δ, and ant1Δ cells revealed that all these mutants formed the typical Pex11–GFP breakdown product (data not shown). Surprisingly, pexophagy is fully functional in pex14Δ cells (Figure 1B).
We also used GFP–PTS1 expressed from the oleate-inducible promoter of the peroxisomal catalase gene (CTA1) to monitor pexophagy (Figure 1C). The decrease in fluorescent puncta and the appearance of a vacuolar GFP signal, as seen above for Pex11–GFP and HcRed–PTS1, is indicative of pexophagy in WT cells. Peroxisomes remained intact in atg1Δ and atg36Δ cells, and also in pep4-3, prb1-1122 cells, in which many of the vacuolar hydrolases fail to become activated. The identity of the vacuolar compartment was confirmed with FM4-64 (Figure 1D) (Vida and Emr, 1995). Therefore, pexophagy as assayed by western blot correlates with pexophagy as assayed by microscopy.
As described above, peroxisomes were more numerous in post-log phase glucose-grown atg36Δ cells compared with WT cells, suggesting that peroxisome degradation in post-log cells also depends on Atg36. To test this, we grew cells expressing Pex11–GFP in either glucose, glycerol or oleate medium for 3 days continuously and monitored pexophagy at 24 h intervals by immunoblotting. We found pexophagy occurred under all these conditions, and in all cases was Atg36-dependent (Figure 1E).
Atg36 is not required for mitophagy, the Cvt pathway or non-specific macroautophagy
To test whether the requirement for Atg36 is specific to pexophagy, we examined the functionality of other types of autophagy in atg36Δ cells.
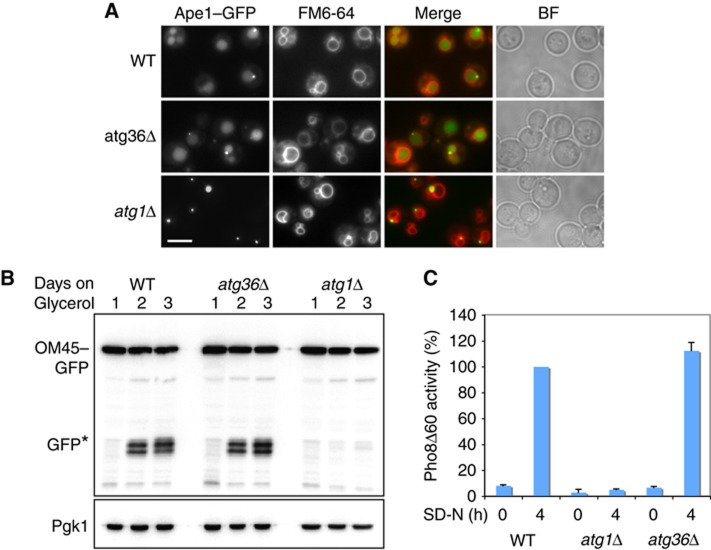
The Cvt pathway was tested by the accumulation of the marker Ape1–GFP in the vacuole. This pathway is functioning normally in atg36Δ cells (Figure 2A). Mitophagy was measured by degradation of the mitochondrial outer membrane marker OM45–GFP after growing cells for up to 72 h in glycerol (Kanki and Klionsky, 2008; Okamoto et al, 2009). The appearance of free GFP indicates vacuolar degradation of OM45–GFP. As shown in Figure 2B, mitophagy occurs in atg36Δ cells after 2 days growth, similar to WT cells. In contrast, atg1Δ cells are unable to degrade OM45–GFP. Non-selective autophagy induced by nitrogen starvation was monitored by measuring the uptake of a cytosolic form of alkaline phosphatase into vacuoles. As shown in Figure 2C, this pathway is fully functional in atg36Δ cells.
Figure 2.
Atg36 is not required for the Cvt pathway, mitophagy or non-specific autophagy. (A) Cvt pathway activity was assessed by endogenous expression of Ape1–GFP in WT, atg36Δ and atg1Δ cells. Cells were grown for 18 h in glucose medium and vacuoles were stained with FM4-64. Accumulation of GFP in the vacuole indicates normal function of pathway. Bar, 5 μm. (B) Mitophagy of WT, atg36Δ and atg1Δ cells was assayed by western blot analysis of OM45–GFP after culturing cells for up to 3 days in glycerol medium. Samples were taken at 24 h intervals. (C) WT, atg36Δ and atg1Δ cells were assayed for non-specific autophagy by alkaline phosphatase assay. Cells were grown in YPD and shifted to SD-N medium for 4 h. Samples were collected and processed for Pho8Δ60 activity. The results represent the mean and s.d. of three experiments. WT 4 h starvation is set at 100%.
We conclude that Atg36 is a new autophagy factor required specifically for pexophagy.
Atg36 localises to peroxisomes and its overexpression induces pexophagy
To examine the localisation of Atg36, we tagged it in the genome with GFP. The signal was very low under most conditions tested (Supplementary Figure S2). However, a faint punctate pattern became apparent in post-log glucose or oleate-grown cells that was absent in pex3Δ cells. Coexpression in WT cells with HcRed–PTS1 confirmed colocalisation with peroxisomes. Peroxisomes labelled with Atg36–GFP are clustered rather than dispersed throughout the cell. This clustering is also observed when Atg36 is tagged with monomeric GFP (Supplementary Figure S2), indicating that dimerisation of GFP is not the cause of the clustering.
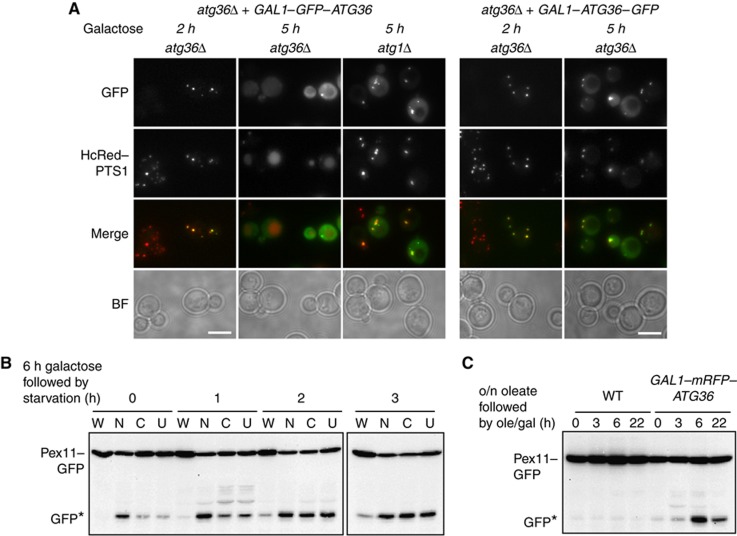
We expressed Atg36 from a centromeric plasmid either untagged or with an N- or a C-terminal GFP or mRFP tag under control of the GAL1 promoter (Figure 3). We noticed that at early time points after growth in galactose, Atg36 could be seen colocalising with the peroxisomal marker HcRed–PTS1 (Figure 3A). After 5 h induction of GFP–Atg36, the punctate pattern of HcRed–PTS1 had disappeared and labelling of the vacuole became apparent. The autophagic nature of this degradation was confirmed by overexpressing GFP–Atg36 in atg1Δ cells, where Atg36-labelled peroxisomes, but no degradation occurred (Figure 3A). Autophagic degradation of peroxisomes was less evident at this time point when overexpressing C-tagged Atg36 (right hand panel). We compared the galactose-inducible pexophagy activity of the various tagged versions with pexophagy by endogenous Atg36 in WT cells grown for 6 h on galactose, then switched to starvation conditions (Figure 3B). WT cells contained either empty plasmid (W), GAL1–mRFP–Atg36 (N), GAL1–Atg36–mRFP (C) or GAL1–Atg36 (U) (Figure 3B). Cells overexpressing any of the three versions of Atg36 induce pexophagy above endogenous level in WT cells. The N-tagged version is much more active than the C-tagged or untagged versions, and its activity is less dependent on switching to starvation conditions. Interestingly, the C-tagged and untagged versions are activated to the level of the N-tagged version by growth in starvation medium. Furthermore, the N-tagged version induced pexophagy even under peroxisome proliferation conditions (Figure 3C). This suggests that N-tagged Atg36 is constitutively active. The N- and C-tagged versions are expressed at similar levels after 6 h galactose induction (Supplementary Figure S3B), and this level is ∼50- to 100-fold the endogenous level under the same growth conditions (Supplementary Figure S3B and C). Although the tagged versions are enormously overexpressed, pexophagy is only moderately induced, suggesting that the level of Atg36 on its own does not determine the magnitude of the pexophagy response (see also below).
Figure 3.
Atg36 localises to peroxisomes and induces their autophagic degradation. (A) N- (left) and C- (right) GFP-tagged ATG36 under control of the GAL1 promoter was induced in atg36Δ or atg1Δ cells. Early (2 h) and later (5 h) time points after induction are shown. Pexophagy is assessed using the peroxisomal marker HcRed–PTS1 expressed from the constitutive HIS3 promoter. (B) Pex11–GFP western blot showing pexophagy in WT cells grown 6 h in galactose medium (0) then switched to starvation medium for 1, 2 or 3 h as indicated. (W), WT (empty plasmid); (N), GAL1–mRFP–ATG36; (C), GAL1–ATG36–mRFP; (U), GAL1–ATG36. WT shows pexophagy by endogenous Atg36. (C) Pex11–GFP pexophagy of WT cells containing either empty plasmid (WT) or GAL1–mRFP–ATG36 grown 18 h in oleate medium (0) then switched to oleate medium containing 2% galactose and harvested at 3, 6 or 22 h as indicated.
Pex3 recruits Atg36 to peroxisomes
Bioinformatic analysis of the Atg36 amino-acid sequence did not reveal any characteristics that indicate it could associate with membranes or be imported into organelles, as no potential transmembrane region, lipid-binding domain or targeting signal was detected. However, binding of Atg36 to peroxisomes is saturable: Atg36-labelled peroxisomes at early time points after galactose induction, but further expression resulted in cytosolic accumulation of GFP–Atg36 (Figure 3A).
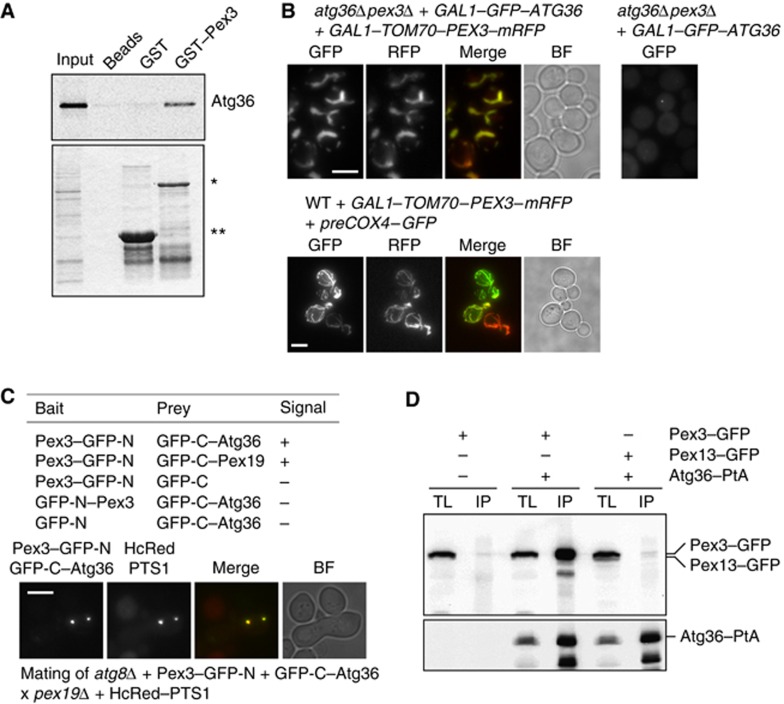
Since we identified Atg36 in a yeast two-hybrid screen using the cytosolic domain of Pex3 as bait, we expected Pex3 to interact physically with Atg36. To test this, we incubated the Escherichia coli-expressed cytoplasmic domain of Pex3 (as a GST fusion immobilised to beads) with a reticulocyte lysate in which Atg36 was produced radioactively. As shown in Figure 4A, Atg36 bound specifically to Pex3.
Figure 4.
Atg36 binds peroxisomes via Pex3. (A) An in vitro quick-coupled transcription/translation assay was performed using E. coli-produced GST–Pex3 (40–441) and GST bound to Glutathione Sepharose beads. After extensive washing of the bound protein, Atg36 synthesised in vitro in the presence of [35S]-Methionine was added. Following further washing, protein was eluted with GSH elution buffer and subjected to SDS–PAGE. Bound fractions and the Atg36 input were analysed by Coomassie staining (bottom panel) and PhosphoImaging (top panel). A beads-only sample was used as a further control. *Represents bound GST–Pex3 (40–441) and **represents bound GST. (B) atg36Δpex3Δ cells were transformed with plasmids encoding GAL1–GFP–Atg36 and a GAL1–Tom70–Pex3–mRFP chimeric protein (left panels, top) or GAL1–GFP–Atg36 (right panel, top). Expression was induced for 3 h. WT cells were transformed with GAL1–Tom70–Pex3–mRFP and preCox4–GFP (Motley et al, 2008) to confirm the mitochondrial localisation of Tom70–Pex3–mRFP. Bar, 5 μm. (C) Split-GFP analysis in atg8Δ cells. Cells expressing GFP halves were grown for 4 h in galactose medium, and scored for presence of fluorescence (−, no signal; +, fluorescent signal). Peroxisomes in cells expressing GFP–C-Atg36 plus Pex3–GFP-N were identified by mating with pex19Δ cells expressing HcRed–PTS1. Multiple fluorescent images were acquired in Z-axis and flattened into a single image. Bright-field image is a single plane. Bar, 5 μm. (D) Coimmunoprecipitation of Pex3 with Atg36. IP was performed in background strains of C13 abyss or C13–Atg36–PtA using plasmids expressing Pex3–GFP or Pex13–GFP under control of their endogenous promoters. Cells grown for 24 h to post-log phase in glucose medium. IgG Sepharose beads were used to immobilise Atg36–PtA from spheroplast yeast lysates. SDS–PAGE gels were probed with anti-GFP and PAP. Yeast lysates represent 5% of the lysate added to the beads and analysed by immunoblotting. TL, total lysate; IP, immunoprecipitate.
We have previously used a Tom70–Pex3–mRFP fusion to show Inp1 binds Pex3 when Pex3 is present on mitochondria (Munck et al, 2009). Although a fusion of the Tom20 mitochondrial targeting signal to Pex3 cytosolic domain was shown to restore peroxisome formation in pex3Δ cells (Rucktaschel et al, 2010), this is not the case for our full-length Tom70 fusion to the cytsolic domain of Pex3. We found that whereas GFP–Atg36 labelled the cytoplasm of pex3Δatg36Δ cells, it bound to mitochondria when coexpressed with Tom70–Pex3–mRFP (Figure 4B). The finding that Atg36 binds Pex3 even when Pex3 is mislocalised to mitochondria indicates that the cytosolic domain of Pex3 is sufficient to localise Atg36 to membranes. Occasional puncta of GFP–Atg36 are present in some cells lacking peroxisomes (Figure 4B). Since they do not colocalise with Atg11, nor are they present on the vacuolar membrane (not shown) we did not investigate them further.
Our results indicate that Pex3 and Atg36 interact both in vitro and in vivo. The interaction was examined further by split-GFP analysis in atg8Δ cells to prevent pexophagy and degradation of the signal we are trying to detect. Colocalisation of the split-GFP interaction with HcRed–PTS1 in mating cells shows that Pex3 and Atg36 interact at the peroxisomal membrane (Figure 4C). We confirmed the Pex3–Atg36 interaction by a coimmunoprecipitation experiment (Figure 4D): Atg36–PtA brings Pex3–GFP down specifically. These observations suggest that Pex3 alone may be responsible for recruiting Atg36 to peroxisomes.
Isolation of pexophagy-specific mutants of PEX3
Pex3 is required for peroxisome segregation in dividing cells via its binding to the inheritance factor Inp1. We have previously used a library of pex3 alleles to identify mutations in PEX3 that give rise to a peroxisome segregation defect phenotype like that of inp1Δ cells (Munck et al, 2009). The pex3-1 allele gives rise to this segregation phenotype and encodes a mutant Pex3 protein that cannot bind Inp1. Based on our finding that Pex3 binds the pexophagy factor Atg36 and on the observations in methylotrophic yeast that implicate Pex3 in pexophagy, we decided to screen the library of pex3 alleles for pexophagy defective mutants.
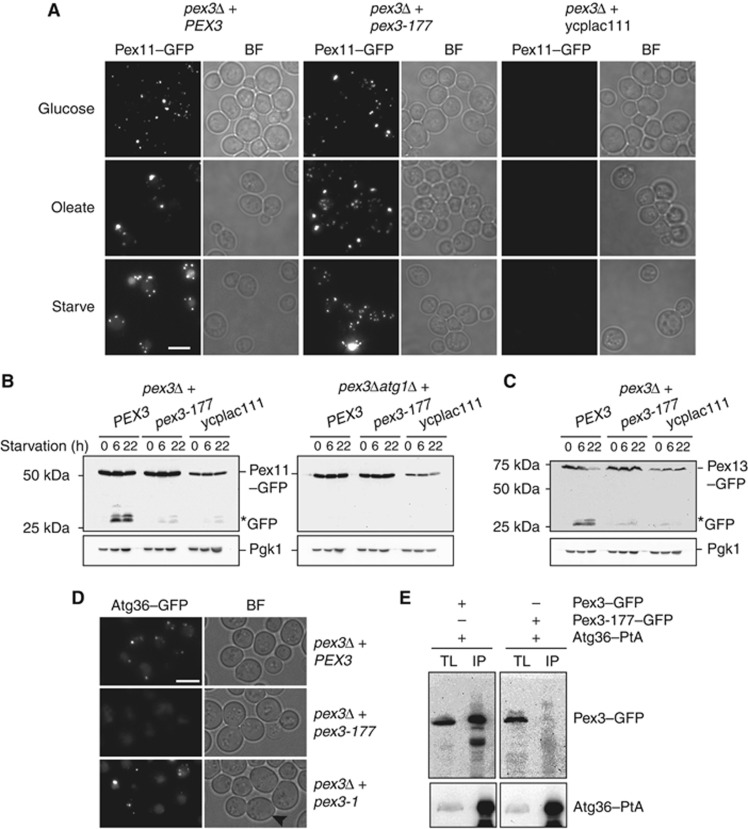
We screened our library of pex3 alleles by microscopy using the GFP–PTS1 pexophagy assay. Most of the alleles show cytoplasmic localisation of GFP–PTS1 with at most a few peroxisomes per cell, that is, Pex3 function is severely compromised. These mutants were not considered. Among the 191 mutants that contain peroxisomes and show no cytoplasmic labelling when grown on oleate medium, we found three pex3 mutants that have a phenotype similar to atg36Δ cells, that is, peroxisomes remain intact and GFP–PTS1 does not accumulate in the vacuole on starvation. All other 188 mutants displayed vacuolar GFP labelling upon starvation. We recovered the PEX3 plasmids from the three mutants and reintroduced them into pex3Δ cells expressing GFP–PTS1. Again these alleles restored peroxisome formation to pex3Δ cells, but failed to support pexophagy (shown for the pex3-177 allele in Figure 5A; Supplementary Figure S4D).
Figure 5.
The pex3-177 allele is affected in pexophagy. (A) Pex11–GFP pexophagy assayed by microscopy in pex3Δ cells and pex3Δatg1Δ cells transformed with plasmids containing either WT PEX3 or pex3-177, or with an empty plasmid ycplac111. Cells were grown in glucose, transferred to oleate medium for 18 h and switched to SD-N medium for 22 h. (B) Western blot analysis of same cells during starvation on SD-N medium. GFP* indicates the relative protease-resistant degradation product and reflects vacuolar breakdown. Time points for western blots indicated. (C) Pexophagy was assayed by Pex13–GFP breakdown in pex3Δ cells carrying plasmids as above. (D) Analysis of Atg36 localisation in pex3Δ cells expressing PEX3, pex3-177 and pex3-1. The mother cells of pex3-1 are devoid of peroxisomes (black arrowhead). (E) Coimmunoprecipitation of Pex3-177 with Atg36. IP was performed in the background strain of C13–Atg36–PtA using a plasmid expressing Pex3-177–GFP under control of its endogenous promoter. Cells grown for 24 h to post-log phase in glucose medium. IgG Sepharose beads were used to immobilise Atg36–PtA from spheroplast yeast lysates. SDS–PAGE gels were probed with anti-GFP and PAP. Yeast lysates represent 5% of the lysate added to the beads and analysed by immunoblotting. TL, total lysate; IP, immunoprecipitate. Comparison to Pex3–GFP IP (left panel).
We investigated the pexophagy phenotype further by following the degradation of Pex11–GFP (Figure 5B; Supplementary Figure S4A). In pex3Δ cells, most PMPs including Pex11 are rapidly broken down, which results in low levels of these PMPs (Hettema et al, 2000). As expected, the level of Pex11–GFP is low in pex3Δ cells grown on oleate (Figure 5B, ycplac111). Subsequent disruption of ATG1 does not stabilise Pex11–GFP (Figure 5B, atg1Δpex3Δ cells, ycplac111), furthermore, the lack of the GFP breakdown product in atg1Δpex3Δ cells indicates that Pex11–GFP is degraded by autophagy-independent pathways (e.g., the ubiquitin/proteasome machinery) in the absence of Pex3 (Figure 5B).
Pex11–GFP is stabilised in pex3Δ cells containing the pex3-177 allele (Figure 5A and B) and is present in peroxisomes (Supplementary Figure S4C). Upon shifting these cells to starvation medium, only a minor fraction of Pex11–GFP is broken down (Figure 5B), showing that this allele is strongly defective in pexophagy. A second peroxisomal membrane marker (Pex13–GFP) confirmed these observations (Figure 5C). Therefore, three independent markers (GFP–PTS1, Pex11–GFP and Pex13–GFP) indicate that peroxisomes in pex3-177 cells are resistant to breakdown under pexophagy conditions. Two further Pex3 alleles were isolated using the microscopic pexophagy screen, and both of these alleles stabilise Pex11–GFP under pexophagy conditions (Supplementary Figure S4A).
We analysed peroxisome distribution between mother cell and bud in pex3Δ cells containing the segregation deficient pex3-1 allele or the pexophagy-deficient pex3-177 allele (Supplementary Figure S4B). In contrast to in pex3-1 cells, we found peroxisome segregation was intact and Inp1–GFP was recruited to peroxisomes in pex3-177 cells (Supplementary Figure S4D).
To test whether the pexophagy phenotype in pex3-177 cells results from an inability of peroxisomes to bind Atg36, we expressed Atg36–GFP on a plasmid from the ATG36 promoter in pex3-177 or pex3-1 cells (Figure 5D). We found Atg36–GFP binding to peroxisomes was undetectable in pex3-177 cells. Furthermore, we show that Pex3-177 does not interact in vivo with Atg36–PtA (Figure 5E).
We conclude that cells containing the pex3-177 allele are able to form peroxisomal structures and segregate them during cell division, but are defective in Atg36 recruitment, and this results in the pexophagy defect. This Atg36-binding function of Pex3 can be separated genetically from that required for peroxisome formation and segregation.
ATG36 expression is induced prior to pexophagy
If Atg36 marks peroxisomes for autophagic degradation, we would expect its expression to be highest prior to degradation of peroxisomes. In order to compare the timing of ATG36 expression with that of pexophagy, we tagged Atg36 in the genome Atg36 at the C-terminus with PtA and analysed its expression under various growth conditions in cells also expressing Pex11–GFP (Figure 6A). We found the level of Atg36 increased on oleate without inducing pexophagy, and started to decrease when cells were switched to starvation medium and pexophagy commenced as seen by Pex11–GFP degradation. Together with the overexpression experiments described in Figure 3 it is clear that there is no direct correlation between the level of Atg36 expression and pexophagy, that is, an increase in the level of Atg36 (either endogenous, or induced) is not sufficient to trigger pexophagy, which occurs only after switching to starvation medium (Figures 3B and 6A, with the exception of the N-tagged version, which appears to be constitutively active). This suggests that Atg36 is activated by switching to starvation medium. Notably, it appears that Atg36–PtA is differentially modified depending on growth conditions: Atg36–PtA migrates as a set of fuzzy bands whose mobility changes upon starvation and which displays differential sensitivity to CIP treatment (Figure 6A; Supplementary Figure S3D). The observation that Atg36 is upregulated in oleate and declines when cells are switched to starvation conditions was examined further in WT cells expressing GFP-tagged Atg36 from its genomic locus (Figure 6B). After 6 h on starvation medium, most Atg36–GFP is cleaved as seen by the appearance of the typical GFP-specific breakdown product that accumulates upon vacuolar entry of GFP fusion proteins, and the appearance of this cleavage is Atg1-dependent. Interestingly, Atg36–GFP disappears also from atg1Δ cells, but since no GFP cleavage product accumulates, this breakdown is not occurring in the vacuole. We conclude that in WT cells, Atg36 is degraded by autophagy after being cotransported with peroxisomes into the vacuole, but in atg1Δ cells, atg36 is degraded via an autophagy-independent process.
Figure 6.
Atg36 expression and cleavage correlates with Pex11–GFP degradation. (A) WT cells expressing genomic Atg36–PtA and Pex11–GFP on a plasmid were grown in glucose for 22 h, transferred to oleate medium, and then shifted to SD-N medium. Samples were taken at the times indicated and processed for western blot with peroxidase anti-peroxidase complex to detect Atg36–PtA (top panel) or anti-GFP to detect Pex11–GFP pexophagy (bottom panel). GFP* indicates the relative protease-resistant degradation product and reflects vacuolar breakdown. (B) Western blot analysis of genomic Atg36–GFP under oleate and starvation conditions for the indicated time points in the strains indicated. (C) Western blot analysis of genomic Atg36–GFP in WT, pex14Δ, atg1Δ and pex3Δ cells grown for up to 3 days in oleate medium. Samples were taken at 24 h intervals. (D) Pex3 level in total lysate was determined from WT cells grown in glucose medium for 22 h, transferred to oleate medium and then shifted to SD-N medium. Samples were taken at the times indicated and processed for western blot with anti-Pex3. A pex3Δ glucose 22 h sample was used to determine the specificity of the antibody. **Indicates non-specific band. (E) Western blot analysis of pex3Δ and atg36Δpex3Δ cells expressing Pex3–GFP from its own promoter after growth in glucose, oleate-containing medium and after shifting to SD-N medium for the times indicated.
We investigated the degradation of Atg36–GFP in cells grown continuously for 3 days in oleate medium (Figure 6C). As was seen in Figure 1E, pexophagy commences on day 2. Whereas Atg36–GFP is broken down and the protease-resistant GFP fragment appears during pexophagy in WT cells, it does not appear in pex3Δ or atg1Δ cells (Figure 6C). Atg36–GFP is cleaved in pex14Δ cells, which is in line with our finding that pexophagy is intact in these cells. We conclude that Atg36 enters the vacuole with peroxisomes via an autophagy process, as Atg36–GFP is not cleaved in pex3Δ or atg1Δ cells.
Bellu et al (2002) have reported that H. polymorpha Pex3 is rapidly removed from peroxisomes when pexophagy was induced and degraded prior to vacuolar uptake of the whole organelle. To investigate whether this is the case in S. cerevisiae, we followed stability of endogenous Pex3 (Figure 6D) and Pex3–GFP (Figure 6E) in cells grown on glucose, oleate and starvation medium. We found that the time course of both Pex3 and Pex3–GFP degradation is comparable to that of Atg36–PtA and Pex11–GFP (Figure 6A), with most degradation occurring for all three proteins between 3 and 6 h on starvation medium. The finding that Pex3 is degraded with the same kinetics as the peroxisomal marker Pex11 and Atg36 suggests that S. cerevisiae Pex3 is not removed from peroxisomes and degraded prior to pexophagy, in contrast to what has been found for H. polymorpha Pex3 (Bellu et al, 2002). Furthermore, H. polymorpha Pex3 is degraded even in cells blocked in autophagy, whereas in S. cerevisiae, Pex3–GFP remains intact in atg36Δ cells (Figure 6E). This clearly indicates a mechanistic difference between pexophagy in H. polymorpha and S. cerevisiae.
Atg36 is required to link peroxisomes to the autophagy apparatus
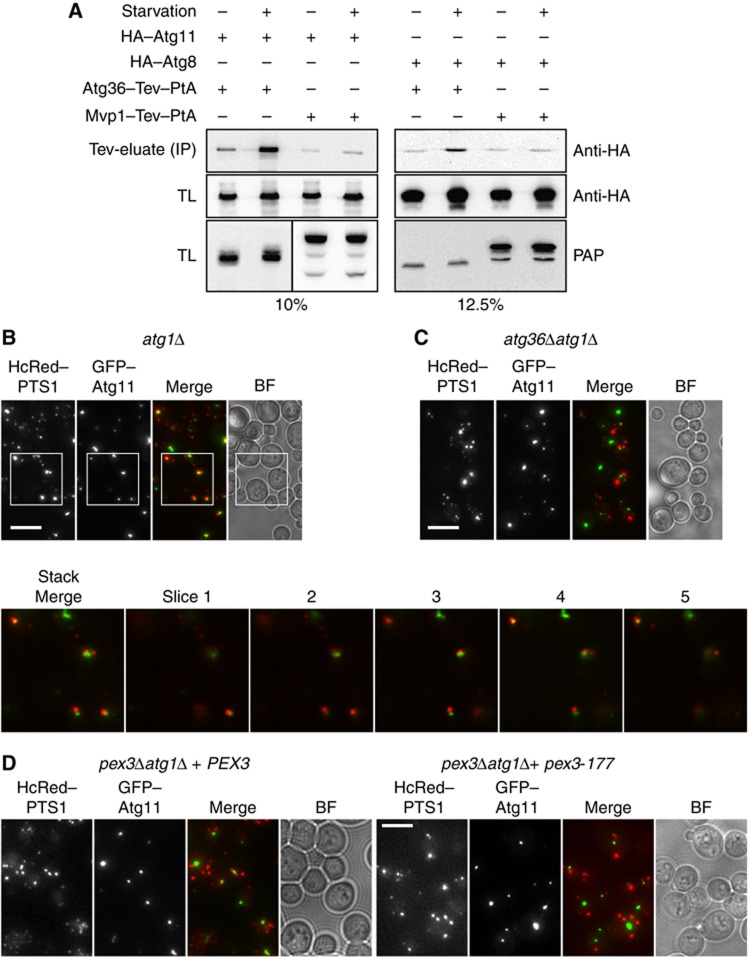
Atg11 has been proposed to act as an adaptor protein that links receptors for selective cargo to the core autophagy machinery. For instance, the receptors for mitophagy and the Cvt pathway in S. cerevisiae (Atg32, Atg19, respectively) and pexophagy in P. pastoris (PpAtg30) bind Atg11 (Yorimitsu and Klionsky, 2005a; Farre et al, 2008; Kanki et al, 2009; Okamoto et al, 2009). Atg32 has also been shown to bind Atg8 (Okamoto et al, 2009) and in mammalian cells, many selective receptors interact with LC3, the mammalian Atg8 orthologue (Johansen and Lamark, 2011). We therefore sought to determine whether Atg11 and Atg8 interact with Atg36. We expressed Atg36–PtA tagged from its chromosomal locus in cells coexpressing either HA–Atg11 or HA–Atg8. Cells were grown under oleate and starvation conditions. Atg36–PtA was immunoprecipitated with IgG sepharose beads and copurified proteins were eluted by Tev protease cleavage. The eluate was tested for the presence of HA–Atg11 and HA–Atg8 by immunoblotting (Figure 7A). Both HA–Atg11 and HA–Atg8 copurified with Atg36–PtA from lysate prepared from starved cells. A low level of binding was also observed under oleate conditions but a similar level of HA–Atg11 and HA–Atg8 copurified with the negative control Mvp1–PtA. We conclude that Atg36 interacts with both Atg11 and Atg8 and that this interaction is induced under pexophagy conditions.
Figure 7.
Atg11 and Atg8 bind Atg36. (A) Coimmunoprecipitation of Atg11 and Atg8 with Atg36. IP was performed in either C13–Atg36–PtA atg1Δ or C13–Mvp1–PtA atg1Δ cells containing plasmids expressing either HA–Atg11 or HA–Atg8 under control of the TPI1 promoter. Cells were grown for 24 h to post-log phase in glucose medium followed by growth in oleate medium for a further 18 h. Cells were then grown under non-starvation or starvation conditions for a further 2 h. IgG Sepharose beads were used to immobilise Atg36–PtA or Mvp1–PtA from spheroplast yeast lysates. After extensive washing, bound material was eluted by Tev protease cleavage. SDS–PAGE gels (percentage indicated) were probed with anti-HA (monoclonal 12CA5) and PAP. TL, total lysate; IP, immunoprecipitate (Tev eluate). (B–D) GFP–Atg11 was expressed in strains as indicated and cells were grown to post-log phase in glucose medium. Inset shows magnification plus individual slices of stack from atg1Δ cells. Bar, 5 μm.
Many cargo receptors have been shown to contain an AIM/LIR that links them to Atg8/LC3. Analysis of Atg36 reveals eight amino-acid sequences weakly resembling the AIM/LIR consensus (Supplementary Figure S5). Mutational analysis shows that although one of these sequences is required for Atg36 function, it is not conserved and does not constitute a functional AIM (Supplementary Figure S5).
We then examined the requirement for Atg36 to link peroxisomes to Atg11-positive structures in cells where autophagy is blocked (atg1Δ cells): we expressed GFP–Atg11 and quantified the degree of colocalisation with peroxisomes in the presence (Figure 7B) or absence (Figure 7C) of ATG36. We used atg1Δ cells as they assemble PAS containing Atg proteins but are blocked in the further formation of autophagosomes (Suzuki et al, 2007). Most post-log atg1Δ cells (>65%) expressing both markers showed close proximity or colocalisation of GFP–Atg11 puncta with peroxisomes, whereas in atg1Δ atg36Δ cells, only a minority (<15%) of cells showed close proximity between these two markers. This confirms that Atg36 is required to bring peroxisomes to the autophagy structure marked by GFP–Atg11, and indicates that in atg1Δ cells, peroxisomes, like mitochondria, associate with Atg11-positive structures, most likely to be the PAS (Kanki et al, 2009).
In a similar experiment, we found the degree of proximity between peroxisomes and Atg11 is reduced to 30% in pex3Δatg1Δ cells containing pex3-177 compared with 75% in pex3Δatg1Δ cells containing PEX3 (Figure 7D).
Mitochondria-targeted Pex3 can drive mitophagy
We have shown above that Pex3 recruits GFP–Atg36 to membranes, even when Pex3 is mislocalised to mitochondria. We have also shown that induction of mRFP–Atg36 causes pexophagy, even under conditions that normally stimulate peroxisome proliferation. We hypothesised that overexpression of GFP–Atg36 may cause mitophagy if Atg36 is targeted to mitochondria by coexpression of Tom70–Pex3–mRFP.
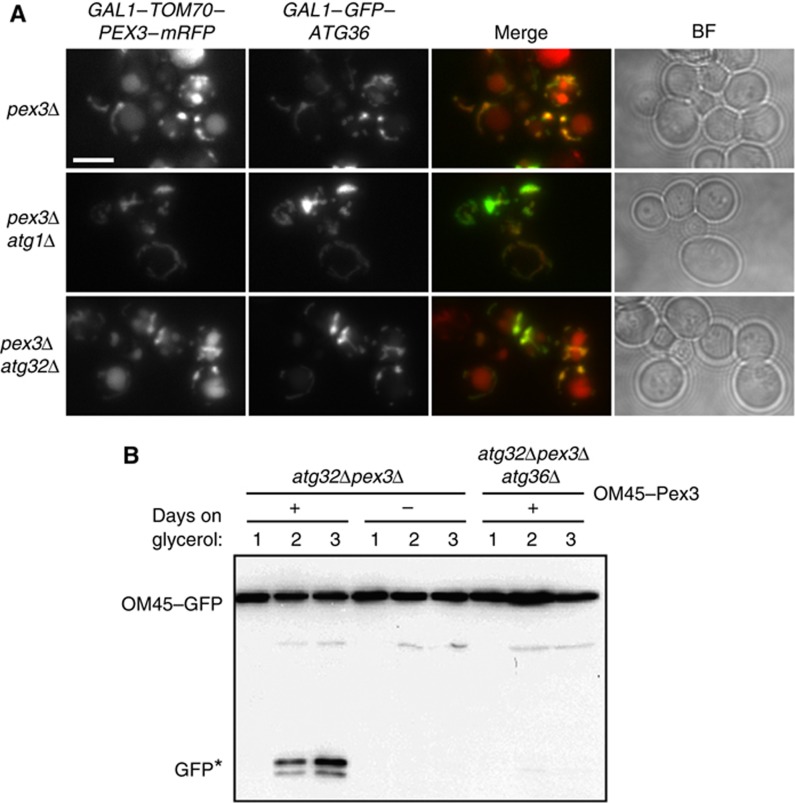
We found indeed that prolonged (22 h) expression of GFP–Atg36 in pex3Δ cells resulted in vacuolar localisation of Tom70–Pex3–mRFP, and this effect was enhanced when cells were shifted to starvation medium (Figure 8A). This effect of GFP–Atg36 expression occurred only when GFP–Atg36 and Tom70–Pex3–mRFP were expressed together, and was Atg1-dependent. This effect of overexpression was also seen in atg32Δ pex3Δ cells, which are otherwise mitophagy-deficient. This suggests that Tom70–Pex3–mRFP and Atg36 can function together as a transplantable module to bring about autophagic degradation of mitochondria.
Figure 8.
Atg36 drives mitophagy when directed to mitochondria. (A) pex3Δ, pex3Δatg1Δ and pex3Δatg32Δ cells were grown for 6 h in galactose medium to express GAL1–Tom70–Pex3–mRFP and GAL1–Atg36–GFP. Cells were shifted to SD-N medium and imaged after 12 h. Multiple fluorescent images were acquired in Z-axis and flattened into a single image. Bright-field image is a single plane. Bar, 5 μm. (B) OM45 was tagged with GFP in the genome of atg32Δpex3Δ and atg32Δpex3Δatg36Δ cells, which were transformed with an empty plasmid (−) or a plasmid encoding OM45–Pex3 (+). The cells were grown for up to 3 days in glycerol medium and samples were taken at 24 h intervals and processed for anti-GFP western blotting. GFP* indicates the relative protease-resistant degradation product and reflects vacuolar breakdown.
We investigated this observation further by testing whether endogenous Atg36 can restore mitophagy in cells lacking the mitophagy receptor Atg32. This was assessed by OM45–GFP breakdown during growth on glycerol for 3 days (Figure 8B). These growth conditions induce mitophagy (Figure 2B) and pexophagy (Figure 1E) starting at day 2. Endogenous Atg36 was directed to mitochondria of atg32Δ pex3Δ cells by expressing OM45–Pex3, which for these experiments comprised a plasmid encoding an OM45–Pex3 fusion protein under control of the OM45 promoter. This fusion does not rescue peroxisome formation in pex3Δ cells (data not shown). Mitophagy was measured by immunoblotting of OM45–GFP.
As can be seen in Figure 8B, free GFP indicative of mitophagy becomes evident only in atg32Δ pex3Δ cells expressing OM45–Pex3. Disruption of ATG36 in atg32Δ pex3Δ cells abolished the ability of OM45–Pex3 to rescue mitophagy, which confirms that it is indeed endogenous Atg36 that is responsible for the mitophagy activity. We conclude that Atg36 acts as a tag to link its cargo to the autophagy machinery, and can even replace Atg32 if it is targeted to mitochondria.
Discussion
We set out to understand the role of Pex3 in peroxisome turnover and have isolated a pex3 allele (pex3-177) that is blocked specifically in autophagic breakdown of peroxisomes. This allele fails to recruit a novel autophagy factor, Atg36, that links peroxisomes to the autophagy machinery. These observations support a model whereby Pex3 acts as an anchor for a pexophagy-specific factor that tags peroxisomes for degradation.
Pex3 recruitment of Atg36 is required for pexophagy
We show that pex3 is required for pexophagy as we isolated three pex3 alleles (including pex3-177) that are affected in pexophagy. We identified a novel Pex3-interacting protein, Atg36. Pex3 is both sufficient and necessary to recruit Atg36 from the cytosol to peroxisomes as Pex3 and Atg36 interact both in vitro and in vivo. If we redirect the cytosolic domain of Pex3 to mitochondria, Atg36 follows. The pex3 pexophagy-deficient alleles have mutations in their cytosolic domain and we show that one of these alleles, pex3-177, fails to bind Atg36 in vivo and consequently does not recruit Atg36 to peroxisomes. Comparison of the mutant alleles did not reveal a mutation hotspot. This suggests that rather than disrupting a linear binding motif, the mutations are more likely to affect the tertiary structure of Pex3 and thereby inhibit binding of Atg36. The mutations still, however, allow binding of Inp1 and Pex19, as pex3-177 is active in peroxisome formation and segregation. Our data show that atg36Δ cells are not affected in peroxisome formation, segregation or functioning. The increase in peroxisome number under most growth conditions is a consequence of a defect in peroxisome turnover.
Atg36 was shown to interact with Pex34 and Inp1 in genome-wide two-hybrid screens (Ito et al, 2001; Yu et al, 2008). These two proteins are involved in peroxisome multiplication and segregation, respectively. We have found no requirement for Inp1 or Pex34 in pexophagy, neither did we find a requirement for Atg36 in peroxisome multiplication nor segregation. Furthermore, the pex3 alleles that are affected in pexophagy or segregation are selectively deficient in peroxisomal recruitment of Atg36 or Inp1, respectively. This implies that these factors are recruited to peroxisomes by Pex3 independently of each other. Pex34 is an integral membrane protein that travels via the ER to peroxisomes (Tower et al, 2011). When we redirect Pex3 to mitochondria, Atg36 follows where it stimulates mitophagy. The simplest interpretation of these results is that Pex34 is not required for Atg36 recruitment or activity. Therefore, the relevance of the observed two-hybrid interactions in genome-wide screens is unclear.
What is the role of Atg36 in pexophagy?
First, we established that Atg36 is specifically required for pexophagy and not for a variety of other autophagic processes including non-selective autophagy, mitophagy or Ape1 targetting to the vacuole via the Cvt pathway.
We propose that Atg36 is the pexophagy receptor in S. cerevisiae based on the following observations. When expressing GFP-tagged Atg36 from the GAL1 promoter, we see its binding to peroxisomes precedes pexophagy, and that continued overexpression of Atg36 induces pexophagy. Fungal receptors for selective autophagy link their cargoes via Atg11 to the core autophagy machinery (Yorimitsu and Klionsky, 2005a; Farre et al, 2008; Kanki et al, 2009; Okamoto et al, 2009). The mitophagy and Cvt receptors Atg32, and Atg19 and Atg34, respectively also interact with Atg8 (Shintani et al, 2002; Okamoto et al, 2009; Suzuki et al, 2010), and in vivo mutational studies have revealed the AIM sequences of Atg19 and Atg34 are crucial for activity of the Cvt pathway (Noda et al, 2008; Suzuki et al, 2010). In mammalian cells, Atg11 appears to be absent but the Atg8-family orthologues, LC3/GABARAP, interact with cargo receptors for selective autophagy via an LIR, and these interactions have been shown to be crucial for function (Pankiv et al, 2007; Noda et al, 2008; Kirkin et al, 2009; Novak et al, 2010; Wild et al, 2011). However, mutation of the AIM of Atg32 does not abolish its in vivo interaction with Atg8, as determined by coimmunoprecipitation, and this mutation has only a mild effect on mitophagy (Okamoto et al, 2009; Kondo-Okamoto et al, 2012). In contrast, interaction of Atg32 with Atg11 is crucial for mitophagy (Kondo-Okamoto et al, 2012). The authors propose that ‘other protein–protein interfaces could contribute to the interaction between Atg32 and Atg8 in vivo’. Likewise, the interaction between P. pastoris Atg11 and Atg30 is crucial for pexophagy but no canonical AIM is present in this protein and no binding to Atg8 has been described.
We show that Atg36 interacts in vivo with both Atg11 and Atg8. Although we identified eight sequences in Atg36 that weakly resemble the AIM/LIR consensus, disruption of seven of them did not affect pexophagy (Supplementary Figure S5). Further analysis of Y191/L194 that is required for Atg36 activity suggests that it is not an AIM, however, as it is not conserved, and it is missing a serine or threonine at −1 which has been found in all yeast AIMs identified to date (Supplementary Figure S5). Furthermore, it lacks the negatively charged residues characteristic of AIM/LIRs at positions X1/X2 or X−1/X−3 (Johansen and Lamark, 2011), and substitution of the D at −2 to A did not affect function. Finally, the finding that a single substitution of Y191 with L (which is present at this position in most yeast Atg36 orthologues (Supplementary Figure S5)) did not affect pexophagy confirms that Y191/L194 is not an AIM. Whether Atg8 interacts with Atg36 via a different motif, or whether these motifs are redundant, remains to be tested. We were unable to detect an interaction between Atg8 and Atg36 in vitro or with yeast two-hybrid (data not shown). However, we do find an interaction in vivo (Figure 7A), and it is possible that Atg8 does not bind Atg36 directly.
In atg1Δ cells, most Atg proteins accumulate at the PAS (Suzuki et al, 2007), but later steps in the autophagic process are blocked. We show that peroxisomes frequently localise to Atg11-containing structures in atg1Δ cells, and that this colocalisation requires Atg36 recruitment to peroxisomes, as atg36Δ cells and pex3-177 cells have reduced colocalisation of peroxisomes with Atg11. All these observations qualify Atg36 as a receptor for pexophagy that links peroxisomes to autophagosomal membranes. This was further tested and confirmed by our transplantation experiment where we directed Pex3 to mitochondria and restored the mitophagy defect of atg32Δ. This restoration was dependent upon Atg36.
An important question that is raised by our observations relates to how pexophagy is regulated. At this stage, we can only hypothesise as to how Atg36 activates pexophagy. We propose that Atg36 does this in a two-step process, that is, first accumulation of Atg36 on the peroxisomal surface followed by an activation event. When cells are grown on oleate, Atg36 levels on peroxisomes increase but this does not correlate with pexophagy. This indicates a second event is required. Pexophagy is activated when cells are subsequently shifted to starvation medium, and western blot analysis shows that this correlates with a shift in mobility of Atg36, implying that Atg36 is post-translationally modified. Under these conditions, we also observe an increased interaction of Atg36 with Atg11 and Atg8. The nature of the modification is complex. Phosphatase treatment suggests that Atg36 is phosphorylated both during growth on oleate and during starvation. However, phosphatase treatment before and during starvation does not result in the same mobility of Atg36. This indicates that Atg36 is differentially modified under these conditions and that this modification may stimulate interactions with the autophagy machinery. A precedent for this is the phosphorylation dependent interaction of Atg11 with ScAtg32 and PpAtg30 (Farre et al, 2008; Aoki et al, 2011). Furthermore, phosphorylation of a serine residue at position −1 of the LIR of optineurin stimulates binding to ATG8-family orthologues and selective autophagy of Salmonella (Wild et al, 2011). There are frequently negative charges or serines or threonines adjacent to position 1 of the AIM/LIR (Noda et al, 2010; Supplementary Figure S5) and it has been proposed that phosphorylation of LIRs is a common mechanism for regulation of autophagy receptors (Wild et al, 2011). Currently, we can only correlate the appearance of the modified forms of Atg36 with induction of pexophagy but are not able to test their relevance. We found that overexpression of Atg36 induces pexophagy even under non-starvation conditions. Since overexpression is at least 50- to 100-fold, it seems plausible that under these conditions the second step in the regulation process can be bypassed. Of particular interest is the effect of ectopically expressed N-tagged Atg36, which seems to be constitutively active, that is, not starvation-dependent.
As discussed above, Atg36 can replace Atg32 in mediating mitophagy, and the timing of Atg36-mediated mitophagy mirrors that of Atg32-mediated mitophagy and Atg36-mediated pexophagy. Whether the artificial Atg36-dependent mitophagy is dependent upon the pexophagy-specific regulatory mechanism is not clear and awaits further investigation. Recently, the MAPKs Slt2 and Hog1 were implicated in selective autophagy. Both Hog1 and Slt2 have been implicated in mitophagy and pexophagy although the data are conflicting for a role for Hog1 in pexophagy (Manjithaya et al, 2010a; Aoki et al, 2011; Mao and Klionsky, 2011; Mao et al, 2011). Their autophagic targets, however, are unknown. Both level and migration pattern of Atg36 in slt2Δ and hog1Δ cells was unaffected compared with WT cells (data not shown). This suggests that neither Slt2 nor Hog1 act at the level of Atg36. This is in agreement with the observations that suggest that Slt2 acts at a late stage of pexophagy, after pexophagosome formation (Manjithaya et al, 2010a).
S. cerevisiae pexophagy differs mechanistically from that of methylotropic yeast
Atg36 homologues can be identified only in closely related species, and no homologue is present in P. pastoris. Likewise, PpAtg30 is not conserved in S. cerevisiae. Our study shows many features are common between PpAtg30 and ScAtg36, including the induction of pexophagy upon overexpression, post-translational modification during conditions of nitrogen starvation and interaction with Pex3 and Atg11 (Farre et al, 2008). Although there are many similarities, there are major differences between pexophagy in P. pastoris and S. cerevisiae. Pex14 is required for recruitment of PpAtg30 to peroxisomes in P. pastoris and is essential for pexophagy induced by PpAtg30 overexpression. A role for Pex3 was suggested as it interacts with PpAtg30, but since PpAtg30 does not localise to Pex3-containing structures in pex14Δ cells, the exact role of Pex3 in P. pastoris pexophagy remains unclear.
Bellu et al (2002) have reported that in the related methylotrophic yeast H. polymorpha, Pex3 is rapidly removed from peroxisomes under pexophagy inducing conditions, prior to sequestration and degradation of peroxisomes in vacuoles. It is also reported that removal of Pex3 is a prerequisite for peroxisome degradation and occurs independently of autophagy (Bellu et al, 2002; van Zutphen et al, 2011). In contrast to this, we find that Pex3 is degraded in parallel with peroxisomes and is not removed from peroxisomes prior to their degradation in the vacuole in S. cerevisiae. Furthermore, in contrast to in H. polymorpha, ScPex3 is not degraded under starvation conditions when autophagy is blocked. Our data strongly support a role for ScPex3 in coupling peroxisomes via Atg36 to the autophagy machinery. This role may be fulfilled by Pex14 in methylotropic yeasts, as P. pastoris Pex14 is required for PpAtg30 to localise to peroxisomes (Farre et al, 2008) and Pex14 is also required for pexophagy in H. polymorpha (Bellu et al, 2001). In mammalian cells, Pex14 has been shown to interact via tubulin to LC3 and thereby has been suggested to play a role in pexophagy (Hara-Kuge and Fujiki, 2008). In contrast, we show that S. cerevisiae Pex14 is completely dispensable for pexophagy (Figure 1B).
We conclude that ScPex3 acts in (at least) three different processes and that these processes can be separated genetically. Pex3 acts as docking factor for Pex19 in peroxisome formation, and it also recruits Inp1 to peroxisomes as part of its role in peroxisome segregation. Now we show that Pex3 recruits the pexophagy-specific protein, Atg36, to peroxisomes and that this recruitment is required for pexophagy. A model emerges whereby Pex3 coordinates the biogenesis and maintenance of the peroxisomal compartment by recruiting process-specific factors. How this is achieved awaits further investigation.
Materials and methods
Yeast strains, media and growth conditions
The yeast strains used in this study are listed in Supplementary Table S1. Gene tagging and disruptions were performed by homologous recombination and strains were checked by PCR. For all experiments, cells were grown overnight in defined selective glucose medium. For analysis of phenotypes by microscopy, cells were subsequently diluted to 0.1 OD600 in fresh selective glucose medium and grown for two to three cell divisions (4–6 h), prior to imaging. Where the induction of a reporter protein was required, cells were transferred to selective galactose medium at 0.1 OD600 and grown for the time indicated in the figures and text. Yeast cells were growth at 30°C in either of the following mediums: rich YPD media (1% yeast extract, 2% peptone, 2% glucose), minimal media (YM2) for the selection of the uracil prototrophic marker (carbon source, 0.17% yeast nitrogen base without amino acids and ammonium sulphate, 0.5% ammonium sulphate, 1% casamino acids) or minimal media (YM1) for the selection of all prototrophic markers (carbon source, 0.17% yeast nitrogen base without amino acids and ammonium sulphate, 0.5% ammonium sulphate). Regarding the carbon sources, glucose and galactose were added to 2% (w/v) and glycerol 3% (v/v). For peroxisome induction, cells were transferred to oleate medium (YM2 oleate: YM2 plus 0.12% oleate (v/v), 0.2% Tween-40® (v/v), 0.1% yeast extract) at a 1/10 overnight dilution. Pexophagy was induced by transferring cells to starvation medium lacking a nitrogen source (SD-N; 0.17% yeast nitrogen base without amino acids and ammonium sulphate, 2% glucose) (Hutchins et al, 1999; Manjithaya et al, 2010a). The appropriate amino-acid stocks were added to minimal media as required. In all, 5–10 OD600 units were collected at selected time points as indicated in the figures and text. Cells were either analysed by immunoblotting or by fluorescence microscopy. For mitophagy induction, mid-log phase yeast cells were grown for 3 days in YM2 glycerol medium (Okamoto et al, 2009). This method was also used as a measure of pexophagy substituting YM2 glycerol for YM2 oleate or YM2 glucose. In all, 5–10 OD600 units were collected at selected time points as indicated in the figures and text. Oleate plates contained 0.67% yeast nitrogen base without amino acids and ammonium sulphate, 0.1% yeast extract, 0.1% oleate (v/v), 0.25% Tween-40 (v/v), 2% agar and amino acids as needed.
Mating experiments were performed as described previously (Motley and Hettema, 2007). The vacuolar membrane was stained as previously described (Vida and Emr, 1995).
Plasmids
Yeast expression plasmids were based on the parental plasmids ycplac33 and ycplac111 (Gietz and Sugino, 1988). The majority of constructs used in this study were generated by homologous recombination in yeast (Uetz et al, 2000). The ORF of interest was amplified by PCR. The 5′ ends of the primers included 18 nt extensions homologous to plasmid sequences flanking the intended insertion site, to enable repair of gapped plasmids by homologous recombination. For expression of genes under control of their endogenous promoter, 500 nt upstream from the ORF were included. Galactose-inducible constructs contained the GAL1 and GAL10 intragenic region. All yeast constructs contain the PGK1 terminator. 3HA–Atg11 and 3HA–Atg8 expression plasmids were constructed by in-frame fusion of either the ATG11 or ATG8 ORF under control of the TPI1 promoter with N-tagged 3HA. The galactose-inducible Tom70–Pex3–mRFP fusion and constitutive expression constructs for HcRED–PTS1 and GFP–PTS1 have been described previously (Motley and Hettema, 2007; Munck et al, 2009). Oleate-inducible GFP–PTS1 is controlled by the peroxisomal catalase (CTA1) promoter as described in Hettema et al (1998). We used GFPS65T for tagging, with monomeric GFP containing GFP L221K (Snapp et al, 2003). Split-GFP constructs were based on the plasmids designed by (Barnard et al, 2008), with the split-GFP fragments introduced behind the GAL1 promoter into centromeric plasmids to generate a conditional split-GFP system (Munck et al, 2009). For E. coli expression, GST–Pex3 (40–441) has been previously described (Munck et al, 2009).
PEX3 mutant screen
In order to identify pexophagy-deficient PEX3 mutants, we screened a library of PEX3 alleles previously generated in the laboratory (Munck et al, 2009). Essentially, we screened the mutants using the pexophagy assay, as described above, in 96-well plates with fluorescence microscopy. Of the 1000 mutants screened, only 191 showed multiple punctate fluorescent structures and no cytosolic labelling. This indicates that these pex3 alleles support formation of peroxisomes. In the pexophagy assay, most of these 191 mutants lost their peroxisomes and showed vacuolar GFP labelling, but three mutants retained their peroxisomes and vacuolar labelling was absent. We recovered these PEX3 plasmids from the three mutants, sequenced them, and reintroduced them into pex3Δ cells expressing GFP–PTS1. Again, these alleles restored peroxisome formation but failed to support pexophagy. DNA sequence analysis of the alleles revealed the following amino-acid substitutions: pex3-58 (F35S, I170T, D196E, Q285R, D374E, T397S, S429R); pex3-153 (E58D, L166P, K210R, Q284L); pex3-177 (F64S, T74A, H354L).
In-vitro transcription/translation
In-vitro transcription/translation was performed using the TnT quick-coupled rabbit reticulocyte transcription/translation kit (Promega) using a PCR-generated template of ATG36 according to the manufacturer’s instructions. GST–Pex3 (40–441) was produced in E. coli as described previously (Munck et al, 2009). SDS–PAGE gels were stained with Coomassie blue followed by PhosphoImaging.
Immunoblotting
For preparation of extracts by alkaline lysis, cells were centrifuged and pellets resuspended in 0.2 M NaOH and 0.2% β-mercaptoethanol and left on ice for 10 min. Soluble protein was precipitated by addition of 5% TCA for a further 10 min. Following centrifugation (13 000 g, 5 min, 4°C), soluble protein was resuspended in 10 μl 1 M Tris–HCl (pH 9.4) and boiled in 90 μl 1 × SDS–PAGE sample loading buffer for 10 min. Samples (0.25–1 OD600 equivalent) were resolved by SDS–PAGE followed by immunoblotting. Blots were blocked in 2% (w/v) fat-free Marvel™ milk in TBS-Tween-20 (50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.1% (v/v) Tween-20). Tagged proteins were detected using either monoclonal anti-HA 12CA5 (mouse 1:50 diluted hybridoma culture supernatant), monoclonal anti-GFP (mouse; 1:3000; Roche), peroxidase-anti-peroxidase (PAP) (rabbit; 1:2000; Sigma), polyclonal anti-Pex3 (rabbit; 1:3000; gift of Ralf Erdmann and Erdmann Girzalsky) or monoclonal anti-PGK 22C5 (mouse; 1:7000; Invitrogen). Secondary antibody was HRP-linked anti-mouse polyclonal (goat; 1:4000; Bio-Rad) or HRP-linked anti-rabbit polyclonal (goat; 1:10 000; Promega). Detection achieved using enhanced chemiluminescence (Biological Industries) and chemiluminescence imaging.
Coimmunoprecipitation
To fully saturate IgG Sepharose™ 6 Fast Flow beads (GE Healthcare) with genomically expressed Atg36–PtA, 500 ml oleate medium was inoculated with 50 ml of a 24-h glucose medium culture. These were grown with agitation for 18 h at 30°C. Non-starvation condition flasks were left a further 2 h while cells for starvation conditions were harvested (2500 g, 5 min), washed, and resuspended in 50 ml SD-N medium and also left a further 2 h. All cells were harvested, washed and pellets were weighed. Spheroplasts were prepared by treatment with zymolase (MP Biomedicals) in sorbitol buffer (50 mM Kpi (pH 7.4), 1.2 M sorbitol, 2% glucose, 0.17% yeast nitrogen base without amino acids and ammonium sulphate, zymolase (5 mg/g cells), lysine/histidine/methionine, 0.5% ammonium sulphate (to non-starvation)) for 1 h. Spheroplasts were harvested, washed once in 50 mM Kpi (pH 7.4)+1.2 M sorbitol, and resuspended in 10 ml lysis buffer (150 mM KCl, 20 mM Tris–HCl (pH 8.0), 4 mM Pefabloc SC (Roche), 1 mM EDTA (pH 8.0), one complete protease inhibitor cocktail tablet/25 ml (Roche), 5 mM Sodium Vanadate, 25 mM Sodium Fluoride). Whole cell lysates were obtained by dounce homogenisation followed by 10 min incubation on ice with addition of 0.5% (v/v) Triton X-100. After centrifugation (13 000 g, 5 min, 4°C), lysates were incubated with 35 μl IgG Sepharose™ 6 Fast Flow beads, pre-washed in lysis buffer, at 4°C with rotation for 15 min. Beads were subsequently washed three times with lysis buffer containing 0.1% (v/v) Triton X-100. The bound material was eluted by addition of lysis buffer and boiling with 4 × SDS–PAGE buffer at 95°C for 5 min or by AcTEV protease cleavage according to the manufacturer’s instructions (Invitrogen).
Alkaline phosphatase assay
Alkaline phosphatase assay for non-selective autophagy was performed as described previously (Noda et al, 1995). In brief, cells were grown overnight till OD600=0.5 in YPD. The culture was harvested and half of it was transferred to SD-N medium and incubated for 4 h. Cells were washed in water and pellets of 1–2 × 106 cells were frozen down in liquid nitrogen and stored at 80°C until analysed.
Cells were lysed in assay buffer (250 mM Tris–HCl (pH 8.5), 0.4% (v/v) Triton X-100, 10 mM MgSO4, 10 μM ZnSO4, 1 mM PMSF) and vortexed with glass beads. After centrifugation, 10 μl lysate was added to 190 μl assay buffer containing 1 mM p-nitrophenol phosphate (Sigma, N9389) and incubated at 30°C. The increase in OD400 was followed at 2 min intervals using a Mutiscan Ascent (Thermo Labsystems) plate reader. The change in OD400/min/mg protein of WT lysate under starvation conditions was set at 100%. Three independent experiments were performed.
Image acquisition
Live cells were analysed with an Axiovert 200M microscope (Carl Zeiss, Inc.) equipped with Exfo X-cite 120 excitation light source, band-pass filters (Carl Zeiss, Inc. and Chroma), and α Plan-Fluar 100 × /1.45 NA or A-Plan 40 × /0.65 NA Ph2 objective lens (Carl Zeiss, Inc.) and a digital camera (Orca ER; Hamamatsu). Image acquisition was performed using Openlab and Volocity software (Perkin-Elmer). Fluorescence images were routinely collected as 0.5 μm Z-Stacks and merged into one plane after contrast enhancing in Volocity and processed further in Photoshop (Adobe) where only the level adjustment was used. On occasions (as indicated in the text) images were collected as single-plane images. Bright-field images were collected in one plane.
Yeast two-hybrid analysis
Yeast two-hybrid screening was performed by Hybrigenics, S.A., Paris, France (http://www.hybrigenics-services.com). The coding sequence for amino acids 40–441 of S. cerevisiae PEX3 was PCR amplified and cloned into pB27 as a C-terminal fusion to LexA (N-LexA–Pex3-C). The construct was checked by sequencing the entire insert and used as a bait to screen at saturation a highly complex S. cerevisiae fragment genomic library constructed into pP6. pB27 and pP6 derive from the original pBTM116 (Vojtek and Hollenberg, 1995) and pGADGH (Bartel et al, 1993) plasmids, respectively. In all, 63 million clones (11-fold the complexity of the library) were screened using a mating approach with Y187 (MATα) and L40DGAL4 (MATa) yeast strains as previously described (Fromont-Racine et al, 1997). 304 His+ colonies were selected on a medium lacking tryptophan, leucine, histidine and supplemented with 5 mM 3-aminotriazole. The prey fragments of the positive clones were amplified by PCR and sequenced at their 5′ and 3′ junctions.
Supplementary Material
Acknowledgments
We thank Ralf Erdmann and Erdmann Girzalsky for the anti-Pex3 antibody and Joanne Munck for making the OM45–Pex3 plasmid and the pex3 allele library. We also thank Matthew Walsh and Stuart Wilson for reagents and discussions on in-vitro transcription/translation, Rick Rachubinski for the pex28Δpex29Δ and pex30Δpex31Δpex32Δ strains, and Peter Piper, Stefan Milson, Sean Munro, and Yohei Ohashi for stains and helpful suggestions relating to the ALP assay. This work was funded by a Wellcome Trust Senior Research Fellowship in Basic Biomedical Science awarded to EH, WT084265MA.
Author contributions: AMM, JMN and EHH designed the experiments; EHH wrote the paper; JMN, AMM and EHH performed the experiments; AMM and JMN constructed strains and plasmids; AMM performed all the microscopy and JMN all the biochemistry.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aoki Y, Kanki T, Hirota Y, Kurihara Y, Saigusa T, Uchiumi T, Kang D (2011) Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol Biol Cell 22: 3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Takeshige K, Baba N, Ohsumi Y (1994) Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol 124: 903–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard E, McFerran NV, Trudgett A, Nelson J, Timson DJ (2008) Development and implementation of split-GFP-based bimolecular fluorescence complementation (BiFC) assays in yeast. Biochem Soc Trans 36: 479–482 [DOI] [PubMed] [Google Scholar]
- Bartel P, Chien CT, Sternglanz R, Fields S (1993) Elimination of false positives that arise in using the two-hybrid system. Biotechniques 14: 920–924 [PubMed] [Google Scholar]
- Bellu AR, Komori M, van der Klei IJ, Kiel JA, Veenhuis M (2001) Peroxisome biogenesis and selective degradation converge at Pex14p. J Biol Chem 276: 44570–44574 [DOI] [PubMed] [Google Scholar]
- Bellu AR, Salomons FA, Kiel JA, Veenhuis M, Van Der Klei IJ (2002) Removal of Pex3p is an important initial stage in selective peroxisome degradation in Hansenula polymorpha. J Biol Chem 277: 42875–42880 [DOI] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P (2006) Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol 4: e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Thorsness PE (1998) Escape of mitochondrial DNA to the nucleus in yme1 yeast is mediated by vacuolar-dependent turnover of abnormal mitochondrial compartments. J Cell Sci 111: Part 162455–2464 [DOI] [PubMed] [Google Scholar]
- Chang J, Mast FD, Fagarasanu A, Rachubinski DA, Eitzen GA, Dacks JB, Rachubinski RA (2009) Pex3 peroxisome biogenesis proteins function in peroxisome inheritance as class V myosin receptors. J Cell Biol 187: 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve C, Wattiaux R (1966) Functions of lysosomes. Annu Rev Physiol 28: 435–492 [DOI] [PubMed] [Google Scholar]
- Dunn WA Jr, Cregg JM, Kiel JA, van der Klei IJ, Oku M, Sakai Y, Sibirny AA, Stasyk OV, Veenhuis M (2005) Pexophagy: the selective autophagy of peroxisomes. Autophagy 1: 75–83 [DOI] [PubMed] [Google Scholar]
- Farre JC, Manjithaya R, Mathewson RD, Subramani S (2008) PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell 14: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain JC, Legrain P (1997) Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet 16: 277–282 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534 [DOI] [PubMed] [Google Scholar]
- Hara-Kuge S, Fujiki Y (2008) The peroxin Pex14p is involved in LC3-dependent degradation of mammalian peroxisomes. Exp Cell Res 314: 3531–3541 [DOI] [PubMed] [Google Scholar]
- Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ (1996) Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem 271: 17621–17624 [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ (1995) Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol 131: 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, Girzalsky W, van Den Berg M, Erdmann R, Distel B (2000) Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J 19: 223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, Ruigrok CC, Koerkamp MG, van den Berg M, Tabak HF, Distel B, Braakman I (1998) The cytosolic DnaJ-like protein djp1p is involved specifically in peroxisomal protein import. J Cell Biol 142: 421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohfeld J, Veenhuis M, Kunau WH (1991) PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J Cell Biol 114: 1167–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins MU, Veenhuis M, Klionsky DJ (1999) Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J Cell Sci 112: Part 224079–4087 [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, Lamark T (2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7: 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ (2008) Mitophagy in yeast occurs through a selective mechanism. J Biol Chem 283: 32386–32393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ (2009) Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell 17: 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Huang WP, Stromhaug PE, Klionsky DJ (2002) Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem 277: 763–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J (2008) Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci USA 105: 20567–20574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Johansen T, Dikic I (2009) NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy 5: 732–733 [DOI] [PubMed] [Google Scholar]
- Knoblach B, Rachubinski RA (2010) Phosphorylation-dependent activation of peroxisome proliferator protein PEX11 controls peroxisome abundance. J Biol Chem 285: 6670–6680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo-Okamoto N, Noda NN, Suzuki SW, Nakatogawa H, Takahashi I, Matsunami M, Hashimoto A, Inagaki F, Ohsumi Y, Okamoto K (2012) Autophagy-related protein 32 acts as autophagic degron and directly initiates mitophagy. J Biol Chem 287: 10631–10638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M (2008) Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 10: 602–610 [DOI] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 6: 463–477 [DOI] [PubMed] [Google Scholar]
- Lynch-Day MA, Klionsky DJ (2010) The Cvt pathway as a model for selective autophagy. FEBS Lett 584: 1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Agrawal G, Subramani S (2011) Peroxisome assembly: matrix and membrane protein biogenesis. J Cell Biol 193: 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R, Jain S, Farre JC, Subramani S (2010a) A yeast MAPK cascade regulates pexophagy but not other autophagy pathways. J Cell Biol 189: 303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R, Nazarko TY, Farre JC, Subramani S (2010b) Molecular mechanism and physiological role of pexophagy. FEBS Lett 584: 1367–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K, Klionsky DJ (2011) MAPKs regulate mitophagy in Saccharomyces cerevisiae. Autophagy 7: 1564–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K, Wang K, Zhao M, Xu T, Klionsky DJ (2011) Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J Cell Biol 193: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchener JS, Shelburne JD, Bradford WD, Hawkins HK (1976) Cellular autophagocytosis induced by deprivation of serum and amino acids in HeLa cells. Am J Pathol 83: 485–492 [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T (2001) Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol 152: 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley AM, Hettema EH (2007) Yeast peroxisomes multiply by growth and division. J Cell Biol 178: 399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley AM, Ward GP, Hettema EH (2008) Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J Cell Sci 121: 1633–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munck JM, Motley AM, Nuttall JM, Hettema EH (2009) A dual function for Pex3p in peroxisome formation and inheritance. J Cell Biol 187: 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10: 458–467 [DOI] [PubMed] [Google Scholar]
- Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, Fujioka Y, Ohsumi Y, Inagaki F (2008) Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13: 1211–1218 [DOI] [PubMed] [Google Scholar]
- Noda NN, Ohsumi Y, Inagaki F (2010) Atg8-family interacting motif crucial for selective autophagy. FEBS Lett 584: 1379–1385 [DOI] [PubMed] [Google Scholar]
- Noda T, Matsuura A, Wada Y, Ohsumi Y (1995) Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun 210: 126–132 [DOI] [PubMed] [Google Scholar]
- Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dotsch V, Ney PA, Dikic I (2010) Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep 11: 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y (2009) Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell 17: 87–97 [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC (2002) Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet 11: 1107–1117 [DOI] [PubMed] [Google Scholar]
- Rich KA, Burkett C, Webster P (2003) Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol 5: 455–468 [DOI] [PubMed] [Google Scholar]
- Rucktaschel R, Girzalsky W, Erdmann R (2011) Protein import machineries of peroxisomes. Biochim Biophys Acta 1808: 892–900 [DOI] [PubMed] [Google Scholar]
- Rucktaschel R, Halbach A, Girzalsky W, Rottensteiner H, Erdmann R (2010) De novo synthesis of peroxisomes upon mitochondrial targeting of Pex3p. Eur J Cell Biol 89: 947–954 [DOI] [PubMed] [Google Scholar]
- Sakai Y, Oku M, van der Klei IJ, Kiel JA (2006) Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta 1763: 1767–1775 [DOI] [PubMed] [Google Scholar]
- Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ (2001) Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell 7: 1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Huang WP, Stromhaug PE, Klionsky DJ (2002) Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell 3: 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp EL, Hegde RS, Francolini M, Lombardo F, Colombo S, Pedrazzini E, Borgese N, Lippincott-Schwartz J (2003) Formation of stacked ER cisternae by low affinity protein interactions. J Cell Biol 163: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kamada Y, Ohsumi Y (2002) Studies of cargo delivery to the vacuole mediated by autophagosomes in Saccharomyces cerevisiae. Dev Cell 3: 815–824 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kondo C, Morimoto M, Ohsumi Y (2010) Selective transport of alpha-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. J Biol Chem 285: 30019–30025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y (2007) Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12: 209–218 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Ohsumi Y (2010) Current knowledge of the pre-autophagosomal structure (PAS). FEBS Lett 584: 1280–1286 [DOI] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y (1992) Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol 119: 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower RJ, Fagarasanu A, Aitchison JD, Rachubinski RA (2011) The peroxin Pex34p functions with the Pex11 family of peroxisomal divisional proteins to regulate the peroxisome population in yeast. Mol Biol Cell 22: 1727–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- van Zutphen T, Veenhuis M, van der Klei IJ (2008) Pex14 is the sole component of the peroxisomal translocon that is required for pexophagy. Autophagy 4: 63–66 [DOI] [PubMed] [Google Scholar]
- van Zutphen T, Veenhuis M, van der Klei IJ (2011) Damaged peroxisomes are subject to rapid autophagic degradation in the yeast Hansenula polymorpha. Autophagy 7: 863–872 [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128: 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM (1995) Ras-Raf interaction: two-hybrid analysis. Methods Enzymol 255: 331–342 [DOI] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Elazar Z (2011) Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 80: 125–156 [DOI] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I (2011) Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333: 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ (2007) Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ (2005a) Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell 16: 1593–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ (2005b) Autophagy: molecular machinery for self-eating. Cell Death Differ 12: Suppl 21542–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C et al. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.