Abstract
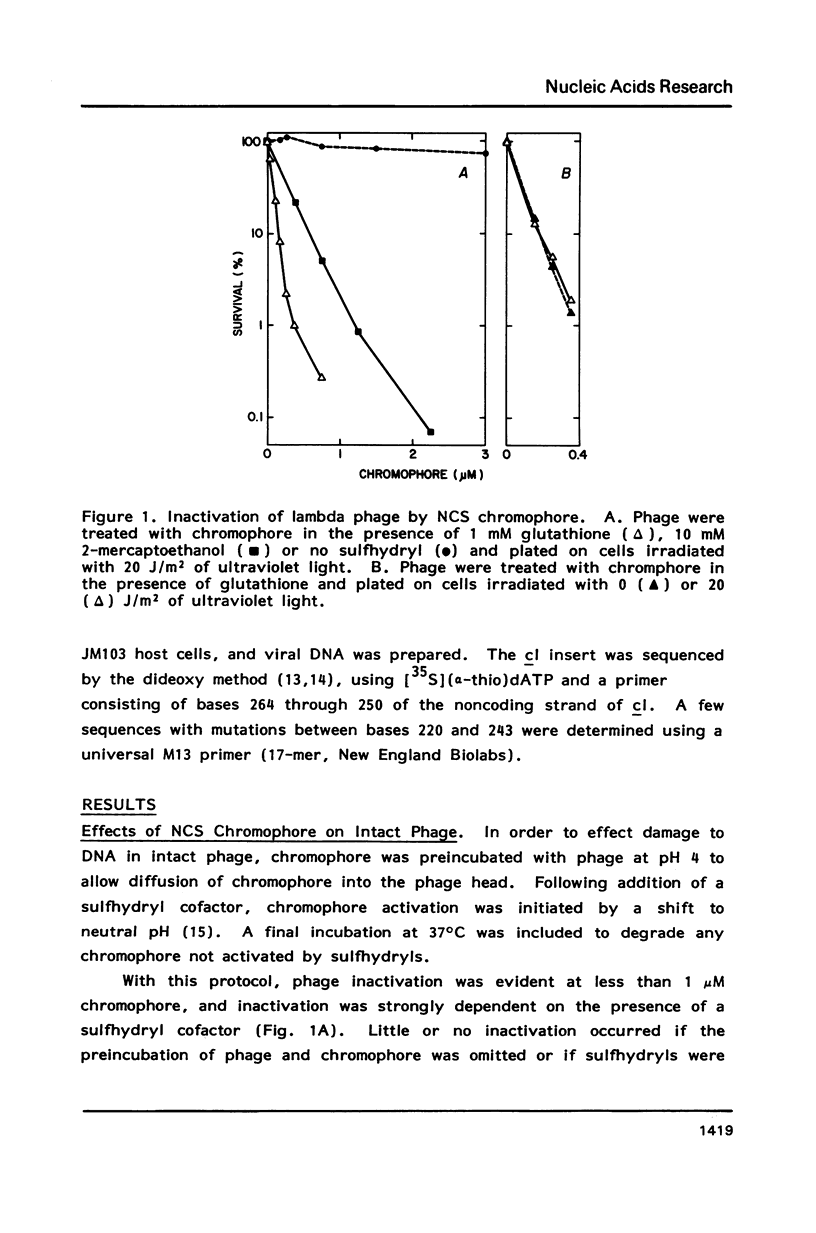
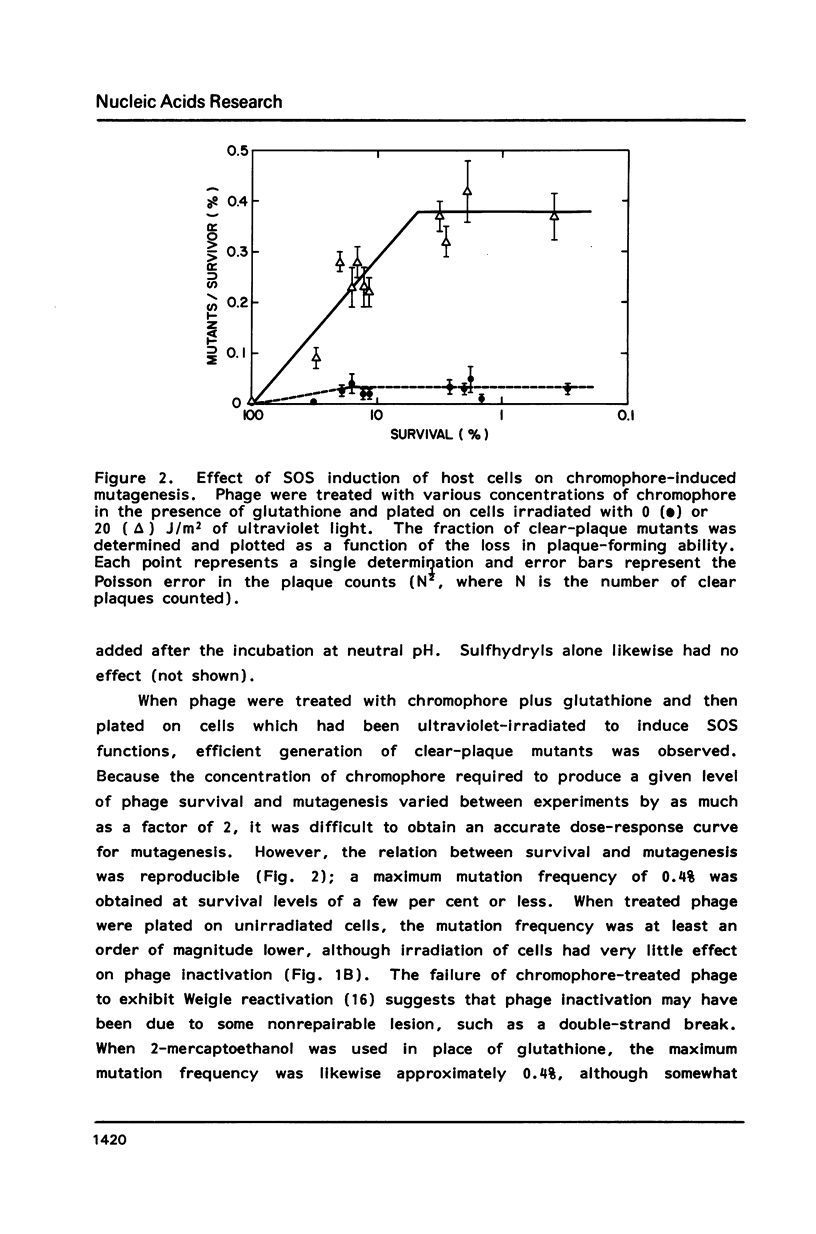
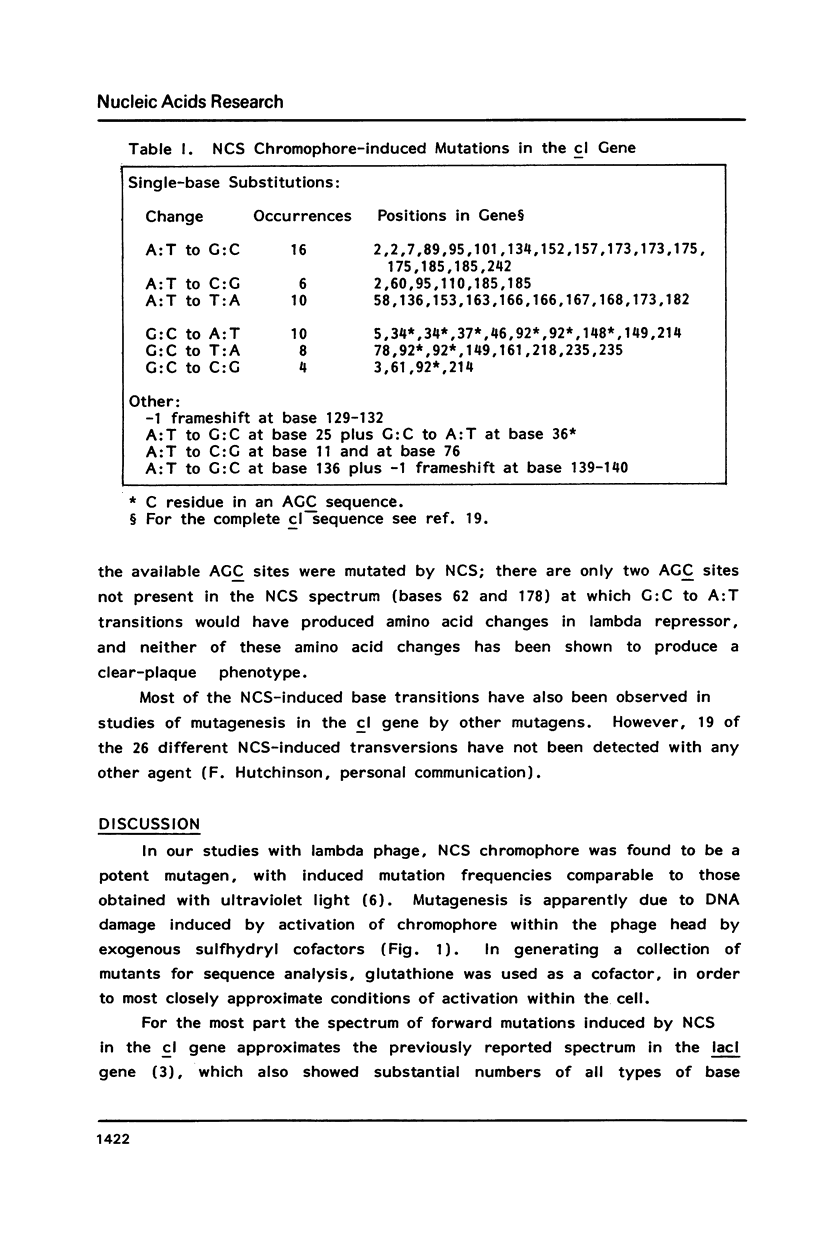
Treatment of intact lambda phage with the nonprotein chromophore of neocarzinostatin resulted in efficient phage inactivation and generation of clear-plaque mutants. Both effects required a preincubation at low pH to allow diffusion of chromophore into the phage head. Chromophore activation was then effected by addition of a sulfhydryl cofactor, followed by a shift to neutral pH. Sequence analysis of mutations mapped to the DNA-binding region of the cI gene revealed that nearly all were single base substitutions. Significant numbers of all possible base changes were found, with A:T to G:C transitions being the most frequent events. Of 11 G:C to A:T transitions, 7 were found at C residues in the trinucleotide sequence AGC, which has previously been shown to be a hotspot for chromophore-induced depyrimidination. This result, as well as the SOS dependence of mutagenesis and the overall distribution of various types of base substitutions, is consistent with the hypothesis that apurinic/apyrimidinic sites are important mutagenic lesions.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belfort M., Noff D., Oppenheim A. B. Isolation, characterization and deletion mapping of amber mutations in the cll gene of phage lambda. Virology. 1975 Jan;63(1):147–159. doi: 10.1016/0042-6822(75)90380-3. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose K. K., Tatsumi K., Strauss B. S. Apurinic/apyrimidinic endonuclease sensitive sites as intermediates in the in vitro degradation of deoxyribonucleic acid by neocarzinostatin. Biochemistry. 1980 Oct 14;19(21):4761–4766. doi: 10.1021/bi00562a007. [DOI] [PubMed] [Google Scholar]
- D'Andrea A. D., Haseltine W. A. Sequence specific cleavage of DNA by the antitumor antibiotics neocarzinostatin and bleomycin. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3608–3612. doi: 10.1073/pnas.75.8.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstadt E., Wolf M., Goldberg I. H. Mutagenesis by neocarzinostatin in Escherichia coli and Salmonella typhimurium: requirement for umuC+ or plasmid pKM101. J Bacteriol. 1980 Nov;144(2):656–660. doi: 10.1128/jb.144.2.656-660.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., Eisenstadt E. Distribution and specificity of mutations induced by neocarzinostatin in the lacI gene of Escherichia coli. J Bacteriol. 1983 Jan;153(1):379–383. doi: 10.1128/jb.153.1.379-383.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatayama T., Goldberg I. H., Takeshita M., Grollman A. P. Nucleotide specificity in DNA scission by neocarzinostatin. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3603–3607. doi: 10.1073/pnas.75.8.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson F., Stein J. Mutagenesis of lambda phage: 5-bromouracil and hydroxylamine. Mol Gen Genet. 1977 Mar 28;152(1):29–36. doi: 10.1007/BF00264936. [DOI] [PubMed] [Google Scholar]
- ISHIDA N., MIYAZAKI K., KUMAGAI K., RIKIMARU M. NEOCARZINOSTATIN, AN ANTITUMOR ANTIBIOTIC OF HIGH MOLECULAR WEIGHT. ISOLATION, PHYSIOCHEMICAL PROPERTIES AND BIOLOGICAL ACTIVITIES. J Antibiot (Tokyo) 1965 Mar;18:68–76. [PubMed] [Google Scholar]
- Isildar M., Schuchmann M. N., Schulte-Frohlinde D., von Sonntag C. gamma-Radiolysis of DNA in oxygenated aqueous solutions: alterations at the sugar moiety. Int J Radiat Biol Relat Stud Phys Chem Med. 1981 Oct;40(4):347–354. doi: 10.1080/09553008114551301. [DOI] [PubMed] [Google Scholar]
- Kappen L. S., Goldberg I. H. Deoxyribonucleic acid damage by neocarzinostatin chromophore: strand breaks generated by selective oxidation of C-5' of deoxyribose. Biochemistry. 1983 Oct 11;22(21):4872–4878. doi: 10.1021/bi00290a002. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Mutational specificity of depurination. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1494–1498. doi: 10.1073/pnas.81.5.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Povirk L. F., Dattagupta N., Warf B. C., Goldberg I. H. Neocarzinostatin chromophore binds to deoxyribonucleic acid by intercalation. Biochemistry. 1981 Jul 7;20(14):4007–4014. doi: 10.1021/bi00517a009. [DOI] [PubMed] [Google Scholar]
- Povirk L. F., Goldberg I. H. Competition between anaerobic covalent linkage of neocarzinostatin chromophore to deoxyribose in DNA and oxygen-dependent strand breakage and base release. Biochemistry. 1984 Dec 18;23(26):6304–6311. doi: 10.1021/bi00321a003. [DOI] [PubMed] [Google Scholar]
- Povirk L. F., Goldberg I. H. Detection of neocarzinostatin chromophore-deoxyribose adducts as exonuclease-resistant sites in defined-sequence DNA. Biochemistry. 1985 Jul 16;24(15):4035–4040. doi: 10.1021/bi00336a035. [DOI] [PubMed] [Google Scholar]
- Povirk L. F., Goldberg I. H. Endonuclease-resistant apyrimidinic sites formed by neocarzinostatin at cytosine residues in DNA: evidence for a possible role in mutagenesis. Proc Natl Acad Sci U S A. 1985 May;82(10):3182–3186. doi: 10.1073/pnas.82.10.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk L. F., Goldberg I. H. Stoichiometric uptake of molecular oxygen and consumption of sulfhydryl groups by neocarzinostatin chromophore bound to DNA. J Biol Chem. 1983 Oct 10;258(19):11763–11767. [PubMed] [Google Scholar]
- Sagher D., Strauss B. Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry. 1983 Sep 13;22(19):4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer R. T. DNA sequence of the bacteriophage gama cI gene. Nature. 1978 Nov 16;276(5685):301–302. doi: 10.1038/276301a0. [DOI] [PubMed] [Google Scholar]
- Skopek T. R., Hutchinson F. DNA base sequence changes induced by bromouracil mutagenesis of lambda phage. J Mol Biol. 1982 Jul 25;159(1):19–33. doi: 10.1016/0022-2836(82)90029-8. [DOI] [PubMed] [Google Scholar]
- Weigle J. J. Induction of Mutations in a Bacterial Virus. Proc Natl Acad Sci U S A. 1953 Jul;39(7):628–636. doi: 10.1073/pnas.39.7.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood R. D., Hutchinson F. Non-targeted mutagenesis of unirradiated lambda phage in Escherichia coli host cells irradiated with ultraviolet light. J Mol Biol. 1984 Mar 5;173(3):293–305. doi: 10.1016/0022-2836(84)90122-0. [DOI] [PubMed] [Google Scholar]
- Wood R. D., Skopek T. R., Hutchinson F. Changes in DNA base sequence induced by targeted mutagenesis of lambda phage by ultraviolet light. J Mol Biol. 1984 Mar 5;173(3):273–291. doi: 10.1016/0022-2836(84)90121-9. [DOI] [PubMed] [Google Scholar]
- Wu J. C., Kozarich J. W., Stubbe J. The mechanism of free base formation from DNA by bleomycin. A proposal based on site specific tritium release from Poly(dA.dU). J Biol Chem. 1983 Apr 25;258(8):4694–4697. [PubMed] [Google Scholar]



