Abstract
Purpose
Anaplastic lymphoma kinase (ALK) rearrangements, associated with sensitivity to an experimental ALK/MET inhibitor, occur in 3% to 5% of non-small cell lung cancers. Intratumoral fluorescence in situ hybridization (FISH) heterogeneity has been reported. We explored the heterogeneity basis, the requirements for accurately determining ALK FISH positivity, and the effect of enriching the tested population using clinical and molecular factors.
Experimental Design
Lung cancer patients were screened by ALK and MET FISH and for EGFR and KRAS mutations.
Results
Thirteen ALK-positive cases were identified from 73 screened patients. Gene copy number increases occurred together with classic rearrangements. All positive cases were adenocarcinomas, 12 were EGFR/KRAS wild-type, and 1 had a coexistent EGFR exon 20 mutation. No association with MET amplification occurred. ALK positivity was associated with <10-pack-year smoking status (P = 0.0004). Among adenocarcinomas, without KRAS or EGFR mutations, with <10-pack-year history, 44.8% of cases were ALK positive. ALK FISH positivity was heterogeneous, but mean values in tumor areas from ALK-positive patients (54% of cells; range, 22-87%) were significantly higher than in adjacent normal tissue or tumor/ normal areas from ALK-negative patients (mean, 5-7%). Contiguous sliding field analyses showed diffuse heterogeneity without evidence of focal ALK rearrangements. One hundred percent sensitivity and specificity occurred when four or more fields (~60 cells) were counted.
Conclusions
Intratumoral ALK FISH heterogeneity reflects technique, not biology. The clinical activity of ALK/MET inhibitors in ALK-positive patients probably reflects ALK, but not MET, activity. Prescreening by histology, EGFR/KRAS mutations, and smoking status dramatically increases the ALK-positive hit rate compared with unselected series.
Transforming rearrangements of the ALK gene have been reported in a few human hematologic and solid malignancies (1, 2). These in-frame gene rearrangements place the ALK kinase domain under the promoter control of another gene, with its intracellular localization and function potentially influenced by the specific NH2-terminal protein fusion partner expressed (1). In 2007, an ALK gene rearrangement creating an in-frame fusion protein between ALK and echinoderm microtubule-associated protein-like 4 (EML4) was described in non-small cell lung cancer (NSCLC; ref. 3). Since then, several different transforming in-frame fusion variants of EML4-ALK in NSCLC have been described with different EML4 and ALK breakpoints, in addition to other rarer non-EML4 fusion partners, including TFG and KIF5B (3-8).
The incidence of ALK gene rearrangements detected by either real-time PCR (RT-PCR) or fluorescence in situ hybridization (FISH) seems to be low in largely unselected NSCLC series (~3-5%; ref. 9). However, ALK status, at least that determined by FISH, does seem to be extremely important as a specific treatment sensitivity marker with respect to small-molecule inhibitors of ALK (9, 10).
The ability to accurately identify a patient as truly ALK positive or negative using any given screening technique is essential for appropriate therapy selection and outcome assessment. Recent reports of significant intratumoral heterogeneity of EML4-ALK fusions detected by FISH have raised questions about both whether ALK gene rearrangements could easily be missed and, if subclones with and without ALK rearrangements commonly coexist in tumors, whether they may be later rather than primary transforming events in human lung cancers in vivo (11). In addition, in the absence of universal screening, being able to identify, in advance, which patients are most likely to possess ALK gene rearrangements would enable treating physicians to prioritize which patients to refer for ALK screening (e.g., by FISH) and possible enrollment within an ALK inhibitor trial.
Here, we report on a detailed exploration of the heterogeneity of ALK-positive signals across tumor and surrounding normal tissue and the specific cytogenetic characteristics of those who tested positive using FISH break-apart probe technology. We also report on how selecting patients on the basis of histology, EGFR and KRAS mutation status, and smoking history has significantly enriched our numbers testing positive for ALK gene rearrangements compared with previously reported unselected series.
Materials and Methods
Patients
All lung cancer patients tested received treatment or consultation from the University of Colorado Thoracic Oncology Program. Since mid-2008, all new NSCLC patients seen at the University of Colorado have had routine testing of their tumors for epidermal growth factor receptor (EGFR) expression by immunohistochemistry, EGFR gene copy number by FISH, and EGFR, KRAS, and TP53 mutational status done through the on-site Clinical Laboratory Improvement Amendments-certified Colorado Molecular Correlates (CMOCO) Laboratory (12). In addition, to identify patients for entry into defined molecular cohorts treated within the phase I study of crizotinib (PF-02341066), we began to screen tumor biopsies from selected NSCLC patients for MET gene copy number alterations and for ALK gene rearrangements by FISH. An Institutional Review Board-approved protocol approved in early 2009 permits both retrospective and prospective clinical correlates to be made on all in-house patients in whom molecular analyses have been conducted within CMOCO.
CMOCO mutation tests
Mutational analysis was done as previously described (13). Briefly, tumor-rich regions were identified in each block of formalin-fixed, paraffin-embedded (FFPE) tissue. Tissue cores were then obtained from these regions using a microarrayer device and 1-mm arrayer needles. Tissue cores were deparaffinized by soaking in xylene overnight and subsequently digested with protease K. DNA was isolated from the incubation mixture using a QIAcube robotic workstation (Qiagen, Inc.) extraction protocol. EGFR exons 19, 20, and 21 and KRAS exon 2 were then amplified and sequenced using a ABI model 3730 capillary gel sequencer. Mutations were identified by visual inspection of the resulting chromatograms and automated scanning using Mutation Surveyor v3.24.
MET and ALK FISH testing
Unstained 4-μm sections of FFPE tumor tissue were submitted to dual-color FISH assays using two probe sets. The MET/CEP7 probe set was developed in house with a DNA insert encompassing MET (RP 11-95I20 bacterial artificial chromosome clone) labeled with SpectrumRed and the SpectrumGreen CEP7 probe (Abbott Molecular). The LSI ALK (anaplastic lymphoma kinase) Dual Color, Break-Apart Rearrangement Probe (Abbott Molecular) was designed to detect rearrangements in 2p23 encompassing the ALK gene. This probe set includes a 250-kb DNA fragment telomeric to ALK (3′ end) labeled in Spectrum-Orange and a 300-kb fragment centromeric to ALK (5′ end) labeled in SpectrumGreen.
For both probe sets, assays were done according to a protocol previously described (14). Signals were enumerated in at least 50 tumor nuclei per core using an epifluorescence microscope with single interference filters sets for green (FITC), red (Texas red), and blue (4′,6-diamidino-2-phenylindole) as well as dual (red/green) and triple (blue, red, green) band-pass filters.
In the MET FISH assay, specimens were classified as carrying gene amplification when the MET/CEP7 ratio was >2 or when >10% tumor cells carried clusters or >15 copies per cell of MET signals. For documentation, images were captured using a charge-coupled device (Photometrics) and merged using dedicated software (CytoVision, Genetix). In the ALK FISH assay, the native state appears as fusedor adjacent red/green (yellow) signals, whereas a rearrangement with a breakpoint at the 2p23 ALK region produces single red and single green signals. Red and green signals separated by a gap larger than 2 signal diameters were considered split. The occurrence of an ALK rearrangement (ALK FISH positive) was concluded if >15% of tumor cells showed split red and green signals (15) and/or single red signals; otherwise, the specimen was classified as ALK FISH negative.
In the detailed FISH analyses to investigate intratumoral heterogeneity and the presence of the rearrangement in nontumor cells, the whole section was scanned using a contiguous “sliding strategy.” In addition to the 13 positive cases detected among the Colorado CMOCO patients, an additional 4 positive Caucasian cases from Italy identified in other studies were also included in these analyses. Beginning at a selected “starting corner” and using the high-power objective, a tumor area was selected (T1) and ~15 representative tumor cells were analyzed. Then, the microscope stage was moved 2 mm along the Y axis and another tumor area (T2) was chosen for analysis of 15 tumor cells. A similar sequence of steps was then used to reach the third location (T3), and so on. In the event that a selected location included nontumor cells such as lymphocytes, macrophages, or smooth muscle cells, this area was identified as N1, and 15 of these cells were analyzed. When reaching an area without tissue, the microscope stage was moved 2 mm along the X axis and the stage was again moved 2 mm along the Y axis, looking for tumor (T) and normal (N) areas. This strategy guaranteed that specimens were scanned in entirety, that no overlap occurred among different areas, and that no cell was counted in more than one area. Following this schema, from 7 to 20 tumor areas and from 5 to 15 nontumor areas were analyzed in each specimen.
Statistical analyses
Unless otherwise specified, for the analyses of clinical and CMOCO markers on the patient samples, the Fisher’s exact test was used to assess correlation between categorical variables and Student’s t test was used to assess association between the distributions of treatment outcome. All reported P values are two-sided unless otherwise specified, and we considered a test as statistically significant if P < 0.05.
To assess whether ALK rearrangement was a focal event, we used a window of four consecutively scored high-power objective fields generated through the sliding strategy described above (e.g., areas T1-T4, T2-T5, etc.) through all the tumor areas tested for ALK rearrangement in each individual ALK-positive patient. We hypothesized that variability resulting from technical aspects of the FISH assay would produce a diffuse effect, and positive signals (>15% cells positive for rearrangement) would be seen in most fields tested. In contrast, if independent ALK-positive and ALK-negative subclones existed, we would expect to see differences between fields in the same tumor. ALK rearrangement was considered focal if only one of the four areas analyzed was positive, whereas all other combina tions were taken as proof that ALK rearrangement was a diffuse event.
To determine the optimal number of high-power microscope fields and tumor nuclei required to accurately define a patient’s tumor as positive or negative for ALK rearrangement, we randomly selected two to seven tumor areas from ALK-positive and ALK-negative patients in 100 permutations. If the average percent of positive cells in the permutation was >15%, we called that permutation ALK positive, and vice versa for ALK negative. Using large numbers of tumor areas to generate what we considered definitive ALK-positive and ALK-negative status, we could compute the number of true positives (TP) and true negatives (TN), false positives (FP), and false negatives (FN) for permutations with lower numbers of counted fields. We defined TP and TN as the tumor areas that were obtained and correctly determined as ALK positive and negative in patients who, overall, were concluded as ALK positive and negative, respectively. FP was defined as tumor areas sampled from ALK-negative patients but determined as ALK positive because these areas showed >15% of cells positive for rearrangement patterns. FN was defined as tumor areas obtained from ALK-positive patients but wrongly classified as ALK negative. From this permutation test, we could calculate sensitivity [TP/(TP + FN)] and specificity [TN/ (TN + FP)] for the number of high-power microscope fields and number of tumor nuclei required to determine ALK positivity.
Results
Patients tested for ALK gene rearrangements
Between June 2008 and October 2009, a total of 73 lung cancer patients were considered for testing with the ALK FISH break-apart assay within the University of Colorado Thoracic Oncology Program. Of these, four cases could not be analyzed due to insufficient tumor material remaining in the biopsy specimen and three cases could not be analyzed due to assay failure, in one case due to DNA degradation following decalcification of a bone biopsy specimen. From the remaining 66 specimens, 13 cases were positive for ALK gene rearrangements (20%).
Cytogenetic patterns of ALK positivity
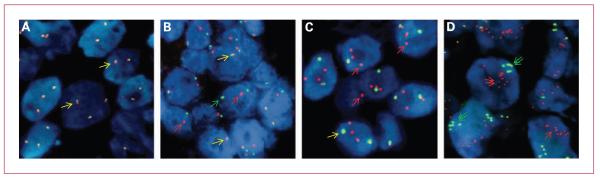
Table 1 shows the descriptive statistics for each of the 13 patients classified as positive for the ALK rearrangement, including the mean copy number per cell (and SD) for the normal signal (fused 3′ red-5′ green ALK), for the split/single 3′ ALK (red), and the split/single 5′ ALK (green) signals, and the distribution of tumor cells according to the possible patterns combining these different signals. Normal signals are frequently seen in cells that also display ALK gene rearrangements. The classic EML4-ALK fusion, as well as translocations between ALK and other genes, should typically show distinct red and green signals separated within the same cell. However, either due to nuclear truncation or true physical deletion of chromosomalsegments, atypical patterns including only single red (3′ end) and only single green (5′ end) signals were also observed. To compile the frequency of positive cells for ALK rearrangements, the sum of cells carrying split red and green and single red (ALK 5′ region) signals was considered, in the presence or absence of normal fused signals. The predominant ALK-positive FISH pattern was fusion and split red and green signals in eight patients (62%), whereas in four (31%) it was fusion and single red, and in one (7%) it was single red and single green signals. Multiple copies per cell of single reds, clusters of red and green signals, and clusters of fusion signals were also seen, suggesting that increases in gene copy number occur in addition to rearrangements. Representative variations in the cytogenetic patterns of ALK rearrangements are shown in Fig. 1.
Table 1.
Descriptive statistics for each of the 13 CMOCO patients classified as positive for the ALK rearrangement, including the mean copy number per cell (and SD) for the normal signal (3′/5′ ALK, fused red/green), for the single 3′ ALK (red) and the single 5′ ALK (green) signals, and the distribution of tumor cells according to the possible patterns combining these three types of signals
| ALK-positive patient |
Fused (3′/5′) |
Single 3′ ALK |
Single 5′ ALK |
Percentage of cells with given patterns | % Cells positive for ALK rearrangement |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||||
| Mean | SD | Mean | SD | Mean | SD | Only 3′/5′ |
Only 3′ |
Only 3′ |
Only 5′ |
3′/5′ and 3′ |
3′/5′ and 5′ |
3′ and 5′ |
3′/5′, 3′ and 5′ |
|
| 1 | 3.63 | 2.68 | 1.18 | 1.41 | 0.06 | 0.24 | 40% | 0% | 0% | 54% | 2% | 0% | 4% | 58% |
| 2 | 2.45 | 1.25 | 0.61 | 0.63 | 0.74 | 0.76 | 40% | 0% | 0% | 3% | 6% | 4% | 47% | 54% |
| 3 | 2.7 | 1.07 | 0.54 | 0.68 | 0.78 | 0.93 | 42% | 0% | 0% | 4% | 12% | 0% | 42% | 46% |
| 4 | 1.69 | 1.07 | 0.42 | 0.59 | 0.38 | 0.6 | 57% | 0% | 0% | 10% | 6% | 9% | 18% | 37% |
| 5 | 1.61 | 0.75 | 0.31 | 0.53 | 0.31 | 0.56 | 69% | 0% | 0% | 5% | 3% | 6% | 17% | 28% |
| 6 | 1.75 | 0.67 | 1.18 | 1.03 | 0.14 | 0.35 | 24% | 0% | 0% | 62% | 7% | 1% | 6% | 69% |
| 7 | 2.4 | 1.21 | 0.79 | 0.86 | 0.79 | 0.88 | 33% | 0% | 0% | 16% | 14% | 3% | 34% | 53% |
| 8 | 0.56 | 0.92 | 2.46 | 2.44 | 1.05 | 1.45 | 17% | 20% | 1% | 6% | 0% | 42% | 14% | 82% |
| 9 | 1.72 | 0.7 | 1.16 | 0.91 | 0.1 | 0.36 | 30% | 2% | 0% | 60% | 0% | 2% | 6% | 70% |
| 10 | 1.6 | 0.86 | 0.88 | 0.82 | 0.91 | 0.87 | 33% | 0% | 0% | 4% | 3% | 8% | 52% | 64% |
| 11 | 1.72 | 0.88 | 0.94 | 0.74 | 0.16 | 0.42 | 24% | 0% | 0% | 62% | 6% | 2% | 6% | 70% |
| 12 | 0.98 | 0.8 | 1.86 | 0.95 | 1.92 | 1.03 | 6% | 0% | 0% | 4% | 0% | 22% | 68% | 94% |
| 13 | 3.1 | 1.25 | 0.5 | 0.61 | 0.64 | 0.72 | 46% | 0% | 0% | 2% | 10% | 0% | 42% | 44% |
Fig. 1.
NSCLCs hybridized with the ALK break-apart FISH probe (Abbott Molecular). Native ALK status (indicated by yellow arrows) shows fusion of the probes adjacent to the 3′ and 5′ ends of the gene, labeled, respectively, with red and green fluorophores. Rearranged ALK is indicated by the presence of split 3′ (red arrows) and 5′ (green arrows) signals, including single red signals. A, ALK FISH-negative specimen. B, “classic” ALK FISH-positive specimen showing split red and green signals. C, ALK FISH-positive specimen showing two or three copies of single red signals per cell. D, ALK FISH-positive specimen showing evidence of gene copy number increase with clusters of multiple single red and single green signals (double arrows).
Details and consequences of heterogeneity in ALK positivity
As depicted in Fig. 2 and Table 2, tumor areas tested in ALK-positive patients had an average of 53.8% of cells with a positive signal, which was significantly higher than in the adjacent normal areas in ALK-positive patients (6.81%; P = 2.2 × 10−8, t test), as well as in both tumor (5.98%; P = 1.9 × 10−8, t test) and adjacent normal areas (5.26%; P = 1.4 × 10−8, t test) in ALK-negative patients. To explore whether intratumoral ALK heterogeneity was a diffuse or focal event, the sliding window technique was used. From 16 to 26 four-area “sliding windows” were available for analysis in each of the 17 ALK-positive specimens and not a single focal event (1 of 4 windows positive) was detected.
Fig. 2.
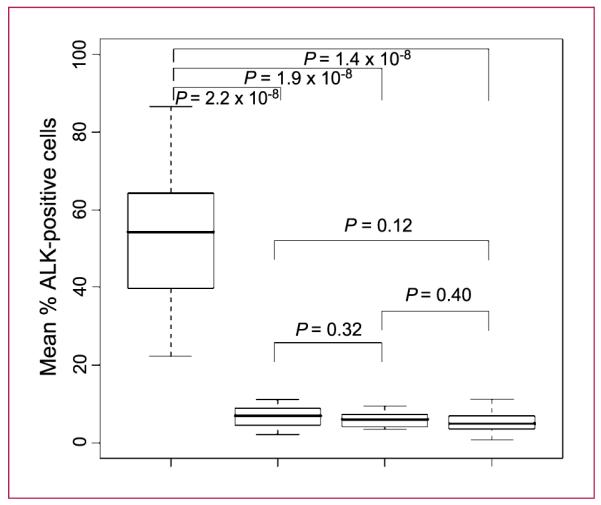
Box-and-whisker plot of ALK-positive and ALK-negative samples in tumor and adjacent normal areas. Seventeen ALK-positive and 15 ALK-negative patients were analyzed to determine the % of ALK-positive cells from various tumor and adjacent normal areas from each individual patient. P values were computed by Welch two-sample t test. The box portion of the box-and-whisker plot includes 50% of the data. The bold line in the box represents the median of the data, and the borders of the box represent the lower quartile (25% percentile) and the upper quartile (75% percentile) of the data. The upper and lower whiskers represent the maximum and minimum % of ALK-positive cells.
Table 2.
ALK-positve and ALK-negative samples in tumor and adjacent normal areas
| Patient category |
||||
|---|---|---|---|---|
| ALK Positive |
ALK negative |
|||
| Histology | Tumor | Nontumor | Tumor | Nontumor |
| No. areas tested (range) | 16 (7-20) | 8 (5-15) | 8 (18-20) | 8 (5-10) |
| Mean no. cells scored (range) | 15 (12-16) | 14 (11-19) | 15 (14-16) | 13 (11-15) |
| % Cells positive for rearrangement (range) | 53.80 (22.25-86.62) | 6.81 (2.14-11.14) | 5.98 (3.51-9.45) | 5.26 (0.71-11.21) |
Table 3 depicts results of statistical simulations on two to seven randomly selected tumor areas (100 permutations) from individual ALK-positive and ALK-negative patients. Large numbers of tumor areas were used to generate “true” ALK-positive and ALK-negative status. The sensitivity and specificity of using different lower numbers of high-power microscope fields and number of tumor nuclei required to determine true ALK positivity are shown. Maximum sensitivity and specificity occurred when four tumor areas (~60 cell nuclei) were analyzed.
Table 3.
Sensitivity and specificity of the ALK FISH assay based on different number of tumor areas/nuclei scored compared with definitive status set by analysis of entire specimen following a contiguous sliding field strategy
| Number of tumor areas |
||||||
|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | 7 | |
| % Sensitivity | 98.6 | 99.3 | 100 | 100 | 100 | 100 |
| % Specificity | 96.6 | 100 | 100 | 100 | 100 | 100 |
| No. nuclei scored | ~30 | ~45 | ~60 | ~75 | ~90 | ~105 |
Clinical and molecular factors associated with ALK positivity
Of the 66 analyzed specimens in our series, 61 were adenocarcinoma, 1 was a small cell lung cancer, 1 was carcinoid, 2 were squamous cancers, and 1 was a large cell neuroendocrine cancer. The primary histology was reviewed in house, and in all 13 ALK-positive cases, the underlying histology was described as adenocarcinoma of the lung. All other analyses described below were only conducted among the adenocarcinoma population.
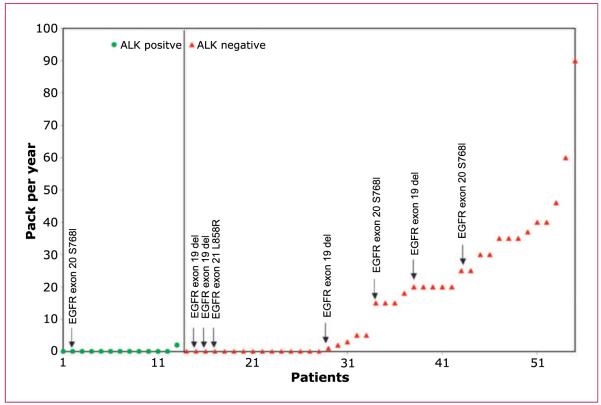
In the first 55 adenocarcinomas screened for ALK gene rearrangements in whom both EGFR and KRAS status was known, 7 of the 42 ALK-negative cases showed EGFR mutations (4 exon 19 deletions, 1 L858R point mutation, and 2 S768I point mutations), whereas among the 13 ALK-positive cases only 1 showed an EGFR mutation (exon 20 point mutation S768I). None of the 55 patients had a KRAS mutation. The smoking status in pack-years of each of these 55 patients and their mutational status are shown graphically in Fig. 3.
Fig. 3.
Scatter plot of patient number [adenocarcinomas of known EGFR and KRAS status (marked)] versus smoking status in pack-years. The vertical line separates ALK-positive from ALK-negative cases. ALK FISH positivity was statistically significantly enriched among never/light smokers (<10 pack-years; P = 0.0004, Fisher’s exact test; proportion positive among never/light smoking adenocarcinomas = 13 of 33, 39%; proportion positive among those with >10–pack-year smoking history = 0 of 22).
Among our 13 ALK FISH-positive cases, MET copy number was also available in 9 (6 with MET high polysomy but no gene amplification and 3 with no or low MET copy number gain). Among the closest comparable group {i.e., ALK-negative never/light smoking adenocarcinomas without either EGFR [tyrosine kinase inhibitor (TKI) sensitivity mutations] or KRAS mutations}, MET information was available in all 16 cases (5 with MET high polysomy but no gene amplification, and 11 negative for any MET-related FISH changes). Taken together, although ALK FISH-positive cases seemed to be significantly correlated with MET high polysomy (P < 0.0001, Fisher’s exact test), they were not correlated with MET gene amplification. No correlation between ALK FISH-positive cases with TP53 mutational status could be established: none of the 11 ALK FISH-positive cases tested for TP53 showed TP53 mutations, whereas 5 of the 41 ALK-negative cases tested were TP53 mutant (P = 0.288, Fisher’s exact test).
Among our 13 ALK-positive patients, the mean and median ages at diagnosis were 55.4 and 53 years, respectively (range, 34-75), and the gender split was 9F:4M. With regard to race, 12 of our ALK-positive patients were Caucasian and 1 was Hispanic. Twelve of the ALK-positive patients were resident in Colorado at the time of diagnosis and 1 was resident in South Carolina. Our referencepopulation was not sufficiently diverse to make any meaningful conclusions about either race or geographic factors on the ALK positivity rate.
Retrospective treatment efficacy results on advanced ALK-positive cases
Progression-free survival (PFS) for patients with advanced disease was calculated from the start of therapy until documented radiographic or clinical progression. Of our 13 ALK-positive cases, advanced disease treatment outcome for platinum-based chemotherapy and for EGFR TKIs were available in 7 (2 ongoing and censored at data gathering) and 3 (none ongoing) patients, respectively. For platinum-based chemotherapy, mean PFS was 3.3 months (range, 2-5 months). For EGFR TKI therapy, mean PFS was 5.3 months (range, 3-9 months). For the ALK-negative cases known to lack EGFR TKI sensitivity mutations and KRAS mutations, treatment outcome for platinum-based chemotherapy and EGFR TKI were available in 22 (4 ongoing and censored at data gathering) and 6 (1 ongoing and censored at data gathering) patients, respectively. For platinum-based chemotherapy, mean PFS was 7.0 months (range, 1-19 months). For EGFR TKI therapy, mean PFS was 6.5 months (range, 1-22 months). Of note, for the ALK-negative patients with EGFR TKI sensitivity mutations, treatment outcome for EGFR TKI therapy was available in 2 (1 ongoing and censored at data gathering) patients, with a mean PFS of 18 months (12 and 24 months). Although numerically ALK-positive patients had a shorter PFS with platinum-based therapy and a comparable PFS on EGFR TKI therapy to ALK-negative EGFR/KRAS wild-type patients (both of these groups appearing to have a much shorter PFS on EGFR TKI therapy than known EGFR mutant patients), given the small sample size of the different subgroups, and the high proportion of patients still on therapy, we did not pursue formal statistical comparisons on this treatment data set.
Discussion
The original identification of EML4-ALK as a potential early driver of lung carcinogenesis came through a screen for transforming cDNAs isolated from a lung adenocarcinoma known to be both EGFR and KRAS wild-type, occurring in a 62-year-old male Japanese with a low smoking history (4.5 pack-years)4 (3). The importance of detecting ALK gene rearranged NSCLCs relates to the apparent high predictive value of the gene rearrangement with regard to sensitivity of the tumor to small-molecule inhibitors of ALK (16). In the first-in-man phase I study of crizotinib (a potent dual inhibitor of ALK and MET), an objective response rate of 53% and a disease control rate at 8 weeks of 79% were reported following its use in advanced NSCLC patients known to harbor ALK gene rearrangements, detected with the break-apart ALK FISH probe (10). Crizotinib is now being explored in ALK FISH-positive advanced NSCLC patients study compared with standard second-line chemotherapy in an international randomized phase III study.
Recently, the apparent intratumoral heterogeneity of ALK gene rearrangements and the presence of rearrangements in nontumor cells have both been raised as areas of concern. Using separate FISH probes for both EML4 and ALK, 2.7% of NSCLCs cases were noted to be EML4-ALK positive in 603 tissue microarray-screened tumors (11). However, rearrangements were only detected in 50% to 100% of cells in positive tumors. Based on this finding, the authors concluded that ALK rearrangements may represent a late, rather than an early, tumorigenic event, resulting in the coproliferation of different clones (both ALK positive and ALK negative) within established tumors. Using RT-PCR, Martelli et al. detected EML4-ALK fusion transcripts in 7.5% of 120 frozen NSCLC samples (17). However, EML4-ALK transcripts were also detected in frozen nontumor lung samples taken at the time of the initial tumor resection in 10 of 67 patients, all of whom were negative for EML4-ALK in their tumors. Among eight paraffin-embedded tumor specimens positive for the RT-PCR transcript in the same study, the frequency of FISH-positive cells ranged from only 1.2% to 9% of cells.
Different ALK assays will undoubtedly have different FP and FN rates. ALK gene rearrangement screening in NSCLC using the FISH break-apart probe described here is technically sophisticated, significantly more so than, for example, FISH that is designed to simple estimate increases in gene copy number. Even among break-apart probe tests, ALK gene rearrangements generally require significant expertise for accurate assessment. When molecular fusions occur between genes mapped in different chromosomes or far distant within the same chromo-some, the sequences at the 3′ and 5′ end of the gene targeted by the break-apart probe will be physically separated and the red and green signals representing these sequences will be easily verified as split. However, the EML4-ALK fusion, which is considered to be the most common ALK gene rearrangement in NSCLC, is a consequence of a paracentric inversion on 2p such that the 3′ and 5′ ALK sequences are only separated by ~12 Mb. In interphase chromatin, the signals generated by these two probes, although split, will therefore usually show as closely “paired” not independently distributed in the nucleus as with other break-apart probes. However, the FISH break-apart probe is the ALK-related marker that has been most extensively explored in relation to clinical activity following ALK inhibitor intervention. We were able to successfully conduct ALK FISH testing in 90% (66 of 73) of our initial screening cohort. The detailed cytogenetic rearrangement patterns seen in ALK-positive tumors (Table 1 and Fig. 1) reveal the potential for activating chromosomal deletions (single red 3′ ALK) and fusion/truncation gene copy number increases, in addition to the classic split signal occurring with the EML4-ALK inversion, to contribute to the transforming potential of ALK in NSCLC. Single green signals (5′ ALK) also occur, but as these do not represent the kinase domain of ALK, they do not contribute to the definition of ALK positivity.
When the signal patterns generated by the break-apart FISH probe in nontumor tissue and in tumor samples from ALK-positive and ALK-negative tumors are compared (Fig. 2), several conclusions can be drawn. Firstly, as previously noted, not all cells within a positive tumor display the positive signal patterns for an ALK rearrangement. In our series, positivity occurred in 22.25% to 86.62% of the cells in ALK-positive tumors. Second, there is a clear “background noise” associated with the assay with up to 11% of cells in both nontumor sections and ALK-negative tumors scoring positive. This “noise” seems to be entirely assay related and not a true biolog ical phenomenon, as the signals in nontumor sections and ALK-negative tumor specimens did not differ significantly from each other, but each did differ significantly from the ALK-positive tumor areas. Importantly, the >15% cut point used in the crizotinib trials falls within the nonoverlapping area of 12% to 21% positivity that seems to accurately differentiate biology from assay variability. FP and FN signals within individual cells are likely to have multiple different etiologies, including nuclear truncation on sectioning (signals are physically lost), aberrant probe hybridization (intended target sequences do not hybridize with the labeled probe and/or the probe does hybridize to nonspecific sequences), background noise (weak specific signals mimic background nonspecific noise), and observer error (nontumor cells included in scoring, adjacent red/green incorrectly classified as fused or split). In further support of assay, rather than biological, variability driving the apparent intratumoral heterogeneity in cell positivity, we found no evidence of discrete foci of ALK gene rearrangements, as might occur if distinct subclones of positive and negative cells truly existed, when a series of four sliding fields were tracked across positive tumor specimens, allowing us to conclude that ALK gene rearrangements are truly diffuse events within ALK-positive tumors. Taken together, these data suggest that variations in the percentage of positive cells within positive tumors are unlikely to represent a true biological phenomenon, and although retrospective clinical data will be required to confirm this, we would predict that the exact percent of positive cells in a positive tumor will probably have little bearing on outcome following treatment with an ALK inhibitor.
Given the propensity for individual cells to falsely read as positive or negative with the break-apart FISH assay,determination of the optimal number of high-power fields and cells to count is imperative for accurate reporting. In Table 2, it is clear that both sensitivity and specificity increase as the number of fields and cells increase. As expected, the worst performance was based on two tumor areas with an average of 30 cells scored, where the decision would be made with 96.6% specificity and 98.6% sensitivity. In contrast, examination of four or more tumor areas including ~60 tumor cells provided 100% sensitivity and specificity in determining ALK positivity, establishing these conditions as the minimum requirements to accurately determine the presence or absence of ALK rearrangements in NSCLC specimens.
Among our initial 66 specimens tested, 13 were positive for ALK gene rearrangements (20%). The reported incidence in unselected series is ~3% to 5%, and our high initial incidence reflects both our routine molecular screening for EGFR and KRAS mutations and an evolving awareness of the clinical and molecular characteristics associated with ALK positivity (specifically, adenocarcinomas known to be EGFR and KRAS wild-type; ref. 9). Together, these factors lead us to adopt a highly focused approach designed to enrich the tested cohort for ALK gene rearranged cancers. Although we can quantify the frequency of ALK positivity within subsets of the group that we have defined, it is important to note that our ALK-negative group for comparisons does not represent an unselected NSCLC population; instead, it represents the negatives seen within the same increasingly preselected group. By taking all EGFR mutations into consideration, our ALK positivity rate in adenocarcinomas known to be EGFR and KRAS wild-type was 12 of 47 (25.5%). The one EGFR mutation noted among our ALK-positive patients was an exon 20 point mutation, which has not been associated with EGFR TKI sensitivity (18).Given that the primary clinical significance of EGFR mutations relates to their association with sensitivity to EGFR TKIs, if we chose to only consider EGFR mutations associated with EGFR TKI sensitivity, our ALK positivity rate became 13 of 50 (26%). Among adenocarcinomas in whom both EGFR and KRAS status was known, there was a trend for ALK-positive cases to be enriched among those known to lack both EGFR (sensitivity mutations) and KRAS mutations (26%; 13 of 50) compared with those with known mutations (0%, 0 of 5; P = 0.245, Fisher’s exact test). Whether this general lack of co-occurrence of EGFR or KRAS mutations with ALK gene rearrangements reflects biology (i.e., lack of a baseline selection pressure for a second oncogenic driver in the presence of an existing one), or simply the low chances of two rare events occurring together, is uncertain. With regard to the first point, although not a TKI sensitivity mutation, the coexisting exon 20 mutation noted in our series may still be a driving mutation. Similarly, within a tissue microarray,coexistence of an ALK gene rearrangement and a classic activating EGFR exon 19 deletion (E746_A750) has also been described (19). How frequently potential molecular codrivers exist when larger data sets are available, how cells with more than one driver will react when treated with specific inhibitors de novo, and whether upregulation or selection of subclones with others drivers could provide a means for acquired resistance following specific inhibitor use, as has, for example, been documented with EGFR mutations and MET gene amplification, remains to be seen (20, 21). Of note, we found no association between ALK gene rearrangements and MET gene amplification, a known preclinical sensitivity factor for MET inhibition, suggesting that the clinical activity seen with crizotinib in ALK-positive cases is likely to reflect the anti-ALK activity of this dual-targeted drug (22). We did see an association with MET high polysomy (P < 0.0001, Fisher’s exact test), although, in the absence of knowing the copy number of other chromosomes, a specific selection for chromosome 7 cannot be distinguished from a more general trend toward increased genetic instability in these cells.
A general trend toward ALK positivity being more common among those with little or no smoking history has been reported (9). In our own series, <10-pack-year smoking status, based on precedents from the EGFR mutation literature, was a highly statistically significant cut point for enriching for ALK-positive patients (Fig. 3; ref. 23). Moreover, when only those known to be wild-type for both EGFR (TKI sensitivity mutations) and KRAS mutations were considered, the effect was even stronger [P = 0.0002, Fisher’s exact test; proportion positive among never/light smoking adenocarcinomas wild-type for both EGFR (TKI sensitivity mutations) and KRAS mutations = 13 of 29, 44.8%; proportion positive among those with >10-pack-year smoking history wild-type for both EGFR (TKI sensitivity mutations) and KRAS mutations = 0 of 21; Fig. 3]. We interpret these data as showing that smokers are more prone to getting other, non-ALK-driven, types of lung cancer, rather than smoking being somehow protective against ALK gene rearrangements.
Taken together, our data suggest that using aspects of histology, EGFR, KRAS, and smoking status to prescreen patients can enrich the population testing positive for an ALK gene rearrangement by ~10-fold (~45% positivity rate) compared with unselected series (9). Because EGFR and KRAS mutational testing is becoming routinely available, in the absence of universal ALK testing, this prescreening approach could significantly increase the chances of finding these rare patients, enabling treating physicians to determine who it would be most appropriate to refer for ALK screening and possible enrollment within an ALK inhibitor trial. However, although the proportions testing positive may increase with this approach, it does not guarantee that ALK-positive patients will not be missed should they exist at lower frequencies outside this group, and until very large data sets are analyzed, we would caution against being too proscriptive in terms of who not to test.
Both age and gender factors have also been reported in association with ALK positivity (9). In series where gender has been documented among adenocarcinomas with ALK gene rearrangements using RT-PCR, the ratios have been 6F:5M (24), 7F:4M (25), and, using the break-apart FISH probe, F8:11M (26). Whereas the first two series were conducted in Japan and China, the third was conducted in the United States. Shaw et al. (26) reported that ALK-positive patients were significantly more likely to be males compared with patients who were either EGFR mutant (P = 0.036) or EGFR wild-type (P = 0.039). However, in the larger data set, taking our own series together with the other U.S. series or all four series together, no dominant gender difference is apparent (United States: 17F:15M; United States + Japan + China: 30F:24M). In series where age has been documented, trends toward ALK-positive patients being younger than the general lung cancer population have been reported (9, 26). Here, our median age at diagnosis was similarly low at 53 years, but because the range is so broad (34-75 in our series), we do not believe that age per se should be used as a preselection criterion for ALK screening for fear of inappropriately excluding patients.
Lung cancer is now being divided into several well-defined molecular subtypes. In addition to helping to identify a significant majority of the patients potentially appropriate for treatment with an ALK inhibitor, in the absence of widespread screening of all lung cancer cases for ALK gene rearrangements, the categorical factors (adeno-carcinoma, EGFR/KRAS wild-type, and <10-pack-year smoking history) we have associated with ALK positivity may also prove a useful first step toward reexploring lung cancer epidemiology at the molecular level. Several nonsmoking-related risk factors, including radon gas exposure and various familial syndromes, have been described for lung cancer in general (27). The specific etiology of ALK-driven lung cancers remains unknown. By looking in different series of patients preselected by the characteristics we describe here to maximally enrich for patients testing positive, any apparent regional and/or national variation in the frequency of ALK positivity may generate hypotheses relating to high-risk genetic and/or environmental factors differing between the series to then be explored in more detail in the future.
Translational Relevance.
Rare ALK gene rearrangements detected by a specific fluorescence in situ hybridization assay are associated with marked sensitivity to a small-molecule inhibitor of ALK currently in clinical development in advanced non-small cell lung cancer. Reports of intratumoral heterogeneity of ALK rearrangements have raised issues about the true importance of ALK in carcinogenesis and the possibility of high false-positive/false-negative screening results. Here, we show that the apparent heterogeneity reflects technique rather than biology, determine the optimal number of fields to count to ensure maximal sensitivity and specificity, and confirm that when >4 fields are counted the >15% cell positivity cut point used in the current trials falls within the range accurately distinguishing positive from negative tumors and from nontumor regions. In addition, we show how a combination of clinical and other molecular factors identifies a population particularly enriched for ALK gene rearrangements that could significantly increase the ALK-positive screening hit rate in the future.
Acknowledgments
We thank Amanda Schubert and Courtney Scott (University of Colorado, Clinical Investigations Core) and Natalie Thomas (CMOCO) for clinical and molecular data tracking on all described patients; University of Colorado Cancer Center Cytogenetics Core for technical assistance with FISH assays.
Grant Support
Italian Association for Cancer Research (AF fellowship) and Colorado Lung Specialized Program of Research Excellence grant P50CA58187.
Footnotes
H. Mano, personal communication.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note: Current address for Q. Zhou: Division of Pulmonary Oncology, Cancer Center, Guangdong General Hospital and Guangdong Academy of Medical Sciences, Guangzhou 510080, China.
References
- 1.Palmer RH, Vernersson E, Grabbe C, Hallberg B. Anaplastic lymphoma kinase: signalling in development and disease. Biochem J. 2009;420:345–61. doi: 10.1042/BJ20090387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mano H. Non-solid oncogenes in solid tumors: EML4-ALK fusion genes in lung cancer. Cancer Sci. 2008;99:2349–55. doi: 10.1111/j.1349-7006.2008.00972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 4.Choi YL, Takeuchi K, Soda M, et al. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68:4971–6. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 5.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 6.Takeuchi K, Choi YL, Soda M, et al. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res. 2008;14:6618–24. doi: 10.1158/1078-0432.CCR-08-1018. [DOI] [PubMed] [Google Scholar]
- 7.Takeuchi K, Choi YL, Togashi Y, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15:3143–9. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 8.Horn L, Pao W. EML4-ALK: honing in on a new target in non-small-cell lung cancer. J Clin Oncol. 2009;27:4232–5. doi: 10.1200/JCO.2009.23.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solomon B, Varella-Garcia M, Camidge DR. ALK gene rearrangements: a new therapeutic target in a molecularly-defined subset of non-small cell lung cancer. J Thorac Oncol. 2009;4:1450–4. doi: 10.1097/JTO.0b013e3181c4dedb. [DOI] [PubMed] [Google Scholar]
- 10.Kwak EL, Camidge DR, Clark J, et al. Clinical activity observed in a phase I dose escalation trial of an oral c-met and ALK inhibitor, PF-02341066 [abstract 3509] J Clin Oncol. 2009;27:15s. [Google Scholar]
- 11.Perner S, Wagner PL, Demichelis F, et al. EML4-ALK fusion lung cancer: a rare acquired event. Neoplasia. 2008;10:298–302. doi: 10.1593/neo.07878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lucas D, Varella-Garcia M, Camidge R, et al. Triple-platform testing (IHC, FISH and mutation) to predict response to targeted therapy in NSCLC; The United States and Canadian Academy of Pathology Annual Meeting; Boston, MA. 2009; Poster 233. [Google Scholar]
- 13.Franklin WA, Haney J, Sugita M, Bemis L, Jimeno A, Messersmith WA. KRAS mutation: comparison of testing methods and tissue sampling techniques in colon cancer. J Mol Diagn. 2010;12:43–50. doi: 10.2353/jmoldx.2010.080131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cappuzzo F, Marchetti A, Skokan M, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol. 2009;27:1667–74. doi: 10.1200/JCO.2008.19.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodig SJ, Mino-Kenudson M, Dacic S, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res. 2009;15:5216–23. doi: 10.1158/1078-0432.CCR-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDermott U, Iafrate AJ, Gray NS, et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008;68:3389–95. doi: 10.1158/0008-5472.CAN-07-6186. [DOI] [PubMed] [Google Scholar]
- 17.Martelli MP, Sozzi G, Henandez L, et al. EML4-ALK rearrangement in non-small cell lung cancer and non tumor lung tissues. Am J Pathol. 2009;174:661–70. doi: 10.2353/ajpath.2009.080755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28(Suppl 1):S24–31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koivunen JP, Mermel C, Zejnullahu K, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–83. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turke AB, Zejnullahu K, Wu YL, et al. Preexistence and clonal selectionof MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelman JA, Jänne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–9. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 22.McDermott U, Sharma SV, Dowell L, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104:19936–41. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatinpaclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 24.Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol. 2009;22:508–15. doi: 10.1038/modpathol.2009.2. [DOI] [PubMed] [Google Scholar]
- 25.Wong DW, Leung EL, So KK, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from non-smokers with wild-type EGFR and KRAS. Cancer. 2009;115:1723–33. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- 26.Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–53. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]