SUMMARY
Mitosis is triggered by the activation of Cdk1-cyclin B1 and its translocation from the cytoplasm to the nucleus. Positive feedback loops regulate the activation of Cdk1-cyclin B1 and help make the process irreversible and all-or-none in character. Here we examine whether an analogous process, spatial positive feedback, regulates Cdk1-cyclin B1 redistribution. Using chemical biology approaches and live cell microscopy, we show that nuclear Cdk1-cyclin B1 promotes the translocation of Cdk1-cyclin B1 to the nucleus. Mechanistic studies suggest that cyclin B1 phosphorylation promotes nuclear translocation and, conversely, nuclear translocation promotes cyclin B1 phosphorylation, accounting for the feedback. Interfering with the abruptness of Cdk1-cyclin B1 translocation affects the timing and synchronicity of subsequent mitotic events, underscoring the functional importance of this feedback. We propose that spatial positive feedback ensures a rapid, complete, robust and irreversible transition from interphase to mitosis and suggest that bistable spatiotemporal switches may be widespread in biological regulation.
INTRODUCTION
Mitosis is one of the most dramatic events in cell biology. At the onset of mitosis a mammalian cell changes from flat to round, condenses its chromosomes, undergoes nuclear envelope breakdown (NEB), and reorganizes its microtubules into a spindle (Morgan, 2007). The transition from interphase to mitosis is temporally abrupt, all-or-none in character, and irreversible.
Mitosis is triggered by the activation and nuclear translocation of Cdk1-cyclin B1 (Heald et al., 1993; Jin et al., 1998; Li et al., 1997; Pines and Hunter, 1991). A priori, none of the individual biochemical processes that culminate in the accumulation of active Cdk1-cyclin B1 in the nucleus would be expected to be all-or-none in character. A HeLa cell, for example, possesses approximately 1,600,000 Cdk1-cyclin B1 complexes (Sun et al., 2010). With such a large number of Cdk1-cyclin B1-complexes, a cell could, in principle, be imbued with an almost continuously graded range of nuclear Cdk1-cyclin B1 activities. Yet mitotic entry is a switch-like, all-or-none process, and interphase and mitosis are qualitatively distinct cellular states.
Likewise, none of the processes that lead to the accumulation of active Cdk1-cyclin B1 in the nucleus are intrinsically irreversible. The phosphorylation of Cdk1 by CAK is opposed by PP2C (Cheng et al., 1999), the Cdk1 activator Cdc25 is opposed by Wee1 (Russell and Nurse, 1986, 1987), and nuclear import of the Cdk1-cyclin B1 complexes is opposed by nuclear export (Hagting et al., 1999; Hagting et al., 1998; Toyoshima-Morimoto et al., 2001; Yang et al., 2001). Nevertheless, mitotic entry is normally irreversible. These observations raise the question of how these graded, reversible signaling proteins collectively produce a switch-like, irreversible response.
Part of the answer lies in the regulation of the kinase activity of Cdk1-cyclin B1 by phosphorylation of kinase subunit in its N-terminal lobe. The phosphorylation reaction, which inactivates the cyclin-dependent kinase, is carried out by the Wee1 and Myt1 kinases (Mueller et al., 1995a; Mueller et al., 1995b), and the dephosphorylation reaction, which activates Cdk1, is carried out by Cdc25 (Atherton-Fessler et al., 1994; Hoffmann et al., 1993; Kumagai and Dunphy, 1992; Solomon et al., 1990; Tang et al., 1993). Cdk1, Cdc25 and Wee are organized in positive and double negative feedback loops. Active Cdk1-cyclin B1 phosphorylates and activates Cdc25, which dephosphorylates and activates Cdk1-cyclin B1; active Cdk1-cyclin B1 phosphorylates and inactivates Wee1 and Myt1, which in turn can phosphorylate Cdk1-cyclin B1. Studies in Xenopus extracts have shown that these feedback loops allow the Cdk1-cyclin B1 system to collectively function as a bistable trigger that flips between alternative stable steady-states (Pomerening et al., 2003; Sha et al., 2003), helping to make the transition from interphase to mitosis switch-like and irreversible in character. Recent work has begun to explore the quantitative behavior of the trigger in mammalian cell lines (Deibler and Kirschner, 2010; Gavet and Pines; Lindqvist et al., 2007; Pomerening et al., 2008), and overall the mechanisms of Cdk1 activation and inactivation appear to be well-conserved evolutionarily.
But the biological activity of Cdk1-cyclin B1 is regulated not only at the level of its biochemical activity, but also at the level of its localization. Cdk1-cyclin B1 must translocate to the nucleus to bring about nuclear envelope breakdown. Work from several labs has implicated the multisite phosphorylation of cyclin B1 (that is, phosphorylation of the cyclin subunit rather than the Cdk subunit) in the regulation of Cdk1-cyclin B1 translocation (Hagting et al., 1999; Hagting et al., 1998; Li et al., 1995, 1997; Toyoshima-Morimoto et al., 2001; Walsh et al., 2003; Yang et al., 2001). In human cyclin B1 there are five mitotic phosphorylation sites: Ser 116, Ser 126, Ser 128, Ser 133, and Ser 147, four of which are conserved in vertebrates from humans through amphibians. Phosphorylation of these sites promotes the nuclear import of Cdk1-cyclin B1 and inhibits its nuclear export (Hagting et al., 1998; Li et al., 1995, 1997; Toyoshima-Morimoto et al., 2001; Walsh et al., 2003; Yang et al., 2001).
Most theoretical models of cell cycle regulation assume that the system is spatially homogeneous (Ferrell et al., 2011). However, this assumption is patently untrue for the mitotic trigger, where the enzymes that control Cdk1-cyclin B1 activation and Cdk1-cyclin B1 localization are localized to specific compartments. A satisfactory model of the trigger requires therefore some consideration of the inhomogeneous localization of the proteins involved. Moreover, the combination of spatial inhomogeneity and regulated protein localization has the potential to give rise to complex systems-level behaviors, including feedback and bistability, just as regulated protein activity does (Ferrell, 1998). This led us to hypothesize that spatial positive feedback in the system that regulates Cdk1-cyclin B1 localization contributes to the all-or-none character of mitosis, to the temporal abruptness of mitotic entry, and to the irreversibility of this cell cycle transition.
Here we experimentally test whether spatial positive feedback is present in the mitotic trigger, using strategies to dynamically control the spatial distribution of Cdk1-cyclin B1. We find that the translocation of Cdk1-cyclin B1 from the cytoplasm to the nucleus promotes the nuclear translocation of additional Cdk1-cyclin B1 complexes, and triggers mitosis. In addition, compromising the abruptness of mitotic entry affects the successful completion of mitosis. Spatial positive feedback depends upon cyclin B1 multisite phosphorylation, and this phosphorylation allows Cdk1-cyclin B1 to bind stably to mitotic chromosomes. Moreover, we show that cyclin B1 phosphorylation is more favorable in the nucleus than in the cytoplasm, so that phosphorylation promotes nuclear localization, which further promotes phosphorylation. Finally, we formulate a simple theoretical model of Cdk1-cyclin B1 translocation, which accounts for the observed spatial positive feedback.
RESULTS
Quantifying the Redistribution of Cyclin B1
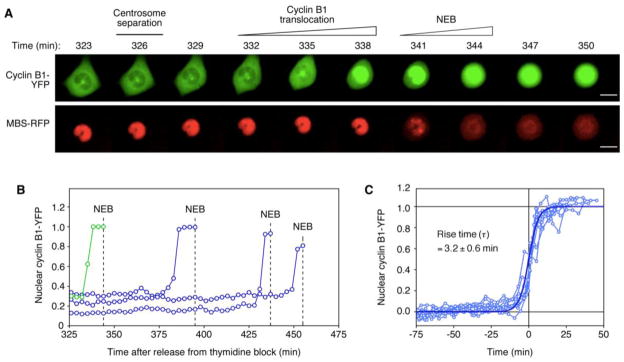
To quantitatively assess the kinetics of cyclin B1 redistribution, we transfected HeLa cells with two fluorescent probes, cyclin B1-YFP and a mitotic biosensor (MBS-RFP) (Jones et al., 2004), and then followed individual synchronized cells into mitosis by real-time fluorescence imaging. In agreement with previous reports (Hagting et al., 1999), cyclin B1-YFP was found to be mostly cytoplasmic during interphase, accumulated on centrosomes late in interphase, and then abruptly moved to the nucleus shortly before NEB (Figure 1A,B and Movie S1). To quantify how rapidly cyclin B1 redistributed, we fitted the time course data to the logistic equation (Figure 1C), used here as a generic sigmoidal curve. The fitted rise time was found to be 3.2 ± 0.6 min (mean ± S.D.). In agreement with previous work (Gavet and Pines, 2010a; Hagting et al., 1999), the redistribution of cyclin B1 occurred prior to the general breakdown in nuclear permeability (Figure S1 and Movie S2).
Figure 1. Abrupt Cyclin B1 Nuclear Translocation Prior to Nuclear Envelope Breakdown.
(A) A representative HeLa cell expressing cyclin B1-YFP (top) and the mitotic biosensor (MBS-RFP) (bottom) after release from G1/S block. Centrosome separation (top), cyclin B1 translocation (top), and NEB (bottom) occur in quick succession. Scale bars: 10 μm.
(B) Quantitation of nuclear accumulation of cyclin B1-YFP after release from thymidine block of the cell shown in (A) (green) and three other representative cells (blue). For each cell, the last data point shown represents the time of NEB.
(C) Translocation kinetics are well-approximated by the logistic equation. Cyclin B1-YFP translocation was measured for more than 100 cells. Individual time courses were b fitted to the logistic equation and were scaled to their fitted maximum and minimum values (b and a respectively) and half-maximal times (t0). Ten scaled time courses are shown (light blue). Rise times (τ), were calculated from the curve fits for all >100 cells and are expressed as means ± S.D. The fitted logistic equation curve with τ = 3.2 min is shown in dark blue. Note that with the logistic equation, y goes from 27% to 73% of its maximal value over a time interval of 2τ. See also Figure S1 and Movies S1 and S2.
Cdk1AF and NLS-Cyclin B1-CFP Promote the Translocation of Cyclin B1-YFP
As a first test of the spatial positive feedback hypothesis, we co-transfected Tet-on HeLa cells with NLS-tagged cyclin B1-CFP plus cyclin B1-YFP (as a reporter of “normal” cyclin B1 localization). In addition, we either transfected wild-type Cdk1-mCherry or a Tet-inducible Cdk1AF-mCherry, where Cdk1AF is a mutant of Cdk1 that cannot be inactivated by Wee1 or Myt1 (Figure S2). We released the cells from a G1/S block, induced Cdk1AF expression with doxycycline, and then followed the cells’ progression into mitosis by time-lapse microscopy. The combination of Cdk1AF and NLS-cyclin B1 has been shown to cause premature activation of H1 kinase activity and premature entry into mitosis (Jin et al., 1998); here we asked whether Cdk1AF plus NLS-cyclin B1 would also cause premature redistribution of cyclin B1-YFP from the cytoplasm to the nucleus just prior to NEB.
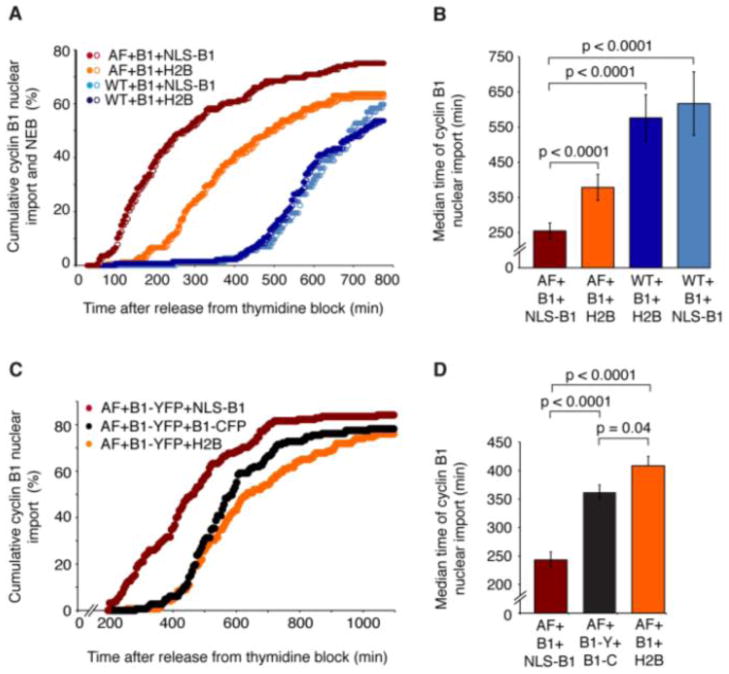
Consistent with previous studies (Jin et al., 1998), cells expressing Cdk1WT and cyclin B1-YFP, plus either histone H2B or NLS-cyclin B1-CFP underwent NEB at approximately 690–720 min post-release (Figure 2A), as is typical for control cells (Gong et al., 2007). The cyclin B1-YFP translocated to the nucleus approximately 5 min before NEB (Figure 2A). Cells transfected with Cdk1AF, cyclin B1-YFP, and H2B underwent NEB more than 3 hours earlier than did cells transfected with Cdk1WT, cyclin B1-YFP, and H2B (Figure 2A, B, orange vs. dark blue). Again the cyclin B1-YFP redistributed to the nucleus prior to NEB (Figure 2A).
Figure 2. In Cdk1AF-Expressing Cells, NLS-Cyclin B1-CFP Expression Promotes Cyclin B1-YFP Translocation.
(A) Cumulative percentage of cells that showed cyclin B1 nuclear translocation (closed circles) and NEB (open circles) after release from G1/S block. Cells were expressing Cdk1AF (AF) or Cdk1-WT (WT), plus cyclin B1-YFP (B1) and either NLS-cyclin B1-CFP (NLS-B1) or histone H2B-CFP (H2B). At least 100 cells were analyzed for each experimental condition. One of three independent experiments yielding similar results.
(B) Median times of nuclear import for the experimental conditions described in (A). In each condition at least 100 cells were analyzed. Error bars represent the median absolute deviations calculated for the first 50% of the cells to accumulate cyclin B1 in the nucleus. p values were calculated by Mann-Whitney and Kolmogorov-Smirnov tests.
(C) Cumulative percentage of cells that showed cyclin B1 nuclear translocation for cells transfected with Cdk1AF (AF) and cyclin B1-YFP (B1-YFP) plus either NLS-cyclin B1-CFP (NLS-B1), cyclin B1-CFP (B1-CFP), or histone H2B-CFP (H2B).
(D) Median times of nuclear import for the experimental conditions described in (C). Error bars and p values were calculated as described in (B). See also Figure S2.
Notably, the combination of Cdk1AF and NLS-cyclin B1 advanced the timing of both NEB and cyclin B1-YFP translocation by an additional ~2 hours (Figure 2A, B, dark red vs. orange). This finding indicates that nuclear Cdk1AF-cyclin B1 is more effective than cytoplasmic Cdk1AF-cyclin B at promoting both NEB and the pre-NEB redistribution of the reporter cyclin B1-YFP to the nucleus.
To see whether the presence of a double-dose of cyclin, rather than the fact that one of the cyclins was nuclear, might account for the acceleration of NEB and cyclin translocation, we compared the nuclear accumulation of cyclin B1-YFP in cells expressing Cdk1AF and cyclin B1-YFP plus either NLS-cyclin B1-CFP, cyclin B1-CFP, or H2B (Figure 2C, D). Cyclin B1-YFP translocation occurred prematurely when the cells expressed cyclin B1-YFP plus NLS-cyclin B1-CFP, but not when they expressed cyclin B1-YFP plus cyclin B1-CFP (Figure 2C, D). Again this suggests that nuclear Cdk1AF-cyclin B1 is more effective than cytoplasmic Cdk1AF-cyclin B1 at promoting the redistribution of the reporter cyclin B1-YFP to the nucleus, consistent with the hypothesis that cyclin B1 nuclear translocation promotes further cyclin B1 translocation.
iRap-Controlled Translocation of Cyclin B1 to the Nucleus
If the spatial positive feedback hypothesis is correct, then acutely inducing one cyclin B1 protein to translocate to the nucleus should promote the translocation of a second distinguishable cyclin B1 protein. To control the translocation of the first cyclin B1, we made use of a small molecule-induced dimerization strategy (Crabtree and Schreiber, 1996; Komatsu et al.). The heterodimerizing protein domains are the FK506 binding protein-12 (FKBP) domain and the mTOR-derived FKBP-rapamycin binding (FRB) domain (Figure 3A and Figure S3), and the dimerization-inducing small molecule is the indole-derivative of rapamycin iRap.
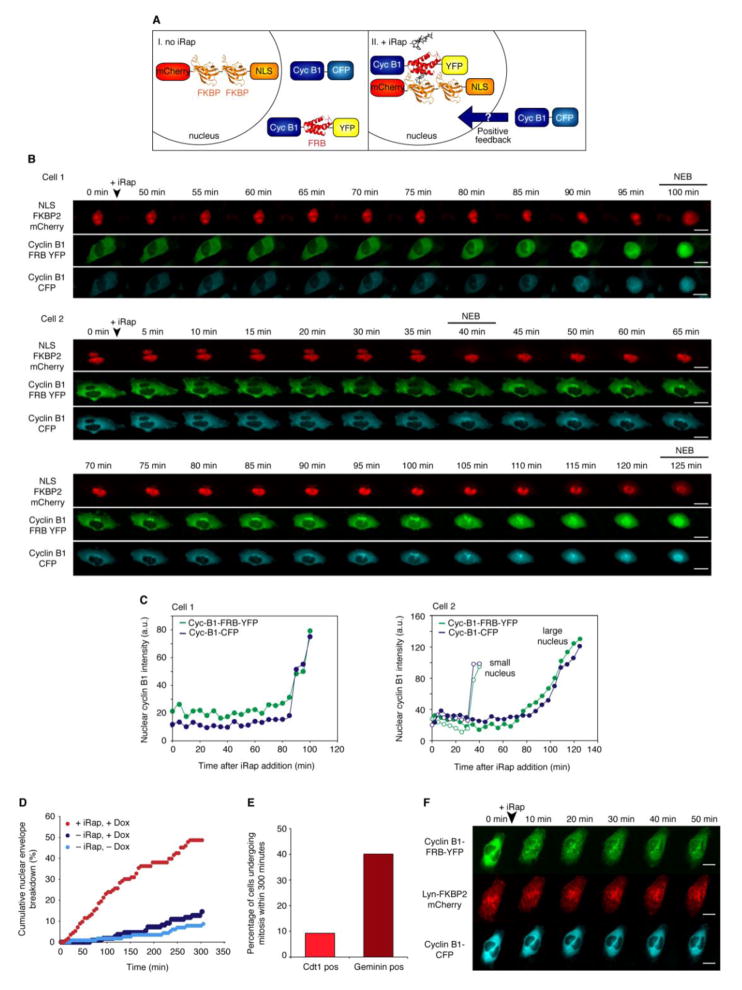
Figure 3. Induced Translocation of Active Cdk1-Cyclin B1 Complexes to the Nucleus Triggers Mitosis and Induces Spatial Positive Feedback.
(A) Schematic of the experimental approach by which the rapamycin-analog iRap causes cyclin B1-FRB to accumulate in the nucleus (see also Figure S3).
(B) Two representative cells expressing mCherry-NLS3-FKBP2 (top), cyclin B1-FRB-YFP (middle), cyclin B1-CFP (bottom) and treated with 5 μM iRap in the presence of 1 μg/ml doxycycline to induce Cdk1AF expression. Scale bars: 10 μm. See also Movies S3, S4.
(C) Quantitation of the accumulation of cyclin B1-FRB-YFP (green) and cyclin B1-CFP (blue) for the cells shown in (B). For the binucleate cells both the import into the smaller nucleus (open points) and the larger nucleus (solid points) are shown.
(D) Cumulative percentage of cells that underwent NEB. Cells were treated with 5 μM iRap and/or 1 μg/ml doxycycline as indicated. More then 100 cells were analyzed for each experimental condition.
(E) Percentage of Cdt1-cerulean positive cells (G1/early S ) and Geminin-cerulean (Late S/G2) positive cells that underwent mitosis within 300 minutes of iRap treatment.
(F) Representative images of one cell ectopically expressing cyclin B1-FRB-YFP (top), cyclin B1-CFP (bottom) and Lyn-FKBP2-mCherry (middle) treated with 5 μM iRap. Scale bars: 10 μm. See also Figure S3 and Movie S5.
To test the induced dimerization system, we transfected cells with a constitutively nuclear NLS-FKBP2-mCherry construct plus a cytoplasmic FRB-YFP construct, and treated the transfected cells with iRap. As shown in Figure S3B, iRap induced the redistribution of FRB-YFP from the cytoplasm to the nucleus on a time scale of ~100 min.
We then used the same strategy to induce the translocation of cyclin B1-FRB-YFP from the cytoplasm to the nucleus, and assessed the consequences in unsynchronized Tet-On cells expressing inducible Cdk1AF. As shown in Figure 3B,C, the FRB-tagged cyclin B1-YFP protein was mostly cytoplasmic prior to iRap treatment, and iRap caused it to move to the nucleus (Figure 3B,C and Movies S3 and S4). Strikingly, a reporter cyclin B1-CFP, which lacks the FRB tag and should therefore be insensitive to iRap, translocated to the nucleus concomitantly with the cyclin B1-FRB-YFP. Two typical cells are shown in Figure 3B,C. In the first cell, the FRB-tagged cyclin B1-YFP began to move to the nucleus about 90 min after iRap treatment, and the untagged cyclin B1-CFP followed within a few minutes (Figure 3B,C). In the second cell (a binucleate cell), the two cyclin proteins first accumulated in the smaller nucleus followed by breakdown of its nuclear envelope. The cyclins then accumulated in the larger nucleus within a few minutes of each other, followed again by NEB (Figure 3B,C). Similar results were seen in other cells (Figure S3C). No redistribution was seen when NLS-FKBP2-mCherry/cyclin B1-YFP-expressing cells were treated with iRap (not shown). Thus, inducing nuclear translocation of YFP-tagged cyclin B1-FRB molecules promoted the redistribution of cyclin B1-CFP molecules to the nucleus.
Strikingly, the redistribution of the cyclin B1 proteins was accompanied by entry of the cells into mitosis. During a five hour time interval, about 50% of the iRap-treated cells entered mitosis, whereas less than 10% of the cells that were not treated with iRap did (Figure 3D). These findings indicate that, in the presence of Cdk1AF, the movement of cyclin B1 to the nucleus can be sufficient to trigger mitosis. To determine what phases of the cell cycle contributed to these iRap-induced mitoses, we transfected cells with the Fucci sensors Cdt1-cerulean and Geminin-cerulean (Sakaue-Sawano et al., 2008). As shown in Figures 3E and S3, the S/G2-phase (Geminin-positive) cells were more likely to enter mitosis in response to iRap than were the G1/early S-phase (Cdt1-positive) cells. However, a few Cdt1-positive cells did enter mitosis (Figure 3E and S3D), suggesting that the time when cells acquire the ability to enter mitosis in response to cyclin B1 translocation is late G1 phase or early S phase.
Next we tested whether some unanticipated physical interaction between cyclin B1-CFP and cyclin B1-FRB-YFP might account for the iRap-induced translocation of cyclin B1-CFP to the nucleus. We targeted the FKBP domain to the plasma membrane and internal membranes rather than the nucleus, using a Lyn-FKBP2-mCherry chimera. iRap caused the rapid translocation of cyclin B1-FRB-YFP to the Lyn-FKBP2-mCherry foci, but had no apparent effect on the distribution of the reporter cyclin B1-CFP protein (Figure 3F and Movie S5). This control experiment argues against the possibility that cyclin B1-FRB-YFP can “drag” cyclin B1-CFP to wherever it is localized.
Compromising the Abruptness of Cdk1-Cyclin B1 Activation and Import Affects Progression through Mitosis
Like mitotic onset, the traversal of Start and the initiation of anaphase in S. cerevisiae are thought to be initiated by bistable triggers (Eser et al., 2011; Holt et al., 2008; Skotheim et al., 2008), and in both of these cases, manipulations that compromise the temporal abruptness of the trigger adversely affect subsequent cell cycle progression (Holt et al., 2008; Skotheim et al., 2008). We therefore asked whether we could alter the abruptness of Cdk1-cyclin B1 translocation and, if so, whether this affected mitotic progression.
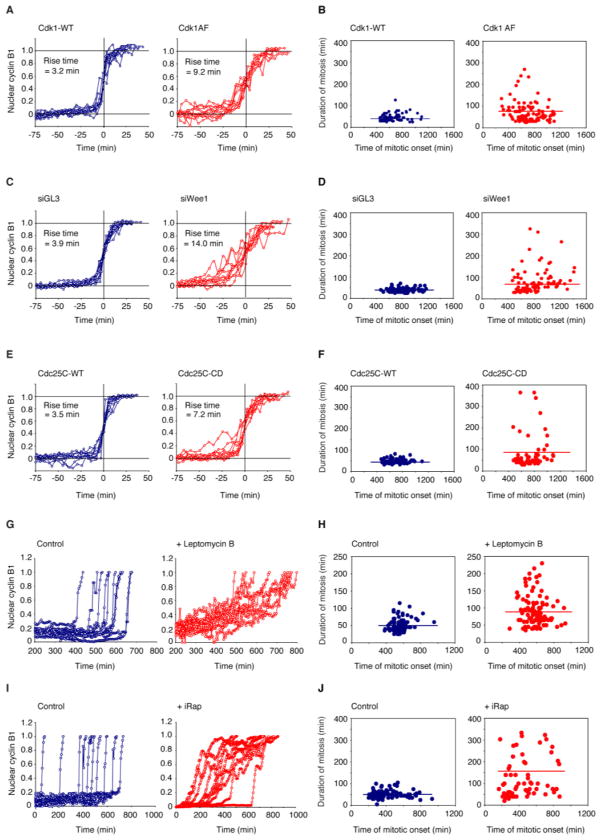
Since Cdk1-cyclin B1 translocation depends upon Cdk1-cyclin B1 activation (Gavet and Pines, 2010a), it seemed plausible that compromising the positive feedback loops that regulate Cdk1 activity might make cyclin B1-YFP translocation less switch-like. To test this idea, we transfected cells with tetracycline-inducible Cdk1AF, a phosphorylation site mutant that cannot be inhibited by Wee1 or Myt1 and thus short-circuits the positive feedback loops that regulate Cdk1 activity (Pomerening et al., 2008), and induced Cdk1AF expression while releasing the cells from a G1/S double-thymidine block (Figures S2 and S4C,D). As previously noted (Jin et al., 1998), Cdk1AF expression had little effect on the timing of NEB (Figure 4B). However, it did cause the rise time for cyclin B1-YFP translocation to increase from 3.2 to 9.2 min (Figure 4A). It also caused cells to take longer to complete mitosis, presumably because it took longer for the cells to satisfy the spindle assembly checkpoint and/or activate APC-Cdc20, and caused the duration of mitosis to become more variable (Figure 4B). There was no correlation between how long it took for cells to enter mitosis (a measure of the durations of S-phase plus G2-phase) and how long it took for cells to complete mitosis (Figure 4B), emphasizing the independence of these events.
Figure 4. Timely Completion of Mitosis Requires Abrupt Activation and Redistribution of Cdk1-Cyclin B1.
(A, B) Cdk1AF expression makes cyclin B1 translocation more graded and makes the duration of mitosis longer and more variable. As a proxy for the duration mitosis we measured the time of completion of cyclin degradation minus the time of cyclin B1 translocation. Rise times were calculated by fitting traces of ~100 cells to the logistic equation as described in Figure 1. Ten individual traces are shown here for each condition. The individual shown for the Cdk1-WT-transfected cells are the same as those shown in Figure 1C.
(C, D) Wee1 knockdown makes cyclin B1 translocation more graded and makes the duration of mitosis longer and more variable. Ten individual traces are shown here for each condition. See also Figure S4.
(E, F) Cdc25-CD expression makes cyclin B1 translocation more graded and makes the duration of mitosis longer and more variable. Ten individual traces are shown here for each condition. See also Figure S4.
(G, H) Leptomycin B treatment makes cyclin B1 translocation more graded and makes the duration of mitosis longer and more variable. Ten individual traces are shown here for each condition. Timing of events was monitored for approximately 100 cells for each condition. See also Figure S4 and Movie S6.
(I, J) iRap-mediated induction of cyclin B1 translocation in the absence of Cdk1AF makes cyclin B1 translocation more graded and makes the duration of mitosis longer and more variable. Ten individual traces are shown here for each condition.
In each panel the timing of events was monitored for approximately 100 cells for each condition.
We also compromised positive feedback in the Cdk1-cyclin B1 activation circuit by knocking down Wee1 with siRNAs (Figure 4C,D and S4A) and by expressing a putative dominant negative form of Cdc25C (the catalytically-inactive C377S mutant) (Figure 4E,F). In both cases, the rise time for cyclin B1-YFP translocation increased (Figure 4C,E and S4C,D) and mitosis became longer and more variable (Figure 4D,F and S4).
To change the abruptness of cyclin B1 nuclear import without directly affecting the process of Cdk1 activation, we treated unsynchronized cells with the nuclear export inhibitor leptomycin B. Leptomycin B caused a gradual accumulation of nuclear cyclin B1, sometimes followed by a more rapid late phase of accumulation (Figure 4G and S4B and Movie S6). Mitosis in the leptomycin B-treated cells was slower and more variable than in control cells (Figure 4H and S4C,D).
Finally, we made use of the iRap-medicated cyclin B1 nuclear accumulation strategy to induce a gradual cyclin B1 nuclear import. In these experiments, unsynchronized cells were transfected with cyclin B1-FRB-YFP, NLS-FKBP-mCherry and cyclin B1-CFP but no Cdk1AF. Upon treatment of cells with iRap, cyclin B1 accumulated gradually in the nucleus (Figure 4I). Again, mitosis was slower and more variable then in control cells (Figure 4J). Moreover, 19% of the iRap-treated cells underwent apoptosis after a prolonged mitosis. These data support the hypothesis that abrupt Cdk1-cyclin B1 import is important for timely completion of mitosis and for keeping mitotic events properly synchronized.
Cyclin B1 Phosphorylation is Required for Spatial Positive Feedback
The multisite phosphorylation of cyclin B1 occurs at the time of mitosis, depends upon the activity of Cdk1 (Figure S6 and S7), and has the potential to regulate the nuclear import and export of Cdk1-cyclin B1. This argues that they are likely to cause the pre-NEB shift of Cdk1-cyclin B1 from the cytoplasm to the nucleus, and in support of this hypothesis, it has been reported that a human cyclin B1 protein with all five mitotic phosphorylation sites mutated to alanines (cyclin B1-5SA) only enters the nucleus once NEB has begun (Hagting et al., 1999). However, a more recent study found that cyclin B1-WT and cyclin B1-5SA enter the nucleus simultaneously (Gavet and Pines, 2010a), indicating that something other than cyclin B1 phosphorylation determines the timing of Cdk1-cyclin B1 redistribution at the onset of mitosis. Since this issue is critical for the question of the mechanism underpinning the spatial positive feedback observed here, we revisited the question of whether or not cyclin B1-WT and cyclin B1-5SA enter the nucleus simultaneously.
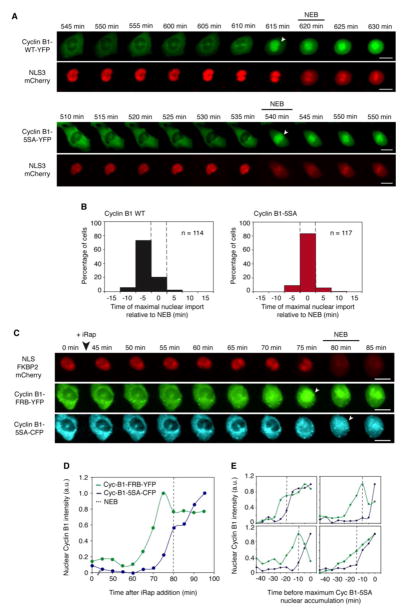
HeLa cells were transfected with cyclin B1-WT-YFP, cyclin B1-5SA-YFP, or cyclin B1-5SE-YFP, plus a nuclear marker (NLS3-mCherry), and were then released from a double thymidine block. We then measured the time of NEB (with the nuclear marker) and the time of maximal cyclin B1 translocation. Representative cells are shown in Figure 5A; binned timings from more than 100 cells are shown in Figure 5B. Cyclin B1-WT-YFP was generally found to translocate ~5 min prior to NEB (Figure 5B), whereas cyclin B1-5SA-YFP translocated concomitantly with NEB. This result is consistent with the hypothesis that cyclin B1 phosphorylation is required for pre-NEB translocation (Hagting et al., 1999).
Figure 5. Cyclin B1 Phosphorylation is Required for the Pre-NEB Translocation of Cyclin B1 to the Nucleus and for Spatial Positive Feedback.
(A, B) Cyclin B1-WT-YFP generally translocates to the nucleus ~5 min prior to NEB. In contrast, cyclin B1-5SA translocates to the nucleus concomitantly with NEB. Panel A shows representative cells. The arrows show when cyclin B1 translocation was maximal. Scale bar represents 10 μm. Panel B shows binned translocation timings for more than 100 cells.
(C–E) Cyclin phosphorylation is required for spatial positive feedback. Cyclin B1-FRB-YFP was induced to translocate to the nucleus by iRap treatment. Panel C shows one representative cell, where cyclin B1-5SA-CFP translocation lags substantially behind that of cyclin B1-FRB-YFP. The arrows show when cyclin B1 translocation was maximal. Scale bar represents 10 μm. Panel D quantifies the translocation shown in panel C. Panel E shows four other cells. See also Figure S5 and Movie S7.
We next asked whether cyclin B1 phosphorylation was required for spatial positive feedback. We repeated the iRap experiment shown in Figure 3, transfecting cells with NLS-FKBP2-mCherry, Cdk1AF, and cyclin B1-FRB-YFP, plus cyclin B1-5SA-CFP, and asked whether iRap would cause the cyclin B1-5SA-CFP to translocate to the nucleus prior to NEB. As shown in Figure 5C–E (see also Movie S7), the cyclin B1-5SA-CFP lagged significantly behind the cyclin B1-FRB-YFP, and moved to the nucleus at the time of NEB rather than prior to NEB. Cdc25C, another mitotic regulator reported to translocate to the nucleus at the onset of mitosis (Dalal et al., 1999; Graves et al., 2000; Kumagai and Dunphy, 1999; Yang et al., 1999), did not accompany cyclin B1-FRB-YFP into the nucleus in response to iRap treatment (Figure S5). These findings indicate implicate cyclin B1 phosphorylation in the observed spatial positive feedback.
Phosphorylation-Dependent Binding of Cyclin B1 to Mitotic Chromosomes
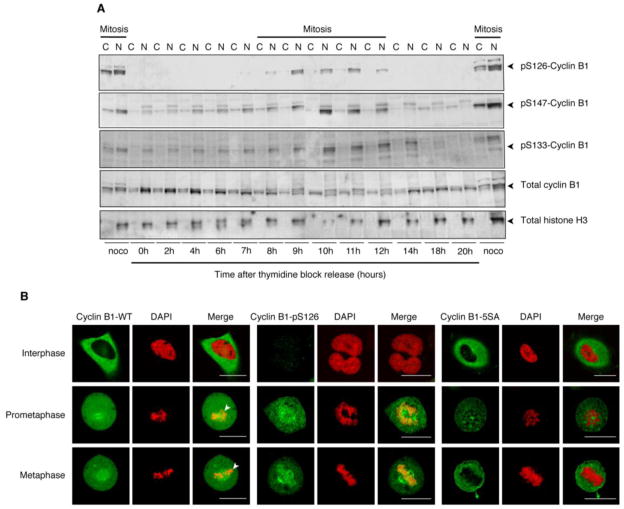
In fractionation studies, almost all of the phosphorylated cyclin B1 is found in the (nuclear) low speed pellet, even when most of the cells are mitotic and intact nuclei are not present (Figure 6A and S6). This suggests that cyclin B1 phosphorylation not only regulates the transport of Cdk1-cyclin B1 through the nuclear pore, but also promotes the stable association of Cdk1-cyclin B1 with easily-pelleted structures like the mitotic chromosomes. To test this idea further, we fixed and stained cells and examined the localization of cyclin B1 by confocal fluorescence microscopy. In interphase cells, cyclin B1 was present in the cytoplasm and excluded from the nucleus (Figure 6B). In prometaphase and metaphase cells it was found throughout the cell and concentrated around the mitotic spindle (Figure 6B). Importantly, phosphospecific cyclin B1 antibodies to Ser 126, the main autophosphorylation site (Hagting, A 1999 Current Bio, Borgne, A. 1999 JBC and Jackman, M 2003) (see Figure S6 and S7) stained only mitotic cells, with the spindle and the mitotic chromosomes staining most heavily (Figure 6B). In contrast, transfected cyclin B1-5SA localized everywhere except on the chromosomes during mitosis (Figure 6B). Taken together, these findings indicate that phosphorylated cyclin B1 stably associates with mitotic chromosomes, and that Cdk1 activity is required for this association.
Figure 6. Cyclin B1 Phosphorylation Is Required for the Stable Association of Cyclin B1 with Mitotic Chromosomes.
(A) Cell fractionation. Cells were released from a double thymidine block and lysed at various times. Mitotic cells were obtained by releasing double thymidine blocked cells into nocodazole for 12 hours. Lysates were separated into a low speed pellet, N, and a low speed supernatant, C, and were blotted for total cyclin B1, phosphorylated cyclin B1, and histone H3 (a nuclear marker).
(B) Fluorescence confocal microscopy. Cells were synchronized by double thymidine block and release. Non-transfected cells were stained with cyclin B1 or pS126-cyclin B1 antibodies (green) plus DAPI (red). Cyclin B1-5SA-YFP (green) transfected cells were stained with DAPI (red). Scale bar represents 10 μm. See also Figure S6.
Nuclear Localization of Cyclin B1 Promotes Cyclin B1 Phosphorylation
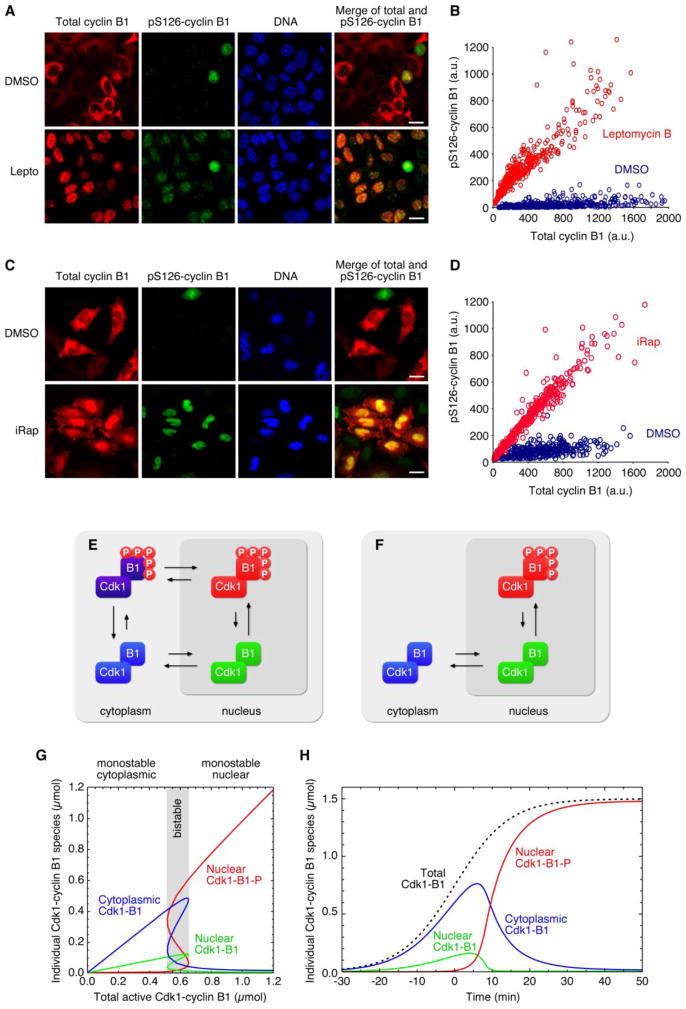
Given that cyclin B1 phosphorylation promotes the translocation of Cdk1-cyclin B1 to the nucleus, if it were also true that the nuclear translocation of Cdk1-cyclin B1 promoted cyclin B1 phosphorylation, the result would be a positive feedback loop. To test this idea, we arrested (non-transfected) HeLa cells by double thymidine block and released them in the presence or absence of leptomycin B. We fixed the cells at a time (8 h post-release) when most of the cells were expected to be in late S phase or G2 phase, and examined the levels and localization of total cyclin B1 and pS126-cyclin B1 by immunofluorescence. Most of the control DMSO-treated cells were in interphase. Their cyclin B1 was cytoplasmic and the cells showed little pS126-staining (Figure 7A, top). The leptomycin B-treated cells were also mostly in interphase, but their cyclin B1 had accumulated in the nucleus and had acquired pS126 staining (Figure 7A, bottom).
Figure 7. Translocation Promotes Cyclin B1 Phosphorylation, Completing the Positive Feedback Loop.
(A) Control (DMSO-treated) HeLa cells (top) and leptomycin B-treated HeLa cells (bottom) 8 h post release from a double thymidine block. Cells were stained for total cyclin B1 (red), pS126-cyclin B1 (green), and DNA (blue).
(B) Quantitation of the cyclin B1 and pS126-cyclin B1 staining in interphase cells treated with DMSO (blue) or leptomycin B (red).
(C) Control (DMSO-treated) HeLa cells (top) iRap-treated HeLa cells (bottom) after 5 hours. Cells were expressing stained for total cyclin B1 (red), pS126-cyclin B1 (green), and DNA (blue).
(D) Quantitation of the cyclin B1 and pS126-cyclin B1 staining in interphase cells treated with DMSO (blue) or iRap (red).
(E) Schematic model of Cdk1-cyclin B1 translocation and autophosphorylation.
(F) A simplified, three-species model of Cdk1-cyclin B1 translocation and autophosphorylation.
(G) Steady-state responses of the model. Parameters were: kimp = 0.25; kexp = 1; kphos = 100; kdephos = 0.2; and, for the Hill function, K = 1 and n = 4. The steady-state responses were solved numerically using Mathematica 7.0 (Wolfram). (H) Dynamical response of the model. The ODEs were solved numerically using Mathematica 7.0. The dashed line is , with τ = 7.2 min. See also Figure S7.
Figure 7B shows the cyclin B1 staining and pS126 for approximately 300 interphase cells. For the DMSO-treated cells, there was a wide range of levels of total cyclin B1, and in all of these cells the cytoplasmic cyclin B1 staining exceeded the nuclear cyclin B1 staining and the pS126 cyclin B1 staining was low. In contrast, the nuclear cyclin B1 staining of the leptomycin B-treated cells exceeded the cytoplasmic cyclin B1 staining, and the pS126 cyclin B1 staining was high. The slope of the pS126-cyclin B1 vs. cyclin B1 plots can be taken as a measure of how favorable cyclin phosphorylation is, and from the data in Figure 7B, pS126 phosphorylation appears to be ~19x more favorable in the nucleus than in the cytoplasm. These findings indicate that moving cyclin B1 from cytoplasm to the nucleus promotes its phosphorylation, completing a positive feedback loop where cyclin B1 phosphorylation promotes nuclear localization, which then promotes further cyclin B1 phosphorylation.
Note that while many of the leptomycin B-treated cells did eventually enter mitosis, most of the pS126-positive leptomycin B-treated cells were in interphase (Figure 7A). This suggests that the amount of Cdk kinase activity required to phosphorylate cyclin B1 at S126 is low relative to the amount required to bring about nuclear envelope breakdown. Alternatively, kinases in addition to Cdk1 may be required for the latter but not the former.
We carried out similar experiments in cells transfected with nuclear NLS-FKBP2-mCherry plus a FRB-cyclin B1-YFP and inducible Cdk1AF. In response to iRap, the FRB-cyclin B1-YFP moved to the nucleus, and this movement was invariably accompanied by an acquisition of nuclear pS126 reactivity (Figure 7C). As was the case with leptomycin B-treated cells, many of the iRap-treated cells eventually entered mitosis, but the majority of the pS126-positive iRap-treated cells were in interphase. The slope of the pS126-cyclin B1 vs. cyclin B1 plot was steeper for the iRap-treated cells than for the control DMSO-treated cells (Figure 7D). Again these findings indicate that cyclin B1 translocation promotes S126 phosphorylation.
An ODE Model for Spatial Positive Feedback in the Mitotic Trigger
We next set out to determine whether a positive feedback loop like that described above could plausibly be expected to generate switch-like translocation of active Cdk1-cyclin B1 from the cytoplasm to the nucleus. To this end we formulated a compartmentalized ordinary differential equation (ODE) model of the process. We chose a differential equation model rather than a stochastic model because the number of Cdk1-cyclin B1 complexes in a cell is large (~1.6 × 106; Sun et al., 2010) and we chose an ODE model rather than a partial differential equation model because we expect the spatial equilibration of Cdk1-cyclin B1 throughout the cytoplasm and the nucleus to be rapid (Smith et al., 2002). To keep any positive feedback in Cdk1-cyclin B1 localization separate from the positive feedback in Cdk1-cyclin B1 activation (the well studied Cdk1/Cd25C/Wee1 system), we included only fully active Cdk1-cyclin B1 in the model.
Initially there were four Cdk1 species in the model: phosphorylated and non-phosphorylated cytoplasmic Cdk1-cyclin B1, and phosphorylated and non-phosphorylated nuclear Cdk1-cyclin B1 (Figure 7E). These species are interconverted by import/export reactions (Figure 7E, horizontal arrows) and phosphorylation/ dephosphorylation reactions (Figure 7E, vertical arrows). The relative sizes of the arrows depicts the fact that the ratio of import to export is higher for phosphorylated Cdk1-cyclin B1, and the ratio of phosphorylation to dephosphorylation is higher for nuclear Cdk1-cyclin B1.
We can simplify the model further without changing its essential behavior if we assume that the phosphorylation of cyclin B1 subunit in the cytoplasm is negligible (see Figure 7A, C; see also Figure S7D, E) and that transport of cyclin-phosphorylated form of Cdk1-cyclin B1 out of the nucleus is also negligible (Figure 7E, F; see also Figure 6A, S6). We also assume that Cdk1-cyclin B1 activation by Cdc25 occurs in the cytoplasm before the complex translocates to the nucleus. We are then left with three Cdk1-cyclin B1 species (Figure 7F), whose interconversion reactions are described by three ordinary differential equations and a small number of kinetic parameters:
where xcyt, xnuc,and xpnuc represent the amounts (not concentrations) of Cdk1-cyclin B1 in the cytoplasm, in the nucleus but non-phosphorylated, and in the nucleus and phosphorylated, respectively. Because we are using units of quantity rather than concentration, a simple conservation equation holds:
The main unknown in the model is the response function Φ that describes how the multisite phosphorylation of nuclear Cdk1-cyclin B1 depends upon the amount of Cdk1-cyclin B1 present in the nucleus. As shown in the Supplemental Information, measurements of the initial rate of Cdk1-cyclin B1 autophosphorylation in vitro are compatible with an intermolecular rather than intramolecular mechanism (Figure S7). Given that multiple phosphorylations on cyclin B1 contribute to the regulation of Cdk1-cyclin B1 nuclear translocation, we have assumed that the Φ a Hill function with a Hill coefficient (or Hill exponent) of 4:
The steady-state response of the three ODE model for various assumed amounts of total active Cdk1-cyclin B1 is shown in Figure 7G. When the total amount of active Cdk1-cyclin B1 present is low, the system is monostable and the Cdk1-cyclin B1 is predominantly cytoplasmic. When the active Cdk1-cyclin B1 levels are high, the system is monostable and the Cdk1-cyclin B1 is predominantly nuclear. At intermediate levels of Cdk1-cyclin B1 (0.51 < xtot < 0.66), the system is bistable, with two alternative stable steady-states and one unstable steady-state (Figure 7G). Thus the positive feedback in the system has produced a spatial toggle switch, provided that the rate constants in the model are set to appropriate values (see legend to Figure 7G).
We can also investigate the dynamic behavior of the system. Using a FRET sensor of Cdk1-cyclin B1 activity, Gavet and Pines showed that Cdk1-cyclin B1 rises just before NEB in a sigmoidal fashion (Gavet and Pines, 2010b). We assumed that Cdk1-cyclin B1 was initially activated in the cytoplasm at a rate comparable to that seen by Gavet and Pines, approximated by a logistic function with a rise time of 7.2 min (Figure 7H, dashed line), and then looked at the modeled dynamics of the three Cdk1-cyclin B1 species. The active Cdk1-cyclin B1 is initially predominantly cytoplasmic, and then abruptly switches to become nuclear (Figure 7H). Thus, a simple positive feedback model that incorporates the known facts about Cdk1-cyclin B1 regulation (phosphorylation promotes localization, localization promotes phosphorylation, and phosphorylation occurs through an intermolecular mechanism with non-linear kinetics) accounts for the experimentally-observed, abrupt redistribution of Cdk1-cyclin B1 from the cytoplasm to the nucleus (Figure 1).
DISCUSSION
In summary, the evidence presented here argues that spatial positive feedback is a key trigger mechanism in the initiation of mitosis. Not only does the activation of Cdk1-cyclin B1 promote Cdk1-cyclin B1 activation, but also the presence of those complexes in the nucleus promotes the further recruitment of Cdk1-cyclin B1 complexes to the nucleus (Figures 2, 3). Since both Cdk1-cyclin B1 activation and translocation are critical for the initiation of mitosis, positive feedback in both Cdk1-cyclin B1 activation and translocation can be expected to contribute to the switch-like activation of the mitotic trigger. Manipulations that compromise the abruptness of the trigger also compromise the cell’s ability to complete mitosis in a timely fashion (Figure 4), arguing that the switch-like initiation of mitosis is important for successful completion of mitosis.
The mechanism we propose for this spatial positive feedback centers on cyclin B1 phosphorylation. Consistent with previous studies (Hagting et al., 1998; Li et al., 1995, 1997; Toyoshima-Morimoto et al., 2001; Walsh et al., 2003; Yang et al., 2001), we found that cyclin B1 phosphorylation regulates Cdk1-cyclin B1 translocation and is required for the abrupt pre-NEB redistribution of Cdk1-cyclin B1 from the cytoplasm to the nucleus, and for spatial positive feedback (Figure 5). While previous work has shown that cyclin B1 phosphorylation regulates its interaction with nuclear import and export proteins, here we have shown that cyclin B1 phosphorylation promotes its stable association with mitotic chromosomes, an association that persists after the nuclear envelope has broken down (Figure 6). We also found that moving cyclin B1 from the cytoplasm to the nucleus, through leptomycin B-treatment, promotes its phosphorylation (Figure 7), completing a spatial positive feedback loop where cyclin B1 phosphorylation promotes nuclear localization and nuclear localization promotes cyclin B1 phosphorylation. Computational modeling showed that a loop like this can convert a graded increase in cyclin B1 concentration into a switch-like, temporally abrupt redistribution of Cdk1-cyclin B1 from the cytoplasm to the nucleus (Figure 7).
We propose that the combined, interlinked positive feedback in both Cdk1-cyclin B1 activation and Cdk1-cyclin B1 localization underlies the abruptness of the onset of mitosis, which we have shown is essential for successful mitotic progression. We envision that the combined loops may be more robust and reliable in triggering a well-coordinated mitotic entry than the individual loops would be by themselves (Brandman et al., 2005; Ferrell, 2008).
Our model of spatial positive feedback is built on the observation that the enzymatic processes that cyclin B1 phosphorylation is more favorable in the nucleus than in the cytoplasm (Figure 7). This suggests that the kinase and phosphatases that regulate cyclin B1 phosphorylation are spatially segregated. While both Plk1 and Cdk1 are distributed throughout the cell, cyclin A2-Cdk1, a candidate cyclin B1 kinase, is concentrated in the nucleus (Pines and Hunter, 1991). In addition, since the volume of the nucleus is ~10 times smaller than the volume of the cytoplasm, the rate of intermolecular cyclin B1-Cdk1 autophosphorylation can be much higher in the nucleus than in the cytoplasm. Less is known about the phosphatases that dephosphorylate cyclin B1, but one plausible candidate is PP2A-B55δ, which is found primarily in the cytoplasm and centrosome ((Schmitz et al.), also see S7D). Any or all of these asymmetries could contribute to the enhanced phosphorylation of cyclin B1 in the nucleus. In addition, while we have treated cyclin B1-Cdk1 localization separately from cyclin B1-Cdk1 activation, for simplicity, the two levels of regulation are tightly linked in vivo (Gavet and Pines, 2010a). Since cyclin B1 phosphorylation depends upon cyclin B1-Cdk1 activation, spatial segregation of the regulators of cyclin B1-Cdk1 activity could also contribute to the enhanced phosphorylation of cyclin B1 in the nucleus. In this light, it is noteworthy that the Cdk1 activators CAK and Cdc25A are predominantly nuclear (Kallstrom et al., 2005; Tassan et al., 1994) (as is the Cdk1 inactivator Wee1), and the Cdk1 inhibitor Myt1 is cytoplasmic (Kornbluth et al., 1994). Consistent with this idea, biochemical fractionation experiments show that active Cdk1 is enriched in the nuclear pellet (Figure S7E).
One of the most important steps forward in our understanding of the systems biology of cellular regulation has been the appreciation that there are specific, recurrent motifs that define how genes and proteins regulate each other’s activities (Alon, 2007; Shen-Orr et al., 2002). However protein function is regulated not only through temporal regulation of activity, but also through the regulation of localization (Mochly-Rosen, 1995; Ptashne and Gann, 1997). The present work shows that positive feedback, a motif found in many signaling pathways, can also be an important design principle in the control of protein localization. Moreover, since freely-diffusing proteins can redistribute very quickly over the distance scales of a typical cell, a bistable trigger making use of spatial positive feedback has the potential to switch rapidly between discrete spatial states. Given how common it is for regulatory proteins to rapidly translocate from the cytoplasm to the nucleus in response to stimuli (e.g. NF-AT, NF-κB, Hog1), it seems likely that spatial positive feedback may prove to be a recurring theme in cellular regulation.
METHODS
Cell Culture, Synchronization, and Transfection
HeLa cells (ATCC) and Tet-On Advanced HeLa cells (Invitrogen) were cultured at 37°C, 5% CO2 in DMEM (Invitrogen) supplemented with 10% FCS, penicillin (100 U/ml), streptomycin (100 μg/ml) and G418 (100 μg/ml, for Tet-On Advanced HeLa cells). Cells were synchronized at the G1/S phase of the cell cycle by double thymidine block. Transfection of cDNAs was typically performed using either Fugene 6 (Roche) (for DNA transfection) or Gene Silencer (Genlantis) (for small interfering RNAs, siRNA, transfection or double DNA/siRNA transfection) according to the manufacturers’ instructions. Transfection of 50 μM dextran-70-kDa-Texas Red was done by suspended drop electroporation (SDE) as described in (Guignet and Meyer, 2008).
Microscopy and Data Analysis
Time-lapse live cell imaging was performed on either an ImageXpress Micro inverted epifluorescence microscope (Molecular Devices) or on a Nikon Eclipse TI with perfect focus, both equipped with temperature, humidity and CO2 control. ImageXpress images were acquired with 20x or 40x plan fluorescence objectives. Excitation (Ex) and emission (Em) filters sets (Chroma Technology Corporation) were as follows: CFP, 427-10nm (Ex), 483-32 nm (Em); YFP, 504-12 nm (Ex), 542-27 nm (Em); mCherry, 589-15 nm (Ex), 632-22 nm (Em). Nikon Eclipse TI images were acquired using either 40x NA 1.3 Plan Flour or 60x NA 1.3 Plan Apo objectives. Excitation (Ex) and emission (Em) filters sets were as follows: CFP, 430-24 nm (Ex), 470-24 nm (Em); YFP, 500-20 nm (Ex), 535-30 nm (Em); mCherry, 572-35 nm (Ex), 632-60 nm (Em). Micromanager 1.3 was used for acquisition of time-lapse images.
A typical experiment would monitor cells from 15–20 h with images taken every 3–5 min. Confocal microscopy was performed on a Leica TCS SP2 ABOS equipped with a 63x 1.3 NA oil immersion objective. DAPI was excited using a 405 nm diode laser; Alexa 488 or YFP were excited with a 488 nm or 514 nm Ar laser and Alexa 647 was excited with a 633 nm HeNe laser line.
All data analysis was done with scripts written in Matlab (Mathworks) or using Cell Profiler (Broad Institute) and ImageJ (National Institutes of Health).
Cdk1-Cyclin B1 Autophosphorylation
For cyclin B1 auto-phosphorylation experiments where we varied the concentration of Cdk1-cyclin B1, purified human Cdk1-cyclin B1 protein complex was incubated at the indicated concentrations at 25°C for 1, 2 or 5 min with kinase buffer (20 mM HEPES/NaOH pH 7.4, 10 mM MgCl2, 1 mM DTT), 15 μM ATP and phosphatase inhibitor cocktail (Sigma) in a total volume of 20 μl. For Cyclin B1 auto-phosphorylation experiments where we varied kinase reaction volumes, purified human Cdk1-cyclin B1 protein complex was incubated at 25°C for 1, 2 or 5min with kinase buffer (20 mM HEPES/NaOH pH 7.4, 10 mM MgCl2, 1 mM DTT), 15 μM ATP and phosphatase inhibitor cocktail (Sigma) in a total volume of 10, 20, 40, 80 or 160 μl. Reactions were stopped by addition of Laemmli sample buffer and analyzed by gel electrophoresis and blotting followed by phospho-imaging.
Nuclear/Cytoplasmic Cell Fractionation
HeLa cell nuclear and cytoplasmic fractions were isolated with NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific) according to the manufacturer’ instructions. Total protein amount in the nuclear and the cytosolic lysates was quantified by BCA assay for protein quantitation. Typically, 15–20 μg or 100–200 μg of total protein for each fraction was used for western blot analysis or immunoprecipitation, respectively. For Cdk1-cyclin B1 nuclear/cytosolic kinase assays, 100 μg of total protein was used to immunoprecipitate cyclin B1 using anti-cyclin B1 antibody (Santa Cruz). Kinase assays were performed using purified histone H1 (Upstate) as a general Cdk1-cyclin B1 substrate and the resulting 32P-labeled histone was detected and quantified by SDS gel electrophoresis and blotting followed by phosphorimaging.
Supplementary Material
Article Highlights.
The onset of mitosis relies on spatial positive feedback.
Cyclin B1 phosphorylation promotes nuclear translocation of Cdk1-cyclin B1.
Cdk1-cyclin B1 nuclear translocation promotes cyclin B1 phosphorylation.
Cyclin B1 phosphorylation allows it to bind to chromatin.
Spatial feedback is important for the decisiveness and irreversibility of mitotic entry
Acknowledgments
We thank S. Pearlman and P. Beltrao for help with analysis scripts; M. Teruel, S. Bandara and T. Wandless for iRap; O. Rocks and S. Spenser for reagents; J. Wu from the Stanford High-Throughput Bioscience Center for technical assistance; and J. Skotheim, P. Beltrao, S. Collins, A. Hayer, A. Salmeen S. Carrasco, J Chang and the Meyer and Ferrell labs for helpful discussions and comments on the manuscript. This work was supported by a grant from the National Institutes of Health (GM061276). S.D.M.S. is a Human Frontier Science Program Fellow (HFSP) and an EMBO fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Atherton-Fessler S, Liu F, Gabrielli B, Lee MS, Peng CY, Piwnica-Worms H. Cell cycle regulation of the p34cdc2 inhibitory kinases. Mol Biol Cell. 1994;5:989–1001. doi: 10.1091/mbc.5.9.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Ferrell JE, Jr, Li R, Meyer T. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 2005;310:496–498. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Ross KE, Kaldis P, Solomon MJ. Dephosphorylation of cyclin-dependent kinases by type 2C protein phosphatases. Genes Dev. 1999;13:2946–2957. doi: 10.1101/gad.13.22.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree GR, Schreiber SL. Three-part inventions: intracellular signaling and induced proximity. Trends Biochem Sci. 1996;21:418–422. doi: 10.1016/s0968-0004(96)20027-1. [DOI] [PubMed] [Google Scholar]
- Dalal SN, Schweitzer CM, Gan J, DeCaprio JA. Cytoplasmic localization of human cdc25C during interphase requires an intact 14-3-3 binding site. Mol Cell Biol. 1999;19:4465–4479. doi: 10.1128/mcb.19.6.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deibler RW, Kirschner MW. Quantitative reconstitution of mitotic CDK1 activation in somatic cell extracts. Mol Cell. 2010;37:753–767. doi: 10.1016/j.molcel.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eser U, Falleur-Fettig M, Johnson A, Skotheim JM. Commitment to a cellular transition precedes genome-wide transcriptional change. Mol Cell. 2011;43:515–527. doi: 10.1016/j.molcel.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE., Jr How regulated protein translocation can produce switch-like responses. Trends Biochem Sci. 1998;23:461–465. doi: 10.1016/s0968-0004(98)01316-4. [DOI] [PubMed] [Google Scholar]
- Ferrell JE., Jr Feedback regulation of opposing enzymes generates robust, all-or-none bistable responses. Curr Biol. 2008;18:R244–245. doi: 10.1016/j.cub.2008.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE, Jr, Tsai TY, Yang Q. Modeling the cell cycle: why do certain circuits oscillate? Cell. 2011;144:874–885. doi: 10.1016/j.cell.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Gavet O, Pines J. Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J Cell Biol. 2010a;189:247–259. doi: 10.1083/jcb.200909144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell. 2010b;18:533–543. doi: 10.1016/j.devcel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D, Pomerening JR, Myers JW, Gustavsson C, Jones JT, Hahn AT, Meyer T, Ferrell JE., Jr Cyclin A2 regulates nuclear-envelope breakdown and the nuclear accumulation of cyclin B1. Curr Biol. 2007;17:85–91. doi: 10.1016/j.cub.2006.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves PR, Yu L, Schwarz JK, Gales J, Sausville EA, O’Connor PM, Piwnica-Worms H. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J Biol Chem. 2000;275:5600–5605. doi: 10.1074/jbc.275.8.5600. [DOI] [PubMed] [Google Scholar]
- Guignet EG, Meyer T. Suspended-drop electroporation for high-throughput delivery of biomolecules into cells. Nat Methods. 2008;5:393–395. doi: 10.1038/nmeth.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagting A, Jackman M, Simpson K, Pines J. Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal. Curr Biol. 1999;9:680–689. doi: 10.1016/s0960-9822(99)80308-x. [DOI] [PubMed] [Google Scholar]
- Hagting A, Karlsson C, Clute P, Jackman M, Pines J. MPF localization is controlled by nuclear export. Embo J. 1998;17:4127–4138. doi: 10.1093/emboj/17.14.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, McLoughlin M, McKeon F. Human wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated Cdc2 kinase. Cell. 1993;74:463–474. doi: 10.1016/0092-8674(93)80048-j. [DOI] [PubMed] [Google Scholar]
- Hoffmann I, Clarke PR, Marcote MJ, Karsenti E, Draetta G. Phosphorylation and activation of human cdc25-C by cdc2--cyclin B and its involvement in the self-amplification of MPF at mitosis. Embo J. 1993;12:53–63. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Krutchinsky AN, Morgan DO. Positive feedback sharpens the anaphase switch. Nature. 2008;454:353–357. doi: 10.1038/nature07050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Hardy S, Morgan DO. Nuclear localization of cyclin B1 controls mitotic entry after DNA damage. J Cell Biol. 1998;141:875–885. doi: 10.1083/jcb.141.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JT, Myers JW, Ferrell JE, Meyer T. Probing the precision of the mitotic clock with a live-cell fluorescent biosensor. Nat Biotechnol. 2004;22:306–312. doi: 10.1038/nbt941. [DOI] [PubMed] [Google Scholar]
- Kallstrom H, Lindqvist A, Pospisil V, Lundgren A, Rosenthal CK. Cdc25A localisation and shuttling: characterisation of sequences mediating nuclear export and import. Exp Cell Res. 2005;303:89–100. doi: 10.1016/j.yexcr.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Komatsu T, Kukelyansky I, McCaffery JM, Ueno T, Varela LC, Inoue T. Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat Methods. 2010;7:206–208. doi: 10.1038/nmeth.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornbluth S, Sebastian B, Hunter T, Newport J. Membrane localization of the kinase which phosphorylates p34cdc2 on threonine 14. Mol Biol Cell. 1994;5:273–282. doi: 10.1091/mbc.5.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Regulation of the cdc25 protein during the cell cycle in Xenopus extracts. Cell. 1992;70:139–151. doi: 10.1016/0092-8674(92)90540-s. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev. 1999;13:1067–1072. doi: 10.1101/gad.13.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Meyer AN, Donoghue DJ. Requirement for phosphorylation of cyclin B1 for Xenopus oocyte maturation. Mol Biol Cell. 1995;6:1111–1124. doi: 10.1091/mbc.6.9.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Meyer AN, Donoghue DJ. Nuclear localization of cyclin B1 mediates its biological activity and is regulated by phosphorylation. Proc Natl Acad Sci U S A. 1997;94:502–507. doi: 10.1073/pnas.94.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A, van Zon W, Karlsson Rosenthal C, Wolthuis RM. Cyclin B1-Cdk1 activation continues after centrosome separation to control mitotic progression. PLoS Biol. 2007;5:e123. doi: 10.1371/journal.pbio.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochly-Rosen D. Localization of protein kinases by anchoring proteins: a theme in signal transduction. Science. 1995;268:247–251. doi: 10.1126/science.7716516. [DOI] [PubMed] [Google Scholar]
- Morgan DO. The Cell Cycle: Principles of Control. London UK: New Science Press Ltd; 2007. [Google Scholar]
- Mueller PR, Coleman TR, Dunphy WG. Cell cycle regulation of a Xenopus Wee1-like kinase. Mol Biol Cell. 1995a;6:119–134. doi: 10.1091/mbc.6.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller PR, Coleman TR, Kumagai A, Dunphy WG. Myt1: a membrane-associated inhibitory kinase that phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science. 1995b;270:86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- Pines J, Hunter T. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerening JR, Sontag ED, Ferrell JE., Jr Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nature Cell Biol. 2003;5:346–351. doi: 10.1038/ncb954. [DOI] [PubMed] [Google Scholar]
- Pomerening JR, Ubersax JA, Ferrell JE., Jr Rapid cycling and precocious termination of G1 phase in cells expressing CDK1AF. Mol Biol Cell. 2008;19:3426–3441. doi: 10.1091/mbc.E08-02-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Russell P, Nurse P. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 1986;45:145–153. doi: 10.1016/0092-8674(86)90546-5. [DOI] [PubMed] [Google Scholar]
- Russell P, Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 1987;49:559–567. doi: 10.1016/0092-8674(87)90458-2. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Schmitz MH, Held M, Janssens V, Hutchins JR, Hudecz O, Ivanova E, Goris J, Trinkle-Mulcahy L, Lamond AI, Poser I, et al. Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat Cell Biol. 12:886–893. doi: 10.1038/ncb2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha W, Moore J, Chen K, Lassaletta AD, Yi CS, Tyson JJ, Sible JC. Hysteresis drives cell-cycle transitions in Xenopus laevis egg extracts. Proc Natl Acad Sci U S A. 2003;100:975–980. doi: 10.1073/pnas.0235349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AE, Slepchenko BM, Schaff JC, Loew LM, Macara IG. Systems analysis of Ran transport. Science. 2002;295:488–491. doi: 10.1126/science.1064732. [DOI] [PubMed] [Google Scholar]
- Solomon MJ, Glotzer M, Lee TH, Philippe M, Kirschner MW. Cyclin activation of p34cdc2. Cell. 1990;63:1013–1024. doi: 10.1016/0092-8674(90)90504-8. [DOI] [PubMed] [Google Scholar]
- Sun T, Yang X, Wang W, Zhang X, Xu Q, Zhu S, Kuchta R, Chen G, Liu X. Cellular abundance of Mps1 and the role of its carboxyl terminal tail in substrate recruitment. J Biol Chem. 2010;285:38730–38739. doi: 10.1074/jbc.M110.177642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Coleman TR, Dunphy WG. Two distinct mechanisms for negative regulation of the Wee1 protein kinase. Embo J. 1993;12:3427–3436. doi: 10.1002/j.1460-2075.1993.tb06017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassan JP, Schultz SJ, Bartek J, Nigg EA. Cell cycle analysis of the activity, subcellular localization, and subunit composition of human CAK (CDK-activating kinase) J Cell Biol. 1994;127:467–478. doi: 10.1083/jcb.127.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima-Morimoto F, Taniguchi E, Shinya N, Iwamatsu A, Nishida E. Polo-like kinase 1 phosphorylates cyclin B1 and targets it to the nucleus during prophase. Nature. 2001;410:215–220. doi: 10.1038/35065617. [DOI] [PubMed] [Google Scholar]
- Walsh S, Margolis SS, Kornbluth S. Phosphorylation of the cyclin B1 cytoplasmic retention sequence by mitogen-activated protein kinase and Plx. Mol Cancer Res. 2003;1:280–289. [PubMed] [Google Scholar]
- Yang J, Song H, Walsh S, Bardes ES, Kornbluth S. Combinatorial control of cyclin B1 nuclear trafficking through phosphorylation at multiple sites. J Biol Chem. 2001;276:3604–3609. doi: 10.1074/jbc.M008151200. [DOI] [PubMed] [Google Scholar]
- Yang J, Winkler K, Yoshida M, Kornbluth S. Maintenance of G2 arrest in the Xenopus oocyte: a role for 14-3-3-mediated inhibition of Cdc25 nuclear import. Embo J. 1999;18:2174–2183. doi: 10.1093/emboj/18.8.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.