Abstract
The adaptors IRS1 and IRS2 link growth factor receptors to downstream signaling pathways that regulate proliferation and survival. Both suppress factor-withdrawal-induced apoptosis and have been implicated in cancer progression. However, recent studies suggest IRS1 and IRS2 mediate differential functions in cancer pathogenesis. IRS1 promoted breast cancer proliferation, while IRS2 promoted metastasis. The role of IRS1 and IRS2 in controlling cell responses to chemotherapy is unknown. To determine the role of IRS1 and IRS2 in the sensitivity of cells to chemotherapy, we treated 32D cells lacking or expressing IRS proteins with various concentrations of chemotherapeutic agents. We found that expression of IRS1, in contrast to IRS2, enhanced the sensitivity of 32D cells to chemotherapy-induced apoptosis. When IRS2 was expressed with IRS1, the cells no longer showed enhanced sensitivity. Expression of IRS1 did not alter the expression of pro- and anti-apoptotic proteins; however, 32D-IRS1 cells expressed higher levels of Annexin A2. In 32D-IRS1 cells, IRS1 and Annexin A2 were both located in cytoplasmic and membrane fractions. We also found that IRS1 coprecipitated with Annexin A2, while IRS2 did not. Decreasing Annexin A2 levels reduced 32D-IRS1 cell sensitivity to chemotherapy. These results suggest IRS1 enhances sensitivity to chemotherapy in part through Annexin A2.
Keywords: insulin receptor substrate, signaling, annexin A2, chemotherapy, cell death
Introduction
Insulin receptor substrate (IRS) proteins 1-6 are large cytoplasmic docking proteins that are important for receptor signaling. IRS1 and IRS2 are widely expressed, while the other family members (IRS3-6) have restricted expression patterns [1]. Most hematopoietic cells express IRS2, but not IRS1 [2, 3]. Other cell types, including many cancer cells express IRS1, IRS2, or both [4, 5]. IRS3 is restricted to mouse adipose tissue while IRS4 is restricted to human brain, liver, kidney, and thymus. IRS5 and IRS6 have less clear roles [1, 6]. IRS1 and IRS2 are encoded by single exons on human chromosome 2q36-37 (mouse chromosome 1), and human chromosome 12q341 (mouse chromosome 8), respectively. These proteins function as large adaptors and have many protein binding motifs that allow them to interact with other proteins. IRS1 and IRS2 have a well conserved pleckstrin homology (PH) domain and a phosphotyrosine binding (PTB) domain, which interact with phospholipids and phosphorylated receptors respectively. Furthermore, they both have a large COOH-terminal region containing many potential sites for tyrosine and serine/threonine phosphorylation [7-9].
IRS1 and IRS2 have been shown to mediate signaling from activated cell surface receptors including the receptors for insulin, insulin like growth factor 1 (IGF-1), prolactin, growth hormone (GH), vascular endothelial growth factor (VEGF), and several cytokine receptors including those for interleukin (IL)-4, -9, -13, and -15 [1, 4, 7, 10]. When these receptors are activated by ligand binding, the IRS proteins are recruited to the receptors by interaction of their PTB domains with phosphorylated residues within the receptor tails. IRS1 and IRS2 are then phosphorylated on multiple residues by receptor associated kinases [7, 11-15]. IRS1 and IRS2 have ~20 potential tyrosine phosphorylation sites in their COOH-terminal region. Once phosphorylated, IRS1 and IRS2 can recruit additional signaling molecules and activate downstream signaling pathways. Phosphorylated IRS1 and IRS2 have been shown to interact with SHP2, Fyn, Grb-2, Nck, Crk, and p85, the regulatory subunit of phosphoinositide-3-kinase (PI-3-K) [6, 14, 16-19]. IRS1 and IRS2 have four and two possible p85 binding sites, respectively. The p85 subunit contains an N-terminal SH3 domain and C-terminal SH2 domain that mediate its interaction with the p110 catalytic subunit. The p110 subunit can phosphorylate membrane lipids, which are involved in the activation of several kinases, including AKT (also known as protein kinase B), that play an important role in cell survival. The p110 subunit can also phosphorylate, by a less well understood mechanism, Ser/Thr protein residues, including those of IRS1 [14, 15].
While the tyrosine phosphorylation of certain residues leads to enhanced recruitment of downstream pathways, phosphorylation of IRS1 has also been shown to reduce certain protein interactions. IRS1 has been shown to interact with the DNA repair protein RAD51 via its PTB domain; tyrosine phosphorylation induced by IGF-1 suppressed this interaction [9]. Furthermore, IRS1 Ser/Thr phosphorylation by multiple kinases, including JNK, mTOR, and PKC, has been shown to interrupt the interaction of IRS1 with activated receptors and may promote its interaction with 14-3-3 proteins. Phosphorylated serines may also sterically prevent other proteins from binding to IRS1. Activation of Ser/Thr kinase activity may create a negative feedback loop to regulate IRS1 function [4, 15, 20-24]. IRS proteins are also regulated by ubiquitination, which can be mediated by Ser/Thr phosphorylation, leading to proteasomal degradation [1, 23-25]. On the other hand, in some cases serine phosphorylation of IRS1 may actually promote cell signaling. For example, in human prostate cancer cells the serine phosphorylation of IRS1 inhibited its tyrosine phosphorylation and promoted cell adhesion while decreasing cell motility because of enhanced interactions with α5β1 integrins [26].
Chemotherapy involves the use of anti-cancer drugs to treat advanced or metastatic cancers systemically. Chemotherapeutic agents can induce double-strand breaks in DNA causing proliferating cells to growth arrest and then undergo apoptosis [9, 27-32]. Many patients show inherent or acquired resistance to chemotherapy and in some cases this is due to alterations in the levels of pro-apoptotic and anti-apoptotic proteins [28, 29, 31]. Despite similar structure and function, IRS1 and IRS2 have also been implicated in cancer progression by differentially regulating cell survival, growth, proliferation, and motility [4, 6, 7, 15, 33-36]. It has been postulated that differences in functions of IRS1 and IRS2 in cancer progression may be due to difference in tissue distribution, subcellular localization, or protein recruitment [19, 34]. However, the roles of IRS1 and IRS2 in regulating responses to chemotherapy are unclear. Therefore, to determine the role of IRS1 and IRS2 in the protection of cells from chemotherapy induced cell death, we utilized the 32D cell model system. The 32D cell line is an IL 3 dependent myeloid progenitor line that lacks expression of IRS1 and IRS2. These cells have been used extensively as models for growth factor receptor signaling, proliferation, and regulation of apoptosis [3, 34, 37-40]. We found that IRS1, but not IRS2, expression sensitized 32D cells to chemotherapy induced caspase-mediated cell death. Although this was not due to changes in expression of pro- or anti-apoptotic proteins, we did find that cells expressing IRS1 expressed higher levels of Annexin A2. In 32D-IRS1 cells IRS1 and Annexin A2 were both located in cytoplasmic and membrane fractions. We also found that IRS1 coprecipitated with Annexin A2, while IRS2 did not. Decreasing Annexin A2 levels reduced 32D-IRS1 cell sensitivity to chemotherapy. These results showed that enhanced expression of IRS1 led to enhanced responses to chemotherapy and suggest that Annexin A2 may participate in this response.
Materials and Methods
Materials
32D cells were obtained from Dr. J. H. Pierce [37]. Recombinant IL-4 (rIL-4) was purchased from R &D Systems and used at a final concentration of 10 ng/ml. The caspase inhibitor, Q-VD-OPh, was also purchased from R & D Systems and used at a final concentration of 10 μM. Neomycin (G418) and tetramethylrhodamine (TMR) were purchased from Invitrogen. Vincristine, etoposide, taxol, and daunorubicin were purchased from Sigma-Aldrich. Taxol was also purchased from LC Laboratories. Propidium Iodide (PI) was purchased from from Sigma-Aldrich. Antibodies for BCL-xL, pBAD(Ser136), BAD, BAK, GAPDH, Sp1, and Annexin II were purchased from Santa Cruz Biotechnology. Antibodies for pAKT(473), AKT, BAX, and BCL-2(50E3) were purchased from Cell Signaling Technologies. Antibody for alpha tubulin was purchased from eBioscience. Antibody for HSP90 was purchased from Stressgen. Antibodies for IRS1 and IRS2 were purchased from Upstate Biotechnology. Imperial protein stain was purchased from Thermo Scientific.
Cell Culture and Reagents
32D cells transfected with either IRS1 or IRS2 were maintained in complete RPMI 1640 (Lonza) supplemented with 10% fetal bovine serum (FBS), 1% glutamine, 1% pen-strep, plus 5% WEHI-3B supernatant as a source of IL-3 [3, 37]. 32D cells transfected with both IRS1 and IRS2 were maintained in complete RPMI plus 5% WEHI-3B supernatant and 800 μg/ml neomycin (Invitrogen). WEHI-3B supernatant was prepared by growing WEHI-3B cells to terminal density in RPMI 1640 (Lonza) containing 15% fetal bovine serum (FBS), 1% glutamine, and 1% pen strep as described [37].
Transfection Experiments
32D cells expressing IRS1 (clone 484.2 and clone 260.3) were previously described [37]. 32D cells expressing IRS2 were generated similarly. To express both IRS1 and IRS2 in 32D cells, 1 × 107 cells were washed and resuspended in 0.3 ml of phosphate buffered saline (PBS). The cells were mixed with 10 g of IRS1 cDNA in a vector containing a neomycin resistance marker and 10 g of IRS2 cDNA in a vector. The mixture was subjected to electroporation using a Bio-Rad gene-pulsar set on 200 V and 960 F. The cells were returned to ice for 20 minutes and then cultured overnight in culture media before selection with 800 g/ml neomycin. Neomycin resistant cells were tested for the expression of IRS1 and IRS2 by western immunoblotting using an IRS1- or IRS2-specific antibody. The phosphorylation of IRS1 and IRS2 after IL-4 stimulation was tested by immunoprecipitation with an IRS1- or IRS2-specific antibody and then western immunoblotting using a phosphotyrosine specific antibody.
Knock-down of Annexin A2 in 32D-IRS1 Cells
32D-IRS1 (clone 260.3) cells were plated to 80% confluency in 2 mls of culture media. Various amounts of Annexin A2 shRNA lenti-viral supernatant (Sigma TRCN0000110696) were used to infect the cells. Two days after infection, cells were selected in 700 ng/ml puromycin. Puromycin-resistant cells were tested for the expression of Annexin A2 by western immunoblotting using an Annexin A2-specific antibody. Two Annexin A2-depleted cell lines were generated and characterized (101-2W3 and 101-2W4). Non-target shRNA control lentiviral supernatant (Sigma SHC002V) was also found to knock-down Annexin A2.
Western Immunoblotting
Cells were washed and deprived of serum for two hours. Cells were treated in the presence or absence of IL-3, supplied as 5% WEHI-3B supernatant, or 10 ng/ml IL-4 (R&D) for 30 minutes. Cells were washed with ice cold PBS and lysed with lysis buffer (50 mM HEPES at pH 7.5, 0.5% NP-40, 5 mM EDTA, 50 mM NaCl, 10 mM Na pyrophosphate, 50 mM NaF, fresh 1 mM NaOV, fresh 1mM PMSF, fresh 1X protease cocktail, and fresh 100 nM calyculin A). In some experiments a protein subcellular fractionation kit (Pierce Biotechnology) was used to obtain lysates. Protein concentrations were determined using a BCA Kit (Pierce Biotechnology). Equal amounts of protein were loaded on the gel, separated by SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis), and transferred to a nitrocellulose membrane (Millipore). The blot was blocked with 3% BSA in TBST (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% Tween-20) wash buffer for 1 hour. The appropriate primary antibody was added for either 1 hour or overnight. The blot was then washed and the secondary antibody conjugated with horseradish peroxidase was added for 1 hour. The blot was washed again and chemiluminescence reagents (Denville Scientific) were used to visualize the signal on x-ray film. In some experiments the membranes were stripped with a 37° Celsius stripping solution (Millipore) for 30 minutes at room temperature. These membranes were then used for immunoblotting with another antibody. For immunoprecipitation experiments, specific antibodies were added to lysates containing equal, normalized amounts of total cellular protein. Protein A/G-agarose beads were used to pull down antibody-bound proteins prior to analysis by SDS-PAGE and western blotting.
Protein Identification and Sequence Alignment
Cell lysates were prepared from 32D and 32D-IRS1 cells, loaded on a gel, and separated by SDS-PAGE. The proteins were stained and visualized with Imperial™ protein stain overnight and then de stained in distilled water. Bands that showed a change in staining intensity compared to the control lysate were excised for further analysis. Proteins were digested in gel with trypsin. Subsequently, the extracted peptides were analyzed by liquid chromatography, tandem mass spectrometry using an LCQ Advantage ion trap mass spectrometry and nano spray ion source equipped with a 105 mm by 0.030 mm stainless steel emitter. Proteins were identified using X!Tandem where the NCBI Refseq (mouse) database was used to identify peptides and correlate those peptide hits to protein identifications [41]. Protein sequence alignment was performed using the NCBI site.
Nuclear DNA Content Assay
Cells were treated with chemotherapy agents as described [9]. Cells were cultured in RPMI 1640 media containing IL-3 supplied as 5% WEHI-3B supernatant which contains 0.75% serum. Various concentrations of chemotherapeutic drugs were added for 24 hours. Cells were centrifuged and washed twice with PBS (Lonza). Nuclear DNA was quantified with propidium iodide stain (0.1% Sodium Citrate, 0.2% Triton, 50 g/ml RNase, 50 μg/ml propidium iodide, in PBS). The cells were analyzed by FACS as described in detail by Zamorano et al. [3]. Based on multiple biochemical and morphological parameters, cells with < 2N DNA content (hypodiploid) were considered apoptotic [3, 42].
PI Uptake Assay
Cells were pretreated in the presence or absence of 10 μM Q-VD-OPh, a broad spectrum cell-permeant caspase inhibitor, for 1 hour and then treated in the presence or absence of 4 g/ml etoposide, for 20 hours. The cells were then washed in HEPES Buffered Saline (HBS, pH 7.4), containing 25 mM HEPES and 137 mM NaCl and then resuspended in PBS containing 5 μg/ml Propidium Iodide (PI). FACS analysis was used to measure the percentage of PI+ cells.
Mitochondrial Membrane Potential Assay
Cells were pretreated in the presence or absence of 10 μM Q-VD-OPh, a broad spectrum cell-permeant caspase inhibitor, for 1 hour and then treated in the presence or absence of 4 g/ml etoposide, for 20 hours. Mitochondrial membrane potential (MMP) was measured by labeling cells with 80 nM tetramethylrhodamine (TMR) for 30 minutes as previously described [43]. FACs analysis was used to measure the percentage of TMR low cells.
Results
Expression of IRS proteins in 32D cells
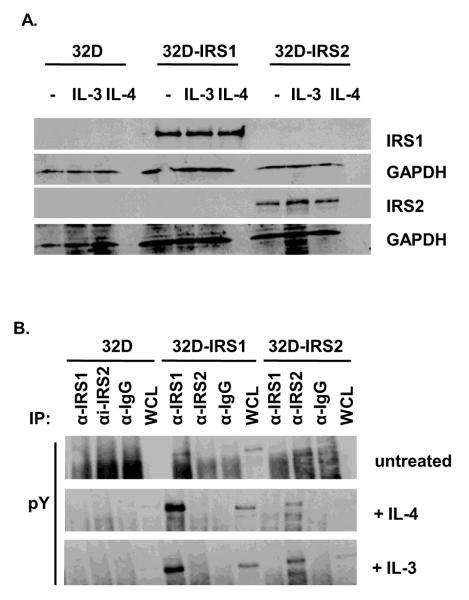
32D cells expressing IRS1 as a result of transfection have been described previously [3, 37]. We generated 32D cells expressing IRS2 using a similar strategy. The cDNA containing vector also encoded for a neomycin resistance marker that was used for positive clone selection. Positive clones were selected in 800 g/ml of G418. We verified the expression levels of IRS proteins in these transfectants in the presence or absence of cytokines (Figure 1A). The expression levels of the IRS proteins were not affected by the addition of recombinant IL-4 or the cognate growth factor for 32D cells, IL-3, supplied as 5% WEHI-3B supernatant. When we analyzed phosphorylation of these proteins, we observed basal IRS1 tyrosine phosphorylation that was increased by the addition of IL-4 or WEHI-3B supernatant which contains IL-3 and 0.75% serum (Figure 1B).
Figure 1. Expression of IRS proteins in 32D cells.
A. 32D cells and cells transfected with eithera IRS1 or IRS2 cDNA were treated in the presence or absence of IL-3 (5% WEHI-3B supernatant) or IL-4 (10 ng/ml) for 30 minutes. Cell lysates were prepared and the western blot was probed with anti IRS1, anti IRS2, or anti GAPDH antibodies. B. Cells were treated in the presence or absence of IL-3 (5% WEHI-3B supernatant) or IL-4 (10 ng/ml) for 30 minutes. Cell lysates were prepared and immunoprecipitated with antibodies specific for IRS1, IRS2, or an isotype control antibody (IgG). The western blot was probed with an anti phosphotyrosine (pY) antibody.
IRS1 expression sensitizes 32D cells to vincristine-induced cell death
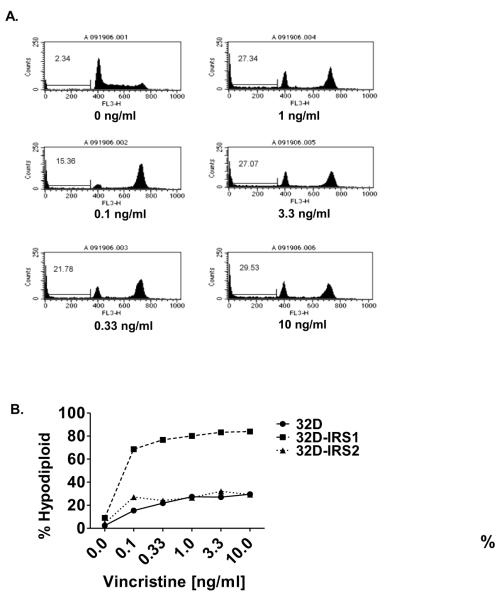
We previously reported that IRS1 expression protected cell lines from factor-withdrawal-induced death [3]. In tumor models, IRS1 has been shown to regulate cell proliferation, while IRS2 is primarily thought to control metastasis [4, 35]. However, the role of IRS1 and IRS2 in modulating chemotherapy induced cell death is unknown. Therefore, this study examined the effect of IRS1 and IRS2 expression on sensitivity of 32D cells to chemotherapy. We first examined the effect of vincristine on the 32D parental cells. Vincristine inhibits microtubule assembly by binding tubulin and inducing aggregate formation [31]. It arrests the cell cycle in the G2/M phase and induces apoptosis. To test for sensitivity to chemotherapy without inducing cytokine withdrawal, we treated the cells with various concentrations of vincristine in the presence of 5% WEHI-3B supernatant. Nuclear DNA content was measured by propidium iodide staining followed by FACS analysis. As previously described, this approach identifies hypodiploid, apoptotic cells [3, 42]. Vincristine treatment of 32D cells increased the percentage of hypodiploid, apoptotic cells in a dose dependent manner and increased the percentage of cells in the G2 phase of the cell cycle (Figure 2A). Vincristine also induced cell death in the 32D cells expressing IRS1 or IRS2; however, expression of the IRS proteins differentially affected the sensitivity of 32D cells to vincristine (Figure 2B). Specifically, 32D cells expressing IRS1 were substantially more sensitive to vincristine than were the parental 32D cells or IRS2 expressing 32D cells. Expression of IRS1 also increased the sensitivity of 32D cells to other chemotherapeutics tested including taxol, camptothecin, etoposide, and daunorubicin (unpublished observations).
Figure 2. IRS1 sensitizes 32D cells to vincristine induced cell death.
A. 32D cells were treated with various concentrations of vincristine, a microtubule assembly inhibitor, in the presence of IL-3, supplied as 5% WEHI-3B supernatant which contains 0.75% serum, for 24 hrs. The percentage of cells with <2N DNA content (hypodiploid) was measured by propidium iodide staining followed by FACs analysis. Representative histograms are shown. The percentage of sub diploid cells is indicated. B. 32D cells lacking or expressing IRS1 (clone 484.3) or IRS2 were treated with various concentrations of vincristine in the presence of IL-3, supplied as 5% WEHI-3B supernatant which contains 0.75% serum. The percentage of hypodiploid cells was measured by propidium iodide staining of nuclear DNA content followed by FACS analysis. The percentage of sub-diploid cells is indicated.
32D cells expressing both IRS1 and IRS2 are resistant to chemotherapy
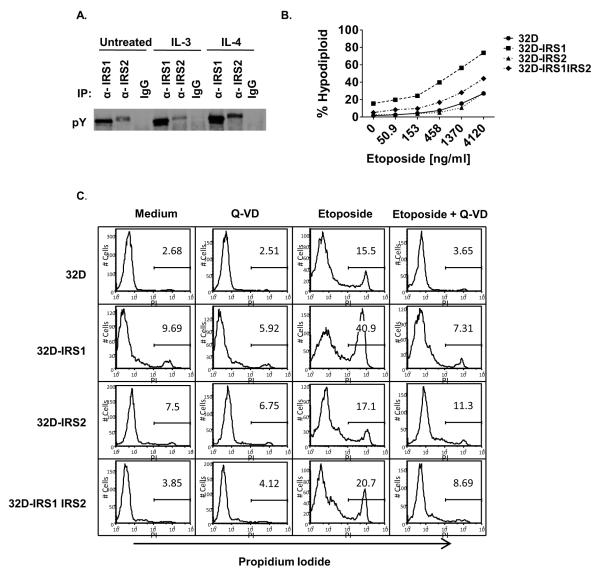
Other cell types, including cancer cells, can co-express both IRS1 and IRS2; therefore, we transfected both IRS1 and IRS2 into the 32D parental cells to examine the effects of the expression of both IRS1 and IRS2 on chemotherapy-induced cell death [4, 5]. We obtained a double-expressing clone that showed modest basal phosphorylation of both IRS1 and IRS2; the tyrosine phosphorylation of both IRS1 and IRS2 was increased in response to IL-4 while IL-3, supplied as 5% WEHI-3B supernatant, predominantly induced the tyrosine phosphorylation of IRS1 (Figure 3A). As mentioned above, WEHI-3B supernatant provides low serum (0.75%) which may contain additional growth factors that signal via IRS1. Etoposide binds topoisomerase II and arrests the cell cycle in S/G2 phase prior to undergoing apoptosis. To determine the effect of IRS1 on etoposide sensitivity, the cells were treated with various concentrations of etoposide. The percentage of hypodiploid, apoptotic cells was measured by propidium iodide staining of nuclear DNA content followed by FACS analysis (Figure 3B). While the 32D-IRS1 cells were persistently sensitive to chemotherapeutic-induced cell death, the 32D-IRS1IRS2 cells were resistant to etoposide induced cell death. Results were similar for death by vincristine and daunorubicin as well (unpublished observations). These results suggest that expression of IRS2 modulates the effect of IRS1 on sensitivity to chemotherapy.
Figure 3. IRS2 expression prevents IRS1 from sensitizing cells to chemotherapy induced cell death.
A. 32D cells were transfected with both IRS1 and IRS2. Clones were selected and the cells were treated in the presence or absence of IL-3 (5% WEHI-3B supernatant) or IL-4 (10 ng/ml) for 30 minutes. Cell lysates were prepared and immunoprecipitated with antibodies specific for IRS1, IRS2, or an isotype control antibody (IgG). The western blot was probed with an anti phosphotyrosine (pY) antibody. B. 32D cells lacking or expressing IRS1 (clone 260.3) or IRS2, or both IRS1 and IRS2 were treated with various concentrations of etoposide in the presence of IL-3, supplied as 5% WEHI-3B supernatant which contains 0.75% serum. The percentage of hypodiploid cells was measured by propidium iodide staining of nuclear DNA content followed by FACS analysis. The percentage of sub diploid cells is shown. C. 32D cells and transfectants were pretreated in the presence or absence of 10 μM Q VD OPh for 1 hr and then treated in the presence or absence of 4 g/ml etoposide, for 20 hrs. Cells were analyzed by FACS to determine the percentage of PI positive cells. Representative histograms are shown.
32D-IRS1 etoposide-induced cell death is caspase mediated
As a second measure of cell death, we evaluated membrane integrity using propidium iodide uptake in the presence or absence of the broad spectrum cell permeant caspase inhibitor Q-VD (Figure 3C). In the presence of WEHI-3B supernatant only, the percentage of PI+ cells was low, ranging from ~3-10%. The addition of Q-VD made a negligible impact on this percentage. Addition of etoposide increased the percentage of PI+ cells, with the 32D IRS1 cells showing a larger percentage (40.9% PI+) than 32D (15.5%), 32D-IRS2 (17.1%), or 32D IRS1IRS2 (20.7%). Concomitant treatment with Q-VD suppressed the etoposide-induced uptake of PI in all cases. These results demonstrate that etoposide induced death is caspase-dependent and confirms that 32D cells expressing IRS1 are more sensitive to etoposide.
IRS1 expression does not alter the levels of pro- or anti-apoptotic proteins
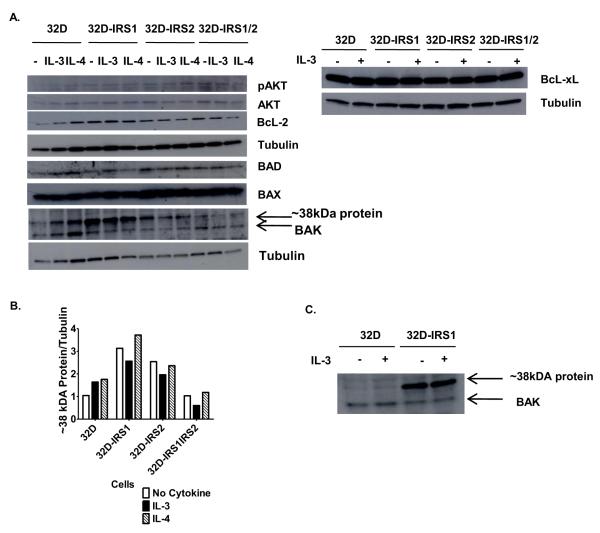
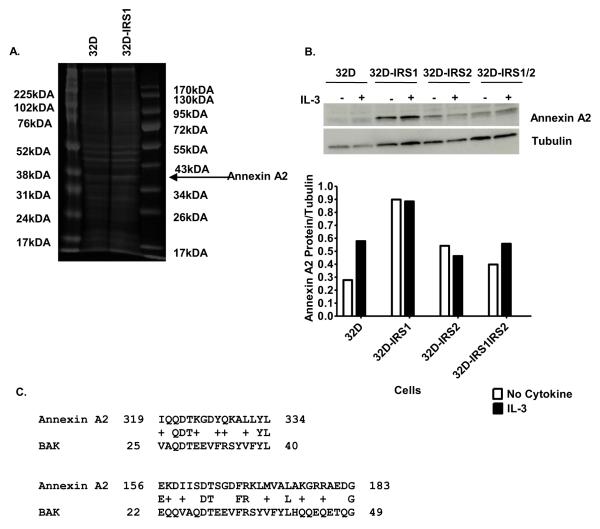
Since etoposide-induced cell death was caspase-mediated, we next tested whether IRS1 expression would alter the levels of various pro- or anti-apoptotic proteins. 32D cells and transfectants were treated in the presence or absence of IL-3, supplied as WEHI 3B supernatant, or with IL-4 for 30 minutes and then lysed. Western blotting and densitometric analyses showed equal levels of phosphorylated AKT (pAKT), AKT, BCL-xL, BAD, BAX, and BAK in the cell lines in the presence or absence of cytokines (Figure 4A and unpublished observations). No pBAD was detected. We noted that, BCL-2 levels were increased in all of the transfected cell lines as compared to 32D parental cells (Bcl2/tubulin ratio = 0.32, 0.82, 0.94, 0.91 for parental, IRS1-, IRS2-, and IRS1,IRS2-expressing 32D, respectively). This increase was not specific to 32D cells expressing IRS1 and therefore cannot explain the differential response observed when only IRS1 is expressed. Interestingly, the anti BAK antibody detected a ~38kDa protein that was expressed at higher levels in 32D cells expressing IRS1 (Figure 4A-C).
Figure 4. IRS1 expression does not alter the levels of pro and anti-apoptotic proteins.
A. 32D cells lacking or expressing IRS1 and/or IRS2 were serum starved and treated in the presence or absence of IL-3, supplied as WEHI-3B supernatant which contains 0.75% serum, or IL-4 (10 ng/ml) for 30 minutes. Cell lysates were prepared and the western blot was probed with anti-pAKT, anti-AKT, anti-BCL-2, anti-BCL-xL, anti-BAD, anti-BAD, anti BAX, anti-BAK and anti-tubulin antibodies. Blots were stripped and re probed no more than once. B. Densitometry was used to quantify bands. C. Lysates from 32D cells lacking or expressing IRS1 treated in the presence or absence of IL-3 were probed with anti BAK antibody.
Annexin A2 is increased in 32D cells expressing IRS1
In order to identify the 38 kDa protein, lysates were prepared from 32D and 32D-IRS1 cells and separated by SDS-PAGE. The proteins were stained and visualized with Imperial™ protein stain overnight and then destained in distilled water (Figure 5A). Bands that showed a change in staining intensity compared to the control lysate were excised for further analysis by mass spectrometry. Digests of proteins found in the excised 38 kDa band isolated from the 32D-IRS1 sample contained 16 unique peptides that correlated to the Annexin A2 amino acid sequence covering 44% of the amino acid sequence. These peptides were not found in the digests of proteins obtained from the excised band isolated from the 32D control sample. The identity of this protein was confirmed by western blotting with the use of an Annexin A2 specific antibody. Expression of Annexin A2 was greater in 32D cells expressing IRS1 (Figure 5B). BAK is a multi-domain BCL-2 family member that creates pores in the mitochondrial membrane resulting in cytochrome c release [44]. To better understand why the anti-BAK antibody detected Annexin A2, a protein sequence alignment was performed. The sequence alignment demonstrates that there are 2 regions of mouse BAK and mouse Annexin A2 that are very similar. Amino acids 156-183 of Annexin A2 are 25% identical and 43% similar to amino acids 22-49 of BAK. Amino acids 319-334 of Annexin A2 are 31% identical and 63% similar to amino acids 25-40 of BAK (Figure 5C). This similarity is seen in human BAK and human Annexin A2 as well (unpublished observations).
Figure 5. 32D-IRS1 cells express higher levels of Annexin A2.
A. 32D cells lacking or expressing IRS1 were treated with IL-3 for 30 minutes. Cell lysates were prepared and loaded on a gel, separated by SDS-PAGE, and stained with Imperial™ protein stain overnight. The region of the gel corresponding to ~38kDa was excised for both samples. Proteins were eluted and analyzed by tandem mass spectrometry. Peptide representation was compared between the 2 samples. B. 32D cells lacking or expressing IRS1 and/or IRS2 were treated in the presence or absence of IL-3, supplied as 5% WEHI-3B supernatant which contains 0.75% serum. Cell lysates were prepared and the western blot was probed with anti Annexin A2. Densitometry was used to quantify bands. C. Amino acid sequences for mouse Annexin A2 (NP_031611.1) and mouse BAK (CAA73684.1) were aligned using the NCBI site. Annexin A2 contains 339 amino acids and BAK contains 208 amino acids.
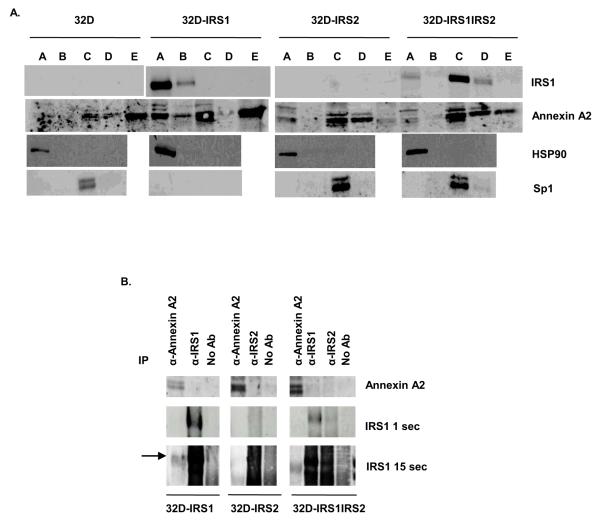
Annexin A2 and IRS1 are both found in the cytoplasmic and membrane fractions of 32D IRS1 cells
Both Annexin A2 and IRS1 have been observed at the membrane and in the nucleus of cells [5, 45]. In order to determine the subcellular location of Annexin A2 and IRS1, we performed subcellular fractionation. Soluble extracts of the fractions were separated by SDS-PAGE. Western blotting of 32D-IRS1 fractions showed that IRS1 was predominantly located in the cytoplasmic fraction with some protein also present in the membrane fraction (Figure 6A). Annexin A2 was mainly present in the cytoplasmic, soluble nuclear, and cytoskeletal fractions, with lower levels in the membrane and chromatin-bound nuclear fractions. Thus, both Annexin A2 and IRS1 were located in the cytoplasmic and membrane fractions. In cells expressing both IRS1 and IRS2, IRS1 and Annexin A2 localized to different fractions of the cell. IRS1 was mainly expressed in the soluble nuclear fraction and weakly expressed in the cytoplasmic and chromatin bound nuclear fractions. Annexin A2 was mainly expressed in the soluble nuclear, chromatin bound nuclear, and cytoskeletal fractions, and weakly expressed in the cytoplasmic fraction. This suggests that expression of IRS2 altered the localization of IRS1. Control blots demonstrated that HSP90 was predominantly located in the cytoplasmic fraction of all cells as expected, while Sp1 was limited to the soluble nuclear fraction. We noted that 32D-IRS1 cells lacked expression of Sp1. Importantly, none of the nuclear fractions were positive for HSP90.
Figure 6. Annexin A2 and IRS1 are expressed in overlapping subcellular compartments and weakly co precipitate in 32D-IRS1 cells.
A. 32D, 32D-IRS1, and 32D-IRS1IRS2 cells were lysed using a subcellular fractionation kit. The subcellular fractions were prepared and analyzed by western blotting with anti-IRS1 and anti-Annexin A2 antibodies. A= cytoplasmic extract, B= membrane extract, C= soluble nuclear extract, D= chromatin bound nuclear extract, E= cytoskeletal extract. The images for 32D cells and 32D-IRS1 cells were derived from the same exposure of the same film. Antibodies to HSP90 and Sp1 were used as positive controls for the cytoplasmic and soluble nuclear fraction, respectively. B. 32D cells expressing IRS1, IRS2, or both were treated with IL-3 for 30 minutes. Cell lysates were prepared and immunoprecipitated with anti-IRS1, anti IRS2, or anti-Annexin A2. Lysates were loaded on a gel and separated by SDS-PAGE. Western blots were probed with anti-Annexin A2 or anti IRS1 antibodies. Several exposures of anti-IRS1 blot are shown. The images for each cell line were derived from the same exposure of the same film.
Annexin A2 and IRS1 weakly interact in 32D-IRS1 cells
Because we found that IRS1 and Annexin A2 localize to similar fractions of the cell we wanted to determine if Annexin A2 interacts with IRS1. We performed immunoprecipitations with anti-Annexin A2 and anti-IRS1 antibodies followed by western blotting. We found that low amounts of IRS1 coprecipitated with anti-Annexin A2 in 32D-IRS1 cells, but not in 32D-IRS1IRS2 cells suggesting a weak interaction between Annexin A2 and IRS1 (Figure 6B). No interaction was detected between Annexin A2 and IRS2 in 32D-IRS2 cells (unpublished observations). Although we did not detect Annexin A2 in the anti IRS1 precipitates, it is possible that the anti Annexin A2 antibody binds a site that is concealed during the Annexin A2 and IRS1 interaction.
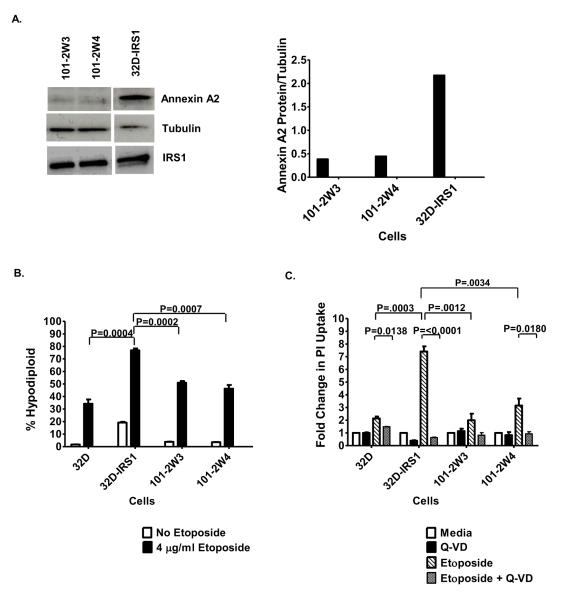
Decreasing Annexin A2 reduces 32D-IRS1 cell sensitivity to chemotherapy
To determine if the ability of IRS1 to sensitize 32D cells to etoposide was mediated by Annexin A2, we used shRNA to decrease Annexin A2 levels in 32D-IRS1 cells. Two independent cell lines were generated (101-2W3 and 101-2W4). We verified the expression levels of Annexin A2 and IRS1 by western blotting and densitometry. While the Annexin A2 levels were decreased in the transduced clones, the IRS1 levels were not affected (Figure 7A). Using both the nuclear DNA-content assay (Figure 7B) and the PI uptake assay (Figure 7C), we found that 32D-IRS1 cells were significantly less sensitive to cell death induced by etoposide when Annexin A2 levels were decreased. The cell death was more similar to what was obtained for 32D cells. Similar results were obtained when cells were treated with vincristine (unpublished observations). In all cases, treating the cells with Q-VD suppressed the etoposide-induced cell death. Together these results suggest that the ability of IRS1 to sensitize cells to chemotherapy-induced death is, at least partially, mediated by Annexin A2.
Figure 7. Decreasing Annexin A2 reduces 32D-IRS1 cell sensitivity to chemotherapy.
A. 32D-IRS1 (260.3) cells were infected with Annexin A2 shRNA and two cell lines were generated (101-2W3 and 101-2W4). Cell lysates were prepared and the western blot was probed with anti-Annexin A2, anti-IRS1 or anti-Tubulin. The images for each cell type were derived from the same exposure of the same film. Densitometry was used to quantify bands. B. 32D cells and transfectants were treated with 4 g/ml etoposide, in the presence of IL-3, supplied as 5% WEHI-3B supernatant which contains 0.75% serum, for 24 hours. The percentage of hypodiploid cells was measured by propidium iodide staining of nuclear DNA content followed by FACS analysis. The percentage of sub-diploid cells is shown. Two-tailed, unpaired, t-test was used to determine statistical significance. C. 32D cells and transfectants were pretreated in the presence or absence of 10 μM Q-VD-OPh for 1 hr and then treated in the presence or absence of 4 g/ml etoposide, for 20 hrs. Cells were analyzed by FACS to determine the percentage of PI positive cells. The percentage of PI positive cells was expressed as fold change relative to media control. Two-tailed, unpaired, t-test was used to determine statistical significance.
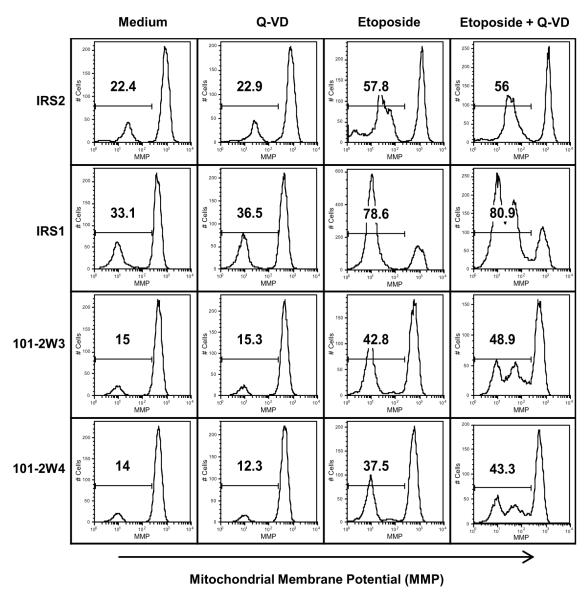
IRS1 and Annexin A2 influence the etoposide-induced loss of mitochondrial membrane potential
Previous studies suggested that chemotherapeutic drugs induce apoptosis by disrupting mitochondrial membrane potential [32, 44, 46]. In order to determine whether expression of IRS1 and Annexin A2 influenced drug-induced changes in mitochondrial membrane potential (MMP), transfectant cells were labeled with tetramethylrhodamine (TMR), a fluorescent probe that binds mitochondrial membranes in the presence of high mitochondrial membrane potential. The cells were then incubated in the presence or absence of etoposide and Q-VD. In the presence of WEHI-3B supernatant only, the percentage of cells with reduced mitochondrial membrane potential (TMR-low) ranged from ~14-33.1%, with the 32D-IRS1 cells showing the highest background (Figure 8). The addition of Q-VD made a negligible impact on this percentage. Addition of etoposide increased the percentage of TMR-low cells, with the 32D IRS1 cells showing a greater percentage (78.6%) than 32D IRS2 (57.8%). 32D-IRS1 cells with reduced expression of Annexin A2, 101 2W3 and 101 2W4, demonstrated reduced percentages of TMR low cells (42.8 and 37.5%, respectively) in response to etoposide. Concomitant treatment with Q-VD had little effect on the overall percentage of TMR low cells observed after etoposide treatment. However, all 3 IRS1 expressing cells (32D-IRS1, 101-2W3, and 101-2W4) demonstrated 2 peaks in the TMR-low fraction. These results suggest that IRS1 expression sensitizes cells to loss of mitochondrial membrane potential upstream of caspase activation and that this is in part dependent on Annexin A2.
Figure 8. Decreasing Annexin A2 reduces 32D-IRS1 loss of mitochondrial membrane potential.
32D cells that express IRS2 or IRS1 or that have reduced Annexin A2 levels were pretreated in the presence or absence of 10 μM Q-VD-OPh for 1 hr and then treated in the presence or absence of 4 g/ml etoposide, for 20 hrs. To measure mitochondrial membrane potential (MMP) cells were labeled with 80 nM tetramethylrhodamine (TMR) for 30 mins. FACS analysis was used to measure the percentage of TMR low cells. Representative histograms from FACs analysis are shown.
Discussion
Previous studies from our laboratory have implicated both IRS1 and IRS2 in protection from factor-withdrawal-induced apoptosis [3]. However, analysis in cancer models found that IRS1 and IRS2 have differential roles in cellular proliferation, growth, motility, metastasis, and survival [4, 6, 7, 15, 33-36]. The role of IRS1 and IRS2 in protecting cells from chemotherapy-induced cell death was previously unknown. In this study, we utilized the well described 32D model system to express IRS proteins [3, 34, 37-40]. We found that IRS1 expression enhanced the sensitivity of 32D cells to chemotherapy-induced death, while IRS2 expression did not. When examining the effects of the expression of both IRS1 and IRS2 on chemotherapy-induced cell death, we found that 32D cells expressing both IRS1 and IRS2 were less sensitive to chemotherapy-induced death suggesting that the presence of IRS2 abrogates the IRS1 effect.
In order to evaluate sensitivity to chemotherapy without inducing IL-3 withdrawal, our experiments were performed in the presence of 5% WEHI-3B supernatant, the standard growth supplement for 32D cells. Based on previously published studies, we maintained low serum (0.75%) conditions during the chemotherapy treatment protocols [9]. As shown in Figures 1 and 2, these conditions were sufficient to modestly induce tyrosine phosphorylation of IRS1 although not to the same level as IL-4. Interestingly, in some experiments we found that 32D-IRS1 cells had a higher basal level of apoptosis as compared to the other cells suggesting that they were more dependent on other growth and survival factors present in serum. Indeed, we found that in the presence of 10% fetal bovine serum, the 32D-IRS1 cells became less sensitive to chemotherapy and responded similarly to the control and IRS2-expressing cells at the same drug concentrations used in the other experiments (unpublished observations). This resistance is likely due to the presence of higher amounts of factors found in serum including insulin and IGF-1which could activate cell survival signaling pathways dependent on the tyrosine phosphorylation of IRS1 and IRS2 [9].
In the absence of tyrosine phosphorylation, IRS1 interacts with the DNA repair protein RAD51 in the perinuclear region of the cell preventing RAD51 from localizing to the nucleus; RAD51 is not known to interact with IRS2 [9]. RAD51 is a key enzyme in homologous recombination-directed DNA repair (HRR). Proliferating cells repair their DNA by HRR or non-homologous end joining (NHEJ), while quiescent cells favor NHEJ [9]. Under low serum conditions, the 32D cells and transfectants were not proliferating (unpublished observations) and therefore may not depend on RAD51 for DNA repair. However, under high serum conditions, 32D cells proliferate rapidly and may utilize HRR to repair DNA in the presence of chemotherapy. Signaling via growth factors in serum can lead to the tyrosine phosphorylation of IRS1 thereby inhibiting its interaction with RAD51 and promoting DNA repair and cell survival in the presence of chemotherapeutic agents [9]. Thus, one possible explanation for our observations is that under low serum conditions high cytoplasmic IRS1 suppresses the nuclear translocation of DNA repair proteins such as Rad51. Other studies have shown that IRS1 is able to interact with proteins that induce its nuclear localization and interaction with Rad51. In medulloblastoma, proteins such as estrogen receptor beta and JC virus T-antigen have been shown to facilitate the IRS1 and RAD51 interaction in the nucleus. Furthermore, in this way JC virus T-antigen sensitized cancer cells to cisplatin [47, 48]. In our studies, co expression of IRS2 caused the localization of IRS1 to become primarily nuclear rather than cytoplasmic and reduced the IRS 1 mediated sensitivity to chemotherapy. Thus, we do not believe the nuclear interaction of IRS1 with Rad51 will explain our findings. The interaction between IRS1 and RAD51 in the cytoplasm and the nucleus will be explored in future studies.
In addition to strongly activating anti-apoptotic pathways, growth factors have been shown to alter the ability of IRS1 to interact with other proteins. Serine phosphorylation of IRS1 prevented its ability to bind to the p85 subunit of PI3K and SHP-2 [4, 15, 49]. In cells expressing both IRS1 and IRS2, IRS2-dependent signaling resulted in the serine phosphorylation of IRS1, and thus inactivation of IRS1 creating a negative feedback loop [4]. This type of regulation could explain why expressing both IRS1 and IRS2 in 32D cells de sensitizes the cells to chemotherapy induced cell death. It would be informative in the future to analyze the phosphorylation status of IRS1 using proteomic approaches and to determine whether serine phosphorylation induces its nuclear localization and inhibits the ability of IRS1 to sensitize cells to chemotherapy.
In testing whether IRS1 expression would alter the levels of pro-apoptotic or anti-apoptotic proteins, a ~38 kDa protein, later identified as Annexin A2, was detected by the anti BAK antibody and showed increased expression in 32D cells expressing IRS1. Similarly, using an anti Bim antibody, Liu et al. identified a ~36 kDa protein that was identified as Annexin A2 by mass spectrometry [50]. Bim is a BH3-only pro-apoptotic BCL-2 family member protein [44]. Bim interacts with other BCL-2 family members like the anti-apoptotic BCL-2 protein and other effector molecules. These data suggest that Annexin A2 shares similarity with multiple pro apoptotic BCL-2 family members [50].
Annexin A2 is a calcium-dependent phospholipid binding protein. It has an N-terminus that contains a nuclear export signal and multiple phosphorylation sites. In various cell-types Annexin A2 has been shown to play a role in exocytosis, endocytosis, lipid raft formation, cell cell adhesion, proliferation, cell surface fibrinolysis, angiogenesis, osteoclast formation and bone resorption [51-54]. Annexin A2 is also expressed in tumors, but at variable levels depending on the tumor type. Annexin A2 is increased in pancreatic, breast, brain, gastric, kidney, liver, and lung cancers; as well as acute promyelocytic leukemias. Conversely, Annexin A2 is decreased in esophageal cancer, and osteosarcomas. In prostate carcinomas, and head and neck carcinomas, Annexin A2 expression results are conflicting; some have reported Annexin A2 to be increased, while others have reported Annexin A2 to be decreased. In tumors, Annexin A2 has been shown to play diverse roles in cell proliferation, adhesion, invasion and metastasis, and apoptosis [50-52, 54, 55].
Our results showed that increased Annexin A2 correlated with increased IRS1 expression and that in 32D-IRS1 cells both Annexin A2 and IRS1 were located in the cytoplasmic and membrane fractions. In the cytoplasm IRS1 acts as a docking protein allowing it to possibly recruit Annexin A2 to the membrane, either directly or indirectly, in order to facilitate signaling that promotes apoptosis or inhibits cell survival. Expression of IRS2 changed the localization of IRS1 causing IRS1 to be mainly expressed in the nucleus, where Annexin A2 was also expressed. Because 32D-IRS1IRS2 cells were much less sensitive to chemotherapy induced death than 32D-IRS1 cells, we propose that the localization of either IRS1 and/or Annexin A2 in the cytoplasm and/or membrane of the cell is needed for the increased sensitivity to chemotherapy. Since Annexin A2 shares similarity with BAK and BIM, pro apoptotic BCL-2 family members, future studies should determine whether Annexin A2 similarly localizes to the mitochondria under certain conditions and also if Annexin A2 has some functions similar to the pro apoptotic BCL-2 family members.
We obtained some evidence that IRS1 and Annexin A2 may interact, although the impact of this interaction is currently unknown. This is an interesting finding because Ueno et al. showed that both IRS1 and IRS2 can interact with BCL-2 in the perinuclear region and regulate BCL-2 phosphorylation and anti-apoptotic function [56]. IRS1 can also interact with BCL XL, another anti-apoptotic protein, but it does not interact with the apoptotic proteins BAX or BIK. Moreover, although IRS-1, via its BCL-2 interaction, could protect Ba/F3 from IL-3 withdrawal, IRS1 could not protect the cells from apoptosis in the absence of serum [56]. More exhaustive analyses with various antibodies and buffer formats will be needed to further explore IRS1 interaction with Annexin A2 and BCL-2 family members.
Annexin A2 has multiple functions; one of its functions in the nucleus is to participate in a primer recognition protein complex that enhances DNA polymerase activity thus playing a role in DNA synthesis and cell proliferation [45]. IRS1 is also found in the nucleus interacting with several other proteins that aid in its translocation, including viral oncoproteins, the upstream binding factor 1 (UBF1), β-catenin, and hormone receptors. In addition, IRS1/β-catnein complexes were found bound to promoter sequences of multiple genes, including c-myc and cyclin D1, and shown to regulate their transcription [5, 9, 57-60] Thus, IRS1 has roles in gene transcription, nuclear localization, and DNA repair. It is possible that IRS1 and Annexin A2 could interact in the nucleus and together regulate transcription of genes that influence apoptosis.
Studies have reported that Annexin A3, Annexin A4, and Annexin A11 were implicated in chemoresistance [54]. Takano et al. suggested that Annexin A2 was associated with rapid recurrence of human pancreatic cancers and shorter overall survival of patients receiving adjuvant chemotherapy [61]. Chuthapisith et al. showed that human breast cancers with enhanced expression of Annexin A2, together with decreased expression of Annexin A1, had a poor response to neoadjuvant chemotherapy. Importantly those results also showed that neither Annexin A2 nor Annexin A1 alone was a good predictor of the response to this therapy [62]. Furthermore, our results showed that decreasing Annexin A2 made 32D-IRS1 cells less sensitive to chemotherapy with reduced lost of mitochondrial membrane potential suggesting a link between Annexin A2 and chemosensitivity. Annexin A2 can undergo post-translational modifications and also proteolysis which result in various forms of the protein [54]. This, as well as differences in cell types, and localization of Annexin A2 in the cell may explain the conflicting results. Future experiments should determine whether over-expressing Annexin A2 in 32D cells that lack IRS1 will be sufficient to sensitize the cells to chemotherapy or if IRS1 is necessary for this response.
These results, along with our other results described herein, suggest that high IRS1 expression, together with high Annexin A2 expression, could be a good indicator of favorable responses to chemotherapy. These studies also suggest that antagonizing IRS2 may provide a way to more efficiently inhibit cancer progression and to enhance the efficacy of chemotherapy in vivo. Such enhancement is desirable since current chemotherapy treatments for cancer patients have many side effects. Avoiding treatments that a patient will not respond to, and instead focusing on treatments that are more likely to result in a better response is critical.
Conclusions
In conclusion, our results show that IRS1 enhanced the sensitivity of 32D cells to chemotherapy induced caspase mediated cell death. This response is partially mediated by Annexin A2 and abrogated by expression of IRS2. Understanding the molecular programming of a cell may allow for the better prediction of a patient’s response to treatment.
Highlights.
IRS1 enhanced the sensitivity of 32D cells to chemotherapy induced apoptosis.
This sensitivity is abrogated by the expression of IRS2.
Expressing IRS1 in 32D cells increased levels of Annexin A2.
Both IRS1 and Annexin A2 were located in cytoplasmic and membrane fractions.
Decreasing Annexin A2 in 32D-IRS1 cells abated their sensitivity to chemotherapy.
Acknowledgments
We acknowledge Brian Hampton for his technical expertise with the tandem mass spectrometry in the CVID Protein Analysis Lab. This work was supported in part by the Transfusion Medicine Research Training Program T32 HL007698, NRSA NIH (F31) Predoctoral Diversity Fellowship Grant 1F31CA142165-01A1 (to H.A.P), NIGMS Initiative for Maximizing Student Development Grant R25-GM55036, Procter and Gamble, NIH Grant R01AI038985-12-17 (to A.D.K), NCI grant K22CA128882 (to G.B.C) and NCI grant UH2CA158689 (to G.B.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests: The authors declare that they have no competing interests.
References
- [1].Lee YH, White MF. Insulin receptor substrate proteins and diabetes. Arch Pharm Res. 2004;27:361–370. doi: 10.1007/BF02980074. [DOI] [PubMed] [Google Scholar]
- [2].Keegan AD, Zamorano J. Regulation of gene expression, growth, and cell survival by IL 4: contribution of multiple signaling pathways. Cell Res. 1998;8:1–13. doi: 10.1038/cr.1998.1. [DOI] [PubMed] [Google Scholar]
- [3].Zamorano J, Wang HY, Wang LM, Pierce JH, Keegan AD. IL 4 protects cells from apoptosis via the insulin receptor substrate pathway and a second independent signaling pathway. J Immunol. 1996;157:4926–4934. [PubMed] [Google Scholar]
- [4].Ma Z, Gibson SL, Byrne MA, Zhang J, White MF, Shaw LM. Suppression of insulin receptor substrate 1 (IRS 1) promotes mammary tumor metastasis. Mol Cell Biol. 2006;26:9338–9351. doi: 10.1128/MCB.01032-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mardilovich K, Pankratz SL, Shaw LM. Expression and function of the insulin receptor substrate proteins in cancer. Cell Commun Signal. 2009;7:14. doi: 10.1186/1478-811X-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Withers DJ, White M. Perspective: The insulin signaling system a common link in the pathogenesis of type 2 diabetes. Endocrinology. 2000;141:1917–1921. doi: 10.1210/endo.141.6.7584. [DOI] [PubMed] [Google Scholar]
- [7].White MF. The IRS signalling system: a network of docking proteins that mediate insulin action. Mol Cell Biochem. 1998;182:3–11. [PubMed] [Google Scholar]
- [8].Hotamisligil GS, Peraldi P, Budavari A, Ellis R, White MF, Spiegelman BM. IRS 1 mediated inhibition of insulin receptor tyrosine kinase activity in TNF alpha and obesity induced insulin resistance. Science. 1996;271:665–668. doi: 10.1126/science.271.5249.665. [DOI] [PubMed] [Google Scholar]
- [9].Trojanek J, Ho T, Del Valle L, Nowicki M, Wang JY, Lassak A, Peruzzi F, Khalili K, Skorski T, Reiss K. Role of the insulin like growth factor I/insulin receptor substrate 1 axis in Rad51 trafficking and DNA repair by homologous recombination. Mol Cell Biol. 2003;23:7510–7524. doi: 10.1128/MCB.23.21.7510-7524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yenush L, White MF. The IRS signalling system during insulin and cytokine action. Bioessays. 1997;19:491–500. doi: 10.1002/bies.950190608. [DOI] [PubMed] [Google Scholar]
- [11].Sun XJ, Rothenberg P, Kahn CR, Backer JM, Araki E, Wilden PA, Cahill DA, Goldstein BJ, White MF. Structure of the insulin receptor substrate IRS 1 defines a unique signal transduction protein. Nature. 1991;352:73–77. doi: 10.1038/352073a0. [DOI] [PubMed] [Google Scholar]
- [12].Araki E, Haag BL, 3rd, Kahn CR. Cloning of the mouse insulin receptor substrate 1 (IRS 1) gene and complete sequence of mouse IRS 1. Biochim Biophys Acta. 1994;1221:353–356. doi: 10.1016/0167-4889(94)90261-5. [DOI] [PubMed] [Google Scholar]
- [13].Sun XJ, Pons S, Wang LM, Zhang Y, Yenush L, Burks D, Myers MG, Jr., Glasheen E, Copeland NG, Jenkins NA, Pierce JH, White MF. The IRS 2 gene on murine chromosome 8 encodes a unique signaling adapter for insulin and cytokine action. Mol Endocrinol. 1997;11:251–262. doi: 10.1210/mend.11.2.9885. [DOI] [PubMed] [Google Scholar]
- [14].Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL 4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol. 1999;17:701–738. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- [15].Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- [16].Sun XJ, Pons S, Asano T, Myers MG, Jr., Glasheen E, White MF. The Fyn tyrosine kinase binds Irs 1 and forms a distinct signaling complex during insulin stimulation. J Biol Chem. 1996;271:10583–10587. doi: 10.1074/jbc.271.18.10583. [DOI] [PubMed] [Google Scholar]
- [17].Beitner Johnson D, Blakesley VA, Shen Orr Z, Jimenez M, Stannard B, Wang LM, Pierce J, LeRoith D. The proto oncogene product c Crk associates with insulin receptor substrate 1 and 4PS. Modulation by insulin growth factor I (IGF) and enhanced IGF I signaling. J Biol Chem. 1996;271:9287–9290. doi: 10.1074/jbc.271.16.9287. [DOI] [PubMed] [Google Scholar]
- [18].Lee CH, Li W, Nishimura R, Zhou M, Batzer AG, Myers MG, Jr., White MF, Schlessinger J, Skolnik EY. Nck associates with the SH2 domain docking protein IRS 1 in insulin stimulated cells. Proc Natl Acad Sci U S A. 1993;90:11713–11717. doi: 10.1073/pnas.90.24.11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Saltiel AR, Pessin JE. Insulin signaling pathways in time and space. Trends Cell Biol. 2002;12:65–71. doi: 10.1016/s0962-8924(01)02207-3. [DOI] [PubMed] [Google Scholar]
- [20].Li Y, Soos TJ, Li X, Wu J, DeGennaro M, Sun X, Littman DR, Birnbaum MJ, Polakiewicz RD. Protein Kinase CÎ Inhibits Insulin Signaling by Phosphorylating IRS1 at Ser1101. Journal of Biological Chemistry. 2004;279:45304–45307. doi: 10.1074/jbc.C400186200. [DOI] [PubMed] [Google Scholar]
- [21].Ozes ON, Akca H, Mayo LD, Gustin JA, Maehama T, Dixon JE, Donner DB. A phosphatidylinositol 3 kinase/Akt/mTOR pathway mediates and PTEN antagonizes tumor necrosis factor inhibition of insulin signaling through insulin receptor substrate 1. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4640–4645. doi: 10.1073/pnas.051042298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gao Z, Hwang D, Bataille F, Lefevre M, York D, Quon MJ, Ye J. Serine phosphorylation of insulin receptor substrate 1 by inhibitor kappa B kinase complex. J Biol Chem. 2002;277:48115–48121. doi: 10.1074/jbc.M209459200. [DOI] [PubMed] [Google Scholar]
- [23].Rui L, Fisher TL, Thomas J, White MF. Regulation of insulin/insulin like growth factor 1 signaling by proteasome mediated degradation of insulin receptor substrate 2. J Biol Chem. 2001;276:40362–40367. doi: 10.1074/jbc.M105332200. [DOI] [PubMed] [Google Scholar]
- [24].White MF. Regulating insulin signaling and beta cell function through IRS proteins. Can J Physiol Pharmacol. 2006;84:725–737. doi: 10.1139/y06-008. [DOI] [PubMed] [Google Scholar]
- [25].Sun XJ, Goldberg JL, Qiao LY, Mitchell JJ. Insulin induced insulin receptor substrate 1 degradation is mediated by the proteasome degradation pathway. Diabetes. 1999;48:1359–1364. doi: 10.2337/diabetes.48.7.1359. [DOI] [PubMed] [Google Scholar]
- [26].Reiss K, Wang JY, Romano G, Tu X, Peruzzi F, Baserga R. Mechanisms of regulation of cell adhesion and motility by insulin receptor substrate 1 in prostate cancer cells. Oncogene. 2001;20:490–500. doi: 10.1038/sj.onc.1204112. [DOI] [PubMed] [Google Scholar]
- [27].Beere HM, Chresta CM, Alejo Herberg A, Skladanowski A, Dive C, Larsen AK, Hickman JA. Investigation of the mechanism of higher order chromatin fragmentation observed in drug induced apoptosis. Mol Pharmacol. 1995;47:986–996. [PubMed] [Google Scholar]
- [28].Wang MY, Chen PS, Prakash E, Hsu HC, Huang HY, Lin MT, Chang KJ, Kuo ML. Connective tissue growth factor confers drug resistance in breast cancer through concomitant up regulation of Bcl xL and cIAP1. Cancer Res. 2009;69:3482–3491. doi: 10.1158/0008-5472.CAN-08-2524. [DOI] [PubMed] [Google Scholar]
- [29].Honma K, Iwao Koizumi K, Takeshita F, Yamamoto Y, Yoshida T, Nishio K, Nagahara S, Kato K, Ochiya T. RPN2 gene confers docetaxel resistance in breast cancer. Nat Med. 2008;14:939–948. doi: 10.1038/nm.1858. [DOI] [PubMed] [Google Scholar]
- [30].Wang TH, Popp DM, Wang HS, Saitoh M, Mural JG, Henley DC, Ichijo H, Wimalasena J. Microtubule dysfunction induced by paclitaxel initiates apoptosis through both c Jun N terminal kinase (JNK) dependent and independent pathways in ovarian cancer cells. J Biol Chem. 1999;274:8208–8216. doi: 10.1074/jbc.274.12.8208. [DOI] [PubMed] [Google Scholar]
- [31].Zhou J, Giannakakou P. Targeting microtubules for cancer chemotherapy. Curr Med Chem Anticancer Agents. 2005;5:65–71. doi: 10.2174/1568011053352569. [DOI] [PubMed] [Google Scholar]
- [32].Makin G, Hickman JA. Apoptosis and cancer chemotherapy. Cell Tissue Res. 2000;301:143–152. doi: 10.1007/s004419900160. [DOI] [PubMed] [Google Scholar]
- [33].Bruning JC, Winnay J, Cheatham B, Kahn CR. Differential signaling by insulin receptor substrate 1 (IRS 1) and IRS 2 in IRS 1 deficient cells. Mol Cell Biol. 1997;17:1513–1521. doi: 10.1128/mcb.17.3.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zamorano J, Wang HY, Wang R, Shi Y, Longmore GD, Keegan AD. Regulation of cell growth by IL 2: role of STAT5 in protection from apoptosis but not in cell cycle progression. J Immunol. 1998;160:3502–3512. [PubMed] [Google Scholar]
- [35].Byron SA, Horwitz KB, Richer JK, Lange CA, Zhang X, Yee D. Insulin receptor substrates mediate distinct biological responses to insulin like growth factor receptor activation in breast cancer cells. Br J Cancer. 2006;95:1220–1228. doi: 10.1038/sj.bjc.6603354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shi B, Sepp Lorenzino L, Prisco M, Linsley P, deAngelis T, Baserga R. Micro RNA 145 targets the insulin receptor substrate 1 and inhibits the growth of colon cancer cells. J Biol Chem. 2007;282:32582–32590. doi: 10.1074/jbc.M702806200. [DOI] [PubMed] [Google Scholar]
- [37].Wang LM, Myers MG, Jr., Sun XJ, Aaronson SA, White M, Pierce JH. IRS 1: essential for insulin and IL 4 stimulated mitogenesis in hematopoietic cells. Science. 1993;261:1591–1594. doi: 10.1126/science.8372354. [DOI] [PubMed] [Google Scholar]
- [38].Pierce JH, Ruggiero M, Fleming TP, Di Fiore PP, Greenberger JS, Varticovski L, Schlessinger J, Rovera G, Aaronson SA. Signal transduction through the EGF receptor transfected in IL 3 dependent hematopoietic cells. Science. 1988;239:628–631. doi: 10.1126/science.3257584. [DOI] [PubMed] [Google Scholar]
- [39].Di Fiore PP, Segatto O, Taylor WG, Aaronson SA, Pierce JH. EGF receptor and erbB 2 tyrosine kinase domains confer cell specificity for mitogenic signaling. Science. 1990;248:79–83. doi: 10.1126/science.2181668. [DOI] [PubMed] [Google Scholar]
- [40].Sun H, Baserga R. Deletion of the pleckstrin and phosphotyrosine binding domains of insulin receptor substrate 2 does not impair its ability to regulate cell proliferation in myeloid cells. Endocrinology. 2004;145:5332–5343. doi: 10.1210/en.2004-0668. [DOI] [PubMed] [Google Scholar]
- [41].Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- [42].Davidson WF, Haudenschild C, Kwon J, Williams MS. T cell receptor ligation triggers novel nonapoptotic cell death pathways that are Fas independent or Fas dependent. J Immunol. 2002;169:6218–6230. doi: 10.4049/jimmunol.169.11.6218. [DOI] [PubMed] [Google Scholar]
- [43].Carey GB, Semenova E, Qi X, Keegan AD. IL 4 protects the B cell lymphoma cell line CH31 from anti IgM induced growth arrest and apoptosis: contribution of the PI 3 kinase/AKT pathway. Cell Res. 2007;17:942–955. doi: 10.1038/sj.cr.2007.90. [DOI] [PubMed] [Google Scholar]
- [44].Chipuk JE, Green DR. How do BCL 2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 2008;18:157–164. doi: 10.1016/j.tcb.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Liu J, Vishwanatha JK. Regulation of nucleo cytoplasmic shuttling of human annexin A2: a proposed mechanism. Mol Cell Biochem. 2007;303:211–220. doi: 10.1007/s11010-007-9477-7. [DOI] [PubMed] [Google Scholar]
- [46].Bouillet P, Strasser A. BH3 only proteins evolutionarily conserved proapoptotic Bcl 2 family members essential for initiating programmed cell death. J Cell Sci. 2002;115:1567–1574. doi: 10.1242/jcs.115.8.1567. [DOI] [PubMed] [Google Scholar]
- [47].Trojanek J, Croul S, Ho T, Wang JY, Darbinyan A, Nowicki M, Del Valle L, Skorski T, Khalili K, Reiss K. T antigen of the human polyomavirus JC attenuates faithful DNA repair by forcing nuclear interaction between IRS 1 and Rad51. J Cell Physiol. 2006;206:35–46. doi: 10.1002/jcp.20425. [DOI] [PubMed] [Google Scholar]
- [48].Urbanska K, Pannizzo P, Lassak A, Gualco E, Surmacz E, Croul S, Del Valle L, Khalili K, Reiss K. Estrogen receptor beta mediated nuclear interaction between IRS 1 and Rad51 inhibits homologous recombination directed DNA repair in medulloblastoma. J Cell Physiol. 2009;219:392–401. doi: 10.1002/jcp.21683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Gibson SL, Ma Z, Shaw LM. Divergent roles for IRS 1 and IRS 2 in breast cancer metastasis. Cell Cycle. 2007;6:631–637. doi: 10.4161/cc.6.6.3987. [DOI] [PubMed] [Google Scholar]
- [50].Liu JW, Shen JJ, Tanzillo Swarts A, Bhatia B, Maldonado CM, Person MD, Lau SS, Tang DG. Annexin II expression is reduced or lost in prostate cancer cells and its re expression inhibits prostate cancer cell migration. Oncogene. 2003;22:1475–1485. doi: 10.1038/sj.onc.1206196. [DOI] [PubMed] [Google Scholar]
- [51].Bao H, Jiang M, Zhu M, Sheng F, Ruan J, Ruan C. Overexpression of Annexin II affects the proliferation, apoptosis, invasion and production of proangiogenic factors in multiple myeloma. Int J Hematol. 2009;90:177–185. doi: 10.1007/s12185-009-0356-8. [DOI] [PubMed] [Google Scholar]
- [52].Wang CY, Lin YS, Su WC, Chen CL, Lin CF. Glycogen synthase kinase 3 and Omi/HtrA2 induce annexin A2 cleavage followed by cell cycle inhibition and apoptosis. Mol Biol Cell. 2009;20:4153–4161. doi: 10.1091/mbc.E09-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sharma MC, Sharma M. The role of annexin II in angiogenesis and tumor progression: a potential therapeutic target. Curr Pharm Des. 2007;13:3568–3575. doi: 10.2174/138161207782794167. [DOI] [PubMed] [Google Scholar]
- [54].Lokman NA, Ween MP, Oehler MK, Ricciardelli C. The role of annexin A2 in tumorigenesis and cancer progression. Cancer Microenviron. 2011;4:199–208. doi: 10.1007/s12307-011-0064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mohammad HS, Kurokohchi K, Yoneyama H, Tokuda M, Morishita A, Jian G, Shi L, Murota M, Tani J, Kato K, Miyoshi H, Deguchi A, Himoto T, Usuki H, Wakabayashi H, Izuishi K, Suzuki Y, Iwama H, Deguchi K, Uchida N, Sabet EA, Arafa UA, Hassan AT, El Sayed AA, Masaki T. Annexin A2 expression and phosphorylation are up regulated in hepatocellular carcinoma. Int J Oncol. 2008;33:1157–1163. [PubMed] [Google Scholar]
- [56].Ueno H, Kondo E, Yamamoto Honda R, Tobe K, Nakamoto T, Sasaki K, Mitani K, Furusaka A, Tanaka T, Tsujimoto Y, Kadowaki T, Hirai H. Association of insulin receptor substrate proteins with Bcl 2 and their effects on its phosphorylation and antiapoptotic function. Mol Biol Cell. 2000;11:735–746. doi: 10.1091/mbc.11.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chen J, Wu A, Sun H, Drakas R, Garofalo C, Cascio S, Surmacz E, Baserga R. Functional significance of type 1 insulin like growth factor mediated nuclear translocation of the insulin receptor substrate 1 and beta catenin. J Biol Chem. 2005;280:29912–29920. doi: 10.1074/jbc.M504516200. [DOI] [PubMed] [Google Scholar]
- [58].Sun H, Tu X, Prisco M, Wu A, Casiburi I, Baserga R. Insulin like growth factor I receptor signaling and nuclear translocation of insulin receptor substrates 1 and 2. Mol Endocrinol. 2003;17:472–486. doi: 10.1210/me.2002-0276. [DOI] [PubMed] [Google Scholar]
- [59].Tu X, Batta P, Innocent N, Prisco M, Casaburi I, Belletti B, Baserga R. Nuclear translocation of insulin receptor substrate 1 by oncogenes and Igf I. Effect on ribosomal RNA synthesis. J Biol Chem. 2002;277:44357–44365. doi: 10.1074/jbc.M208001200. [DOI] [PubMed] [Google Scholar]
- [60].Wu A, Chen J, Baserga R. Nuclear insulin receptor substrate 1 activates promoters of cell cycle progression genes. Oncogene. 2008;27:397–403. doi: 10.1038/sj.onc.1210636. [DOI] [PubMed] [Google Scholar]
- [61].Takano S, Togawa A, Yoshitomi H, Shida T, Kimura F, Shimizu H, Yoshidome H, Ohtsuka M, Kato A, Tomonaga T, Nomura F, Miyazaki M. Annexin II overexpression predicts rapid recurrence after surgery in pancreatic cancer patients undergoing gemcitabine adjuvant chemotherapy. Ann Surg Oncol. 2008;15:3157–3168. doi: 10.1245/s10434-008-0061-5. [DOI] [PubMed] [Google Scholar]
- [62].Chuthapisith S, Bean BE, Cowley G, Eremin JM, Samphao S, Layfield R, Kerr ID, Wiseman J, El Sheemy M, Sreenivasan T, Eremin O. Annexins in human breast cancer: Possible predictors of pathological response to neoadjuvant chemotherapy. Eur J Cancer. 2009;45:1274–1281. doi: 10.1016/j.ejca.2008.12.026. [DOI] [PubMed] [Google Scholar]