Abstract
DNA methylation is an epigenetic modification that is essential for the development and mature function of the central nervous system. Due to the relevance of this modification in the transcriptional control of gene expression, it is often necessary to examine changes in DNA methylation patterns with both gene and single nucleotide resolution. Here, we describe in depth a basic protocol for direct bisulfite sequencing of DNA isolated from brain tissue, which will permit direct assessment of methylation status at individual genes as well as individual cytosines within a genomic region. This method yields analysis of DNA methylation patterns that are robust, accurate, and reproducible, thereby allowing insights into the role of alterations in DNA methylation in brain tissue.
Keywords: Bisulfite sequencing, DNA methylation, epigenetics, central nervous system
Introduction
Methylation of cytosine nucleotides in DNA is a powerful epigenetic modification that dramatically influences the transcriptional potential of individual genes. This modification, which is conserved across vertebrate and invertebrate species, occurs predominantly at cytosine-guanine dinucleotides (CpGs) within the genome, and involves the addition of a methyl group to the 5′ carbon atom in cytosine’s pyrimidine ring (Holliday and Pugh, 1975). The result is a carbon-carbon bond which is incredibly strong and requires a prohibitive amount of energy to be broken enzymatically (Wolffe et al., 1999). Methylation of cytosines in DNA is catalyzed by DNA methyltransferases (DNMTs), which use S-adenosyl methionine as the methyl donor (Klose and Bird, 2006). Interestingly, there are two types of DNMTs – those that methylate DNA de novo, and those that preferentially methylate DNA that is already methylated on one strand (and are therefore termed maintenance DNMTs). Thus, even when methylation of one DNA strand is lost, enzymatic reactions are in place to recognize and re-methylate the corresponding DNA strand (Day and Sweatt, 2010). The result is that DNA methylation can be an incredibly stable and long-lasting molecular modification. In vertebrate systems, cytosine methylation generally occurs at cytosine-guanine dinucleotides (so-called “CpG” sites), which are underrepresented in the genome, but tend to cluster around the promoter region of around 60% of genes (Jaenisch and Bird, 2003). Methylation at these clusters, termed CpG islands, has long been appreciated for its role as a master regulator of gene transcription. Thus, cytosine methylation tends to be at low levels in actively transcribed genes, and occurs at higher levels in gene that are repressed.
Due to its role as a potent modulator of gene transcription, DNA methylation is a critical component of cellular and organismal development. In fact, it is via changes in DNA methylation that individual cells establish and maintain gene programs that ultimately determine cell fate and perpetuate a given cellular phenotype across the lifespan and across cell division. Additionally, recent studies have led to an appreciation for a role of DNA methylation in both dividing and non-dividing cells within the central nervous system (Miller and Sweatt, 2007; Lubin et al., 2008; Ma et al., 2009b; Feng et al., 2010; Ma et al., 2010; Miller et al., 2010). DNA methylation status in the brain is dynamically altered in response to a number of different forms of behavioral experience, and this phenomenon generally occurs at genes that are known to control neuronal plasticity (Weaver et al., 2004; Weaver et al., 2005; Miller and Sweatt, 2007; Lubin et al., 2008; Roth et al., 2009; Gupta et al., 2010; Miller et al., 2010; Penner et al., 2010; Guo et al., 2011c). Although DNA methylation was once thought to be almost irreversible once established in non-dividing cells, it is now clear from these studies that methylation of cytosines in DNA can both increase and decrease in response to neuronal activity. Critically, a series of recent studies have identified a demethylation pathway that may contribute to active decreases in methylation in the adult brain (Ma et al., 2009a; Guo et al., 2011a, 2011b). Thus, far from being a static process, DNA methylation patterns are highly dynamic in response to environmental stimuli within the central nervous system.
Given the importance of DNA methylation for a number of cellular processes, it is not surprising that a number of different techniques have been developed over the last 10 years to examine methylation levels in a given tissue. The most basic of these examine changes in DNA methylation at a global level, permitting analysis of overall methylation differences between unique types of tissue or between experimental and control samples. However, as noted above, DNA methylation changes in the brain (as well as other organs) appear to occur at specific gene targets, meaning that methods that can only assay genome-wide methylation changes are inherently limited. Thus, additional techniques have been developed to examine DNA methylation changes at specific gene targets. One example is methylation dependent DNA immunoprecipitation (MeDIP), a technique that involves incubation of DNA with antibodies that recognize methylated cytosines followed by gene-specific PCR amplification (Weber et al., 2005). This technique has been used successfully with brain tissue in a number of studies (Miller et al., 2010; Vucetic et al., 2010; Vucetic et al., 2011), and is useful for interrogating altered DNA methylation within the amplified DNA region. However, it is increasingly being understood that DNA methylation changes at individual CpG sites can have robust consequences for the transcription of certain genes (Lister and Ecker, 2009; Lister et al., 2009). Indeed, studies with brain tissue have reported that DNA methylation changes following a behavioral manipulation can be incredibly site-specific even within relatively short stretches of a single gene (Weaver et al., 2004; Lubin et al., 2008; Roth et al., 2009; Gupta et al., 2010; Guo et al., 2011c). These observations have led to the suggestion that DNA methylation patterns may constitute a kind of methylation “code” in which individual modifications interact to form a complex methylation landscape that interact with other epigenetic and non-epigenetic factors to regulate gene transcription (Turner, 2007; Day and Sweatt, 2011).
Based on the CpG-specific nature of DNA methylation, sequencing-based methods are increasingly being used to probe how this modification changes following experience (Harris et al., 2010; Guo et al., 2011c). In some cases, these techniques expand upon existing methodologies involving immunoprecipitation or methylation-selective DNA restriction enzymes, and are amenable to gene-targeted approaches as well as massively parallel next-generation sequencing. Another approach, which we will detail here, involves bisulfite treatment of DNA, which converts unmethylated cytosines in DNA to uracil in a sulfonation-deamination-desulfonation reaction, while leaving methylated cytosines unmodified (Frommer et al., 1992). In part due to the high fidelity of this reaction as well as the single-nucleotide information that it allows, bisulfite sequencing has been referred to as the “gold standard” of DNA methylation assessment techniques (Lister and Ecker, 2009; Chen et al., 2010; Reed et al., 2010). Indeed, direct bisulfite sequencing of DNA purified from brain tissue has been performed repeatedly in our laboratories, and represents a reliable and relatively low-cost method for examining changes in DNA methylation with gene and CpG-site specific resolution. The protocol described below provides a basic outline for conducting direct bisulfite sequencing with DNA purified from brain tissue.
Strategic Planning
Prior to bisulfite conversion and sequencing, the concentration and purity of starting DNA should be determined using a conventional spectrophotometer, Nanodrop, or similar system. Starting DNA should be high quality, with the total amount of DNA ranging from 100ng – 5 μg. The volume (μl) of DNA required from each sample can be computed given the concentration. PCR tubes should be labeled and a thermal cycler should be set as outlined in the bisulfite kit in preparation for the start of bisulfite treatment. Care should be taken in advance to perform appropriate in silico analysis required for primer design and gene amplicon selection, based on the guidelines outlined below.
Direct Bisulfite Sequencing of DNA from Brain Tissue in Order to Determine DNA Methylation Levels at Gene Specific Targets
Direct bisulfite sequencing for analysis of DNA methylation levels is a technique that was developed in order to determine CpG site-specific changes (Frommer et al., 1992). Determination of DNA methylation changes is fundamental to understanding control of gene expression within the brain and how these changes affect neuronal activity. Neuronal changes within the brain will inevitably be associated with prior changes in DNA methylation; thus, DNA methylation may be an important control mechanism for all neuronal activity (Dong et al., 2008; Guo et al., 2011c).
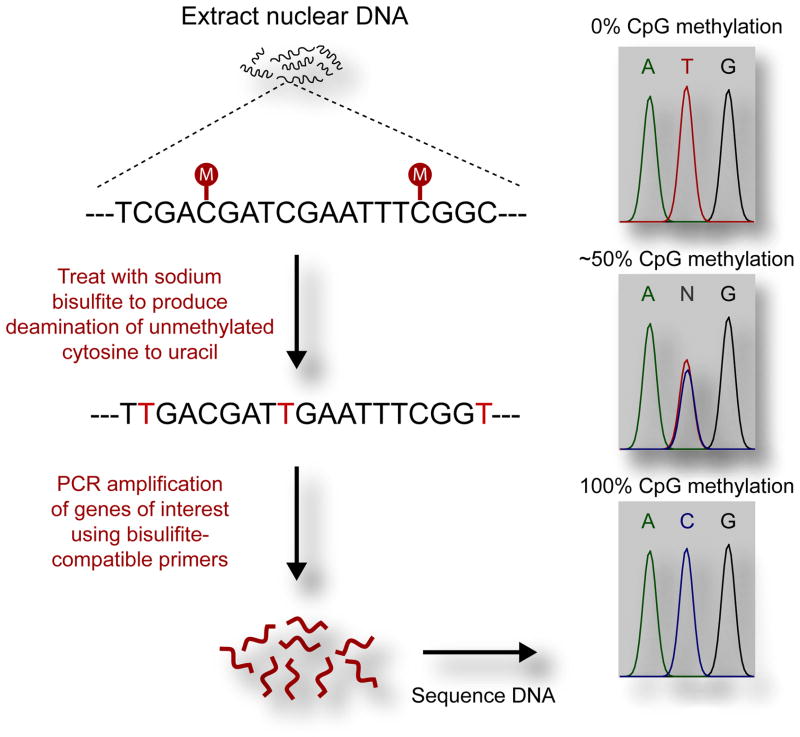
Direct bisulfite sequencing is an effective way to determine DNA methylation changes within brain tissue, providing accurate and relatively quick results. Direct bisulfite sequencing starts when DNA is treated with sodium bisulfite, which conserves methylated cytosine while converting unmethylated cytosine to uracil. Following bisulfite treatment, PCR amplification can be performed with bisulfite-compatible primers which target a specific genomic region. During PCR amplification, cytosine bases which were converted to uracil during bisulfite treatment will be converted to thymine. The PCR product is then purified, and the resulting products are prepared for sequencing. A general schematic diagram of the protocol is shown in Figure 1.
Figure 1.
Schematic diagram of direct bisulfite sequencing workflow. DNA is first bisulfite treated to conserve methylated cytosine while unmethylated cytosine is converted to uracil. During PCR, uracil is converted to thymine and product is amplified. PCR product is then sequenced. The methylation level of each CpG in the amplicon will result in differing peak sizes for cytosine and thymine as read on the chromatogram.
Materials
Isolated, purified DNA
Qiagen EpiTect Bisulfite Kit (Catalog no. 59104)
Thermal cycler
SYBR Green Supermix by Bio-Rad (Catalog no. 170-8882), or similar master mix
Nuclease-free water
Forward and Reverse primers to amplify genomic of region of interest
ExoSAP-IT by Affymetrix (Part no. 78200)
Ethanol
Pipets and pipet tips
PCR tubes
1.5 mL microcentrifuge tubes
Microcentrifuge
Agarose
Gel electrophoresis system
Power supply for gel electrophoresis
Access to an ABI 3730 DNA analyzer
Standard DNA samples from EpigenDX (Cat no. 80-8060-PreMix)
Perform bisulfite conversion of DNA
-
1
Using fresh-frozen brain tissue, extract and purify DNA. The purification of DNA from brain tissue can be accomplished using a number of accepted methods, including the use of silica spin-columns and phenol-chloroform extraction. The method and kit used for this procedure should be adjusted based on the amount of starting tissue and expected yield. However, the Qiagen MicroAmp DNA extraction kit has generally yielded good results in our laboratory. A concentration between 500 ng to 1 μg of starting DNA for bisulfite conversion per sample has worked well for our lab. However, bisulfite treatments can work effectively at concentrations from ranges of 1 ng to 2 μg. Concentration of DNA should remain consistent across all samples.
-
2
Bisulfite conversion can be performed using a number of commercially available kits, although the Qiagen EpiTect Bisulfite Kit has worked well for our lab. The first step of the kit requires thermal cycling that requires approximately 5 hours (this is where the bisulfite conversion is occurring), which we have found best to perform overnight, as overnight holding in the thermal cycler can be done without loss of effectiveness. In addition to using a kit such as the Qiagen EpiTect Bisulfite kit, it is also possible to develop a bisulfite treatment protocol of DNA within your own lab. A basic bisulfite conversion of DNA occurs by incubation of DNA at high temperature with a low PH. The DNA should be incubated with a high bisulfite salt concentration. It is important to note that although many protocols are available for bisulfite conversion of DNA, reliability of conversion of cytosine to uracil can be a problem with many bisulfite conversion protocols. Furthermore, a significant amount of DNA degradation may also occur during bisulfite conversion, while also being time consuming. Many resources are available on the limitations of bisulfite conversion protocols and how to perform your own bisulfite treatment of DNA (Yang et al., 2006; Genereux et al., 2008). Following incubation, the DNA must undergo purification and elution to remove salts and other impurities due to bisulfite treatment. Purification and elution of the converted DNA can be performed by using any standard DNA purification or clean-up protocol.
-
3
Following the thermal cycling to perform the bisulfite conversion, the second part of the Qiagen EpiTect Bisulfite Kit kit involves washing, desulfonation of cytosines (the final step in the conversion to uracil), and purification of the bisulfite converted DNA for downstream PCR. This is accomplished with an EpiTect spin column. After purification, the bisulfite converted DNA samples can then be stored for 24 h at 2–8 °C or at −20°C for at least 3 years without loss of product.
Amplification of region of interest
-
4
Primer design rules for bisulfite sequencing PCR is similar to primer design for other PCR reactions, with a few variations (see “Anticipated Results and Troubleshooting” section for additional discussion of this topic). For primers specific to bisulfite-treated DNA, your DNA region of interest can be analyzed using a free web program called MethPrimer (found at http://www.urogene.org/methprimer/index1.html). MethPrimer will determine the presence and location of CpG islands within a given DNA sequence. Often CpG islands will be the targets of interest for bisulfite sequencing given that the effects of CpG island methylation on gene transcription are well understood. It is important that you verify that you do not have any CpG sites within your primer regions, as the presence of CpG sites within the primer can disrupt primer binding and lead to amplification that is biased towards either methylated or unmethylated DNA. The optimal product size is around 200 base pairs. Once designed, primers for bisulfite-treated DNA are used no differently than any other primer sets. Both forward and reverse primers should be used at a 10 μM concentration.
-
5
Using a thermal cycler, amplify the region of interest using standard PCR. One μL of bisulfite treated DNA per sample has worked well for us. Prepare a master mix of 10 μL of SYBR Green Supermix (by Bio-Rad) with 8 μL of nuclease-free water, plus 0.5 μL each of the forward and reverse primers, for a total of 19 μL of mater mix per reaction. Add 1 μL of bisulfite-treated DNA to this master mix, for a total 20 μL per reaction. A standard thermal cycler protocol that has worked well for us with most primers is as follows: a denaturation cycle for 5 min at 95°C; 50 cycles of denaturation for 1 min at 95°C, annealing for 1 min at 60°C, and extension for 1 min at 72°C; followed by a final extension cycle of 5 min at 72°C and termination at 4°C. Post-PCR products can then be purified or stored at −20°C for later use.
Verification and purification of product
-
6
The size of the amplicon resulting from PCR amplification should be verified prior to proceeding to ensure the correct size. For verification of product, 0.5 μL of post-PCR product should undergo electrophoretic separation on a 2% agarose gel, as with any standard PCR amplification. One strong band with the anticipated product size should be present. Samples containing several bands or primer dimers should be excluded and re-processed.
-
7
PCR products can then be purified using a number of different techniques. The main consideration is removal of any unwanted salts remaining from the PCR reaction, as well as removal of remaining primers and free nucleotides. This can most easily be accomplished using ExoSAP-IT by Affymetrix. Treatment with ExoSAP-IT will require at least 5 μL of post-PCR DNA product and will remove unused primers and nucleotides. See “Support Protocols” for further consideration of other PCR cleanup methods. Once the DNA from PCR amplification is purified, the samples are prepared for sequencing.
Sequencing and analysis of region of interest
-
8
Prepared samples should then be sequenced, with each sample being sequenced in triplicate. A concentration range of 30–100 ng is generally suitable for dye-terminator sequencing. Our samples are sequenced on an Applied Biosystems 3730 DNA Analyzer, which should be available to most researchers. Samples can be sequenced with either the forward or reverse primers of your region of interest at a concentration of 5 μM. If the reverse primer is used for sequencing, the resulting sequencing read will actually be the compliment of the intended DNA sequence.
-
9
Sequenced samples from the ABI 3730 DNA Analyzer will be returned as .ab1 files. AB1 files can be analyzed using an number of different software programs, including either Chromas (for PC, available at http://www.technelysium.com.au/chromas.html), ABI Sequence Scanner (for PC, available at http://www.appliedbiosystems.com/absite/us/en/home.html), and 4Peaks (for Mac, available at http://www.mekentosj.com/science/4peaks). Each of these software programs automatically interprets the chromatogram data and makes base calls determined by the results from the DNA sequencing data. Additionally, each program allows the user to observe DNA sequences and calculate the peak height of individual bases in the DNA sequence (see Fig. 2).
-
10
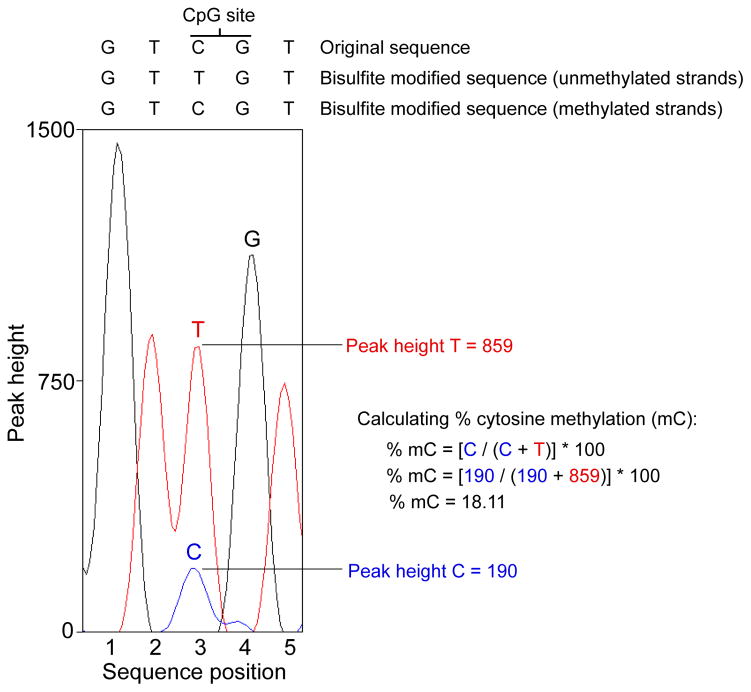
The sequence file should contain the exact same DNA sequence and bp length as the intended PCR amplicon, except for locations at which methylation of cytosines can occur. In these cases, cytosine methylation will yield two unique chromatogram peaks that correspond to the presence of cytosine at that position or the presence of thymine at that position (see Figure 2). As noted above, because bisulfite treatment does not convert methylated cytosines to uracil, the cytosine signal at each CpG location will correspond to the degree of methylation at that site within a given sample. Likewise, the thymine signal will correspond to the degree of cytosines that were not methylated (and were therefore converted to uracil during the bisulfite conversion process). Further, the relative size of these peaks will be proportionally related to the total percentage of cytosine bases that were methylated. Therefore, methylation levels for each CpG site within the DNA amplicon can be quantified by measuring the ratio between peak height values of cytosine (C) and thymine (T), yielding the basic equation for the methylation percentage to be (C/(C+T)*100). Note that this only applies in cases where the forward primer was used for DNA sequencing. If the reverse primer was used, the guanine (G) and adenine (A) peak heights should be used instead, yielding the equation (G/(G+A)*100). An example of this analysis is shown in Figure 2. In our experience, sequencing with the reverse primer results in a cleaner chromatogram and more consistent analysis of DNA methylation.
Figure 2.
An example of how to perform data analysis of a single CpG site with use of the forward primer for sequencing. Chromatogram peaks for thymine (representing unmethylated cytosines that were converted to uracil during bisulfite treatment) and cytosine (representing methylated cytosines that were spared during bisulfite treatment) are compared to determine the average level of methylation for each site within a given sample. The same analysis should be applied when using the reverse primers for sequencing except methylation levels will be derived using the ratio of adenine (unmethylated) to guanine (methylated) instead of thymine and cytosine.
Confirmation of accurate sequencing
-
11
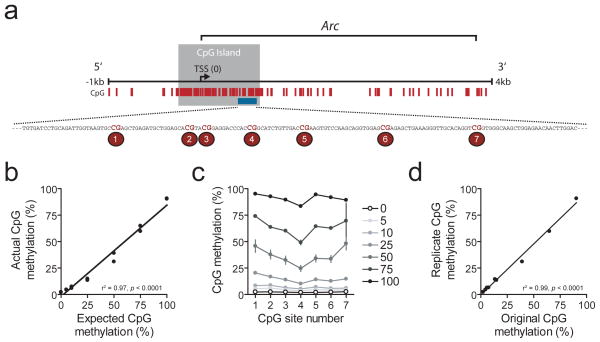
Each primer set used for bisulfite sequencing should be independently verified, using DNA methylation standards to confirm the accuracy of your sequencing data. Standard DNA samples can be ordered from commercial sources (EpigenDX Cat no. 80-8060-PreMix for rat DNA), or created in the lab using available CpG methyltransferase enzymes. Standard DNA samples from EpigenDX come premixed at methylation levels of 0, 5, 10, 25, 50, 75, and 100 percent. The standards should undergo steps 1–10 alongside the samples. It is critical that a standard curve be derived for each gene target of interest prior to interpretation of experimental samples. As shown in Figure 3, direct bisulfite sequencing can provide very accurate results, but must be verified for each set of designed primers.
Figure 3.
Verification of bisulfite sequencing using DNA methylation standards. A) Schematic of a region of the Arc gene with known CpG sites (tick marks), and CpG island (gray box). The 7 CpG sites enlarged are the sites used to determine efficiency of direct bisulfite sequencing at the Arc gene. B) Accuracy of direct bisulfite sequencing. Graph shows actual average methylation versus expected average methylation of known quantities of methylated to unmethylated DNA (using EpigenDx premixed DNA standards). As outlined in the text, bisulfite sequencing resulted in DNA methylation levels that bore a significant linear relationship to expected methylation levels. C) Actual CpG methylation for each site in the amplified region from A, shown for each level of expected methylation (0–100%). Results indicate accuracy of direct bisulfite sequencing across sites. D) Reliability of direct bisulfite sequencing. Independent DNA standard samples were processed for bisulfite sequencing. Results indicate a highly significant and robust linear relationship between two independent replicates – an original (abscissa) and a replicate (ordinate).
Support Protocols
Alternatives for bisulfite conversion of DNA
The Qiagen EpiTect Bisulfite Kit has worked very well for us. However, many other kits are available for bisulfite treatment of DNA as well as designing your own bisulfite conversion protocol. Kits for bisulfite conversion are available from companies such as Applied Biosystems, Invitrogen, Epigentek, SigmaAldrich, and Zymo among many others.
Alternatives for purification of PCR products
PCR products can also be purified by separating the entire product by gel electrophoresis on a 2% agarose gel and then performing a gel extraction of the product. Alternatively, PCR products can be purified with any standard PCR clean-up kit or your own protocol for DNA purification. It is important to note that in order to have a readable, clean chromatogram sequence, no salt or unused products from the PCR reaction should remain in the DNA samples that are being sequenced.
COMMENTARY
Background Information
Direct bisulfite sequencing is a powerful and effective way to determine percent methylation of individual CpG sites, along with percent methylation across multiple CpG sites. This method allows an investigator to start determining how DNA methylation may affect expression of a target gene of interest. Although control of gene expression is very complex, changes in DNA methylation have been shown to correlate to changes in gene expression within the hippocampus in vitro and in vivo due to stimuli (Levenson et al., 2006; Lubin and Sweatt, 2007; Dong et al., 2008; Day and Sweatt, 2010). DNA methylation is another molecular mechanism that allows neuroscientists to further understand how molecular changes affect synaptic plasticity.
Direct bisulfite sequencing can reliably determine changes in DNA methylation within a DNA region of interest. Nevertheless, direct bisulfite sequencing is not without limitations. For example, direct bisulfite sequencing is not able to elucidate the difference between DNA 5-Methylcytosine and 5-Hydroxymethylcytosine. However, to our knowledge there are no available techniques able to resolve the difference between these DNA modifications with single nucleotide resolution. Another limitation of applying direct bisulfite sequencing to brain tissue collected from model organisms is that a number of different cell types are present in the CNS. The investigator is not only sequencing DNA from different types of neurons but also glial cells, so it is not possible to examine cell-type specific changes using direct bisulfite sequencing unless the cells are dissociated and sorted prior to DNA extraction.
Many other techniques for determining DNA methylation following bisulfite conversion of DNA are available and will be discussed briefly; however, the specifics are beyond the scope of this paper. A few other common techniques for determination of site specific changes in DNA methylation are cloning with DNA sequencing and pyrosequencing. Cloning with DNA sequencing is only a slight modification from the protocol provided above, with the main difference being that the region of interest is bacterially cloned prior to sequencing. An advantage to cloning prior to sequencing is that each site from each clone will provide either 100% or 0% methylation, making analysis fairly easy and unambiguous. However, cloning is an extensive process that can be expensive, and in cloning tissue from the brain it is uncertain how many of the clones are DNA from glia as opposed to neurons. Nevertheless, cloning with sequencing is another effective way to determine DNA methylation. Pyrosequencing is another efficient and cost-effective way to determine site-specific DNA methylation levels using bisulfite-treated DNA. Pyrosequencing has been shown to have near equivalent accuracy to bisulfite sequencing; however, one disadvantage to pyrosequencing is the need for multiple sequencing primers, as you can typically only pick up between 2 to 4 CpG sites per each sequencing primer (Ronaghi, 2001; Reed et al., 2010). Pyrosequencing can also be performed alongside direct bisulfite sequencing to confirm accuracy of your DNA sequencing data. Other available techniques which will not be discussed for determination of DNA methylation following bisulfite conversion of DNA include Methylation-sensitive single-strand conformation analysis (MS-SSCA), High resolution melting analysis (HRM), Methylation-sensitive single-nucleotide primer extension (MS-SnuPE), Base-specific cleavage/MALDI-TOF, and Methylation-specific PCR (MSP).
Anticipated Results and Troubleshooting
Bisulfite sequencing should enable reproducible, highly accurate interrogation of the methylation status of individual CpG sites within a gene amplicon of interest. For example, Figure 3 shows representative data obtained by bisulfite sequencing of a region of the Arc gene, which is involved in AMPA receptor trafficking and synaptic plasticity in multiple brain structures. We examined the DNA methylation status of 7 different CpG sites in this gene via bisulfite sequencing using commercially available DNA methylation standards from rat DNA. Figure 3a shows the location of the individual CpG sites in the Arc gene relative to the gene’s transcription start site. As is evident, the selected amplicon is located within a CpG island that straddles the start site of the gene. The resulting analysis demonstrates that bisulfite sequencing at this site is accurate both in terms of the average methylation level across the amplicon (Figure 3b) as well as individual CpG sites within the amplicon (Figure 3c). Furthermore, these results are highly reproducible (Figure 3d). However, certain problems in sequencing results can arise due to a number of potential complications, which may be related to primer selection, DNA purity, amplicon of interest, as well as sequencing interpretation. These will be considered below.
As with all PCR reactions, care must be taken to design primers that are optimal for your gene of interest. Although standard primer design troubleshooting for bisulfite-treated DNA applies as with any other PCR process, there are a number of factors that are especially relevant to primers designed for use with bisulfite sequencing. For example, given that the bisulfite conversion process will result in the conversion of most cytosines into thymines, the base complexity of the resulting DNA will be reduced as compared to unaltered DNA. Due to this fact, extra care must be taken to ensure that the primer sequence is specific for the target of interest and does not bind to and amplify a second site in the DNA template. Further, extra care must be taken to ensure that the primer does not contain excessively long adenine or thymine repeats, which will reduce bond strength between the primer and DNA template. Due to these factors, investigators should verify that the sequencing results actually represent the region that was intended to be sequenced. Although the DNA product size may be confirmed by electrophoresis on an agarose gel, it is possible that the sequence might not be the intended product. Thus, it will be critical to verify the sequence correspondence between the chromatogram and intended amplicon. Furthermore, when using the reverse primer for sequencing, the start of the amplicon sequence will be at the end of the chromatogram trace.
One important consideration when targeting specific genomic regions for PCR amplification of bisulfite treated DNA is to avoid long mononucleotide repeats within the amplicon (such as poly As and poly Ts). Long repeats in the amplicon can cause polymerase slippage during DNA amplification which can shift the resulting sequence and make it difficult to interpret. A shift in the chromatogram can make locating your CpG sites difficult, and can make quantification of DNA methylation challenging and unreliable.
It is also essential to ensure that the chromatogram resulting from bisulfite sequencing has clean, well defined nucleotide base calls. If the chromatogram of the samples is not clean and well defined, it is most likely due to either 1) lack of complete purification of the PCR product or 2) insufficient DNA concentration in sequencing reaction. These problems can be addressed by using more rigorous purification methods and measuring the concentration of DNA prior to sequencing. Incomplete purification can lead to excessively large dye peaks, short DNA trace lengths, or a failed sequencing reaction. Additionally, it is also possible to use a concentration that is too high for dye-terminator sequencing and therefore causes capillary overload when using capillary electrophoresis with sequencing.
In addition, our lab has had success with direct analysis of chromatograms as described above and demonstrated in Figure 2 (without any normalization or data correction). However, it is important to note that other groups have developed algorithms that help to compensate for possible incomplete bisulfite conversion and that can also normalize the signal noise of the chromatogram (Lewin et al., 2004). The application of these algorithms may help to reduce variability between individual replicates and between samples, and has been used with success in a number of publications (Eckhardt et al., 2006; Cortese et al., 2008). Normalization of the DNA sequencing results can be a necessary step for some primers that do not provide optimal standard curves when DNA methylation standards are sequenced.
Time Consideration
Initial preparation and setup before start of bisulfite treatment requires about 15 minutes. Bisulfite treatment and purification of bisulfite-treated samples requires about a day and a half, as we recommend performing the thermal cycle run for bisulfite conversion overnight. Following bisulfite treatment and purification, PCR amplification along with PCR product purification requires about 3 hours. Samples are then prepared for sequencing. If sending the samples to a sequencing core, time requirements may vary, but 3 days should be allotted for sequencing by a core facility. Data analysis of sequencing outcomes can be very time consuming, depending on the amount of samples and amount of CpG sites per sample. However, allow about 20 minutes per each sequenced run for data analysis.
References
- Chen PY, Cokus SJ, Pellegrini M. BS Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese R, Krispin M, Weiss G, Berlin K, Eckhardt F. DNA methylation profiling of pseudogene-parental gene pairs and two gene families. Genomics. 2008;91:492–502. doi: 10.1016/j.ygeno.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70:813–829. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Nelson M, Grayson DR, Costa E, Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc Natl Acad Sci U S A. 2008;105:13614–13619. doi: 10.1073/pnas.0805493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, Haefliger C, Horton R, Howe K, Jackson DK, Kunde J, Koenig C, Liddle J, Niblett D, Otto T, Pettett R, Seemann S, Thompson C, West T, Rogers J, Olek A, Berlin K, Beck S. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genereux DP, Johnson WC, Burden AF, Stoger R, Laird CD. Errors in the bisulfite conversion of DNA: modulating inappropriate- and failed-conversion frequencies. Nucleic Acids Res. 2008;36:e150. doi: 10.1093/nar/gkn691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011a;10:2662–2668. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011b;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, Zhang K, Ming GL, Gao Y, Song H. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011c;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. Histone methylation regulates memory formation. J Neurosci. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, Olshen A, Ballinger T, Zhou X, Forsberg KJ, Gu J, Echipare L, O’Geen H, Lister R, Pelizzola M, Xi Y, Epstein CB, Bernstein BE, Hawkins RD, Ren B, Chung WY, Gu H, Bock C, Gnirke A, Zhang MQ, Haussler D, Ecker JR, Li W, Farnham PJ, Waterland RA, Meissner A, Marra MA, Hirst M, Milosavljevic A, Costello JF. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Lewin J, Schmitt AO, Adorjan P, Hildmann T, Piepenbrock C. Quantitative DNA methylation analysis based on four-dye trace data from direct sequencing of PCR amplificates. Bioinformatics. 2004;20:3005–3012. doi: 10.1093/bioinformatics/bth346. [DOI] [PubMed] [Google Scholar]
- Lister R, Ecker JR. Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 2009;19:959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Sweatt JD. The IkappaB kinase regulates chromatin structure during reconsolidation of conditioned fear memories. Neuron. 2007;55:942–957. doi: 10.1016/j.neuron.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Guo JU, Ming GL, Song H. DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation. Cell Cycle. 2009a;8:1526–1531. doi: 10.4161/cc.8.10.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Marchetto MC, Guo JU, Ming GL, Gage FH, Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat Neurosci. 2010;13:1338–1344. doi: 10.1038/nn.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009b;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, Sweatt JD, Worley PF, Barnes CA. Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol Aging. 2010 doi: 10.1016/j.neurobiolaging.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K, Poulin ML, Yan L, Parissenti AM. Comparison of bisulfite sequencing PCR with pyrosequencing for measuring differences in DNA methylation. Anal Biochem. 2010;397:96–106. doi: 10.1016/j.ab.2009.10.021. [DOI] [PubMed] [Google Scholar]
- Ronaghi M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 2001;11:3–11. doi: 10.1101/gr.11.1.3. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM. Defining an epigenetic code. Nat Cell Biol. 2007;9:2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- Vucetic Z, Kimmel J, Reyes TM. Chronic high-fat diet drives postnatal epigenetic regulation of mu-opioid receptor in the brain. Neuropsychopharmacology. 2011;36:1199–1206. doi: 10.1038/npp.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucetic Z, Kimmel J, Totoki K, Hollenbeck E, Reyes TM. Maternal high-fat diet alters methylation and gene expression of dopamine and opioid-related genes. Endocrinology. 2010;151:4756–4764. doi: 10.1210/en.2010-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, Szyf M. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Jones PL, Wade PA. DNA demethylation. Proc Natl Acad Sci U S A. 1999;96:5894–5896. doi: 10.1073/pnas.96.11.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I, Park IY, Jang SM, Shi LH, Ku HK, Park SR. Rapid quantification of DNA methylation through dNMP analysis following bisulfite-PCR. Nucleic Acids Res. 2006;34:e61. doi: 10.1093/nar/gkl257. [DOI] [PMC free article] [PubMed] [Google Scholar]