Abstract
Examining calcium dynamics within the neural crest (NC) has the potential to shed light on mechanisms that regulate complex cell migration and patterning events during embryogenesis. Unfortunately, typical calcium indicators are added to culture media or have low signal to noise after microinjection into tissue that severely limits analyses to cultured cells or superficial events. Here, we studied in vivo calcium dynamics during NC cell migration and patterning, using a genetically encoded calcium sensor, GCaMP3. We discovered that trunk NC cells displayed significantly more spontaneous calcium transients than cranial NC cells, and during cell aggregation versus cell migration events. Spontaneous calcium transients were more prevalent during NC cell aggregation into discrete sympathetic ganglia (SG). Blocking of N-cadherin activity in trunk NC cells near the presumptive SG led to a dramatic decrease in the frequency of spontaneous calcium transients. Detailed analysis and mathematical modeling of cell behaviors during SG formation showed NC cells aggregated into clusters after displaying a spontaneous calcium transient. This approach highlights the novel application of a genetically encoded calcium indicator to study subsets of cells during ventral events in embryogenesis.
Keywords: chick, neural crest, cell migration, calcium transients, GCaMP3, time-lapse imaging, confocal, sympathetic ganglia
INTRODUCTION
During embryogenesis, beautiful discrete structures arise from complex cell behaviors that include long distance cell migration and cell aggregation. Complex events such as these require rapid signaling mechanisms to regulate the timing of cells to move and coalesce such that discrete structures develop at precise locations. Calcium transients or spontaneous increases of intracellular calcium represent one example of rapid signaling within cells. Calcium transients have been observed in complex embryonic events such as convergent extension (Wallingford et al., 2001) and early neuronal differentiation and patterning (Ciccolini et al., 2003; Gu and Spitzer, 1995; Spitzer, 2006). Advances in live imaging are providing a means to better visualize and quantitate cell movements within embryos (Bower et al., 2011; Supatto et al., 2009). However, two of the remaining roadblocks to in vivo calcium imaging are the lack of fluorescence indicators that are easily deliverable into embryos and a means to quantitatively correlate calcium transients with specific cell behaviors and morphogenetic events.
Visualization of calcium transients in embryos has been challenging due to the limited number of available fluorescence indicators and methods of delivery (Hires et al., 2008). Typical calcium indicators, such as Fluo-4 AM (Heidenreich et al., 2008) require invasive delivery into embryos or can be soaked into cells and tissue in vitro by external application. This severely limits the ability to accurately deliver calcium indicators to discrete cell subpopulations within the developing embryo during precise times of complex morphogenetic events. Genetically encoded calcium indicators have emerged as an exciting set of tools to overcome this roadblock. Specifically, GCaMP3, has been developed and applied to monitor complex cell behavioral events in several adult model systems (Tian et al., 2009; Xiang et al., 2010). Additionally, GCamp3 is non-toxic to embryos and can be transfected into cells or delivered by electroporation to specific subpopulations of cells. Thus, genetically encoded calcium indicators provide a means to study calcium transients in vivo in a targeted manner and have the potential to advance our knowledge of complex cell behavioral events during embryogenesis.
One of the prominent examples in embryogenesis where long distance cell migration and cell aggregation give rise to discrete structures is the formation of the peripheral nervous system (PNS) (Kulesa and Gammill, 2010; Kulesa et al., 2009). During formation of the PNS, trunk neural crest (NC) cells travel along ventral migratory pathways and aggregate into discrete cell clusters of the dorsal root (DRG) and sympathetic ganglia (SG) in a repeating pattern along the vertebrate axis (Gammill et al., 2006; Kasemeier-Kulesa et al., 2006; Kasemeier-Kulesa et al., 2005). One of the major questions of PNS development is how trunk NC cell behaviors are regulated in space and time to produce the pattern of the DRG and SG. Surprisingly, trunk NC cell behaviors are more complex than originally thought. The formation of the primary SG anlagen occurs after NC cells travel to the dorsal aorta (DA) in a discrete migratory stream, then disperse along the dorsal aorta before re-aggregating into a discrete cluster (Kasemeier-Kulesa et al., 2005). Thus, it is clear that insights into calcium dynamics could shed light on the cell migration and patterning events during PNS development.
In this study, we examined the calcium dynamics during chick NC cell migration using GCaMP3 and confocal time-lapse imaging. The GCaMP3 vector was electroporated into pre-migratory NC cells and calcium transients were visualized in vivo in whole chick embryos and in trunk sagittal slice explants. We found differences in the spatiotemporal pattern of fluorescence activity of GCaMP3-labeled cranial and trunk NC cells. We characterized the differences in NC cell calcium transient dynamics, especially during phases when NC cells stopped and coalesced into discrete structures of the SG. We analyzed changes in calcium transients after blocking N-cadherin activity in trunk NC cells during SG formation. Lastly, we constructed a mathematical model, based on detailed NC cell behavioral measurements,that quantitatively correlated calcium transients with NC cell aggregation into discrete SG. Our approach provides a powerful tool for analyzing in vivo NC cell calcium dynamics during cell migratory and patterning events in embryogenesis.
EXPERIMENTAL PROCEEDURES
Embryo labeling
Fertilized white leghorn chicken eggs (Phil's Fresh Eggs, Forreston, IL) were incubated at 38°C in a humidified incubator until the appropriate stages. Eggs were rinsed with 70% ethanol and 3 ml of albumin was removed before windowing the eggshell and staging (Hamburger and Hamilton, 1951). A solution of 10% India ink (Pelikan Fount; www.mrart.com, Houston, TX) in Howard Ringer's solution was injected below the area opaca to visualize each embryo. Embryos were injected and electroporated between 6 and 8 somites for cranial labeling; and 9 and 11 somites for trunk NC cell labeling (Kasemeier-Kulesa et al., 2006; Stark and Kulesa, 2007) with a cocktail of DNA plasmids appropriate for each experiment to fluorescently label premigratory NC cells located in the dorsal neural tube in the trunk. Eggs were resealed with adhesive tape and incubated at 38°C until the appropriate stage. After incubation, we evaluated each embryo prior to manipulation for brightness and uniformity of GCaMP3 labeling using a FITC filter and selected embryos that were developing normally.
GCaMP3 under the control of a CAG promoter was a kind gift from L. Looger. Either a monomeric red fluorescent protein mCherry (gift of R. Lansford) was co-injected for a cytoplasmic label, or to aid in cell tracking, we co-injected a nuclear localized H2B-mCherry (gift from R. Krumlauf lab) both under a CMV promoter. All constructs were used at initial concentration of 5ug/ul.
Embryo Mounting for Imaging
For cranial studies, whole embryos were harvested at HH Stage 11 and mounted dorsal side up on a Millicell culture insert (Millipore, Billerica, MA) inside a glass bottom culture dish (Mattek Corp, Ashland, MA) with approx 1.5mL Neurobasal Media (Invitrogen, Carlsbad, CA).
For trunk imaging, embryos were allowed to incubate 2-3 days post electroporation then selected for imaging at HH Stages 17-20. Sagittal slice explants were selected at the brightest axial level, and between the forelimb and hindlimb level to consistently acquire data on trunk NC cell migration and patterning. Tissue was prepared and mounted as in (Kasemeier-Kulesa et al., 2007) and allowed 5 minutes of rest at 37°C before imaging. Briefly, an approximately 6-8 somite long sections of the trunk were excised down the neural tube midline and gently placed on a Millicell culture insert in a Mattek dish with Neurobasal media. The culture dish was then sealed with parafilm to prevent evaporation. For N-cadherin antibody experiments, the antibody (AbCam, ab11340, Cambridge, MA) was diluted to 300ug/mL in a solution of Howard Ringer's and a very small amount of Fast Green FCF (Sigma-Aldrich, St. Louis, MO) was added to visualize injection into the tissue. We microinjected into the mounted tissue sample lateral to NC cells dispersed along the anterior-posterior axis adjacent to the dorsal aorta, similar to (Kasemeier-Kulesa et al., 2006), and all along the tissue length instead of just one somite length. The culture chamber was sealed and time-lapse imaging was performed immediately after injection.
For in vitro studies, embryos were electroporated between 6 and 8 somites and allowed to re-incubate for approximately 2 hours. Well-labeled and healthy embryos were harvested and the cranial (mid-rhombomere (r3-r5)) region of the neural tube was excised and mounted on a fibronectin and poly-L lysine coated Mattek dish (McKinney et al., 2011). The explanted cells were incubated for 12 hours to allow the cells to move away from the neural tube before imaging.
Time-Lapse Imaging
An LSM5 Pascal or LSM-510 (Zeiss, Thornwood, NY) were used to collect single plane images in 2 channels (GCaMP3 and mCherry) of the labeled NC cells every 30 seconds. The confocal pinhole was adjusted to 2.5 Airy units to collect more fluorescence in the single plane image so that frequent images could be collected without damaging the tissue with as much laser light exposure as in a z-stack. A chamber was constructed around the microscope with a heater to sustain 38°C while imaging (Kasemeier-Kulesa et al., 2006). The GCaMP3 was excited with a 488nm laser line and imaged with a 505-530nm bandpass filter and the mCherry was excited using 543nm laser light and collected with a 560-615nm bandpass filter. Either a 10x/ 0.45NA Plan-Apochromat (Zeiss) or a 10x/0.5NA Fluar (Zeiss) objective was used. All time-lapse imaging sessions were performed for at least 4 hours and up to 10 hours.
Data Analysis
Images were collected in AIM software (Zeiss) and processed in Imaris (Bitplane, South Windsor, CT). Calcium transients were located in Imaris by hand with the “spots” function and statistical information was exported to analysis software in Matlab (Mathworks Inc., Natick, MA). The spot size in Imaris was created so that the measurements did not extend out of the cell body to contain background fluorescence or in the case of H2B-mCherry labeling, only include the nuclear area. ΔF/F0 was calculated for each calcium transient by (It – I0)/I0 where It was the mean intensity of the cell during a calcium transient within the spot created in Imaris and I0 was the mean intensity in the frame preceding the calcium transient. This value was calculated for both GCaMP3 and mCherry channels. To eliminate the possibility of changes in GCaMP3 intensity due to cell movement rather than calcium concentration changes confusing the results, cells that had increases in ΔF/F0 in the green channel that were accompanied by increases in ΔF/F0 in the red channel were excluded from analysis.. Boundaries of the tissue regions were determined from the confocal images and used to automatically assign every calcium transient to a presumptive tissue position.
Mathematical Model and Simulations
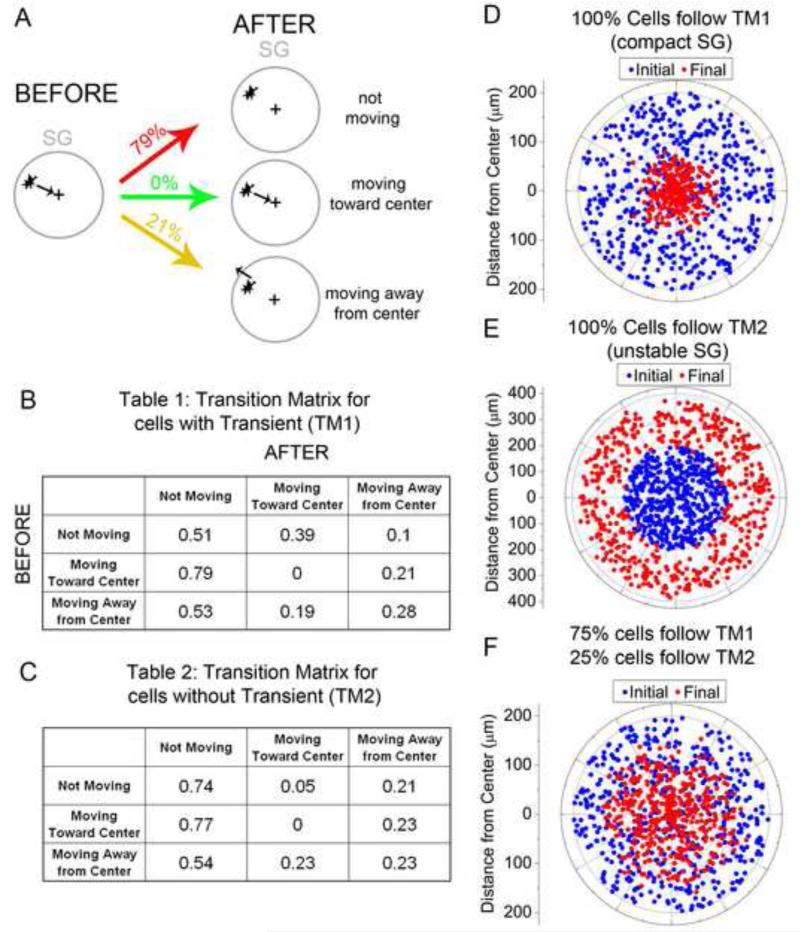
Cells with calcium transients were examined in Imaris before and after each calcium transient. Only cells near and in the SG region were examined for directional movement. The average direction of cell motion before and after each calcium transient was determined by hand in general terms as being either toward or away from the center of the sympathetic ganglia or not moving. 87 cells with calcium transients were examined to tally the number of transitions between states. Probabilities were calculated for transition matrix 1. Calcium transients were not counted if the density of the SG prevented faithful tracking of the cell or if multiple cells were observed to have a calcium transient at once, in case a coordinated calcium signaling event heralded a different behavior from the cells. Non-calcium signaling neighbors of the cells that exhibited calcium transients were also examined for cell behavior within the same time window to create transition matrix 2 for cells without calcium transients.
A simulation framework was created in Matlab where 500 randomly positioned cells in a circle of 150um radius were assigned a state of not moving, moving toward the center or moving away from the center. The number of cells assigned to each initial state was proportional to the number of cells that were counted in each state before a calcium transient. At each time step, the cells moved according to their assigned state either 5um toward the center, away from the center or no movement. Then each cell chose a new state based on a weighted sample from the transition matrix. The simulation was run for 2880 time steps for each transition matrix. For the combination experiment, different fractions of the population were randomly chosen at each time step to follow either transition matrix 1 or 2 until a ratio was found that resulted in a cluster of cells 150um in diameter, typical of a primary SG anlagen.
RESULTS
Calcium transients in migratory neural crest cells lasted between 200ms and 60sec
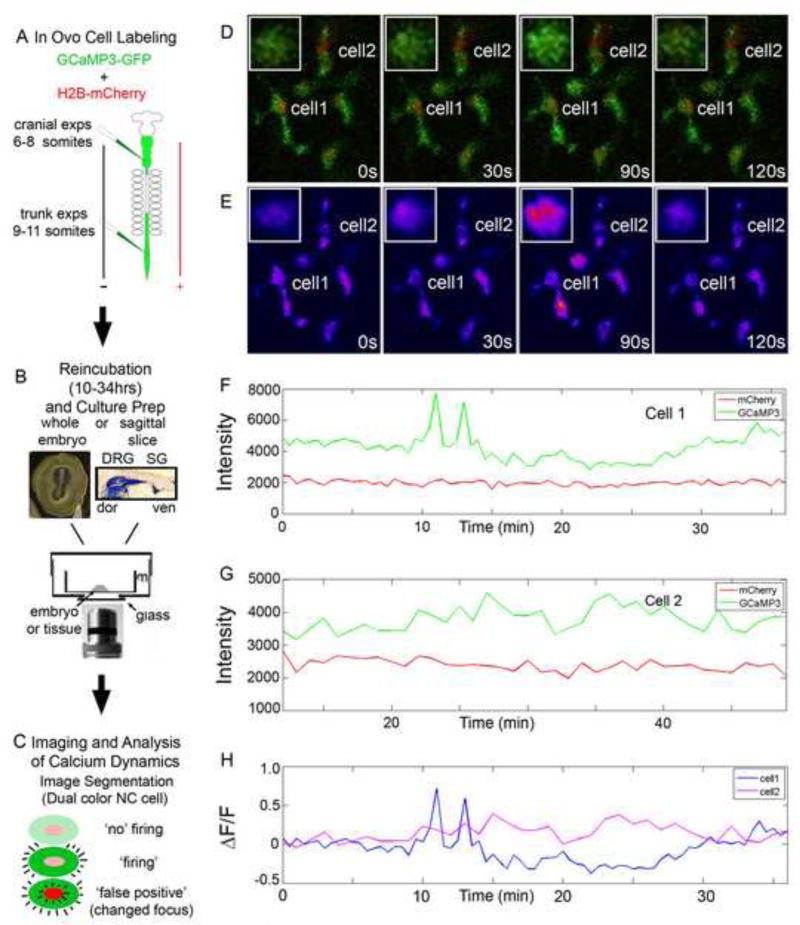
We were able to determine a time window for a typical calcium transient observed in migratory GCaMP3-labeled NC cells (Fig. 1). Laser scanning confocal microscopy captured one image (field of view) every 30sec, which allowed us to collect single scans in 200ms of individual NC cells (average cell diameter of 20um). One image field of view required approximately 7.5 seconds to collect and the tissue was allowed 7.5 seconds of rest in the dark. Imaging schemes using faster time resolution were attempted, but resulted in no observable spontaneous calcium transients and often the cells retracted their filopodia and ceased migration. In a typical time series, we scanned the GCaMP3 channel and when a calcium transient was observed, the fluorescence intensity was high throughout the entire NC cell (753/754 cells; Fig. 1, Sup Movie 1). We very seldom (1/754 cells) observed calcium transients where a subregion of the NC cell had higher fluorescence intensity. Therefore, a NC cell calcium transient lasted at least 200ms (Fig. 1). In 91% of cases, a NC cell remained bright for only 1 frame, however in 9% of cases the calcium transients lasted for 2 frames. Typical calcium transients lasted less than 60sec, but could be sustained for greater than 90sec. We did not observe a NC cell in the firing state for more than 90sec. Thus, NC cell calcium transients typically lasted from 200ms and up to 60sec.
Figure 1. In vivo confocal time-lapse of GCaMP3 labeled NC cells.
A) Embryos were labeled by injection of a cocktail of DNA constructs into the neural tube and electroporation to label the premigratory NC cells. 6-8 somite embryos were used for cranial NC experiments while 9-11 somite embryos were used for trunk NC experiments. B) Embryos were incubated until the proper stage was reached (10 hours for cranially labeled, up to 34 for trunk). Whole embryos were mounted for cranial measurements while a sagittal section was created and mounted for trunk experiments on a culture insert. C) Confocal time-lapse imaging was performed on samples and the calcium transients were segmented out. The calcium transient was verified by constant mCherry fluorescence intensity. D) Selected images from a time-lapse imaging session of NC cells in the forming SG. Inset: magnified image of Cell 1. E) Heat map display of GCaMP3 intensity of same images as in D. F and G) Intensity plots of Cell1 and 2 in both GCaMP3 and mCherry channels showing the difference between a cell with a calcium transient and a neighboring cell without a calcium transient. H) ΔF/F0 for Cell 1 and 2 calculated from the intensity plots in F and G.
Migratory trunk neural crest cells displayed significantly more calcium transients than cranial neural crest cells
NC cells follow stereotypical migratory pathways throughout the head and trunk (Kulesa and Gammill, 2010). Although NC cells give rise to a diverse set of derivatives that vary depending on axial level, discrete NC cell migratory streams share a common feature of traveling as loosely connected individual cells. To determine whether there were differences in calcium transient activities in migratory cranial and trunk NC cells, we analyzed changes in GCaMP3 fluorescence in both sub-regions of the developing chick embryo (Fig. 2). In the head, we examined the calcium transients in the cranial NC cell migratory stream emerging lateral to rhombomere 4 (r4). We compared these observations with calcium transients in the trunk NC cell migratory streams that emerge between the forelimb and hindlimb. In both cases, we analyzed calcium transients in the early emerging NC cells as streams of cells propagated from the neural tube to the periphery (first 10-12 hrs of migration; Fig. 1). We examined calcium transient activity in both whole chick embryos (head) and sagittal slice explants (trunk) and only focused on migratory NC cells that had delaminated from the neural tube (Fig. 2A, B). Time-lapse experiments were conducted identically for the head and trunk (Fig. 1B, C).
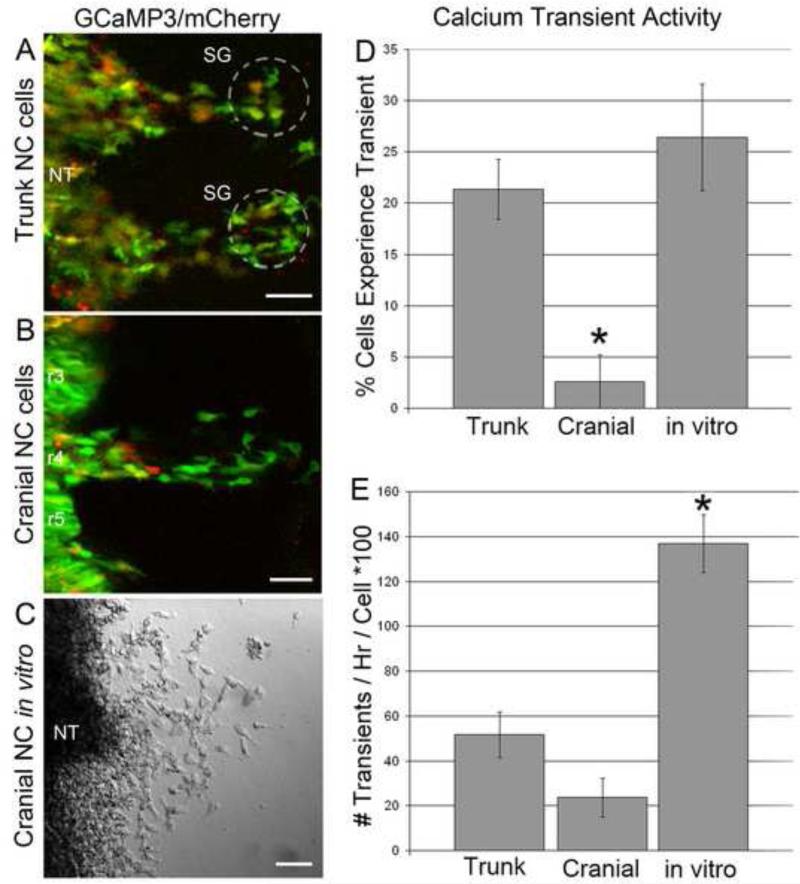
Figure 2. NC cells in the trunk are more likely to exhibit a calcium transient.
Example images from confocal time-lapse series from A) trunk NC cells, B) cranial NC cells and C) cranial cells in a neural tube explant culture in vitro with structural landmarks labeled. D) The percentage of cells excluding NT cells that experienced a calcium transient during a time-lapse for each of the three scenarios. p = 0.028. C) The number of calcium transients per hour for cells that exhibited at least one calcium transient for each of the three scenarios. p = 0.010.
We found a significantly higher percentage of trunk NC cells exhibiting calcium transients (21%; n=2021) compared to migratory NC cells in the head (2%; n=170). Cranial NC cells cultured in vitro in neural tube explants cultures displayed a higher number of calcium transients (26%; n=207). This calcium transient behavior mimicked in vivo trunk NC cell calcium activity (Fig. 2D). Thus, there was a clear difference in calcium transient activity that depended on the axial level of NC cells. Examining the frequency of calcium transients in the trunk, we found that the calcium transients occurred at a rate of 0.52 transients/hr/cell in the cells that experienced a calcium transient. Similarly, cranial NC cells in vivo showed calcium transients at a rate of 0.24 transients/hr/cell (Fig. 2E). In the head, time-lapse images were also acquired after the NC cells reached their target destination in the second branchial arch (BA2), but there were no calcium transients observed in BA2 at the stages we examined (data not shown). We did not perform time-lapse imaging at later developmental stages during cranial ganglia formation, since heartbeat vibration and tissue opacity obscure single cell resolution. Regardless of axial level, spontaneous calcium transients were observed in the entire cell, not localized to one sub-region of the cell or only observed for a partial number of scan lines through the image.
In contrast, in vitro the cells that experienced a calcium transient had a much higher frequency of calcium transients at a rate of 1.37 transients/hr/cell. The cranial NC cells in vitro (Fig. 2C-E) had a percentage of active cells similar to the trunk but they were much more active than either the trunk or the cranial cells in vivo. Therefore, the NC cell behaviors in vitro may not have adequately represented calcium transient behaviors observed in vivo.
NC cells near the sympathetic ganglia were more likely to exhibit a calcium transient
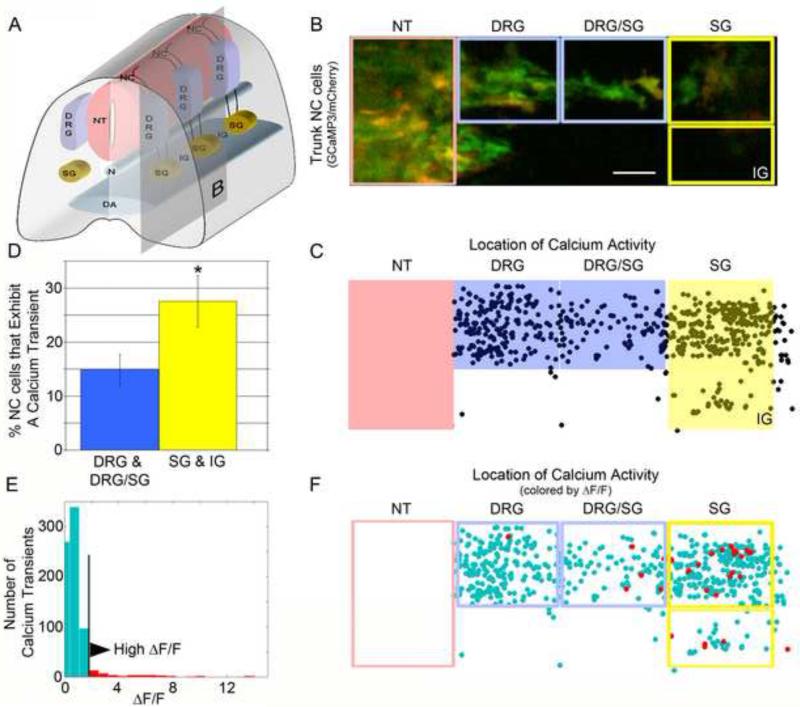
When we examined the NC cells near the dorsal aorta, including the SG and inter-somitic region (IG), we found that 28% of NC cells had at least one calcium transient during the course of a typical time-lapse (Fig. 3A, B). In contrast, NC cells located further from the dorsal aorta, including the presumptive DRG and region between the DRG and SG showed that only 15% had at least one calcium transient during a typical time-lapse session (Fig. 3D). We also examined the propensity of a migratory trunk NC cell to have a calcium transient versus a NC cell that was located within its target site. We compared the population of cells located within the DRG and SG regions with the DRG/SG and IG regions since the cells in these in-between regions were still migrating towards a destination. Cells located within the DRG and SG were observed in proximity to their final positions. There was no significant difference between these two populations in the percentage of cells showing spontaneous calcium transients (Sup Fig 1). These two sets of measurements suggest that NC cells near the dorsal aorta have an environmental influence on their calcium fluctuations that cells within the developing DRG do not.
Figure 3. NC cells near DA are more likely to exhibit at calcium transient and have a larger change in calcium concentration.
A) Schematic of structures in the trunk of the embryo showing the NC derived structures of the DRG and SG. Vertical plane illustrates section imaged in B. B) Example image stream of trunk NC cells with the five sub-structures we divided the stream into. C) Map of all calcium transients from 18 confocal time-lapse series placed onto one template sagittal section. Events in the NT were not counted in analysis since these cells could be NT cells and not NC cells. D) Percentage of cells that exhibited at least one calcium transient in the regions further from the DA (blue boxes) vs the regions closer to the DA (yellow boxes). p = 0.028. E) Distribution of ΔF/F0 values for all calcium transients in the trunk. Top 5% of events are colored red. F) Spatial map of calcium transients with top 5% of events colored red where almost all high calcium events are located near the DA.
The largest changes in neural crest cell calcium concentration occurred during sympathetic ganglia formation
In addition to the frequency of NC cell calcium transients, we also examined the concentration of calcium released into the cell cytoplasm (Fig. 3E, F). We examined the relative GCaMP3 fluorescence intensity in migratory NC cells as a measure of the relative concentration of calcium within the cells, but did not calibrate the GCaMP3 for specific concentration. We measured the quantity ΔF/F0, using the fluorescence intensity during the calcium transient, compared to the NC cell's intensity during a resting period. From these measurements, we determined the distribution of ΔF/F0 values of all calcium transients throughout the trunk (Fig. 3E) and spatial position of all calcium transients in the trunk by their ΔF/F0 value (Fig. 3F). We found that the greatest changes in calcium concentration occurred in the region of the presumptive SG. Clustering of the top 5% of ΔF/F0 values (37 events) revealed that nearly all high ΔF/F0 values were found near the presumptive SG (Fig. 3F).
N-cadherin signaling influenced the propensity of neural crest cell calcium transients
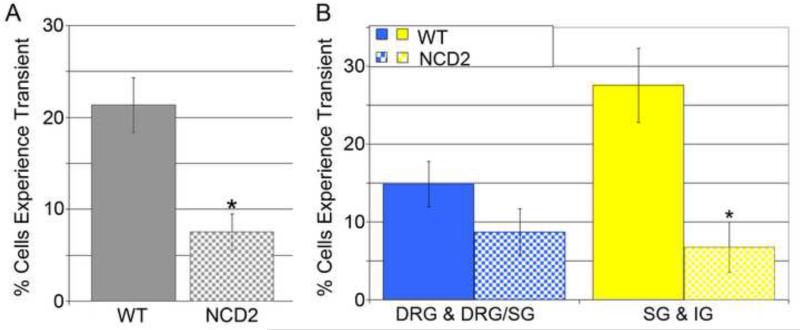
We have previously shown that N-cadherin signaling plays a critical role during chick SG formation (Kasemeier-Kulesa et al., 2006). N-cadherin allows chick NC cells to adhere to each other through the external domain of the N-cadherin protein and thus NC cells coalesce to form a discrete cluster. When N-cadherin was overexpressed, SG formed more compactly (Kasemeier-Kulesa et al., 2006) . In contrast, blocking of N-cadherin signaling led to loosely distributed SG (Kasemeier-Kulesa et al., 2006). Here, we examined whether alterations in the migration and organization of trunk NC cells by perturbation of N-cadherin signaling would affect NC cell calcium transient activity. We found that microinjection of an N-cadherin blocking antibody (NCD2) into the presumptive region of the SG reduced the percentage of NC cells that displayed a calcium transient throughout the trunk (Fig. 5A), most significantly near the DA where the antibody was injected (Fig 5B).
Figure 5. Blocking N-Cadherin near the SG reduces the number of calcium transients.
A) Percentage of cells in the trunk exhibiting at least one calcium transient without and with the blocking antibody treatment. p = 0.005. B) Percentage of cells exhibiting a calcium transient in regions further from the DA and closer to the DA. Comparing cells in the regions of the SG and IG, there is a significant drop in the percentage of cells that experience a calcium transient. p = 0.023.
Mathematical model simulations revealed a correlation in neural crest cell behaviors and calcium transients
We observed more calcium transients during sympathetic ganglia formation. This led us to hypothesize that NC cells that experienced a calcium transient changed their cell behaviors to segregate into the neuronal core and perimeter of the SG. To address this, we examined NC cell movements before and after cells experienced a calcium transient and created a transition matrix (Fig. 4). A cell was examined for approximately 7 minutes (15 frames) before and after a transient and its general direction of migration was scored. NC cells near the SG were examined and only in cases where one NC cell had a calcium transient at a time. Many NC cells were not examined because cells could not be faithfully tracked once cells coalesced into the cluster of cells that defined the SG anlagen. We also did not track NC cells that were a part of a multi-cellular calcium transient event in case these events were associated with a different cellular action. n=87 calcium transients in NC cells near the SG were analyzed and Table 1 (Fig 4B) was created. For example, if a NC cell had a general direction of moving toward the center of the presumptive SG and then expressed a calcium transient, the NC cell could display one of three distinct behaviors afterwards. First, a NC cell could continue to move towards the center of the SG. Second, the NC cell could move away from the SG, or third, the NC cell could stop moving and remain in position. We found that if a NC cell was moving toward the SG center before the calcium transient, 79% of the time the cell stopped moved after a calcium transient. 21% of the time, the NC cell reversed its direction and moved away from the SG center and did not continue moving towards the center (Fig 4A). A cell has the possibility of experiencing multiple calcium transients during a time lapse but the behavior during each transient is not necessarily the same. Therefore each calcium transient is treated as a new event. The transition matrix probabilities for NC cells that started in a non-moving state or moving away from the SG center were also calculated (Table 1, Figure 4B). The data suggested that most often a NC cell stopped moving after experiencing a calcium transient. However, the bias for moving toward the SG center, when starting in a state of non-movement, caused the NC cells to cluster toward the center of the SG.
Figure 4. Modeling cell behavior based on calcium transients.
A) A cell moving towards the center of the SG before a calcium transient has certain probability of each of 3 behaviors after the calcium transient. B) transition matrix for all possible cell migratory behaviors before and after a calcium transient. C) Transition matrix for neighboring non-active cells showing the probability of all possible cell migratory behaviors before and after their neighbor had a calcium transient. D) Simulation results for the case where all cells followed transition matrix 1 at every time step. Cells in their final positions are clustered around the center of the SG. E) Simulation results for the case where all cells followed transition matrix 2 at every time step. Cells in their final positions moved away from the center of the SG. F) Optimal combination of cells following each transition matrix forming an SG that is 150um in diameter. At each time step a random 75% of cells followed transition matrix 1 and the remainder followed transition matrix 2.
To illustrate this point more clearly, we created a mathematical model that simulated NC cell behaviors. That is, each simulation was created where 500 cells were randomly positioned within a circle of 150μm and given random initial states of moving toward, not moving, or moving away from the SG center. The cells changed direction and moved based on the transition matrix probabilities (Figure 4B). The cells were allowed to move 0.45μm at each time step either toward the SG center or away from the center, or nowhere if in a state of not moving. Each time step represented 30 seconds, since our actual time-lapse data was taken in 30 second intervals. After moving, each NC cell chose a new state based on the transition matrix and moved again. Each simulation was run for the equivalent of 24 hours and the initial and final positions of the cells were examined (Fig 4D).
Using the transition matrix created from NC cells that exhibited a calcium transient, the cells in the simulations tended to cluster to the center of the SG. To test the pattern formed from NC cells that did not exhibit a calcium transient, a second transition matrix was created using the behavior of NC cells that were non-active neighbors of the NC cells that exhibited a calcium transient (Table 2, Fig. 4C, n=65). Neighbor cells were chosen based on their proximity to a cell that did exhibit a calcium transient at the same time. The neighbor cells were not biased to be in a state of moving towards, away or not-moving just as in the case of cells experiencing a transient. In similar simulations using the new transition matrix, neighboring cells without calcium transients tended to move away from the SG center (Fig. 4E). Therefore, the model simulations revealed that NC cells that exhibited calcium transients had the tendency to move toward the center of the SG, while NC cells that did not exhibit a calcium transient had the tendency to move away from the center of the SG.
The primary SG is a structure roughly 150μm in diameter, supported by our time-lapse data. In order to create a structure roughly 150μm in diameter in 24 hours, we needed to combine the two populations of NC cells (with and without a calcium transient) to balance the opposing migrational bias of each population. We found that in order to achieve this, we needed to randomly assign 75% of the NC cells to transition matrix 1 and the other 25% to transition matrix 2 at each time step (Fig 4F). Our data indicated that 28% of the cells near the SG experienced a calcium transient (Fig 3D). Therefore, the model simulations predicted we missed roughly 2/3 of the calcium transients due to our 30 second interval time-lapse imaging. Our observed calcium transients had a lower limit of 200ms, so our simulation results were entirely reasonable. However, we were able to draw many conclusions from our data even though we concede we did not detect every calcium transient.
DISCUSSION
We used the chick neural crest (NC) as a model system and a genetically encoded calcium indicator, GCaMP3, to study calcium transients during long distance embryonic cell migration and patterning. The targeted delivery of GCaMP3 and semi-automated imaging and analysis allowed us to overcome the barriers to visualize and quantify in vivo calcium transients during a morphogenetic event deeper within the embryo. Here, our results demonstrated a frequency and pattern of NC cell calcium transients that consisted of several aspects. First, we found that trunk NC cells displayed significantly more calcium transients than cranial NC cells, within a temporal range between 200ms and 60sec. Second, a higher number of calcium transients and the highest changes in calcium concentration occurred in trunk NC cells during sympathetic ganglia (SG) formation. Third, down-regulation of N-cadherin signaling resulted in significant a reduction in calcium transients. Fourth, our mathematical model, constructed from transition matrices of observed NC cell behaviors with and without a calcium transient, predicted that calcium transients are correlated with the segregation of NC cells into a neuronal core and progenitor cell perimeter of the sympathetic ganglia.
GCaMP3 was efficiently delivered into premigratory NC cells and easily visualized in vivo using standard confocal time-lapse microscopy, suggesting genetically encoded calcium indicators are an excellent tool for studying cell patterning events during embryogenesis. Our use of GCaMP3 to study a developmental event extended its range of applications that originally included adult C. elegans, Drosophila, and mouse model systems (Tian et al., 2009). We discovered using our methods that in vivo NC cell calcium transients occurred throughout the entire cell body and as an individual event, rather than in punctate subregions of the cell leading to waves of activity (Fig. 1). Calcium transients in NC cells lasted between 200ms and 60sec (Fig. 1). The duration of our observed NC cell calcium transients (200ms to 60sec) was shorter than previous in vitro NC cell data (approximately 60-180s (Carey and Matsumoto, 1999)). Temporal analysis of NC-derived cells, obtained from cultured mouse neural tubes, showed that NC cells displayed calcium transients in waves and spikes (Carey and Matsumoto, 1999). In their assay, NC cells showed an average of 2-3 spikes per hour, but NC cell calcium transients could go up to 96 spikes per hour (Carey and Matsumoto, 1999). In our in vivo analysis, it is possible that there were faster calcium events that went undetected due to the relatively long scan time (200ms of observation then 30 seconds of rest). Thus, our in vivo temporal measurements of NC cell calcium transients were comparable with recorded measurements in other cell types (Ciccolini et al., 2003; Gu and Spitzer, 1995; O'Donovan, 1999), but had unique dynamics compared to in vitro NC cell analysis. Most spontaneous calcium transients were observed as single-cell events (64%) however many multi-cellular events were also observed (36%). These clusters of cells could be as small as 2 or as many as 9 with an average of 3.2 cells per multi-cellular event. The spontaneous calcium transients did not appear to correlate with cell contact, the location of a cell within the embryo or developmental time. It is possible that our imaging scan speed resulted in missed multi-cellular events so we cannot draw any conclusions as this time to the function of a cluster of cells with calcium transients as opposed to a single cell.
The spatiotemporal pattern of calcium transients revealed more activity when NC cells were near the DA, suggesting correlation with sympathetic ganglia formation (Figs. 3 and 6). We have previously shown that early emerging trunk NC cells reach the DA in discrete migratory streams through the rostral somite halves, under the guidance of CXCR4/CXCL12 signaling (Kasemeier-Kulesa et al., 2010). We were surprised to find only random calcium transients during NC cell migration in the head and trunk, since previous in vitro studies have identified calcium activities in fibroblasts (Wei et al., 2010), NC cell delamination (Newgreen and Gooday, 1985) and attachment to the substrate (Heidenreich et al., 2008), and motor axon guidance (Hanson and Landmesser, 2004). In contrast, the pattern of calcium transients after NC cells reached the DA was exciting (Figs. 3 and 6) and correlated with morphogenetic events we have previously described. For example, chick NC cells spread out along the DA before re-aggregating into discrete SG (Kasemeier-Kulesa et al., 2005). Discrete SG form from a combination of cell repulsion and cell adhesion mechanisms that pattern the NC cells into discrete clusters (Kasemeier-Kulesa et al., 2006). Taken together, these data establish NC cell calcium transient activity with the developmental stages of SG formation.
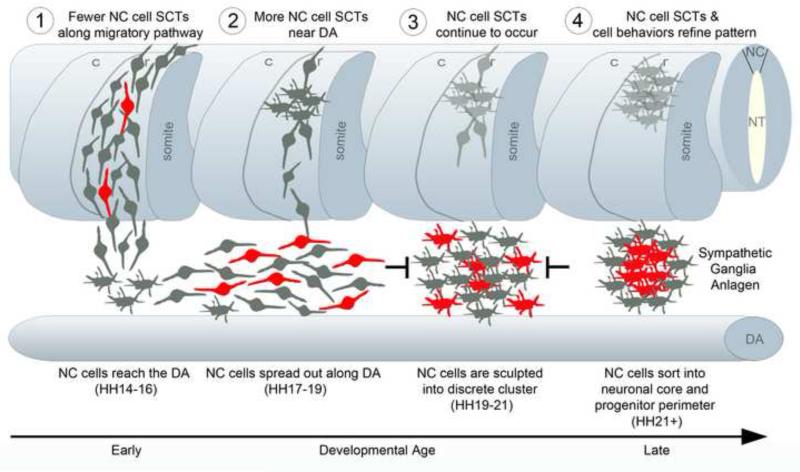
Figure 6. Model of how spontaneous calcium transients refine the substructure of the primary SG anlagen.
Schematic spatiotemporal model of the migration of NC cells forming the SG and the frequency of spontaneous calcium transients (SCT). 1) SCT's occur as NC cells migrate from the NT towards the dorsal aorta (DA) but fewer cells than in later stages exhibit an SCT. 2) NC cells near the DA exhibit SCT's more frequently as they move in an anterior and posterior directions along the DA. 3) NC cells continue to exhibit SCT's as the repulsive forces in the interganglionic regions and attractive forces between NC cells increase. 4) Cells with SCT's promote migration to the center of the cluster forming the neuronal core. Those cells without SCT's move more peripherally forming the progenitor perimeter. Cells in red portray an SCT.
Our data support a model that calcium transients expressed in trunk NC cells are correlated with the refinement of the SG pattern into a neuronal core surrounded by a perimeter of progenitor cells. The primary SG consists of a neuronal core surrounded by a progenitor cell subpopulation identified by differences in TuJ1 and Sox10 expression (Hendershot et al., 2008; Tsarovina et al., 2008). Secreted signals from the DA, in the form of bone morphogenetic proteins (BMPs) initiate a neuronal differentiation program in some trunk NC cells (Goridis and Rohrer, 2002). What has been unclear is how NC cells sort into the neuronal core and progenitor subpopulations of the primary SG anlagen (Kulesa et al., 2009). We observed that the highest calcium concentration changes were in NC cells located all along the DA (Fig. 3), rather than preferentially at the presumptive SG locations. This suggested that a subpopulation of NC cells may respond to differentiation signals expressed uniformly along the DA, but are then segregated into a neuronal core by cell sorting mechanisms. The differentiation program of trunk NC cells has been shown to be dependent on electrical activity (Carey and Matsumoto, 1999; Hao et al., 2010). Thus, we postulate that the sorting of NC cells into the primary SG anlage is also calcium dependent.
We speculate that NC cells that respond to neuronal differentiation signals display calcium transients which alter their behavior to segregate into the neuronal core of the SG. In model simulations, when transition matrices are invoked as a cell's behavioral response to experiencing a calcium transient, firing NC cells cluster into a tight center core while non-firing NC cells scatter outward (Fig. 4). In our observations, NC cells that experienced a calcium transient near the DA either remained stationary or moved toward the center of the presumptive SG with higher probability than moving away (Fig. 4). Local transient calcium elevation has been shown to induce new filopodia from nascent axons or existing filopodia (Lau et al., 1999). In addition, when the leading edges of axonal growth cones sense ligands in the microenvironment, internal calcium release may initiate changes in growth cone direction (Guan et al., 2007). It has also been shown that NC-derived cells that express neuronal markers in the developing enteric nervous system migrate more slowly than non-differentiated enteric NC cells (Hao et al., 2009). These facts support the model that once neuronal differentiation begins, the NC cell's migratory behavior changes and can be related to calcium fluctuations within the cell. It has been shown that when NC cell adhesion was blocked, by microinjection of an N-cadherin signaling blocker (NCD2), primary SG anlage are less compact (Kasemeier-Kulesa et al., 2006) and here we see that NC cells display significantly fewer calcium transients (Fig. 5). Calcium transients in pre-neuronal NC cells could lead to increased cell adhesion and thus neighboring NC cells with calcium transients hold on more tightly to each other thus forming a more compact cluster of cells with calcium transients.
To our knowledge, these data represent the first report of in vivo spontaneous calcium transient dynamics during NC cell migration and patterning. The pattern of significant spontaneous calcium transients appeared to correlate with trunk NC cell formation of the primary SG. Based upon the observed spatiotemporal pattern of spontaneous calcium transients and subsequent trunk NC cell behaviors, we interpret that NC cell spontaneous calcium transient activity refines the pattern of NC cells into the neuronal core and progenitor cell perimeter of a primary SG anlagen. Further experiments will have the challenge to dissect the mechanisms underlying the function of spontaneous calcium transients and refined segregation of NC cells. Details of downstream signaling after spontaneous calcium transients will help shed light on this important NC cell sorting event of sympathetic nervous system development and to other developmental processes that segregate neuronal cell subpopulations. The approach shown here is an exciting step forward in our ability to directly observe and measure in vivo calcium dynamics in subsets of cells during important patterning events deeper within the embryo.
Supplementary Material
Highlights.
first report of in vivo spontaneous calcium transients during patterning of the sympathetic nervous system
neural crest cells in vivo at trunk axial levels more likely to exhibit spontaneous calcium transient than cranial
pattern of spontaneous calcium transients appeared to correlate with formation of the sympathetic ganglia
mathematical model created from cell behaviors indicates calcium transients lead to cluster formation
we interpret that spontaneous calcium transients refine the pattern of the sympathetic ganglia anlagen
ACKNOWLEDGMENTS
We kindly thank Loren Looger (Janelia Farm RC) for the GCaMP3 construct. We also thank lab members Jennifer Kasemeier-Kulesa and Rebecca McLennan for assistance with embryo culture preparation. This work was funded by NIH grant 1R01HD057922 and the Stowers Institute for Medical Research.
REFERENCES
- Bower DV, Sato Y, Lansford R. Dynamic lineage analysis of embryonic morphogenesis using transgenic quail and 4D multispectral imaging. Genesis. 2011 doi: 10.1002/dvg.20754. [DOI] [PubMed] [Google Scholar]
- Carey MB, Matsumoto SG. Spontaneous calcium transients are required for neuronal differentiation of murine neural crest. Dev Biol. 1999;215:298–313. doi: 10.1006/dbio.1999.9433. [DOI] [PubMed] [Google Scholar]
- Ciccolini F, Collins TJ, Sudhoelter J, Lipp P, Berridge MJ, Bootman MD. Local and global spontaneous calcium events regulate neurite outgrowth and onset of GABAergic phenotype during neural precursor differentiation. J Neurosci. 2003;23:103–111. doi: 10.1523/JNEUROSCI.23-01-00103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammill LS, Gonzalez C, Gu C, Bronner-Fraser M. Guidance of trunk neural crest migration requires neuropilin 2/semaphorin 3F signaling. Development. 2006;133:99–106. doi: 10.1242/dev.02187. [DOI] [PubMed] [Google Scholar]
- Goridis C, Rohrer H. Specification of catecholaminergic and serotonergic neurons. Nat Rev Neurosci. 2002;3:531–541. doi: 10.1038/nrn871. [DOI] [PubMed] [Google Scholar]
- Gu X, Spitzer NC. Distinct aspects of neuronal differentiation encoded by frequency of spontaneous Ca2+ transients. Nature. 1995;375:784–787. doi: 10.1038/375784a0. [DOI] [PubMed] [Google Scholar]
- Guan CB, Xu HT, Jin M, Yuan XB, Poo MM. Long-range Ca2+ signaling from growth cone to soma mediates reversal of neuronal migration induced by slit-2. Cell. 2007;129:385–395. doi: 10.1016/j.cell.2007.01.051. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton H. A series of normal stages in the development of the chick embryo. J Morph. 1951;88:49–92. [PubMed] [Google Scholar]
- Hanson MG, Landmesser LT. Normal patterns of spontaneous activity are required for correct motor axon guidance and the expression of specific guidance molecules. Neuron. 2004;43:687–701. doi: 10.1016/j.neuron.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Hao MM, Anderson RB, Kobayashi K, Whitington PM, Young HM. The migratory behavior of immature enteric neurons. Dev Neurobiol. 2009;69:22–35. doi: 10.1002/dneu.20683. [DOI] [PubMed] [Google Scholar]
- Hao MM, Moore RE, Roberts RR, Nguyen T, Furness JB, Anderson RB, Young HM. The role of neural activity in the migration and differentiation of enteric neuron precursors. Neurogastroenterol Motil. 2010;22:e127–137. doi: 10.1111/j.1365-2982.2009.01462.x. [DOI] [PubMed] [Google Scholar]
- Heidenreich DJ, Reedy MV, Brauer PR. Homocysteine enhances cardiac neural crest cell attachment in vitro by increasing intracellular calcium levels. Dev Dyn. 2008;237:2117–2128. doi: 10.1002/dvdy.21644. [DOI] [PubMed] [Google Scholar]
- Hendershot TJ, Liu H, Clouthier DE, Shepherd IT, Coppola E, Studer M, Firulli AB, Pittman DL, Howard MJ. Conditional deletion of Hand2 reveals critical functions in neurogenesis and cell type-specific gene expression for development of neural crest-derived noradrenergic sympathetic ganglion neurons. Dev Biol. 2008;319:179–191. doi: 10.1016/j.ydbio.2008.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hires SA, Tian L, Looger LL. Reporting neural activity with genetically encoded calcium indicators. Brain Cell Biol. 2008;36:69–86. doi: 10.1007/s11068-008-9029-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasemeier-Kulesa JC, Bradley R, Pasquale EB, Lefcort F, Kulesa PM. Eph/ephrins and N-cadherin coordinate to control the pattern of sympathetic ganglia. Development. 2006;133:4839–4847. doi: 10.1242/dev.02662. [DOI] [PubMed] [Google Scholar]
- Kasemeier-Kulesa JC, Kulesa PM, Lefcort F. Imaging neural crest cell dynamics during formation of dorsal root ganglia and sympathetic ganglia. Development. 2005;132:235–245. doi: 10.1242/dev.01553. [DOI] [PubMed] [Google Scholar]
- Kasemeier-Kulesa JC, Lefcort F, Kulesa PM. Sagittal Explant Culture for 3D Confocal Time-Lapse Analysis of Chick Peripheral Nervous System Formation. Cold Spring Harb Protoc. 2007 doi: 10.1101/pdb.prot4791. 2007, pdb.prot4791- [DOI] [PubMed] [Google Scholar]
- Kasemeier-Kulesa JC, McLennan R, Romine MH, Kulesa PM, Lefcort F. CXCR4 controls ventral migration of sympathetic precursor cells. J Neurosci. 2010;30:13078–13088. doi: 10.1523/JNEUROSCI.0892-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesa PM, Gammill LS. Neural crest migration: patterns, phases and signals. Dev Biol. 2010;344:566–568. doi: 10.1016/j.ydbio.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesa PM, Lefcort F, Kasemeier-Kulesa JC. The migration of autonomic precursor cells in the embryo. Auton Neurosci. 2009;151:3–9. doi: 10.1016/j.autneu.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Lau PM, Zucker RS, Bentley D. Induction of filopodia by direct local elevation of intracellular calcium ion concentration. J Cell Biol. 1999;145:1265–1275. doi: 10.1083/jcb.145.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney MC, Stark DA, Teddy J, Kulesa PM. Neural crest cell communication involves an exchange of cytoplasmic material through cellular bridges revealed by photoconversion of KikGR. Dev Dyn. 2011 doi: 10.1002/dvdy.22612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgreen DF, Gooday D. Control of the onset of migration of neural crest cells in avian embryos. Role of Ca++-dependent cell adhesions. Cell and tissue research. 1985;239:329–336. doi: 10.1007/BF00218011. [DOI] [PubMed] [Google Scholar]
- O'Donovan MJ. The origin of spontaneous activity in developing networks of the vertebrate nervous system. Current Opinion in Neurobiology. 1999;9:94–104. doi: 10.1016/s0959-4388(99)80012-9. [DOI] [PubMed] [Google Scholar]
- Spitzer NC. Electrical activity in early neuronal development. Nature. 2006;444:707–712. doi: 10.1038/nature05300. [DOI] [PubMed] [Google Scholar]
- Stark DA, Kulesa PM. An in vivo comparison of photoactivatable fluorescent proteins in an avian embryo model. Dev Dyn. 2007;236:1583–1594. doi: 10.1002/dvdy.21174. [DOI] [PubMed] [Google Scholar]
- Supatto W, McMahon A, Fraser SE, Stathopoulos A. Quantitative imaging of collective cell migration during Drosophila gastrulation: multiphoton microscopy and computational analysis. Nature protocols. 2009;4:1397–1412. doi: 10.1038/nprot.2009.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, Bargmann CI, Jayaraman V, Svoboda K, Looger LL. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsarovina K, Schellenberger J, Schneider C, Rohrer H. Progenitor cell maintenance and neurogenesis in sympathetic ganglia involves Notch signaling. Mol Cell Neurosci. 2008;37:20–31. doi: 10.1016/j.mcn.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Wallingford JB, Ewald AJ, Harland RM, Fraser SE. Calcium signaling during convergent extension in Xenopus. Curr Biol. 2001;11:652–661. doi: 10.1016/s0960-9822(01)00201-9. [DOI] [PubMed] [Google Scholar]
- Wei C, Wang X, Chen M, Ouyang K, Zheng M, Cheng H. Flickering calcium microdomains signal turning of migrating cells. Canadian journal of physiology and pharmacology. 2010;88:105–110. doi: 10.1139/Y09-118. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Yuan Q, Vogt N, Looger LL, Jan LY, Jan YN. Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature. 2010;468:921–926. doi: 10.1038/nature09576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.