The question comes up periodically at conference lunches where scanning probe microscopists are gathered: “Do you think we could image biological macromolecules under a thin layer of water?” It would be wonderful if we could confine biological macromolecules under a thin layer of water or other aqueous solution such that the biomolecules were all close enough to the surface for the atomic force microscope (AFM) tip to image them. AFMs have imaged enzymes in action and biological macromolecules in motion (1–6). Nevertheless, it is always a challenge to find conditions where the macromolecules are bound to a flat surface tightly enough for good AFM imaging and loosely enough for their biological activities to occur. If the macromolecules could be trapped under a thin fluid layer, we might be able to image them without binding them to a surface. The current issue of PNAS presents a step in this direction in the scanning probe microscope (SPM) images of biological macromolecules under a thin layer of aqueous solution (7).
SPMs image sample surfaces by raster scanning a tip back and forth over the sample surface. The tip senses changes in the height or other characteristics of the sample surface. The AFM is a well known member of the SPM family. The AFM uses a tip on a soft cantilever to feel the sample surface, much as a blind person reads Braille or feels a person's face. Much AFM is now done with an oscillating or “tapping” tip, which oscillates up and down as it scans back and forth over the sample surface to map out its surface features. This tapping tip exerts much smaller lateral forces against macromolecules on the sample surface.
Two other SPMs are the scanning tunneling microscope (STM) and the scanning electrochemical microscope (SECM). The SECM measures electrochemical properties of a sample surface under fluid (8). The STM measures the current of electrons that tunnels through the gap between a conductive tip and a conductive sample. The STM was the first SPM to be invented. Remarkably, both the STM and the AFM were invented by the same person, G. Binnig, with collaborators (9–12).
The possibility of doing SPM under a thin fluid layer was tested a few years ago by Dai et al. (13). They prepared an AFM cantilever with a micrometers-long nanotube tip and a sample surface with a sub-micrometer-deep fluid layer. As expected, the cantilever's tapping frequency response was similar to its frequency response in air. The cantilever oscillation amplitude dropped sharply, as it should, when the cantilever approached the surface. However, no images were captured under the thin fluid layer.
Fan and Bard (7) show SECM images under a thin layer of salty aqueous fluid. The SECM has been used previously to detect localized electrochemical reactions at surfaces, including chemical reactions localized to single molecules at the surface (14). Previous SECM has been done with the entire tip submerged in fluid. When submerged in fluid, the sides of the tip must be insulated, so that electrochemical reactivity occurs only at the end of the tip. Such insulated tips tend to have too high a radius of curvature for any good imaging of biomolecules. When imaging in a thin layer of fluid, however, the SECM tip does not need to be insulated, because only the end of the tip is in contact with the fluid. The tip's radius of curvature is, therefore, considerably smaller, and biological macromolecules have been imaged.
With these images, the SECM joins other members of the SPM family that have imaged biological macromolecules at insulating surfaces. The main member of this family is the AFM, images from which are shown in Figs. 1 and 2 a and b. In addition to the SECM, the STM belongs to this family of SPMs that have imaged biomolecules on insulators. The STM was discovered, unexpectedly, to give lovely images of biological macromolecules such as DNA on mica (an insulator) in humid air (ref. 15; Fig. 2c).
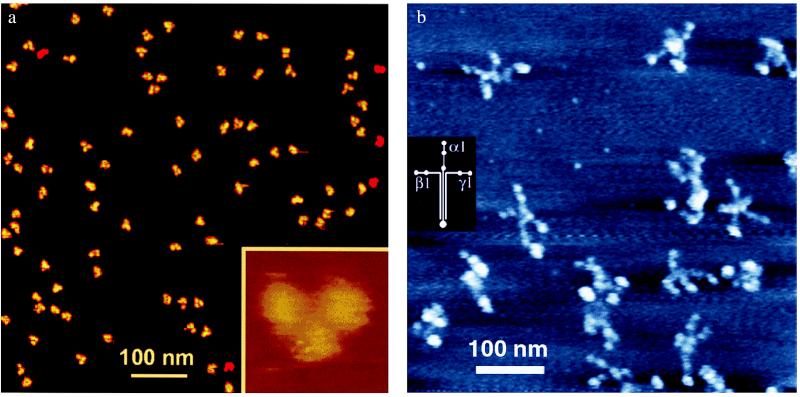
Figure 1.
(a) AFM of IgG antibody molecules under aqueous fluid. IgG molecules were imaged by using the fluid “Tapping Mode” at low amplitude on cantilever resonance. Imaging buffer: 10 mM NaOAc, pH 4.0. Scan size: 700 nm (large field) or 30 nm (Inset). The image was provided by D. Anafi and G.-M. Wu (Amgen Biologicals, Thousand Oaks, CA) and was taken from http://www.di.com (Digital Instruments, Veeco Metrology Group, Santa Barbara, CA). (b) Field laminin molecules in air, taken by AFM. Varied conformations of laminin-1 molecules show the large globular domain at the end of the long arm of the cross and two small globular domains on the ends of many of the short arms. For imaging conditions, see ref. 23. The diagram of laminin-1 chains was derived from electron microscopy, proteolytic degradation, and amino acid sequencing; Molecular mass is 900 kDa (32).
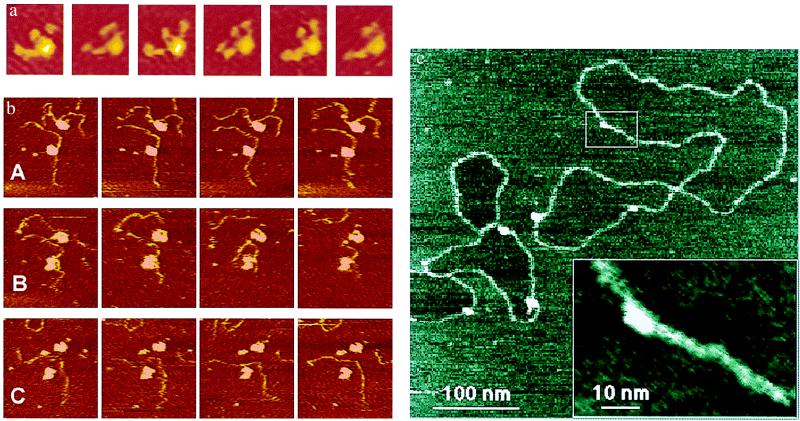
Figure 2.
(a) A laminin molecule moving its arms under fluid, taken by AFM. Six sequential frames from a 58-frame time-lapse movie show the arm movements of a molecule of Laminin-1 on mica. Images are 95 nm × 115 nm. Imaging buffer: 20 mM Mops/5 mM MgCl2/150 mM NaCl, pH 7.4. AFM cantilever was narrow V-shaped silicon nitride, 100-μm-long. See ref. 23. (b) Two active complexes of DNA with RNAP polymerase (RNAP) under fluid in an AFM. Escherichia coli RNAP transcription complexes were prepared with a 1,047-bp DNA template (3, 24). A, B, and C each show a series of four consecutive images at 42-s intervals. (A) DNA strands move near the surface in Zn(II) buffer. (B) These images were taken 3.5–6 min after the last image in A. RNAP transcribes and/or detaches from DNA strands after NTPs are introduced. (C) These images were taken 6–8 min after the last image in B. Zn(II) buffer is reintroduced. DNA images are 310 nm × 330 nm. (c) STM of humid plasmid DNA molecules. Hydration STM shows a high-resolution image of plasmid DNA, prepared on mica (an insulator) and imaged in humid air. The DNA shows a width of only 3 nm along much of its length. This image and other hydration STM images of DNA show an unusual feature: the mica surface inside some of the closed DNA loops appears to be slightly lower than the mica surface outside of closed DNA loops. The box overview marks the area shown in the Inset. This Inset is a cutout of a zoomed-in image taken immediately after the overview image. Imaging conditions: tunneling current, 0.5 pA; sample bias, −7 V; relative humidity, 65%. See ref. 15.
There is some disagreement about the exact mechanism by which STM and SECM can image biomolecules on mica in humid air (16). Because the SECM in humid air and the STM are basically identical microscopes, one would expect them to image in identical ways. STM of DNA on humid mica is interpreted as a tunneling of electrons from the tip to the sample surface (15), where they are dissipated by flowing through a submonolayer of aqueous solution to the electrode. In contrast, SECM of DNA on humid mica is interpreted as a fluid contact between the tip and the sample, such that an electrochemical reaction is occurring (7). The answer to this controversy may be that both mechanisms are correct, because Guckenberger's group (17) has evidence that the mechanism is sometimes electron tunneling and sometimes electrochemistry via a water bridge to the tip, depending on the applied voltage.
The STM's resolution on humid DNA molecules (Fig. 2c) is often so good that it appears to be revealing the major groove of the DNA double helix. This resolution is as good as that of the best AFM, which can detect periodicity the size of the major groove of the DNA double helix only occasionally, either with an unusually fine sharp tip (18) or when the DNA molecules are tightly packed on the surface so that only the very tip of the tip interacts with the DNA molecules during scanning (19).
The AFM images of Figs. 1 and 2 a and b are a few representatives of a rapidly growing body of impressive work. Beautiful images of Y-shaped IgG antibody molecules (Fig. 1a) show a resolution previously achieved only with cryo-AFM (20), even though they were imaged at ambient temperature under aqueous solution. Note that Fig. 1a is a large field of similar but not identical IgG molecules. The IgG molecules have a random rotational orientation on the surface. This feature is important for good SPM imaging—objects should not appear as oriented on a surface when they should be random. It is easy for novices to over-interpret their SPM images, and such over-interpretations have led to a number of published artifacts (21, 22).
The field of larger and more flexible molecules in Fig. 1b shows only a couple of molecules that closely resemble the laminin-1 diagram on the left. On closer inspection, however, several other molecules in Fig. 1b have four arms, stretched or bent in different orientations. Larger clusters of lumps can be classified as laminin dimers or trimers, depending on their volumes as measured from the AFM images. The laminin molecules shown in Fig. 1b were in air, whereas the laminin molecule shown in Fig. 2a was under aqueous solution, with three of its four arms moving in these successive images (23).
At the bleeding edge of biological processes imaged by AFM is the process of DNA being transcribed by RNAP (3, 24). Fig. 2b is an example of this process that occurred too quickly to be captured by a commercial AFM. The two DNA strands in Fig. 2bA move around near the surface in a Zn(II)-containing buffer (25). After addition of the four NTPs (Fig. 2bB), the two RNAPs either transcribe or detach from their two DNA molecules. After Zn(II) buffer is reintroduced (Fig. 2bC), the DNA molecules again become visible—or, rather, the DNA molecule from the lower RNAP complex is visible, whereas the DNA molecule from the upper RNAP complex has moved out of this field of view. Faster AFM imaging is clearly desirable and is now being achieved with new small-cantilever AFMs (26, 27), which have imaged DNA degradation by the DNase I enzyme as fast as 2 s per image (28)—20 times faster than the imaging rate of Fig. 2.
The AFM is clearly a leader among SPMs in biological imaging. The SECM has a strength in imaging that the AFM lacks, however, because the SECM can identify regions of differing electrochemical properties at the sample surface. Perhaps the SECM's niche in biomolecular imaging is to differentiate biomolecules based on their electrochemistry.
Returning to the question of imaging biomolecules in action under a thin layer of fluid—on one hand, this imaging seems to be not only difficult but also undesirable. The out-of-sight DNA strands in Fig. 2bB are probably out of touch with the AFM tip before they are as far above the mica surface at the top of the RNAP molecule. If so, then the thinnest of water layers may already be too thick to keep the macromolecules confined at the surface.
On the other hand, science is filled with seemingly impossible achievements. For example, an AFM tip has recently written words so small that a thousand paragraphs would fit on the head of a pin. The key to this success was the use of a water meniscus, controlled by the relative humidity, to regulate the ink flow (29–31). Clearly the thinnest of water layers has properties that we are only beginning to probe.
Acknowledgments
I thank Hermann Gaub, Deron Walters, James Thompson, and Mario Viani for helpful discussions; Christine Chen for the images of Figs. 1b and 2a; and Scott Hansma for the nanoconvert tiff-conversion software. This work was supported by National Science Foundation Grant MCB 9604566.
Footnotes
See companion article on page 14222 in issue 25 of volume 96.
References
- 1.Drake B, Prater C B, Weisenhorn A L, Gould S A, Albrecht T R, Quate C F, Cannell D S, Hansma H G, Hansma P K. Science. 1989;243:1586–1589. doi: 10.1126/science.2928794. [DOI] [PubMed] [Google Scholar]
- 2.Bezanilla M, Drake B, Nudler E, Kashlev M, Hansma P K, Hansma H G. Biophys J. 1994;67:2454–2459. doi: 10.1016/S0006-3495(94)80733-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasas S, Thomson N H, Smith B L, Hansma H G, Zhu X, Guthold M, Bustamante C, Kool E T, Kashlev M, Hansma P K. Biochemistry. 1997;36:461–468. doi: 10.1021/bi9624402. [DOI] [PubMed] [Google Scholar]
- 4.Shlyakhtenko L S, Potaman V N, Sinden R R, Lyubchenko Y L. J Mol Biol. 1998;280:61–72. doi: 10.1006/jmbi.1998.1855. [DOI] [PubMed] [Google Scholar]
- 5.Lin H, Clegg D O, Lal R. Biochemistry. 1999;38:9956–9963. doi: 10.1021/bi990800q. [DOI] [PubMed] [Google Scholar]
- 6.Goldsbury C, Kistler J, Aebi U, Arvinte T, Cooper G J. J Mol Biol. 1999;285:33–39. doi: 10.1006/jmbi.1998.2299. [DOI] [PubMed] [Google Scholar]
- 7.Fan F-R F, Bard A J. Proc Natl Acad Sci USA. 1999;96:14222–14227. doi: 10.1073/pnas.96.25.14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bard A J, Cliffel D E, Demaille C, Fan F R F, Tsionsky M. Ann Chim. 1997;87:15–31. [Google Scholar]
- 9.Binnig G, Rohrer H, Gerber C, Weibel E. Phys Rev Lett. 1982;49:57–61. [Google Scholar]
- 10.Binnig G, Rohrer H, Gerber C, Wiebel E. Phys Rev Lett. 1983;50:120–123. [Google Scholar]
- 11.Binnig G, Quate C F, Gerber C. Phys Rev Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 12.Binnig G, Gerber C, Stoll E, Albrecht R T, Quate C F. Europhys Lett. 1987;3:1281–1286. [Google Scholar]
- 13.Dai H, Hafner J H, Rinzler A G, Colbert D T, Smalley R E. Nature (London) 1996;384:147–150. [Google Scholar]
- 14.Fan F R F, Bard A J. Science. 1995;267:871–874. doi: 10.1126/science.267.5199.871. [DOI] [PubMed] [Google Scholar]
- 15.Guckenberger R, Heim M, Cevc G, Knapp H F, Wiegrabe W, Hillebrand A. Science. 1994;266:1538–1540. doi: 10.1126/science.7985024. [DOI] [PubMed] [Google Scholar]
- 16.Fan F-R F, Bard A J, Guckenberger R, Heim M. Science. 1995;270:1849–1852. doi: 10.1126/science.270.5243.1849. [DOI] [PubMed] [Google Scholar]
- 17.Heim M, Steigerwald R, Guckenberger R. J Struct Biol. 1997;119:212–221. doi: 10.1006/jsbi.1997.3874. [DOI] [PubMed] [Google Scholar]
- 18.Hansma H G, Bezanilla M, Laney D L, Sinsheimer R L, Hansma P K. Biophys J. 1995;68:1672–1677. doi: 10.1016/S0006-3495(95)80343-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mou J, Czajkowsky D M, Zhang Y, Shao Z. FEBS Lett. 1995;371:279–282. doi: 10.1016/0014-5793(95)00906-p. [DOI] [PubMed] [Google Scholar]
- 20.Han W, Mou J, Sheng J, Yang J, Shao Z. Biochemistry. 1995;34:8215–8220. doi: 10.1021/bi00026a001. [DOI] [PubMed] [Google Scholar]
- 21.Hansma H G, Hoh J. Annu Rev Biophys Biomol Struct. 1994;23:115–139. doi: 10.1146/annurev.bb.23.060194.000555. [DOI] [PubMed] [Google Scholar]
- 22.Leatherbarrow R J, Stedman M, Wells T N. J Mol Biol. 1991;221:361–365. doi: 10.1016/0022-2836(91)80056-z. [DOI] [PubMed] [Google Scholar]
- 23.Chen C H, Clegg D O, Hansma H G. Biochemistry. 1998;37:8262–8267. doi: 10.1021/bi973097j. [DOI] [PubMed] [Google Scholar]
- 24.Guthold M, Zhu X, Rivetti C, Yang G, Thomson N H, Kasas S, Hansma H G, Smith B, Hansma P K, Bustamante C. Biophys J. 1999;77:2284–2294. doi: 10.1016/S0006-3495(99)77067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansma H G, Laney D E. Biophys J. 1996;70:1933–1939. doi: 10.1016/S0006-3495(96)79757-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaeffer T E, Viani M, Walters D A, Drake B, Runge E K, Cleveland J P, Wendman M A, Hansma P K. Proc SPIE. 1997;3009:48–52. [Google Scholar]
- 27.Viani M B, Schaeffer T E, Paloczi G T, Pietrasanta L I, Smith B L, Thompson J B, Richter M, Rief M, Gaub H E, Plaxco K W, et al. Rev Sci Instrum. 1999;70:4300–4303. [Google Scholar]
- 28.Hansma H G, Pietrasanta L I, Golan R, Sitko J C, Viani M, Paloczi G, Smith B L, Thrower D, Hansma P K. In: Biological Structure and Dynamics. Sarma R H, Sarma M H, editors. Albany, NY: Adenine; 2000. , in press. [DOI] [PubMed] [Google Scholar]
- 29.Piner R D, Zhu J, Xu F, Hong S H, Mirkin C A. Science. 1999;283:661–663. doi: 10.1126/science.283.5402.661. [DOI] [PubMed] [Google Scholar]
- 30.Hong S H, Zhu J, Mirkin C A. Science. 1999;286:523–525. doi: 10.1126/science.286.5439.523. [DOI] [PubMed] [Google Scholar]
- 31.Service R F. Science. 1999;286:389–391. [Google Scholar]
- 32.Timpl R, Brown J. Matrix Biol. 1994;14:275–281. doi: 10.1016/0945-053x(94)90192-9. [DOI] [PubMed] [Google Scholar]