Abstract
The Mycobacterium tuberculosis (MTB) Beijing strain is highly virulent, drug resistant, and endemic over Asia. To explore the genetic diversity of this family in several different regions of eastern Asia, 338 Beijing strains collected in Taiwan (Republic of China) were analyzed by mycobacterial interspersed repetitive unit-variable number tandem repeat (MIRU-VNTR) typing and compared with published MIRU-VNTR profiles and by the Hunter-Gaston diversity index (HGDI) of Beijing strains from Japan and South Korea. The results revealed that VNTR2163b (HGDI>0.6) and five other loci (VNTR424, VNTR4052, VNTR1955, VNTR4156 and VNTR 2996; HGDI>0.3) could be used to discriminate the Beijing strains in a given geographic region. Analysis based on the number of VNTR repeats showed three VNTRs (VNTR424, 3192, and 1955) to be phylogenetically informative loci. In addition, to determine the geographic variation of sequence types in MTB populations, we also compared sequence type (ST) data of our strains with published ST profiles of Beijing strains from Japan and Thailand. ST10, ST22, and ST19 were found to be prevalent in Taiwan (82%) and Thailand (92%). Furthermore, classification of Beijing sublineages as ancient or modern in Taiwan was found to depend on the repeat number of VNTR424. Finally, phylogenetic relationships of MTB isolates in Taiwan, South Korea, and Japan were revealed by a minimum spanning tree based on MIRU-VNTR genotyping. In this topology, the MIRU-VNTR genotypes of the respective clusters were tightly correlated to other genotypic characters. These results are consistent with the hypothesis that clonal evolution of these MTB lineages has occurred.
Introduction
Tuberculosis (TB) remains a major infectious and deadly disease in many parts of the world. It is an immense public health problem in Taiwan (Republic of China) despite a steady decrease in both incidence and mortality rates since 1950. Among the most prevalent Mycobacterium tuberculosis (MTB) strains worldwide is the Beijing genotype, which has spread from its origin in Asia to countries of the former Soviet Union, Europe, Africa and the United States, often in association with hypervirulence and multiple drug resistance [1], [2], [3], [4], [5]. The Beijing genotype was first described from isolates in the Beijing region and Mongolia in 1995 [6] and is defined by its highly conserved spoligotype patterns and characteristic IS6110 RFLP profiles.
Beijing strains can be further divided into ancestral, modern and W subfamilies based on the presence or absence of transposon IS6110 insertions in the NTF chromosomal region [7], [8], [9]. The modern subfamily contains a single IS6110 insertion upstream of the NTF region. It is reportedly dominant worldwide, leading to speculation that the modern subfamily has higher virulence and transmissibility than the other two subfamilies [4], [10]. The W subfamily possesses two IS6110 insertions in the NTF region. In contrast to the worldwide prevalence of the modern subfamily, W strains are dominant in the United States, where they have been responsible for outbreaks of multidrug resistant TB [3], [11]. The ancestral subfamily is identified by the absence of IS6110 insertions within the NTF region. Compared with W strains, this subfamily is highly diverse and has caused small-scale outbreaks in the United States [12]. In the last dozen years, discrimination of MTB strains has been achieved by the use of PCR-based methods, including spoligotyping and MIRU-VNTR typing [11], [13].
Our previous studies reported that the Beijing genotypes in the Taipei region of Taiwan mainly belong to the modern subfamily [14], [15], [16]. In contrast, ancient Beijing strains are dominant in Japan [17], [18]. However, it is not known whether the observed variability (modern versus ancient) is due to a selective advantage of particular Beijing strains over other MTB genotypes or to geographical differences among human populations. Here we analyzed a total of 338 Beijing strains collected from four geographical regions in Taiwan by MIRU-VNTR typing and synonymous single nucleotide polymorphism (SNP) typing. We then compared our results against published profiles from South Korea, Japan and Thailand to estimate potential epidemiological linkage in the region.
Results
Discriminatory Power of VNTR Markers in Taiwan
A total of 338 Beijing strains was collected from 5 general hospitals located in four geographical regions of Taiwan, including northern (Taipei Tri-Service General Hospital), eastern (Mennonite Christian Hospital), central (Wan-Chiao Hospital) and southern regions. The copy number of each MIRU-VNTR locus was found to vary from 1 to over 15 (Table 1), and the HGDI values representing the discriminatory power of each locus are shown in Table 2. Among the 24 loci, the discriminatory index for two loci (VNTR424, VNTR2163b) was high (HGDI >0.6), according to the definition of Sola et al. [19]. Seven loci (VNTR154, VNTF3192, VNTR4052, VNTR1955, VNTR4156, VNTR 2996 and VNTR960) were dispersed and had moderate discriminatory power (0.3≤ HGDI ≤0.6). Fifteen loci (VNTR2347, VNTR2059, VNTR2687, VNTR2531, VNTR3007, VNTR580, VNTR2461, VNTR1644, VNTR577, VNTR2401, VNTR3171, VNTR4348, VNTR3690, VNTR2165 andVNTR802) had poor discriminatory power (HGDI<0.3).
Table 1. Frequency of MIRU-VNTR alleles and allelic diversity at each locus in M. tuberculosis isolates from Taiwan.
| MIRU-VNTRlocus | No. of isolates with indicated MIRU allelea | No. AVb | |||||||||||||||
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | >15 | ||
| VNTR2347 (Mtub29) | 1 | 1 | 336 | 3 | |||||||||||||
| VNTR2059 (MIRU20) | 2 | 336 | 2 | ||||||||||||||
| VNTR2687 (MIRU24) | 335 | 3 | 2 | ||||||||||||||
| VNTR2531 (MIRU23) | 1 | 335 | 2 | 3 | |||||||||||||
| VNTR3007 (MIRU27) | 1 | 1 | 334 | 2 | 4 | ||||||||||||
| VNTR580 (MIRU04) | 3 | 2 | 330 | 1 | 1 | 1 | 6 | ||||||||||
| MIRU2461 (ETR-B) | 5 | 328 | 4 | 1 | 4 | ||||||||||||
| VNTR1644 (MIRU16) | 8 | 323 | 7 | 3 | |||||||||||||
| VNTR577 (ETR-C) | 3 | 3 | 321 | 11 | 4 | ||||||||||||
| VNTR2401 (Mtub30) | 18 | 319 | 1 | 3 | |||||||||||||
| VNTR3171 (Mtub34) | 11 | 312 | 13 | 2 | 4 | ||||||||||||
| VNTR4348 (MIRU39) | 3 | 13 | 303 | 19 | 4 | ||||||||||||
| VNTR3690 (Mtub39) | 2 | 2 | 300 | 24 | 9 | 1 | 6 | ||||||||||
| VNTR2165 (ETR-A) | 1 | 39 | 296 | 1 | 1 | 5 | |||||||||||
| VNTR802 (MIRU40) | 14 | 17 | 297 | 9 | 1 | 5 | |||||||||||
| VNTR154 (MIRU02) | 156 | 2 | 10 | 164 | 5 | 1 | 6 | ||||||||||
| VNTR960 (MIRU10) | 6 | 51 | 273 | 5 | 2 | 1 | 6 | ||||||||||
| VNTR2996 (MIRU26) | 2 | 1 | 1 | 9 | 10 | 49 | 244 | 14 | 3 | 4 | 1 | 11 | |||||
| VNTR4156 (QUB4156) | 4 | 58 | 209 | 61 | 5 | 5 | |||||||||||
| VNTR1955 (Mtub21) | 1 | 3 | 12 | 126 | 184 | 3 | 3 | 4 | 1 | 1 | 10 | ||||||
| VNTR4052 (QUB26) | 2 | 3 | 7 | 4 | 20 | 38 | 216 | 33 | 6 | 5 | 4 | 11 | |||||
| VNTR3192 (MIRU31) | 1 | 11 | 175 | 17 | 128 | 4 | 2 | 7 | |||||||||
| VNTR424 (Mtub04) | 5 | 6 | 50 | 125 | 148 | 4 | 6 | ||||||||||
| VNTR2163b (QUB11b) | 1 | 4 | 22 | 31 | 100 | 145 | 26 | 3 | 1 | 2 | 2 | 11 | |||||
N = 338.
AV, allelic variants.
Table 2. HGDI values of M. tuberculosis isolates from Taiwan, Japan and South Korea.
| MIRU-VNTRlocus | Taiwan | Japan [18] | SouthKorea [20] |
| VNTR2347 (Mtub29) | 0.011 | N.D. | 0.118 |
| VNTR2059 (MIRU20) | 0.012 | 0.05 | 0.108 |
| VNTR2687 (MIRU24) | 0.017 | 0 | 0 |
| VNTR2531 (MIRU23) | 0.017 | 0.146 | 0.025 |
| VNTR3007 (MIRU27) | 0.024 | 0.163 | 0.182 |
| VNTR580 (MIRU04) | 0.047 | 0.05 | 0.049 |
| MIRU2461 (ETR-B) | 0.058 | N.D. | 0.025 |
| VNTR1644 (MIRU16) | 0.086 | 0.307 | 0.153 |
| VNTR577 (ETR-C) | 0.097 | 0.028 | 0.049 |
| VNTR2401 (Mtub30) | 0.107 | 0.317 | 0.152 |
| VNTR3171 (Mtub34) | 0.146 | N.D. | 0.0722 |
| VNTR4348 (MIRU39) | 0.192 | 0.183 | 0.277 |
| VNTR3690 (Mtub39) | 0.207 | 0.185 | 0.463 |
| VNTR2165 (ETR-A) | 0.22 | 0.145 | 0.12 |
| VNTR802 (MIRU40) | 0.224 | 0.269 | 0.528 |
| VNTR960 (MIRU10) | 0.325 | 0.436 | 0.178 |
| VNTR2996 (MIRU26) | 0.456 | 0.341 | 0.521 |
| VNTR154 (MIRU02) | 0.552 | 0 | 0.037 |
| VNTR4156 (QUB4156) | 0.554 | 0.622 | 0.628 |
| VNTR1955 (Mtub21) | 0.565 | 0.459 | 0.614 |
| VNTR4052 (QUB26) | 0.566 | 0.675 | 0.707 |
| VNTR3192 (MIRU31) | 0.587 | 0.296 | 0.479 |
| VNTR424 (Mtub04) | 0.651 | 0.474 | 0.698 |
| VNTR2163b (QUB11b) | 0.71 | 0.767 | 0.737 |
Numbers in bold indicate HGDI >0.3.
N.D. = not determined.
Comparison of VNTR and ST profiles from Taiwan, South Korea, Japan and Thailand
To determine whether certain MIRU-VNTR loci could be used to discriminate Beijing strains, we analyzed published MIRU profiles and calculated the HGDI of Beijing strains from two other East Asian countries, Japan [18] and South Korea [20] (Table 2). Among the loci, the discriminatory index of VNTR2163b was greater than 0.6 in Taiwan, Japan and South Korea, whereas the HGDIs of five loci (VNTR 424, VNTR 4052, VNTR1955, VNTR 4156 and VNTR 2996) were higher than 0.3. Furthermore, the HGDIs of three loci (VNTR960, VNTR2401 and VNTR1644) from Japan and two loci (VNTR802 and VNTR 3690) from South Korea were also greater than 0.3.
To better understand the extent of geographic variation of the sequence types in MTB isolate populations, we also analyzed the ST profiles of Beijing strains from Taiwan (present study) (Table S1), Japan [18] and Thailand [21] (Table 3). Eight-hundred and fifty-six Beijing strains from these three studies were classified into 11 ST types, with 402 ancient and 454 modern Beijing isolates (Table 3). The proportion of ST10 was significantly higher in Taiwan (53%) and Thailand (58%) than in Japan (17%). As a group, the three major Beijing sublineages ST10, ST22, and ST19 were found to be prevalent in Taiwan (82%) and Thailand (92%). In contrast, ST3, STK, and ST19 were found to be prevalent in Japan (69%).
Table 3. Distribution features of sequence types of M. tuberculosis isolates from Taiwan, Japan and Thailand.
| Beijingsublineagea | No. (%) strains | ||
| Taiwan | Japan [18] | Thailand [21] | |
| ST11 | 4(1.18) | 4(1.13) | |
| ST26 | 27(7.99) | 28(7.89) | 3(1.84) |
| ST3 | 13(3.85) | 84(23.66) | |
| STK | 4(1.18) | 51(14.37) | 3(1.84) |
| ST19 | 50(14.79) | 111(31.27) | 28(17.18) |
| ST25 | 7(2.07) | 2(0.56) | |
| ST22 | 49(14.50) | 14(3.94) | 28(17.18) |
| ST10 | 180(53.25) | 61(17.18) | 94(57.67) |
| ST8 | 5(3.07) | ||
| STN | 4(1.18, M) | ||
| STF | 2(1.23) | ||
| Ancientb | 88(26.04) | 280(78.87) | 34(20.86) |
| Modern | 250(73.96) | 75(21.13) | 129(79.14) |
| Total | 338 | 355 | 163 |
Sequence type of Beijing sublineages.
The proportion of ancestral or modern Beijing strains was calculated by determining the presence of an IS6110 insertion in the NTF chromosomal region.
Evolution of Beijing Sublineages in Taiwan and Japan
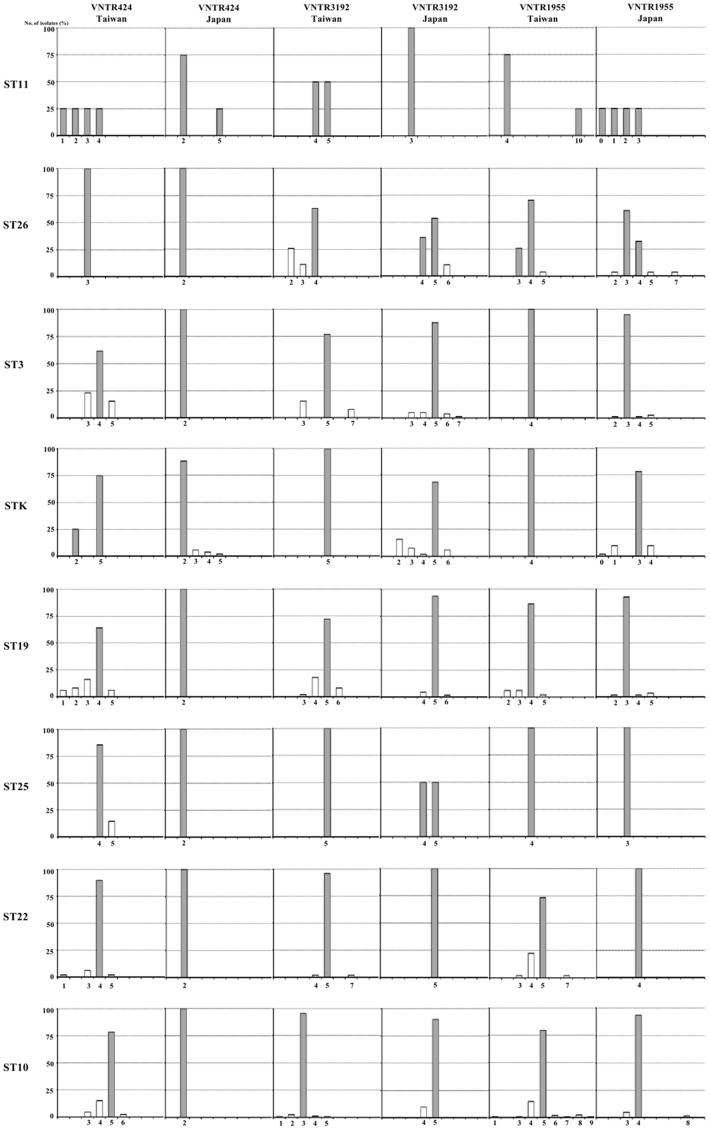
In an attempt to address the evolution of Beijing sublineages, the proportions of each Beijing sublineage were categorized according to the number of repeats of a particular VNTR locus. Three of the loci (VNTR424, VNTR3192 and VNTR1955) were phylogenetically informative (Fig. 1). Evolution of the ancestral or modern Beijing subfamilies, based on the repeat number of VNTRs, was also revealed (ancestral or modern Beijing strains were classified by determining the presence of an RD deletion as described in our previous study [16]). For example, VNTR3192 of Taiwan was characterized mainly by 4 repeats in ST11 and ST26 (ancestral subfamily), but showed almost exclusively 5 repeats in ST3, STK, ST19, ST25, ST22 (modern subfamily) (Fig. 1). VNTR1955 of Japan showed mainly 3 repeats in ST11, ST26, ST3, STK, ST19 and ST25 (ancestral subfamily), but 4 repeats in ST22 and ST10 (modern subfamily) (Fig. 1).
Figure 1. Allelic distribution of three VNTR loci in Taiwan and Japan.
The repeat numbers of each VNTR locus are shown as gray and white bars for the numbers of isolates that comprised >25% and <25% of the proportion, respectively.
Furthermore, the proportions of 3 repeats or 4 and 5 repeats in VNTR424 of Taiwan demonstrated high specificity and sensitivity with respect to the classification of Beijing subfamilies (Fig. 1) (3 repeats distinguished the ancestral Beijing strains: sensitivity 28/31, 90%; specificity 22/307, 93%; 4 and 5 repeats distinguished the modern Beijing strains: sensitivity 272/307, 89%; specificity 1/31, 97%).
Phylogenetic Analysis of Beijing Family Isolates from Taiwan, Japan, and South Korea
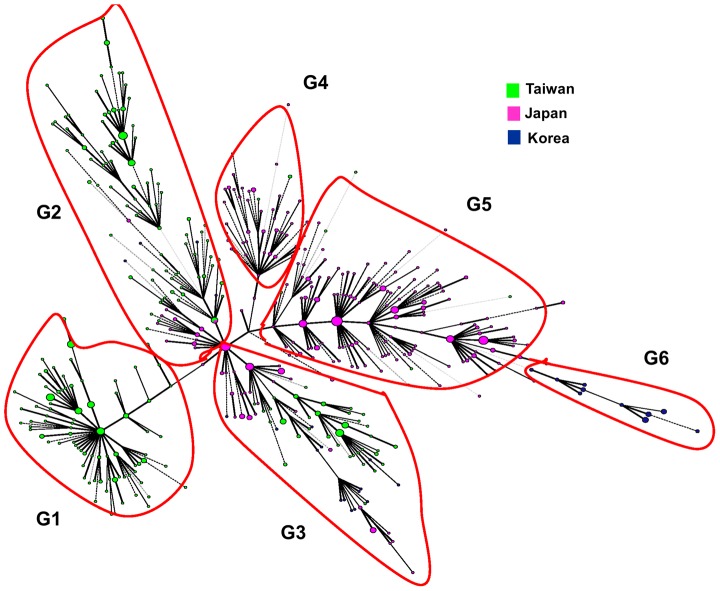
Japan, South Korea and Taiwan are geographically close to mainland China. In addition to geographic nearness, Japan, South Korea, and Taiwan have had historical relationships with mainland China. Business and cultural exchanges among these countries began thousands of years ago. In World War II, Japan colonized Taiwan and Korea. Chinese characters are also used in these three countries. In view of these economic and cultural ties, we compared the MTB strains that have been studied in these three countries. Figure 2 shows the genotype distributions in Taiwan, Japan and South Korea, represented graphically by different colors. Interestingly, the isolates from Taiwan (shown in green) are mostly found in three clusters (G1–G3). We analyzed the allelic diversity and distribution pattern of VNTR loci of the six major phylogenetic sublineages (G1–G6). The G1 sublineage, composed of modern Beijing strains, is predominant in Taiwan. Sublineages G2 and G3, which also contain mainly modern Beijing strains, localize to Taiwan and Japan. Sublineages G4 and G5, composed of predominantly ancient Beijing lineages, are present mainly in Japan. Sublineage G6, also composed predominantly of ancient Beijing strains, is localized in South Korea.
Figure 2. A minimum spanning tree based on 21–MIRU-VNTR genotyping of MTB isolates from Taiwan, Japan, and South Korea.
The sizes of the branches correspond to the number of isolates with a particular genotype. Each country is assigned a different color. MTB isolates from Taiwan, Japan, and South Korea are colored green, pink, and blue, respectively.
Discussion
The Beijing strain is one of the most prominent MTB lineages in East Asia and has spread to other countries worldwide [22], [23]. The reasons for the apparent global success of the Beijing strain are not understood, but could include a variety of host-related factors, such as human population movements [5], selective pressure due to increased worldwide BCG vaccine coverage [7], and ineffective treatment of drug-resistant strains, leading to increased transmission.
Associations of different MTB lineages with different geographic populations have been described [24]. Here we show that geographical clustering is also observed within the Beijing lineage (Fig. 2). The G1, G2 and G3 groups, which contain mainly modern Beijing strains, localize to Taiwan and Japan (Fig. 2). However, G4, G5 and G6, composed predominantly of ancient Beijing strains, are localized in Japan and South Korea (Fig. 2).
As shown in this study, the most frequent strains in Taiwan were ST10 (53.25%), followed by ST19 (14.79%) and ST22 (14.50%), similar to the frequencies in Thailand (ST10, 56.67%; ST22, 17.18%; and ST19, 17.18%) (Table 3). Taiwan is a relatively isolated island. As a similar ST pattern occurred in Taiwan and Thailand, it might be due to frequent travel and immigration between the two countries in the last decade.
The modern Beijing sublineages have been more predominant than the ancestral sublineages throughout other Asian countries, including South Korea, Vietnam, Malaysia and Mongolia [1]. However, Wada and Iwamoto demonstrated that the Beijing family strains in Japan belong mainly to the ancestral subfamily [25]. According to Wada et al. [18], the most frequent strain in Japan was ST19 (31.27%), followed by ST3 (23.66%) and STK (14.37%), which belong to the ancestral subfamily (Table 3). The proportion of ancestral sublineages was similar between Taiwan (26.04%) and Thailand (20.86%) (Table 3), and significantly higher in Japan (78.87%).
Some epidemiological studies have indicated that MTB genotype distribution is highly related to geography, human population and ethnicity [15], [24], [26]. The 12-loci MIRU pattern of one of the strains from our previous investigation was determined to be 223325173533, which is the most predominant pattern for the classical Beijing type (ST1) in Taiwan [14]. Interestingly, this strain was also found in other countries, including Russia, China, and Vietnam [27]. Sequence typing permitted us to further subdivide this major group of strains into ST3, ST10, ST19, ST22 and ST25 in our studies. We believe that these modern Beijing strains, with their high degree of transmissibility, are currently spreading throughout the world. It was previously reported that the BCG vaccination favors the positive selection of modern Beijing strains [28]. Our results support this finding. Moreover, an important study by Reed et al. [29] showed that 80% of the strains from modern Beijing sublineages, but not from ancient sublineages, synthesize relatively high quantities of phenolic glycolipid (PGL), which suppresses proinflammatory cytokines. These findings suggest that modern sublineages may be more pathogenic than ancient sublineages.
The predominance of ancient Beijing lineages in Japan may be because Japan is an island nation and human interaction with foreign countries was probably largely restricted until the 19th century. In fact, TB was not reported as a prevalent infectious disease in Japan until the mid-20th century. But the predominance of modern Beijing sublineages that has occurred recently in countries outside of Japan [30], [31] demonstrates the importance of uncovering the nature of the modern sublineages that has led to their worldwide prevalence and transmission.
The present study also gives the first overview of the evolution of Beijing sublineages in Taiwan based on analyzing the number of repeats of a particular VNTR locus. In our study, three evolutionarily informative loci (VNTR424, VNTR3192 and VNTR1955) were identified. We found that these loci had an increasing trend in the number of repeats (Fig. 1), whereas the remaining VNTRs showed an unvarying number of repeats (data not shown). According to the results, most of the VNTR loci exhibited small variations in the number of repeats (data not shown), which makes it difficult to construct a phylogenetic tree. Notably, the evolution of repeat number in VNTR1955 from Taiwan, Japan and Thailand [21] presented the same trend, which showed a constant repeat number in all STs (principally 4 repeats in Taiwan, 3 repeats in Japan and Thailand) but with increasing numbers in ST22 and ST10 (principally 5 repeats in Taiwan, 4 repeats in Japan and Thailand) (Fig. 1). We propose that the evolutionary history of Beijing isolates in these regions might involve the same events. We also demonstrated the evolutionary pathway of the Beijing strains based on repeat numbers of VNTRs. For example, VNTR424 showed 3 repeats in ST11 and ST26 (ancestral strains), which increased to 4–5 repeats in all other STs that belong to the modern subfamily. VNTR3192 also showed a similar trend (principally 4 repeats in the ancestral sublineages, 5 repeats in the modern sublineages).
Phylogenetic clues in the studied East Asian countries were also revealed (Fig. 2). The NTF and RD analyses in Taiwan indicated that ancient Beijing strains are prevalent among the aboriginals, and modern Beijing strains predominate among military veterans and the general population [15]. The major ethnic population associated with the G1 sublineage of Taiwan, which consists of mainly modern Beijing strains, is Han Chinese (classification by ethnic population in the sampled regions). Sublineage G2, also composed of mainly modern strains, is found predominantly among Han Chinese and military veterans. Taiwan was continuously occupied by the Japanese from 1895 until 1945. The five Taiwanese patients in the G3 sublineage (belonging to the ancient subfamily) were all aboriginals. Therefore, the MTB isolates of the G3 sublineage in Taiwan probably originated in Japan. The minimal spanning tree constructed in this study may constitute a good model with which to study MTB transmission.
In conclusion, this study compares the evolutionary pathway of MTB Beijing isolates from Taiwan against published profiles from three other Asian countries. The classification of MTB Beijing sublineages based on VNTRs was also described. This study also supports the clonal evolution of M. tuberculosis lineages in Taiwan and gives the first overview of the evolution of the Beijing family of MTB from neighboring Asian countries. Our study may be useful in epidemiological studies of global MTB Beijing isolates.
Materials and Methods
Bacterial Strains
A total of 338 Beijing strains was collected during 2003 to 2007 from four geographical regions of Taiwan (north, east, central and south) as described in our previous publication [16]. All 338 isolates were genotyped by spacer oligonucleotide typing (spoligotyping) and 24-locus MIRU-VNTR typing (Table S1). The M. tuberculosis strain H37Rv was used as the control.
DNA Extraction
Mycobacterial chromosomal DNA was extracted by boiling a cultured cell suspension scraped from Lowenstein-Jensen slants in 200 µl distilled water at 85°C for 30 minutes. After centrifugation, the supernatants containing DNA were removed and stored at −20°C until further use.
24-locus MIRU-VNTR Typing
The 12 classical MIRU-VNTR loci (‘12-locus’), 3 exact tandem repeats (ETR A, B and C) and 9 additional loci (Mtub04, Mtub21, Mtub29, Mtub30, Mtub34, Mtub39, QUB11b, QUB26 and QUB4156) were selected and individually amplified in all 338 Beijing isolates as previously described by Supply et al. [32]. The resulting typing pattern from the 24 loci was used to create a 24-digit allelic profile for each isolate.
SNP Typing
PCR procedures and extension primers were designed as described in our previous publication [16]. SNP typing at 10 loci was based on the genome sequences of the H37Rv strain [2], [17].
Statistical Analysis
The Hunter-Gaston diversity index (HGDI) was calculated as described previously [33] and used for comparison of the discriminatory power of MIRU-VNTR typing of Beijing strains from each of the studied East Asian countries. According to this index, the loci were designated as highly (HGDI>0.6), moderately (HGDI 0.3–0.6) and poorly discriminatory (HGDI <0.3) as described in Sola et al. [19].
Supporting Information
24-MIRU-VNTR and SNP profiles of 338 Beijing isloates in this study.
(XLS)
Acknowledgments
All participants of this consortium are acknowledged for valuable discussions. We also thank Dr. Daryl Henderson for his kind help in improving the English of this manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This project was supported by grants from the National Health Research Institutes and National Science Council (NSC 100-3111-B-400 -001), Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bifani PJ, Mathema B, Kurepina NE, Kreiswirth BN. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 2002;10:45–52. doi: 10.1016/s0966-842x(01)02277-6. [DOI] [PubMed] [Google Scholar]
- 2.Filliol I, Driscoll JR, van Soolingen D, Kreiswirth BN, Kremer K, et al. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J Clin Microbiol. 2003;41:1963–1970. doi: 10.1128/JCM.41.5.1963-1970.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glynn JR, Whiteley J, Bifani PJ, Kremer K, van Soolingen D. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg Infect Dis. 2002;8:843–849. doi: 10.3201/eid0808.020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanekom M, van der Spuy GD, Gey van Pittius NC, McEvoy CR, Ndabambi SL, et al. Evidence that the spread of Mycobacterium tuberculosis strains with the Beijing genotype is human population dependent. J Clin Microbiol. 2007;45:2263–2266. doi: 10.1128/JCM.02354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mokrousov I, Ly HM, Otten T, Lan NN, Vyshnevskyi B, et al. Origin and primary dispersal of the Mycobacterium tuberculosis Beijing genotype: clues from human phylogeography. Genome Res. 2005;15:1357–1364. doi: 10.1101/gr.3840605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Soolingen D, Qian L, de Haas PE, Douglas JT, Traore H, et al. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J Clin Microbiol. 1995;33:3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurepina NE, Sreevatsan S, Plikaytis BB, Bifani PJ, Connell ND, et al. Characterization of the phylogenetic distribution and chromosomal insertion sites of five IS6110 elements in Mycobacterium tuberculosis: non-random integration in the dnaA-dnaN region. Tuber Lung Dis. 1998;79:31–42. doi: 10.1054/tuld.1998.0003. [DOI] [PubMed] [Google Scholar]
- 8.Mokrousov I, Narvskaya O, Limeschenko E, Otten T, Vyshnevskiy B. Novel IS6110 insertion sites in the direct repeat locus of Mycobacterium tuberculosis clinical strains from the St. Petersburg area of Russia and evolutionary and epidemiological considerations. J Clin Microbiol. 2002;40:1504–1507. doi: 10.1128/JCM.40.4.1504-1507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mokrousov I, Narvskaya O, Otten T, Vyazovaya A, Limeschenko E, et al. Phylogenetic reconstruction within Mycobacterium tuberculosis Beijing genotype in northwestern Russia. Res Microbiol. 2002;153:629–637. doi: 10.1016/s0923-2508(02)01374-8. [DOI] [PubMed] [Google Scholar]
- 10.Mokrousov I, Jiao WW, Sun GZ, Liu JW, Valcheva V, et al. Evolution of drug resistance in different sublineages of Mycobacterium tuberculosis Beijing genotype. Antimicrob Agents Chemother. 2006;50:2820–2823. doi: 10.1128/AAC.00324-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bifani PJ, Mathema B, Liu Z, Moghazeh SL, Shopsin B, et al. Identification of a W variant outbreak of Mycobacterium tuberculosis via population-based molecular epidemiology. JAMA. 1999;282:2321–2327. doi: 10.1001/jama.282.24.2321. [DOI] [PubMed] [Google Scholar]
- 12.Milan SJ, Hauge KA, Kurepina NE, Lofy KH, Goldberg SV, et al. Expanded geographical distribution of the N family of Mycobacterium tuberculosis strains within the United States. J Clin Microbiol. 2004;42:1064–1068. doi: 10.1128/JCM.42.3.1064-1068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dou HY, Tseng FC, Lin CW, Chang JR, Sun JR, et al. Molecular epidemiology and evolutionary genetics of Mycobacterium tuberculosis in Taipei. BMC Infect Dis. 2008;8:170. doi: 10.1186/1471-2334-8-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dou HY, Tseng FC, Lu JJ, Jou R, Tsai SF, et al. Associations of Mycobacterium tuberculosis genotypes with different ethnic and migratory populations in Taiwan. Infect Genet Evol. 2008;8:323–330. doi: 10.1016/j.meegid.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Chang JR, Lin CH, Tsai SF, Su IJ, Tseng FC, et al. Genotypic analysis of genes associated with transmission and drug resistance in the Beijing lineage of Mycobacterium tuberculosis. Clin Microbiol Infect. 2011;17(9):1391–1396. doi: 10.1111/j.1469-0691.2010.03436.x. [DOI] [PubMed] [Google Scholar]
- 17.Iwamoto T, Yoshida S, Suzuki K, Wada T. Population structure analysis of the Mycobacterium tuberculosis Beijing family indicates an association between certain sublineages and multidrug resistance. Antimicrob Agents Chemother. 2008;52:3805–3809. doi: 10.1128/AAC.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wada T, Iwamoto T, Maeda S. Genetic diversity of the Mycobacterium tuberculosis Beijing family in East Asia revealed through refined population structure analysis. FEMS Microbiol Lett. 2009;291:35–43. doi: 10.1111/j.1574-6968.2008.01431.x. [DOI] [PubMed] [Google Scholar]
- 19.Sola C, Filliol I, Legrand E, Lesjean S, Locht C, et al. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect Genet Evol. 2003;3:125–133. doi: 10.1016/s1567-1348(03)00011-x. [DOI] [PubMed] [Google Scholar]
- 20.Shamputa IC, Lee J, Allix-Beguec C, Cho EJ, Lee JI, et al. Genetic diversity of Mycobacterium tuberculosis isolates from a tertiary care tuberculosis hospital in South Korea. J Clin Microbiol. 2010;48:387–394. doi: 10.1128/JCM.02167-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faksri K, Drobniewski F, Nikolayevskyy V, Brown T, Prammananan T, et al. Genetic diversity of the Mycobacterium tuberculosis Beijing family based on IS6110, SNP, LSP and VNTR profiles from Thailand. Infect Genet Evol. 2011;11:1142–1149. doi: 10.1016/j.meegid.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Hanekom M, Gey van Pittius NC, McEvoy C, Victor TC, Van Helden PD, et al. Mycobacterium tuberculosis Beijing genotype: a template for success. Tuberculosis (Edinb) 2011;91:510–523. doi: 10.1016/j.tube.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Schurch AC, Kremer K, Hendriks AC, Freyee B, McEvoy CR, et al. SNP/RD Typing of Mycobacterium tuberculosis Beijing Strains Reveals Local and Worldwide Disseminated Clonal Complexes. PLoS One. 2011;6:e28365. doi: 10.1371/journal.pone.0028365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wada T, Iwamoto T. Allelic diversity of variable number of tandem repeats provides phylogenetic clues regarding the Mycobacterium tuberculosis Beijing family. Infect Genet Evol. 2009;9:921–926. doi: 10.1016/j.meegid.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Dou HY, Huang SC, Su IJ. Prevalence of Mycobacterium tuberculosis in Taiwan: A Model for Strain Evolution Linked to Population Migration. Int J Evol Biol. 2011;2011:937434. doi: 10.4061/2011/937434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mokrousov I, Valcheva V, Sovhozova N, Aldashev A, Rastogi N, et al. Penitentiary population of Mycobacterium tuberculosis in Kyrgyzstan: exceptionally high prevalence of the Beijing genotype and its Russia-specific subtype. Infect Genet Evol. 2009;9:1400–1405. doi: 10.1016/j.meegid.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Kremer K, van-der-Werf MJ, Au BK, Anh DD, Kam KM, et al. Vaccine-induced immunity circumvented by typical Mycobacterium tuberculosis Beijing strains. Emerg Infect Dis. 2009;15:335–339. doi: 10.3201/eid1502.080795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reed MB, Domenech P, Manca C, Su H, Barczak AK, et al. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 2004;431:84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- 30.Iwamoto T, Fujiyama R, Yoshida S, Wada T, Shirai C, et al. Population structure dynamics of Mycobacterium tuberculosis Beijing strains during past decades in Japan. J Clin Microbiol. 2009;47:3340–3343. doi: 10.1128/JCM.01061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wada T, Fujihara S, Shimouchi A, Harada M, Ogura H, et al. High transmissibility of the modern Beijing Mycobacterium tuberculosis in homeless patients of Japan. Tuberculosis (Edinb) 2009;89:252–255. doi: 10.1016/j.tube.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 32.Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, et al. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000;36:762–771. doi: 10.1046/j.1365-2958.2000.01905.x. [DOI] [PubMed] [Google Scholar]
- 33.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
24-MIRU-VNTR and SNP profiles of 338 Beijing isloates in this study.
(XLS)