Abstract
The mammalian cochlear duct is tonotopically organized such that the basal cochlea is tuned to high frequency sounds and the apical cochlea to low frequency sounds. In an effort to understand how this tonotopic organization is established, we searched for genes that are differentially expressed along the tonotopic axis during neonatal development. Cochlear tissues dissected from P0 and P8 mice were divided into three equal pieces, representing the base, middle and apex, and gene expression profiles were determined using the microarray technique. The gene expression profiles were grouped according to changes in expression levels along the tonotopic axis as well as changes during neonatal development. The classified groups were further analyzed by functional annotation clustering analysis to determine whether genes associated with specific biological function or processes are particularly enriched in each group. These analyses identified several candidate genes that may be involved in cochlear development and acquisition of tonotopy. We examined the expression domains for a few candidate genes in the developing mouse cochlea. Tnc (tenacin C) and Nov (nephroblastoma overexpressed gene) are expressed in the basilar membrane, with increased expression toward the apex, which may contribute to graded changes in the structure of the basilar membrane along the tonotopic axis. In addition, Fst (Follistatin), an antagonist of TGF-β/BMP signaling, is expressed in the lesser epithelial ridge and at gradually higher levels towards the apex. The graded expression pattern of Fst is established at the time of cochlear specification and maintained throughout embryonic and postnatal development, suggesting its possible role in the organization of tonotopy. Our data will provide a good resource for investigating the developmental mechanisms of the mammalian cochlea including the acquisition of tonotopy.
Introduction
The ability of the mammalian cochlea to respond to sounds of different frequencies is attributable to numerous specializations along the longitudinal (tonotopic) axis of the cochlea [1], [2]. Hair cells in the organ of Corti are the mechanosensory receptors that exhibit different sensitivities to specific frequencies depending on their position along the cochlear duct. Their morphological and mechanoelectrical properties gradually differ along the tonotopic axis, which appears to be critical for the frequency tuning [2]. Spiral ganglion neurons, which relay sound information from the hair cells in the form of electrical impulses to the brain, also display different morphological and electrophysiological characteristics along the length of the cochlea [1], [3]. Differences in synaptic structure, function, and number along the tonotopic axis have also been reported [4]. Besides the sensory receptors and neurons, nonsensory structures such as the basilar membrane and tectorial membrane also exhibit graded changes in composition and mechanical features. In fact, the basilar membrane, which vibrates in response to sound, has been considered as a primary determinant that stimulates the hair cells at a specific position with respect to a specific frequency [5]. The tectorial membrane, a viscoelastic structure that covers the stereociliary bundles of hair cells, has also been suggested to play a crucial role in facilitating tonotopic discrimination [6]. Despite numerous structural and cellular specializations in the cochlea that are thought to contribute to tonotopy, the molecular mechanisms that establish these graded specializations during development remain largely unknown.
Several studies have shown that genes and their isoforms are differentially expressed along the base and apex of the cochlea in both mammals and birds. Recently, the microarray technique has been utilized to obtain more comprehensive gene expression profiles along the tonotopic axis of the cochlea [7]–[9]. Sato and colleagues identified genes that are differentially expressed between basal and apical halves of the cochlea in adult mice [7]. In addition, several microarray analyses have compared gene expression levels along the tonotopic axis of the basilar papilla, the avian auditory organ, at hatching or 2-week-old chickens [8], [9]. These gene expression profiles will aid in understanding the tonotopic characteristics of the mature cochlea. However, since these analyses were performed after onset of hearing and the cochlea is thought to be mature already, they may not provide insight into how tonotopy is established.
In mice, the cochlea continues to develop after birth. The hair cells in the organ of Corti undergo dramatic architectural remodeling and achieve mechanosensitivity during the first week of postnatal development [10]–[12]. Also, the stereociliary bundles on the apical surface of the outer hair cells attain mature height and shape, leading to the characteristic organization of a stair-case pattern [10], [12]. The apical surface of the outer hair cells changes progressively from a rounded-hexagon to a non-convex circumference that correlates with the V-shape of their overlying stereociliary bundles [11]. These morphological changes are closely associated with redistribution of cytoskeletal proteins and spatiotemporal expression patterns of genes involved in mechanotransduction, which leads to maturation of the mechanosensitivity of the hair cells [10], [12], [13]. Moreover, the supporting cells in the organ of Corti also undergo major morphological and physiological changes during this period [14]. In particular, progenitor-associated genes such as Notch effectors are downregulated in supporting cells during postnatal development [15], [16]. The loss of Notch signaling may account for diminishment of stem cell features and lack of regeneration potential in the adult mouse cochlea [17], [18]. In contrast, genes associated with supporting cell maturation and function such as the intermediate filament glial fibrillary acidic protein (GFAP) and the glutamate transporter GLAST are upregulated during neonatal development [19], [20].
Interestingly, the morphological and functional maturation progresses gradually from the base of the cochlea toward the apex [10]–[12], [21]. Such changes precede the onset of auditory function, which suggests that the postnatal development may be crucial to achieving the tonotopic characteristics in the mammalian cochlea. Thus, we obtained comprehensive gene expression profiles along the tonotopic axis at postnatal day (P) 0 and P8, and compared expression profiles along the tonotopic axis (base, mid, apex) as well as between the neonatal stages (P0, P8). Our data should provide a platform for future investigations into the developmental mechanisms of the mammalian cochlea.
Results and Discussion
Hair Cell Maturation Along the Tonotopic Axis during Neonatal Cochlear Development
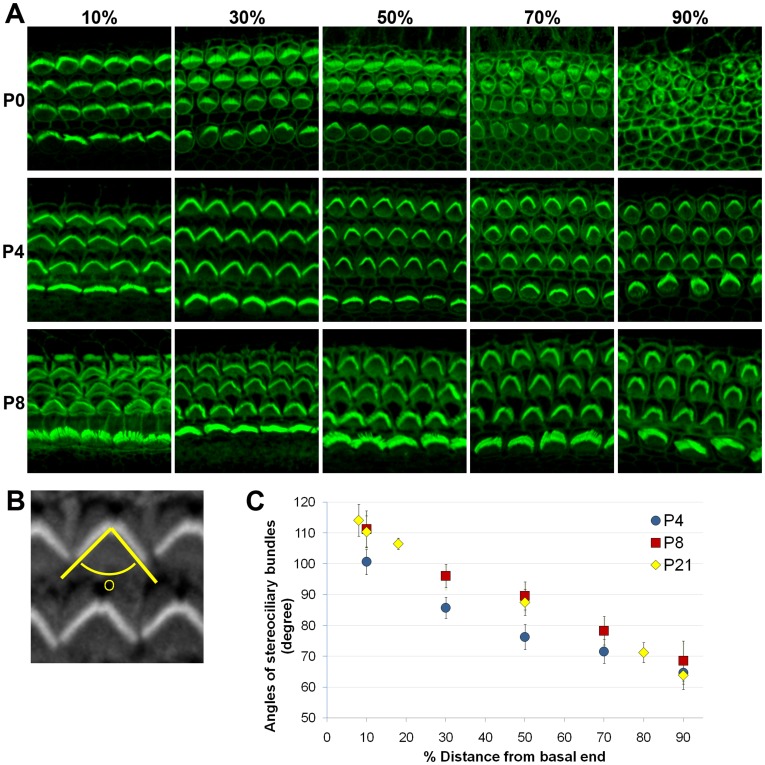
It has been shown that hair cells in the mouse cochlea continue to differentiate after birth [10]–[12]. They acquire mature morphological and mechanoelectrical characteristics during the first postnatal week and hair cell differentiation progresses from the base of the cochlea towards the apex. We examined the maturation process of the hair cells during early postnatal stages in mice (Fig. 1). At P0, the basal half of the cochlea was organized into one row of inner and three rows of outer hair cells with distinct stereociliary hair bundles, while the apical region of the cochlea exhibited multiple rows of unorganized immature hair cells (Fig. 1A), At P4, the entire organ of Corti was in an orderly array with stereocilia of all outer hair cells displaying the characteristic V-shaped (Fig. 1A). The angle of the V-shaped hair bundles was wider at the base and smaller towards the apical cochlea (Fig. 1A). This gradual change in bundle angle is clearly shown in the graph plotted as a function of relative distance from the basal end (Fig. 1B&C). At P8, the morphologies of the hair bundles were almost identical with those of P4, but the angles were generally slightly wider and comparable to those of P21 (Fig. 1C). These results showed that hair cells in the developing cochlea gradually mature in a base-to-apex gradient, and the tonotopic variations along the cochlear duct become evident during neonatal development.
Figure 1. Maturation of hair cells during neonatal cochlear development.
(A) Stereociliary bundles of the mouse cochlea were visualized by staining with Phalloidin at P0, P4 and P8. Pictures were taken from 10%, 30%, 50%, 70% and 90% positions from the basal end of the cochlear duct. (B) Angles of V-shaped hair bundles were measured by drawing two lines along the medial side of the bundles using the Image J program. (C) Angles of the hair bundles are plotted as a function of relative distance from the basal end of the cochlea at P4, P8, and P21.
Gene Expression Profiling Along the Tonotopic Axis during Neonatal Development
Our results and others suggest that neonatal development is crucial for maturation and acquisition of the tonotopic characteristics in the mouse cochlea (Fig. 1) [10], [11]. Genes that play important roles in these processes may be differentially expressed along the cochlear duct during neonatal stages. In order to obtain comprehensive gene expression profiles along the tonotopic axis of the mouse cochlea, we utilized the microarray technique. Approximately eighty temporal bones were dissected from P0 and P8 mouse pups and cochlear tissues containing the organ of Corti and lateral wall were collected. The cochlear tissues were divided into three equal pieces representing the base, middle and apex, and pooled separately (P0-base, P0-middle, P0-apex, P8-base, P8-middle, P8-apex). The same dissection procedure was repeated to obtain a second independent set of pooled cochlear samples from P0 and P8 mice. RNAs were purified from the two biological replicates and were independently subjected to microarray analyses.
The microarray data were analyzed to identify differentially expressed genes along the tonotopic axis at each stage (P0 or P8) by comparing the average expression levels of individual genes. Genes showing at least a 1.5-fold difference (p<0.05) between the base and the apex were considered to be differentially expressed. Interestingly, for most of the genes that were differentially expressed between the base and the apex, expression levels were gradually changed along the cochlear duct, exhibiting either an increasing or decreasing gradient towards the apex. Only a small fraction among the differentially expressed genes (<3%) showed peak expression in the middle cochlear region.
Table 1 shows the top fifty genes that showed higher expression in the base or apex at P0 or P8, ranked by the average fold differences (a complete list of the differentially expressed genes can found in Table S1). The lists contain many genes previously associated with inner ear development or function. For example, Tectb, Isl1, Foxg1, Ush2a, Pcdh15, and Atoh1 showed higher expression in the apex at P0, while Gjb6, Cldn11, and Otoa showed higher expression in the base at P0. At P8, genes showing higher expression in the apex included Tecta, Tectb, Tnc, Fgf10, Smpx, Slitrk6, and Otog, while genes showing higher expression in the base included Crym and Otos. In addition, the lists contain many other genes that have not been previously associated with inner ear or hearing function.
Table 1. Genes differentially expressed along between apex and base of the mouse cochlea at P0 or P8.
| P0 apex high | P0 base high | P8 apex high | P8 base high | ||||
| Gene | Fold diff. apex/base | Gene | Fold diff. apex/base | Gene | Fold diff. apex/base | Gene | Fold diff. apex/base |
| Zpld1 | 4.11 | Atp6v0a4 | 0.24 | Tecta | 5.94 | Tnmd | 0.28 |
| Fst | 3.26 | Pkhd1l1 | 0.28 | Tnc | 5.76 | Hoxa2 | 0.28 |
| Ndst3 | 2.97 | Hoxa2 | 0.32 | Fst | 4.25 | Ifitm5 | 0.35 |
| Clic6 | 2.28 | Fcrlb | 0.33 | Fgf10 | 3.55 | A2m | 0.40 |
| Sema3a | 2.21 | Ucma | 0.33 | Nov | 3.53 | Iyd | 0.41 |
| Tectb | 2.21 | Cyp3a13 | 0.36 | Prkcq | 3.47 | Cntn6 | 0.41 |
| Fam19a4 | 2.17 | A2m | 0.37 | Isl1 | 3.41 | Tmem27 | 0.42 |
| Pcdh15 | 2.16 | Vit | 0.40 | Chrna3 | 3.28 | Tmem196 | 0.42 |
| Zmat4 | 2.16 | Slco1a4 | 0.42 | Zpld1 | 2.98 | Rarres1 | 0.43 |
| Galntl6 | 2.14 | Pkp1 | 0.44 | Glb1l2 | 2.93 | Trdn | 0.44 |
| Rorb | 2.10 | Gjb6 | 0.47 | Syt1 | 2.91 | Fam19a1 | 0.44 |
| Fndc3c1 | 2.07 | Kcna4 | 0.47 | Smpx | 2.90 | Fam198b | 0.45 |
| Isl1 | 2.03 | Cntn5 | 0.48 | Gsbs | 2.81 | Crym | 0.46 |
| Tac1 | 2.01 | Scg2 | 0.48 | Slco5a1 | 2.80 | Pi15 | 0.47 |
| Slco5a1 | 1.98 | Ppp1r1b | 0.50 | Muc15 | 2.75 | Ptgfr | 0.47 |
| Epha3 | 1.92 | Atp6v1c2 | 0.51 | Plch1 | 2.74 | Fgl2 | 0.48 |
| Pcsk2 | 1.85 | Clstn2 | 0.51 | Ccdc148 | 2.73 | Prss35 | 0.49 |
| Hmga2 | 1.82 | Spink5 | 0.54 | Slitrk6 | 2.72 | Pgf | 0.50 |
| Lrrc55 | 1.80 | Wnk4 | 0.54 | Fam40b | 2.70 | Dnase1 | 0.50 |
| Tmem74 | 1.80 | Cfh | 0.54 | Rbm24 | 2.63 | H19 | 0.51 |
| Trpa1 | 1.79 | Rya3 | 0.55 | Cybrd1 | 2.60 | Rgs4 | 0.51 |
| Tyrp1 | 1.78 | Tcfcp2l1 | 0.55 | Sox2 | 2.56 | Myoc | 0.52 |
| Abcg5 | 1.78 | Cldn11 | 0.55 | Adamts19 | 2.56 | Otos | 0.52 |
| P2rx3 | 1.77 | Vcam1 | 0.56 | Kcna10 | 2.53 | Nox3 | 0.52 |
| Pdzrn4 | 1.75 | Mal | 0.56 | Itgb6 | 2.50 | Ppp1r3c | 0.53 |
| Muc15 | 1.73 | Anxa8 | 0.56 | Rassf6 | 2.45 | Smpd3 | 0.53 |
| Cdh4 | 1.72 | Akr1c14 | 0.56 | Cubn | 2.39 | Ctgf | 0.53 |
| Fat3 | 1.71 | Slc38a5 | 0.57 | Otog | 2.38 | Agxt2l1 | 0.54 |
| Angpt1 | 1.70 | Oprk1 | 0.57 | Slitrk3 | 2.38 | Ntn1 | 0.54 |
| Ush2a | 1.70 | Itga4 | 0.57 | Pla2r1 | 2.37 | Ppfia2 | 0.54 |
| Slc39a8 | 1.69 | Kcnj16 | 0.57 | Slc26a5 | 2.36 | Naip1 | 0.54 |
| Dchs2 | 1.68 | Adra2a | 0.58 | Epha3 | 2.33 | Si | 0.55 |
| Rasef | 1.68 | Grin2a | 0.58 | Jag1 | 2.32 | Gpr83 | 0.55 |
| Csmd3 | 1.67 | Lbp | 0.59 | Sfrp2 | 2.32 | Slc38a4 | 0.55 |
| Fam60a | 1.67 | Gabra5 | 0.60 | Frmd3 | 2.30 | Edil3 | 0.55 |
| Foxg1 | 1.66 | Car14 | 0.60 | Xirp2 | 2.29 | Pcolce2 | 0.55 |
| Atoh1 | 1.65 | Wee2 | 0.62 | Il1rapl2 | 2.26 | Kcna4 | 0.55 |
| Robo2 | 1.65 | Cited4 | 0.63 | Spire2 | 2.25 | Sema3d | 0.56 |
| Unc5c | 1.65 | Ndrg1 | 0.63 | Robo2 | 2.25 | Scn7a | 0.56 |
| Chrng | 1.64 | Adm | 0.64 | Tectb | 2.22 | Ly6a | 0.57 |
| Tmem2 | 1.64 | Cntn1 | 0.64 | Mybpc1 | 2.22 | Matn4 | 0.57 |
| Pls1 | 1.64 | Agt | 0.64 | Kcnh8 | 2.22 | Dio2 | 0.57 |
| Mageb16 | 1.63 | Aldh1l1 | 0.64 | Rorb | 2.19 | Mup11 | 0.57 |
| Plch1 | 1.62 | Ano5 | 0.64 | Pla2g4e | 2.19 | Pth1r | 0.57 |
| Prox1 | 1.62 | Cmtm5 | 0.64 | Prox1 | 2.17 | Acan | 0.58 |
| Ar | 1.62 | Ampd3 | 0.64 | Hs6st2 | 2.14 | Fam180a | 0.58 |
| Prkg1 | 1.61 | Tmem38b | 0.64 | Pcsk2 | 2.13 | Mup7 | 0.58 |
| Pappa2 | 1.61 | Itgbl1 | 0.65 | Tsga14 | 2.12 | Ncam2 | 0.58 |
| Scn9a | 1.61 | Otoa | 0.65 | Ush1c | 2.12 | Neto2 | 0.58 |
| Six2 | 1.59 | Slc17a7 | 0.65 | Ppapdc1a | 2.11 | Atp13a5 | 0.59 |
Lists of genes showing >1.5-fold difference (p<0.05) between apex and base at P0 or P8. Shown in the table are fifty genes for each group ranked by the average fold difference Fold difference was determined by comparing expression levels in two biological replicates, each of which were pooled from at least 80 cochlear tissues.
Classification of Gene Profiles Based on Spatial and Temporal Expression Patterns
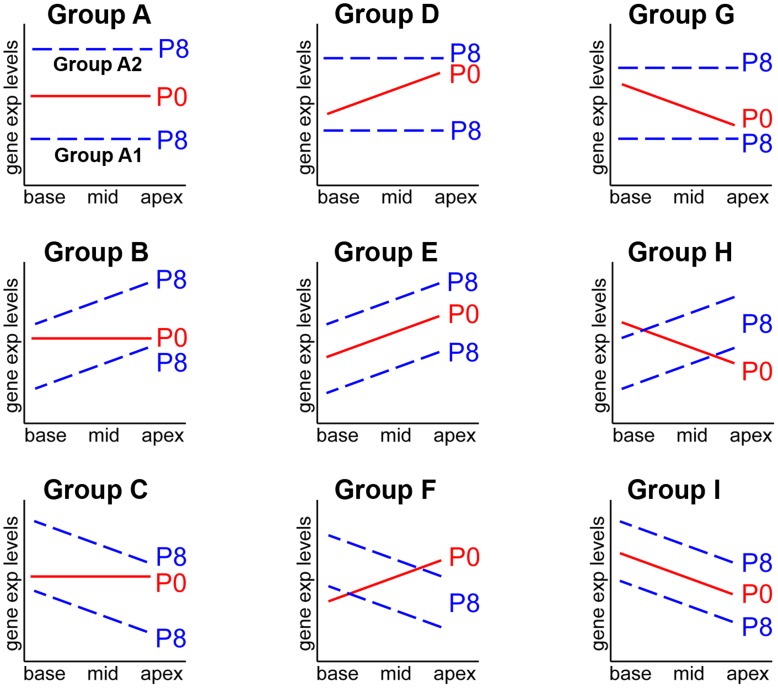
We noticed that there were many genes that exhibited similar or different gradients at P0 and P8 (Table 1 and Table S1). For example, Fst, Otog and Tectb showed higher expression in the apex at both P0 and P8, but Trpa1, P2rx3 and Atoh1 were expressed more in the apex at P0, but no longer differentially expressed along the tonotopic axis at P8. Thus, we further classified the gene profiles according to the spatial (along the tonotopic axis) as well as temporal (between P0 and P8) expression patterns. The entire gene profiles were classified into three groups: 1) genes showing no gradient along the tonotopic axis, 2) genes showing increasing gradient towards the apex and 3) genes showing decreasing gradient towards the apex. Then, the expression patterns were compared between P0 and P8, which yielded nine groups displaying distinct expression patterns (Figure 2).
Figure 2. Schematic representation of distinct gene expression patterns along the tonotopic axis at P0 and P8.
Gene profiles obtained from the microarray were classified based on the spatial (along the tonotopic axis) as well as temporal (between P0 and P8) expression changes. Expression levels of individual genes were compared between base and apex at P0, and classified into three categories: relatively constant along the cochlear duct (average fold-changes <1.5), higher in the apex or base (average fold-changes >1.5). Then, genes in each category were subdivided into three groups base on the expression patterns at P8 (constant, higher in the base or apex), which yielded nine distinct groups. See text for details.
This analysis revealed that a majority of genes (>95%) were expressed relatively constantly along the tonotopic axis at both P0 and P8. Among these genes (no gradient at both P0 and P8), gene showing significant fold-changes between P0 and P8 (fold-change >2, p<0.05) were classified as Group A: some of them were down-regulated (Group A1) or up-regulated (Group A2) during this period. Among the genes showing no gradient at P0, some genes displayed an increasing (Group B) or decreasing (Group C) gradient at P8. Genes showing an increasing gradient toward the apex at P0 were also subcategorized into three groups, comprising genes no longer showing a gradient at P8 (Group D), retaining the increasing gradient (Group E) or showing the opposite (decreasing) gradient at P8 (Group F). Finally, genes showing a decreasing gradient toward the apex at P0 were similarly subcategorized as genes no longer showing a gradient at P8 (Group G), showing the opposite (increasing) gradient (Group H) or retaining the decreasing gradient at P8 (Group I). Examples of genes included in each group were shown in Table 2 (the complete list of the genes in each group can found in Table S2). The possible functional significance of the genes in each group is discussed below.
Table 2. Classification of genes differentially expressed along the tonotopic axis of the mouse cochlea during neonatal development.
| P0 | P8 | P0 | P8 | P0 | P8 | |||||||||
| Group | Gene | P8/P0 | apex/ | apex/ | Group | Gene | P8/P0 | apex/ | apex/ | Group | Gene | P8/P0 | apex/ | apex/ |
| base | base | base | base | base | base | |||||||||
| A1 | Gpr64 | 0.16 | 1.42 | 1.25 | C | Tnmd | 3.95 | 0.86 | 0.28 | E | Fst | 0.72 | 3.26 | 4.25 |
| Dcx | 0.18 | 1.27 | 1.44 | Ifitm5 | 2.30 | 1.02 | 0.35 | Isl1 | 0.50 | 2.03 | 3.41 | |||
| Smoc2 | 0.19 | 1.06 | 1.48 | Iyd | 1.92 | 0.90 | 0.41 | Zpld1 | 0.82 | 4.11 | 2.98 | |||
| Stmn2 | 0.21 | 1.19 | 1.33 | Cntn6 | 1.92 | 0.68 | 0.41 | Slco5a1 | 0.50 | 1.98 | 2.80 | |||
| Bex1 | 0.25 | 1.25 | 0.72 | Rarres1 | 4.54 | 1.27 | 0.43 | Muc15 | 0.92 | 1.73 | 2.75 | |||
| Agtr2 | 0.25 | 0.80 | 0.89 | Trdn | 2.04 | 1.02 | 0.44 | Plch1 | 1.02 | 1.62 | 2.74 | |||
| Cldn6 | 0.26 | 1.10 | 1.27 | Crym | 0.25 | 0.98 | 0.46 | Otog | 0.71 | 1.50 | 2.38 | |||
| Bub1 | 0.28 | 1.41 | 0.97 | Pi15 | 1.22 | 0.76 | 0.47 | Pla2r1 | 0.78 | 1.56 | 2.37 | |||
| Spock3 | 0.29 | 1.04 | 1.16 | Ptgfr | 1.79 | 0.87 | 0.47 | Epha3 | 1.24 | 1.92 | 2.33 | |||
| Igf2bp3 | 0.29 | 1.19 | 0.93 | Fgl2 | 1.75 | 1.11 | 0.48 | Jag1 | 0.53 | 1.57 | 2.32 | |||
| A2 | Clca5 | 12.35 | 0.96 | 1.05 | Prss35 | 0.80 | 1.20 | 0.49 | Robo2 | 0.59 | 1.65 | 2.25 | ||
| Slco1c1 | 12.03 | 0.87 | 0.98 | Pgf | 0.99 | 0.91 | 0.50 | Tectb | 1.74 | 2.21 | 2.22 | |||
| Phex | 9.13 | 1.02 | 0.75 | Mup2 | 2.93 | 0.95 | 0.50 | Rorb | 4.03 | 2.10 | 2.19 | |||
| Emilin2 | 9.11 | 1.15 | 0.87 | Dnase1 | 4.43 | 0.79 | 0.50 | Prox1 | 0.39 | 1.62 | 2.17 | |||
| Hhatl | 8.80 | 0.77 | 0.82 | H19 | 0.58 | 1.08 | 0.51 | Pcsk2 | 0.98 | 1.85 | 2.13 | |||
| Scin | 8.79 | 1.12 | 1.08 | Rgs4 | 2.73 | 0.98 | 0.51 | Ush1c | 1.10 | 1.52 | 2.12 | |||
| Chad | 7.88 | 1.19 | 0.67 | Myoc | 3.67 | 1.09 | 0.52 | Chst15 | 0.20 | 1.54 | 1.99 | |||
| Ces1d | 6.82 | 1.45 | 1.47 | Otos | 6.17 | 0.76 | 0.52 | Ndst3 | 0.56 | 2.97 | 1.78 | |||
| Bhmt | 6.74 | 0.96 | 1.48 | Nox3 | 1.06 | 0.85 | 0.52 | Fam70b | 0.57 | 1.53 | 1.75 | |||
| Prss36 | 5.93 | 1.02 | 0.73 | Smpd3 | 4.97 | 1.17 | 0.53 | Clic6 | 0.64 | 2.28 | 1.74 | |||
| B | Tecta | 0.22 | 1.32 | 5.94 | D | Zmat4 | 0.38 | 2.16 | 1.18 | Foxg1 | 0.47 | 1.66 | 1.71 | |
| Nov | 0.74 | 1.40 | 3.53 | Galntl6 | 0.67 | 2.14 | 1.29 | Sema3a | 0.68 | 2.21 | 1.70 | |||
| Chrna3 | 4.01 | 0.89 | 3.28 | Fndc3c1 | 0.43 | 2.07 | 0.97 | Igf2bp2 | 0.72 | 1.57 | 1.68 | |||
| Glb1l2 | 1.18 | 1.10 | 2.93 | Tac1 | 0.56 | 2.01 | 1.06 | F | No gene is included | |||||
| Syt1 | 0.61 | 1.27 | 2.91 | Trpa1 | 0.53 | 1.79 | 1.15 | G | Atp6v0a4 | 2.92 | 0.24 | 0.76 | ||
| Smpx | 1.12 | 1.30 | 2.90 | Tyrp1 | 0.84 | 1.78 | 0.91 | Pkhd1l1 | 0.71 | 0.28 | 1.20 | |||
| Gsbs | 2.63 | 1.08 | 2.81 | Abcg5 | 0.43 | 1.78 | 1.17 | Ucma | 13.86 | 0.33 | 0.69 | |||
| Ccdc148 | 0.32 | 1.48 | 2.73 | P2rx3 | 0.23 | 1.77 | 1.04 | Slco1a4 | 8.81 | 0.42 | 0.75 | |||
| Slitrk6 | 0.83 | 1.13 | 2.72 | Pdzrn4 | 0.84 | 1.75 | 1.10 | Pkp1 | 1.94 | 0.44 | 0.71 | |||
| Rbm24 | 1.32 | 1.08 | 2.63 | Cdh4 | 0.25 | 1.72 | 1.31 | Gjb6 | 2.70 | 0.47 | 0.93 | |||
| Cybrd1 | 0.51 | 0.97 | 2.60 | Fat3 | 0.56 | 1.71 | 1.01 | Cntn5 | 0.23 | 0.48 | 0.79 | |||
| Sox2 | 0.51 | 1.33 | 2.56 | Angpt1 | 0.63 | 1.70 | 1.35 | Scg2 | 1.32 | 0.48 | 1.24 | |||
| Adamts19 | 0.51 | 1.47 | 2.56 | Slc39a8 | 1.14 | 1.69 | 1.32 | Atp6v1c2 | 2.09 | 0.51 | 1.05 | |||
| Kcna10 | 2.28 | 0.68 | 2.53 | Dchs2 | 1.20 | 1.68 | 1.16 | Clstn2 | 0.95 | 0.51 | 0.75 | |||
| Rassf6 | 1.98 | 1.29 | 2.45 | Csmd3 | 0.54 | 1.67 | 1.02 | H | Fcrlb | 2.58 | 0.33 | 2.02 | ||
| Cubn | 1.66 | 1.24 | 2.39 | Atoh1 | 0.32 | 1.65 | 1.21 | I | Hoxa2 | 1.16 | 0.32 | 0.28 | ||
| Sfrp2 | 0.43 | 1.14 | 2.32 | Unc5c | 0.79 | 1.65 | 0.84 | A2m | 2.87 | 0.37 | 0.40 | |||
| Xirp2 | 0.79 | 0.98 | 2.29 | Chrng | 0.69 | 1.64 | 1.12 | Kcna4 | 1.81 | 0.47 | 0.55 | |||
| Il1rapl2 | 1.73 | 1.25 | 2.26 | Pls1 | 0.54 | 1.64 | 1.48 | Grin2a | 0.76 | 0.58 | 0.65 | |||
| Spire2 | 0.94 | 1.36 | 2.25 | Mageb16 | 0.78 | 1.63 | 1.13 | Ppp1r1b | 2.17 | 0.50 | 0.66 | |||
Genes showing >1.5-fold difference (p<0.05) between apex and/or base (apex/base) were included in the classification. Fold difference between P0 and P8 was calculated with the average values of base, middle, and apex in each age. Shown in the table are examples of genes included in each group. A complete list can be found in the Table S2.
Validation of the Microarray Data with Quantitative RT-PCR
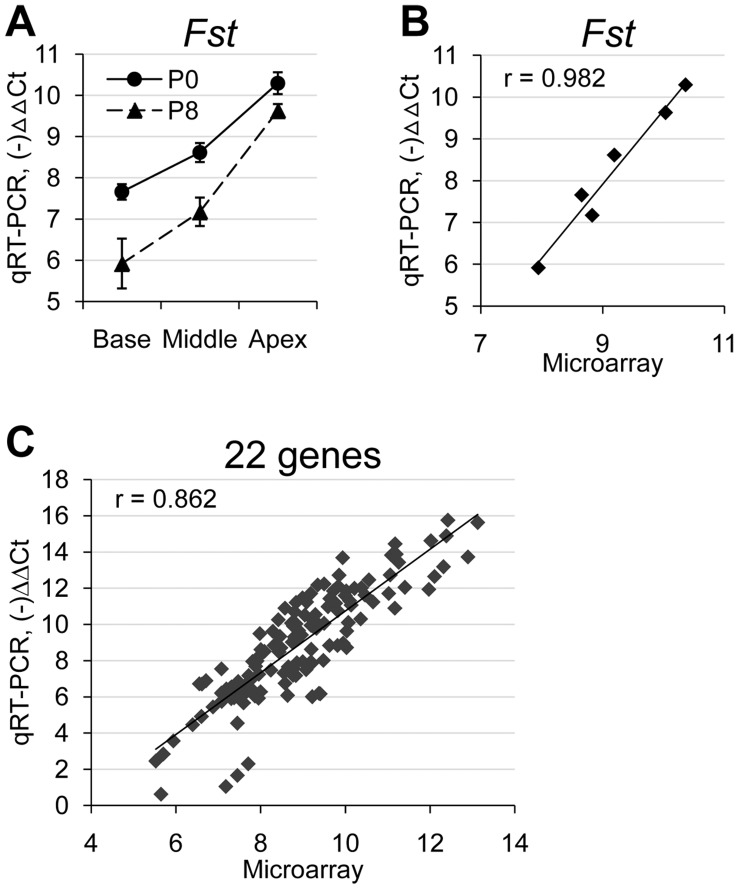
We validated our microarray data by performing quantitative real time PCR (qRT-PCR) on 22 genes selected from various groups (Fig. 3 and Table 3). Genes for qRT-PCR were selected among ones showing higher fold changes in each group such as Stmn2, Tecta, Tnmd, Fst, Atp6v0a4, Hoxa2, and A2m, and also among ones previously shown to be associated with cochlear development or function such as Kcnj10, Kcnq1, Ptprq, Tectb, Otog and Ush1c. As an example, qRT-PCR results for Fst (Follistatin: Group E) showed an increasing gradient toward the apex at both P0 and P8 (Fig. 3A). When the qRT-PCR results were plotted as a function of the values from the microarray, close correlation was clearly shown (Fig. 3B, Pearson correlation coefficient, r = 0.982). Similarly, all 22 selected genes generally showed close correlations between the microarray data and qRT-PCR results (Pearson correlation coefficient, r = 0.862) (Fig. 3C).
Figure 3. Comparison between qRT-PCR and microarray data showed high correlation.
(A) An example of qRT-PCR results for Fst, which showed increasing base-to-apex gradients in expression levels at both P0 and P8. (B) An example showing high correlation between qRT-PCR results and microarray data for Fst (Pearson correlation coefficient, r = 0.982). (C) Comparisons between qRT-PCR results and microarray data for all 22 genes examined showed good correlations (r = 0.862).
Table 3. Validation of microarray data with quantitative RT-PCR.
| Average fold change (Apex/Base) | Average fold change (P8/P0)2 | |||||||
| Microarray | qRT-PCR | Pearson’s Correlation Coefficient | Microarray | qRT-PCR | ||||
| Group | Gene | P0(A/B) | P8(A/B) | P0(A/B) | P8(A/B) | (P8/P0) | (P8/P0) | |
| A1 | Stmn2 | 1.19 | 1.33 | 2.38 | 2.84 | 0.949 | 0.21 | 0.08 |
| Ccnb1 | 1.37 | 0.90 | 1.64 | 0.89 | 0.995 | 0.34 | 0.24 | |
| A2 | kcnj10 | 0.69 | 0.90 | 0.42 | 0.88 | 0.999 | 3.71 | 21.88 |
| Kcnq1 | 0.82 | 0.85 | 0.56 | 0.52 | 0.960 | 2.13 | 3.17 | |
| B | Tecta | 1.32 | 5.94 | 2.20 | 7.56 | 0.957 | 0.22 | 0.03 |
| Ptprq | 0.78 | 2.11 | 0.27 | 2.87 | 0.837 | 1.17 | 1.81 | |
| Chrna10 | 0.84 | 1.92 | 0.55 | 2.96 | 0.796 | 1.11 | 0.83 | |
| Fzd4 | 1.15 | 1.54 | 1.74 | 1.09 | 0.900 | 1.39 | 1.69 | |
| Nrcam | 1.40 | 1.99 | 1.84 | 2.55 | 0.989 | 0.62 | 0.51 | |
| C | Tnmd | 0.86 | 0.28 | 0.81 | 0.12 | 0.981 | 3.95 | 17.79 |
| Crym | 0.98 | 0.46 | 1.48 | 0.24 | 0.982 | 0.25 | 0.12 | |
| D | Pdzrn4 | 1.75 | 1.10 | 2.80 | 1.15 | 0.845 | 0.84 | 0.70 |
| Cdh4 | 1.72 | 1.31 | 2.89 | 1.08 | 0.997 | 0.25 | 0.04 | |
| E | Fst | 3.26 | 4.25 | 6.23 | 13.07 | 0.982 | 0.72 | 0.41 |
| Tectb | 2.21 | 2.22 | 4.86 | 12.86 | 0.953 | 1.74 | 2.87 | |
| Otog | 1.50 | 2.38 | 1.48 | 3.84 | 0.982 | 0.71 | 0.59 | |
| Ush1c | 1.52 | 2.12 | 1.75 | 3.93 | 0.967 | 1.10 | 0.98 | |
| G | Atp6v0a4 | 0.24 | 0.76 | 0.06 | 0.54 | 0.996 | 2.92 | 10.59 |
| Slco1a4 | 0.42 | 0.75 | 0.41 | 0.36 | 0.992 | 8.81 | 13.70 | |
| I | Hoxa2 | 0.32 | 0.28 | 0.06 | 0.01 | 0.942 | 1.16 | 0.85 |
| A2m | 0.37 | 0.40 | 0.21 | 0.16 | 0.997 | 2.87 | 3.95 | |
| Ppp1r1b | 0.50 | 0.66 | 0.26 | 0.35 | 0.969 | 2.17 | 4.95 | |
Fold changes between P0 and P8 (P8/P0) were calculated using the average values of the base, middle, and apex for each age.
In Table 3, we calculated fold changes between the apex and base with the results from qRT-PCR, and compared to those from the microarray. For genes from Group A showing no gradient along the tonotopic axis, average expression levels between P0 and P8 were compared. The results from qRT-PCR appeared to be more sensitive than the microarray data in verifying the changes between the base and the apex. Nevertheless, the expression patterns from the qRT-PCR results were generally consistent with those from the microarray.
Functional Annotation Clustering Analysis
Our microarray analysis yielded an extensive list of genes that are differentially expressed along the tonotopic axis during neonatal development. Analysis of the dynamic expression patterns of individual genes may highlight some key players that are critical to cochlear maturation including the acquisition of tonotopy (Table 2 and Table S2). However, these single-gene analyses may fail to identify important biological processes, which are often regulated collectively by a group of genes involved in common biological function or pathways. Also, biologically significant gene expression changes occurring in a certain group of cells can be missed due to masking by genes expressed in other cell types or inherent noise signals. Thus, in order to evaluate our microarray data at the level of biological function and processes, we performed functional annotation clustering analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatics resources [22]. This analysis identified biological functions and processes specifically enriched in the gene list of each group, which were summarized in Table 4.
Table 4. Functional annotation clustering analysis on the differentially expressed genes.
| Group | Total genes | Enrich. Score1 | Biological annotations | Ratio (%)2 | p-value3 | FDR (%) | Genes |
| A1 | 168 | 8.42 | GO:0022402∼cell cycle process | 13.13 | 1.15E-10 | 1.75E-07 | Bub1 C79407 Ccna2 Ccnb1 Ccnb2 Cdca3 Cks2 Dbc1 Dbf4 Dlgap5 f630043a04rik Hells Kif11 Mki67 Ncaph Plk1 Racgap1 Sgol2 Spag5 Stmn1 Tpx2 |
| 6.86 | GO:0005694∼chromosome | 15.0 | 1.24E-13 | 1.51E-10 | Bub1 C79407 Cenpk F630043a04rik H2afy2 H2afz Hells Hist1h1a Hist1h1b Hist1h2ab Hist1h2af Hist1h2an Hist1h2ao Hist1h2bb Hist1h2bj Hist1h2bl Hist1h2bm Hist1h2bn Hist1h3a Hist1h3b Hist1h3c Hist1h3d Hist1h3e Hist1h3g Hist1h3h Hist1h3i Hist2h3b Hist2h3c1 Hm11277 Hmgb2 Hmgb3 Mki67 Sgol2 Spag5 Tmpo Top2a Zfp57 | ||
| 1.92 | GO:0031012∼extracellular matrix | 6.9 | 3.61E-04 | 0.43868 | Aspn Col6a3 Egfl6 Fbn2 Fras1 Matn1 Oc90 Postn Smoc2 Spock3 Tgfbi | ||
| 1.68 | GO:0022832∼voltage-gated channel activity | 4.4 | 0.00254 | 3.18051 | Cacna1h Kcna6 Kcnh7 Scn2a1 Scn3a Scn3b Scn9a | ||
| 1.51 | GO:0015630∼microtubule cytoskeleton | 10.0 | 7.94E-06 | 0.00968 | Ccnb1 Ckap2 Dlgap5 F630043a04rik Kif2c Kif4 Kif11 Kif15 Kif21b Kif23 Myc Racgap1 Sncg Spag5 Stmn1 Tpx2 | ||
| A2 | 245 | 8.94 | GO:0005578∼proteinaceous extracellular matrix | 9.2 | 4.31E-10 | 5.3E-07 | Abi3bp Chad Chl1 Col4a3 Col4a4 Col8a1 Col24a1 Cpz Ecm2 Emilin2 Entpd2 Fgf1 Gpld1 Kazald1 Kera Lama2 Lgals3bp Npnt Ntn4 Prss36 Spon1 Tnfrsf11b Wnt2b |
| 5.25 | GO:0007155∼cell adhesion | 8.8 | 1.56E-05 | 0.02553 | Anxa9 Atp1b2 Bcam Cd9 Cd36 Cd44 Chl1 Clca5 Cntn4 Col4a3 Col8a1 Col24a1 Emilin2 Itga10 Lama2 Lgals3bp Ninj2 Npnt Sorbs3 Spon1 Tesk2 Wisp1 | ||
| 1.78 | GO:0006811∼ion transport | 8.4 | 0.00113 | 1.82672 | Atp1b2 Atp1b3 Atp2a3 Cacng4 Clca5 Fxyd3 Fxyd5 Glrb Kcne4 Kcnj2 Kcnj10 Kcnj13 Kcnq1 P2rx2 Slc5a5 Slc9a2 Slc10a4 Slc41a2 Slco1c1 Steap3 Tcn2 | ||
| B | 125 | 2.41 | GO:0030182∼neuron differentiation | 0.8 | 0.0015 | 2.28299 | Gata2 Isl2 Lingo1 Mtap2 Nr2e1 Nrcam Slitrk2 Slitrk6 Sox2 Wnt7a |
| 1.88 | GO:0044459∼plasma membrane part | 2.4 | 4.47E-05 | 0.05379 | Apba1 Aqp4 Aqp5 Chrna3 Chrna7 Chrna10 Chrnb4 Cldn9 Cldn23Cubn Cybrd1 Egflam Faim2 Fn1 Kcna10 Kcnb2 Nrcam OtoF Otog Pclo Pla2g4e RgS6 Rgs9 Syt1 Rims2 Six1 Slc6a14 Slc22a3 Sox2 Tecta Tmprss9 Xirp2 | ||
| C | 84 | 2.12 | GO:0005578∼proteinaceous extracellular matrix | 0.9 | 0.00373 | 3.92815 | Acan Crtap Ctgf Nid2 Ntn1 Trf Vcan |
| 1.74 | GO:0006812∼cation transport | 1.2 | 7.95E-04 | 1.13346 | Atp13a5 Kcnh5 Kcnma1 Scn7a Slc4a10 Slc5a3 Slc38a4 Slc39a14 Trf | ||
| D | 46 | 1.87 | GO:0033555∼multicellular organismal response to stress | 7.4 | 7.93E-05 | 0.11043 | P2rx3 Scn9a Tac1 Trpa1 |
| E | 27 | 3.01 | GO:0030182∼neuron differentiation | 28.6 | 9.95E-07 | 0.00138 | Foxg1 Isl1 Jag1 Rorb Robo2 Sema3a Ush1c Ush2a |
| 1.77 | GO:0007423∼sensory organ development | 25.0 | 1.34E-06 | 0.00186 | Foxg1 Jag1 Prox1 Rorb Six2 Ush1c Ush2a | ||
| 1.53 | glycosylation site:N-linked (GlcNAc…) | 46.4 | 0.00117 | 1.44099 | Ndst3 Fst Jag1 Epha3 Pcsk2 Zpld1 Robo2 Chst15 Sema3a Tectb Pla2r1 Muc15 Ush2a | ||
| F | 0 | No gene set was significantly enriched | |||||
| G | 42 | 1.94 | GO:0006811∼ion transport | 21.3 | 7.43E-05 | 0.10516 | Ano5 Atp6v0a4 Atp6v1c2 Cp Gabra5 Kcnj16 Slc17a7 Slco1a4 Tmem38b Wnk4 |
| 1.66 | GO:0007155∼cell adhesion | 17.0 | 5.79E-04 | 0.81684 | Agt Cldn11 Clstn2 Cntn1 Cntn5 Itga4 Pkp1 Vcam1 | ||
| H | 1 | No gene set was significantly enriched | |||||
| I | 5 | No gene set was significantly enriched |
Enrichment scores indicate relative importance (enrichment) of the biological annotations.
Ratio (%) indicates the percentage of the genes included the enriched annotations among the total number of genes in the group.
p-value was calculated with a modified Fisher’s exact test to indicate the significance of the enrichment of the annotations.
In Group A1 (no gradient, down-regulated at P8), genes associated with cell cycle process or chromosomal activity were enriched. Such enrichments indicated that cell proliferation occurring at P0 is generally diminished at P8, which is consistent with active maturation processes observed during neonatal cochlear development. Interestingly, genes associated with microtubule cytoskeleton such as kinesin family members were also shown to be down-regulated at P8, which may be related with a reduced cell cycle and with disassembly of the microtubule-based kinocilium in the hair cells during this period.
On the other hand, Group A2 (no gradient, up-regulated at P8) contained many genes associated with extracellular matrix and cell adhesion such as Col4a3, Col4a4 and Emilin2. Upregulation of these genes may explain the maturation of the cytoarchitecture in the cochlea during the postnatal period. Consistent with our results, Emilin2 was shown to be rapidly accumulated during postnatal development in the extracellular matrix surrounding the collagenous fibers in the basilar membrane [23]. Genes associated with ion transport such as Kcnq1, Kcne1 and Kcnj10 were also enriched. It has been shown that Kcnq1 and Kcne1 are expressed in strial marginal cells and that Kcnj10 in intermediate cells plays a critical role in potassium recycling in the endolymph [24]. Thus, enrichment of such gene sets in P8 cochlea is consistent with the establishment of endocochlear potential during the first postnatal week in the mouse cochlea [25].
In Group B (no gradient at P0, increasing base-to-apex gradient at P8), genes associated with neuron differentiation and plasma membrane were enriched. Several genes involved in cochlear development and function were included in this group such as Tecta, Gata2, Slitrk6, Chrna10, Nrcam, Otof and Otog. The higher expression of these genes in the apex at P8 may be related to the base-to-apex wave of cochlear maturation, since the increasing base-to-apex gradient for some genes resulted from down-regulation at the base by P8. In support of this notion, two genes in this group, Ptprq and Tecta, which are both important for inner ear development, were no longer expressed in the adult cochlea [26], [27]. Interestingly, several genes related to non-sensory structures such as the basilar or tectorial membrane were included in this group. Tecta and Otog, which encode for α-tectorin and otogelin, respectively, that are components of the tectorial membrane [28], were expressed at higher levels in the apex. Tectb, encoding β-tectorin and classified in Group E, also showed higher expression in the apex at P8. The higher expression levels of genes associated with the tectorial membrane in the apex may be related with the decreasing base-to-apex gradient in stiffness of the tectorial membrane, an important factor for tonotopic discrimination [29].
In Group C (no gradient at P0, decreasing base-to-apex gradient at P8), genes related to extracellular matrix were enriched. The biological annotation for proteinaceous extracellular matrix was also shown to be enriched in Group A2 with different sets of genes, suggesting that different types of extracellular matrix components are required at different stages of postnatal cochlear development. Also enriched were genes associated with cation transport. Interestingly, splicing variants of Kcnma1, which encodes for the Maxi-K+ (BK) channel have been shown to be differentially expressed along the tonotopic axis and also during developmental stages in mice and chicken [8], [30]. Kcnma1 expression patterns from our microarray data, showing no gradient at P0 and higher expression in the base at P8, did not closely match with previous reports. This may be because the microarray technique did not distinguish the splicing variants, but displayed a summed expression level of all the variants expressed at the time of analysis. Since differential expressions of specific splice variants have been suggested to contribute to tonotopy [2], it may be necessary to profile the entire transcriptome along the tonotopic axis using deep-sequencing technologies such as the RNA-seq approach [31].
In Group D (increasing base-to-apex gradient at P0, no gradient at P8), enriched genes were related with multicellular organismal responses to stress including P2rx3 and Trpa1. Consistent with our data, P2rx3, encoding the P2X3 subunit of ATP-gated ion channels, was shown to be expressed at higher levels in the apical cochlea at P0, and its expression is gradually decreased and no longer detected after the onset of hearing [32]. Similarly, expression of Trpa1, a subunit for transient receptor potential (TRP) channels, was greatly increased in the apex at around P0 and then decreased to show no gradient by P6 [13]. These expression changes precede the onset of hearing, suggesting their roles in the acquisition of mature cochlear function such as the mechanosensitivity of the hair cells.
In Group E (increasing base-to-apex gradient at both P0 and P8), genes associated with neuron differentiation and sensory organ development were enriched, which is consistent with the ongoing maturation processes in the apical cochlea during neonatal stages (Fig. 1) [10], [11].
In Group G (decreasing base-to-apex gradient at P0, no gradient at P8), genes related to ion transport and cell adhesion were enriched. For most genes in this group, the expression gradient at P0, higher in the base, was lost at P8 due to increased expression of these genes towards the apex during the postnatal development. Since ion transport and cell adhesion molecules are closely associated with cochlear remodeling and function, this expression pattern may be related with the base-to-apex wave of cochlear maturation. Consistently, these gene sets were also shown to be enriched in Group A2, in which genes are up-regulated at P8, showing no gradient.
Finally, in Groups F (higher expression in the apex at P0 and in the base at P8), H (higher expression in the base at P0 and in the apex at P8) and I (higher expression in the base at both P0 and P8), no functionally related genes were significantly enriched.
Confirmation of Expression Patterns for Selected Genes by in Situ Hybridization
We next performed in situ hybridization for selected genes to confirm their differential expression patterns along the tonotopic axis. We selected a few genes previously shown to be expressed in the cochlea such as Chrna10, Ptprq, Kcnj10, Tectb, and Tnc, as well as a few genes that exhibited higher fold changes along the tonotopic axis among the newly identified genes from our analysis such as A2m, Nov, and Fst.
We first performed in situ hybridization on Chrna10, Ptprq, Kcnj10, and Tetcb, whose expression patterns have previously been reported, to confirm our microarray results [26], [27], [33], [34]. Expression domains of these genes within the neonatal cochlea were consistent with the previous reports, and their graded expression patterns along the tonotopic axis generally matched with the microarray data (see Fig. S1 and Fig. S2 for details). Expression pattern of A2m was also consistent with the microarray data (Fig. S2).
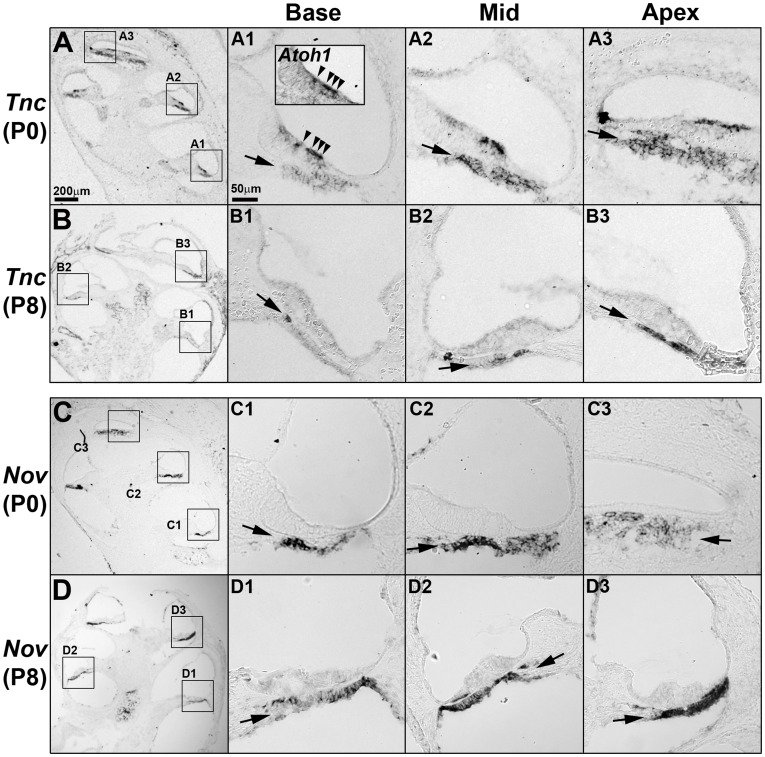
We next examined the expression patterns of the few newly identified genes from our microarray analysis. We found that Nov (nephroblastoma overexpressed gene), identified in Group B, was expressed in the basilar membrane with no obvious gradient at P0 and in an increasing base-to-apex gradient at P8, consistent with the microarray results (Fig. 4C,D, arrows). The Nov (also called as CCN3) gene encodes for a matricellular protein that associates with the extracellular matrix and mediates a variety of cellular functions including proliferation, differentiation, survival, migration and cytoskeleton reorganization [35]. Since the structural organization of the extracellular matrix regulates the mechanical properties of the basilar membrane, gradual changes in Nov expression may contribute to the frequency tuning of the basilar membrane [5]. The expression pattern of Nov in the basilar membrane was compared with that of Tnc, which encodes the glycoprotein tenascin C and has been shown to be expressed in the developing mouse cochlea including the basilar membrane [36]. Tnc was strongly expressed in the hair cells and the basilar membrane along the cochlear duct at P0 (Fig. 4A,B). While Tnc expression in the hair cells was downregulated at P8 (Fig. 4A,B, arrowheads), its expression in the basilar membrane continued in an increasing base-to-apex gradient at P8, consistent with the microarray results (Table 1). The specific expression in the basilar membrane with gradual changes along the cochlear duct of Nov and Tnc suggest that these extracellular matrix proteins may be involved in the developmental regulation of the basilar membrane, specifying differential properties along the tonotopic axis.
Figure 4. Expression patterns of Tnc and Nov in the cochlea during neonatal development.
Expression patterns of Tnc (A,B) and Nov (C,D) were examined by in situ hybridization at P0 (A,C) and P8 (B,D). (A,B) Tnc transcripts were observed in the basilar membrane in an increasing gradient toward the apex at both P0 and P8 (A1–B3, arrows). Tnc expression was also positive in the differentiating hair cells (Atoh1 expression domain) at P0 (inset in A1, arrowheads), but was down-regulated at P8. (C,D) Nov was expressed in the basilar membrane at P0 and P8. Nov expression levels were relatively constant at P0 along the cochlear duct and gradually decreased toward the apex by P8 (D). Scale bar in A (200 µm) applies to B, C, and D; scale bar in A1 (50 µm) applies to A1–A3, B1–B3, C1–C3, and D1–D3.
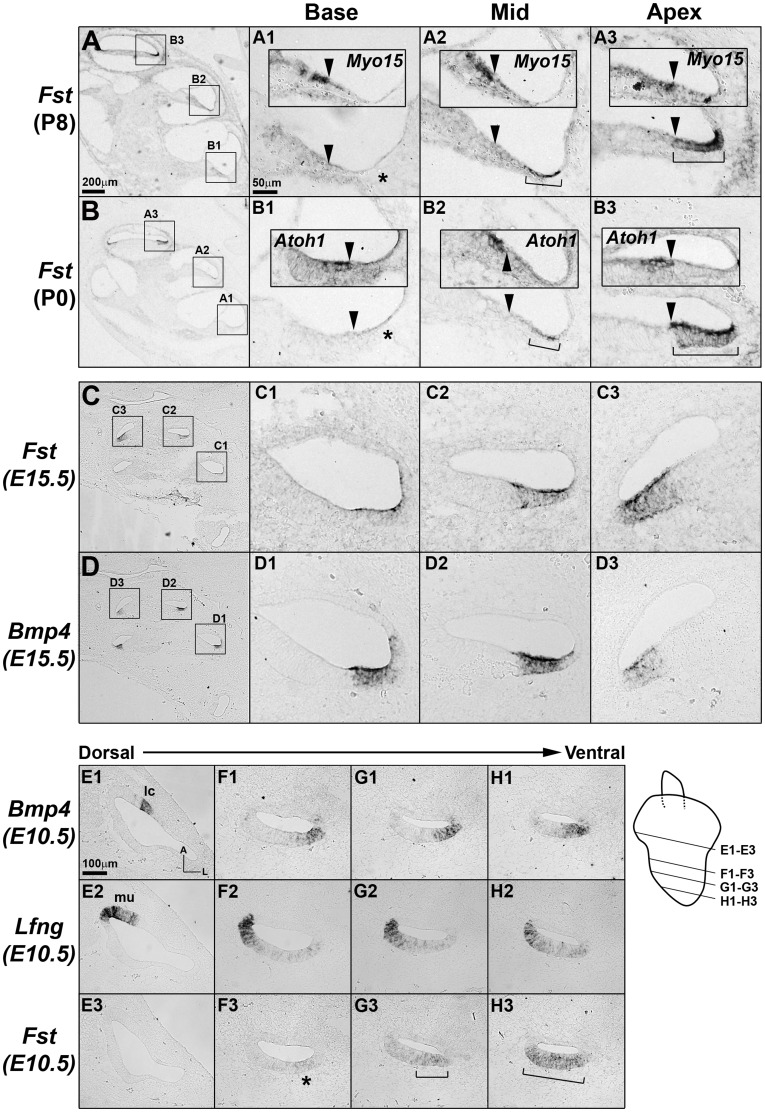
We also examined the expression patterns of Fst (Follistatin), a TGF-β/BMP antagonist, which was identified in Group E and displayed high fold-changes along the tonotopic axis at both P0 and P8 (Table 1 and 2). Fst expression was barely detected in the basal turn of the cochlear duct, began to be observed in the lateral side of the lesser epithelial ridge (LER) in the middle turn and then expanded to the entire LER in the apical turn at both P0 and P8 (Fig. 5A,B, brackets). Since Bmp4 and Bmp7 are also expressed in the LER region (Fig. 5D) [37], the gradual expansion of the Fst expression domain implies gradual decreases of BMP activities toward the apex. We then asked when this graded Fst expression pattern is established. At E15.5 when cochlear morphogenesis is almost complete, the graded expression patterns of Fst were comparable to those observed at P0 and P8 (Fig. 5C). Thus, we examined the inner ears at E10.5, when the primordial cochlear structure just begins to form in the ventral aspect of the otocyst. When examined from dorsal to ventral sections of the otocyst, Fst expression was not detected in the vestibular structures (Fig. 5E1–E3), and began to be faintly observed at the level where Bmp4 and Lfng were expressed in the lateral and medial side of the developing cochlea, respectively (Fig. 5F1–F3) [38]. Fst expression became stronger and broader toward the apical cochlear sections, while Bmp4 and Lfng expression patterns were not changed (Fig. 5G1–H3, brackets). However, at E9.5 when the cochlear anlage has yet to be developed, Fst expression was not detected in the otocyst (data not shown). These results indicate that the longitudinal gradient of Fst expression is established at the time of cochlear specification and maintained throughout cochlear development including embryonic and neonatal stages. Considering the role of BMP signaling in hair cell differentiation and inner ear development [37], [39], [40], the graded expression patterns of Fst suggest a possible role for Follistatin in the specification of gradually different properties along the cochlear duct such as the tonotopy.
Figure 5. Expression patterns of Fst in the cochlea during embryonic and neonatal development.
Fst transcripts were present in the lesser epithelial ridge (LER) region of the cochlea in an increasing base-to-apex gradient during neonatal (A,B) and embryonic development (C-H). (A,B) At P0 and P8, Fst expression was barely detectable in the basal cochlear turns (A1,B1, asterisks), but was evident in the lateral side of the LER in the middle turns (A2,B2, brackets) and its expression domain included the entire LER region in the apical turns (A3,B3, brackets). Insets in A1–B3 show expression domains of Myo15 or Atoh1 in the adjacent sections to indicate the location of hair cells. Arrowheads indicate the lateral border of differentiating hair cells. (C,D) Fst expression patterns at E15.5 were comparable to those observed at P0 and P8. Fst expression domains largely overlapped that of Bmp4 in the LER area. (E1–H3) The increasing gradient of Fst expression patterns in the cochlea was already apparent in the cochlear primordium at E10.5 (F3,G3,H3, asterisk, brackets). Fst expression was not detectable in the presumptive vestibular organs including the lateral crista (E1–E3). Expression domains of Bmp4 (E1,F1,G1,H1) and Lfng (E2,F2,G2,H2) indicated the lateral and medial regions of the developing cochlea, respectively. The schematic diagram indicates the level of sections for the pictures shown in E1–H3. Scale bar in A (200 µm) applies to B-D; scale bar in A1 (50 µm) applies to A1–A3, B1–B3, C1–C3, and D1–D3; Scale bar in E1 (100 µm) applies to E1–H3.
Conclusions
In this study, we applied the microarray technique to identify genes that are differentially expressed along the longitudinal axis of the mouse cochlea during neonatal development. Our analyses identified several candidate genes, which may be involved in cochlear development and acquisition of tonotopy, but have not been appreciated previously. Although the biological relevance of these candidates in cochlear development remains to be determined, the gene expression profiles obtained in this study will be a useful resource for studying various aspects of mammalian cochlear development.
Materials and Methods
Tissue Dissection and RNA Extraction
All animals were handled in accordance with the guidelines for the Care and Use of Laboratory Animals of Yonsei University College of Medicine. The protocol for obtaining cochlear samples was approved by the Committee on Animal Research at Yonsei University College of Medicine (#10-104). Eighty temporal bones were dissected from C57BL/6 mice at P0 and P8. After removing the otic capsule, the cochlear tissues including the organ of Corti, spiral limbus, and lateral wall was dissected in sterile, chilled Hanks’ balanced salt solution under a stereomicroscope (Olympus SZX10). The cochlear tissues were divided into three equal pieces representing the base, middle and apex, and pooled separately (P0-base, P0-middle, P0-apex, P8-base, P8-middle, P8-apex). The pooled cochlear tissues were homogenized with a micro-homogenizer (Microtec, Chiba, Japan), and total RNA was extracted using an RNeasy Mini Kit (Qiagen, Hilden, Germany). The quality and quantity of the extracted total RNA was measured using the Total RNA Nano Assay (Agilent Technologies, Palo Alto, CA, USA). The same procedure was repeated to obtain additional biological replicates for each cochlear sample (P0-base, P0-middle, P0-apex, P8-base, P8-middle, P8-apex).
Microarray Data Analysis
Gene expression profiles were generated using the Affymetrix GeneChip© Mouse Gene 1.0 ST oligonucleotide arrays (Affymetrix, Santa Clara, California). RNA samples isolated from two biological replicates were sent to the local Affymetrix GeneChip© service provider (DNA LINK, Seoul, Korea). Three hundred nano-grams of high quality total RNA was converted into double-stranded cDNA using a random hexamer conjugated with a T7 promoter sequence. Amplified RNA (cRNA) was generated from the double-stranded cDNA using an in vitro transcription (IVT) reaction and then re-converted into cDNA with a mix of dNTPs containing dUTP. This cDNA was then fragmented by uracil-DNA glycosylase (UDG) and APE1 restriction endonucleases and labeled with a biotinylated dideoxynucleotide via a terminal transferase reaction. End-labeled cDNA was hybridized for 16 hours at 45° and 60 rpm according to the Gene Chip Whole Transcript (WT) Sense Target Labeling Assay Manual (Affymetrix). After hybridization, the chips were stained and washed in a Genechip Fluidics Station 450 (Affymetrix) and scanned using a Genechip Array scanner 3000 7G (Affymetrix). Affymetrix Command Console Software version 1.1 was used to process the raw images into CEL files. Gene expression estimates were normalized using the robust multiarray averaging (RMA) method. Microarray data were deposited in the GEO database (#GSE34187) and followed MIAME requirements.
Genes showing an average fold-change greater than 1.5 (p<0.05, unpaired t-test) between the base and the apex and exhibiting a log-transformed gene expression level greater than 6 were considered to be significantly differentially expressed. The differentially expressed genes were further classified into nine groups based on the fold-changes along the tonotopic axis (base vs. apex) and during neonatal development (P0 vs. P8) (Fig. 2). Gene lists in each group were subjected to functional annotation clustering analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/home.jsp). The analysis was performed by following the procedure previously described [22]. Biological annotations with an enrichment score of >1.5, P-value (<0.01) and false discovery rate (FDR, ≤5%) were considered biologically interesting (Table 4).
Quantitative Real-time PCR
Quantitative real-time PCR (qRT-PCR) was performed to confirm the microarray data. RNAs purified from three biological replicates for each of the cochlear samples (two biological replicates used for the microarray analysis and one additional (third) biological replicate collected on a small scale) were used for qRT-PCR. cDNAs were synthesized from each RNA (2 µg) sample using poly (dT)20 and the Superscript III RT kit according to manufacturer’s instructions (Invitrogen). No-RT controls were not included. cDNAs were diluted three times with water and the same amount of cDNAs were used for all PCR reactions. qRT-PCR for 22 selected genes was performed in triplicate wells using SYBR Green PCR Master Mix and the ABI 7500 machine (Applied Biosystems, Carlsbad, California). The samples were denatured at 94°C for 10 minutes and then subjected to 40 cycles of denaturing at 94°C for 15 seconds and annealing and extension at 60°C for 1 minute. We normalized gene expression level (ΔCt) with β-actin (Actb). Relative expression levels were determined by comparative methods (ΔΔCt). Relationships between the microarray data and qRT-PCR were assessed with Pearson correlation coefficient (r) (Table 3). Primers used for qRT-PCR were designed by using the Primer3 program [41]. At least one primer (either forward or reverse) was designed to include an exon-exon junction sequences to avoid amplification of genomic DNA. All the primer sequences are listed in Table S3.
Phalloidin Staining and Measurement for the Angles of Stereociliary Bundles
Stereociliary bundles were visualized by staining the organ of Corti with Alexa Fluor® 488 Phalloidin (Invitrogen) at P0, P4, P8 and P21. Pictures were taken from 10%, 30%, 50%, 70%, and 90% positions from the basal end of the cochlea duct using a confocal microscopy (Zeiss LSM700). The angle formed by two lines drawn along the medial side of the hair bundles of outer hair cells was measured using the Image J program (National Institutes of Health, USA) (Fig. 1B). Angles for at least thirty outer hair cells were measured for each cochlear region and plotted as a function of relative distance from the basal end of the cochlea.
In Situ Hybridization
In situ hybridization was performed as previously described [38]. Probes for Atoh1 [42], Myo15 [43], Bmp4 and Lfng [38] were prepared as previously described. RNA probes for Fst were generated from a 313 base pair (bp) mouse Fst cDNA containing the +36 to +348 coding region (MN_008046); for Tnc, from a 404 bp mouse Tnc cDNA containing the +5835 to +6060 coding region and 178 bp 3′ untranslated region (NM_011607); for Nov, from a 966 bp mouse Nov cDNA containing the 38 bp 5' untranslated region and +1 to +928 coding region (NM_010930); for Chrna10, from a 645 bp mouse Chrna10 cDNA containing the +882 to +1344 coding region and 182 bp 3′ untranslated region (NM_001081424); for Ptprq, from a 455 bp mouse Ptprq cDNA containing the 2 bp 5' untranslated region and +1 to +453 coding region (NM_001081432); for Kcnj10, from a 714 bp mouse Kcnj10 cDNA containing the +749 to +1140 coding region and 322 bp 3′ untranslated region (NM_001039484); for Tectb, from a 406 bp mouse Tectb cDNA containing the +801 to +990 coding region and 216 bp 3′ untranslated region (NM_009348); for A2m, from a 1018 bp mouse A2m cDNA containing the 180 bp 5' untranslated region and +1 to +1000 coding region (NM_175628).
Supporting Information
Expression patterns of Tectb, Chrna10, and Ptprq in the cochlea during neonatal development. Expression patterns of Tectb (A,B), Chrna1 (C,D) and Ptprq (E, F) were examined by in situ hybridization at P0 (A,C,E) and P8 (B,D,F). (A,B) Tectb was classified in Group E showing increasing gradients towards the apex at both P0 and P8. Consistent with the microarray data, Tectb showed higher expression towards the apex at both P0 (A1–A3) and P8 (B1–B3). In the middle turn of P0 cochlea, Tectb expression was found in the lateral margin of greater epithelial ridge, pillar cells, and the third row of Deiter’s cells, consistent with a previous report [26]. Expression domains of Tectb were either diminished in the base or expanded in the apex (A1–A3). At P8, tectb expression in the pillar cells was disappeared and its increasing gradient towards the apex maintained (B1–B3). (C, D) Chrna10 was classified in Group B showing no gradient at P0 and an increasing gradient towards the apex at P8. Consistently, Chrna10 is expressed in the differentiating hair cells relatively constantly along the tonotopic axis at P0 (C1–C3) and in an increasing gradient towards the apex at P8 (D1–D3). (E, F) Ptprq was also classified in Group B showing no gradient at P0 and an increasing gradient towards the apex at P8. However, Ptprq transcripts are observed in the differentiating hair cells with slightly higher expression in the base at P0 (E1–E3, arrowheads), which represents the qRT-PCR results more rather than the microarray data (Table 3). At P8, Ptprq transcripts are observed in an increasing gradient towards the apex at P8, consistent with the microarray and qRT-PCR results (F1–F3). Both Ptprq and Chrna10 were down-regulated in the base at P8, which may reflect the base-to-apex progression of hair cell maturation.
(TIF)
Expression patterns of A2m and Kcnj10 in the cochlea during embryonic and neonatal development. Expression patterns of A2m (A,B) and Kcnj10 (C,D) were examined by in situ hybridization at P0 (A,C) and P8 (B,D). (A,B) A2m was classified in Group I showing decreasing gradients towards the apex at both P0 and P8. Consistent with the microarray data, A2m transcripts are observed in the Reissner’s membrane in a decreasing gradient toward the apex at P0 (A1–A3). Interestingly, A2m expression is no longer detected in the Reissner’s membrane at P8, but is observed in the spiral ligament area retaining the decreasing gradient toward the apex (B1–B3). Kcnj10 was classified in Group A1 showing no gradient at both P0 and P8 with up-regulation at P8. Consistent with the microarray data, Kcnj10 expression was barely detected in the P0 cochlea, but clearly observed in the stria vascularis and the organ of Corti at P8.
(TIF)
Complete list of the differentially expressed genes between apex and base of the mouse cochlear at P0 or P8.
(XLSX)
Complete list of genes in each group (Groups A to H), classified based on spatial and temporal expression patterns.
(XLSX)
Primer sets used for qRT-PCR.
(DOCX)
Acknowledgments
We thank Dr. Doris Wu for critical reading of the manuscript, and Ms. Hyun Joo Kim, Mr. Harinarayana Ankamerddy and Dr. Naga Mahesh Jakka for technical assistance. We also thank Yonsei-Carl Zeiss Advanced Imaging Center, Yonsei University College of Medicine, for technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-0023589 to Dr. Son, 2010-0009623 to Dr. Bok). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Davis RL. Gradients of neurotrophins, ion channels, and tuning in the cochlea. Neuroscientist. 2003;9:311–316. doi: 10.1177/1073858403251986. [DOI] [PubMed] [Google Scholar]
- 2.Mann ZF, Kelley MW. Development of tonotopy in the auditory periphery. Hear Res. 2011;276:2–15. doi: 10.1016/j.heares.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Davis RL, Liu Q. Complex primary afferents: What the distribution of electrophysiologically-relevant phenotypes within the spiral ganglion tells us about peripheral neural coding. Hear Res. 2011;276:34–43. doi: 10.1016/j.heares.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyer AC, Frank T, Khimich D, Hoch G, Riedel D, et al. Tuning of synapse number, structure and function in the cochlea. Nat Neurosci. 2009;12:444–453. doi: 10.1038/nn.2293. [DOI] [PubMed] [Google Scholar]
- 5.Ren T. Longitudinal pattern of basilar membrane vibration in the sensitive cochlea. Proc Natl Acad Sci U S A. 2002;99:17101–17106. doi: 10.1073/pnas.262663699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gummer AW, Hemmert W, Zenner HP. Resonant tectorial membrane motion in the inner ear: its crucial role in frequency tuning. Proc Natl Acad Sci U S A. 1996;93:8727–8732. doi: 10.1073/pnas.93.16.8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato T, Doi K, Hibino H, Kubo T. Analysis of gene expression profiles along the tonotopic map of mouse cochlea by cDNA microarrays. Acta Otolaryngol. 2009. [DOI] [PubMed]
- 8.Frucht CS, Uduman M, Kleinstein SH, Santos-Sacchi J, Navaratnam DS. Gene Expression Gradients along the Tonotopic Axis of the Chicken Auditory Epithelium. J Assoc Res Otolaryngol. 2011;12:423–435. doi: 10.1007/s10162-011-0259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kowalik L, Hudspeth AJ. A search for factors specifying tonotopy implicates DNER in hair-cell development in the chick’s cochlea. Dev Biol. 2011;354:221–231. doi: 10.1016/j.ydbio.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lelli A, Asai Y, Forge A, Holt JR, Geleoc GS. Tonotopic gradient in the developmental acquisition of sensory transduction in outer hair cells of the mouse cochlea. J Neurophysiol. 2009;101:2961–2973. doi: 10.1152/jn.00136.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Etournay R, Lepelletier L, Boutet de Monvel J, Michel V, Cayet N, et al. Cochlear outer hair cells undergo an apical circumference remodeling constrained by the hair bundle shape. Development. 2010;137:1373–1383. doi: 10.1242/dev.045138. [DOI] [PubMed] [Google Scholar]
- 12.Waguespack J, Salles FT, Kachar B, Ricci AJ. Stepwise morphological and functional maturation of mechanotransduction in rat outer hair cells. J Neurosci. 2007;27:13890–13902. doi: 10.1523/JNEUROSCI.2159-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asai Y, Holt JR, Geleoc GS. A quantitative analysis of the spatiotemporal pattern of transient receptor potential gene expression in the developing mouse cochlea. J Assoc Res Otolaryngol. 2010;11:27–37. doi: 10.1007/s10162-009-0193-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roth B, Bruns V. Postnatal development of the rat organ of Corti. II. Hair cell receptors and their supporting elements. Anat Embryol (Berl) 1992;185:571–581. doi: 10.1007/BF00185616. [DOI] [PubMed] [Google Scholar]
- 15.Hartman BH, Basak O, Nelson BR, Taylor V, Bermingham-McDonogh O, et al. Hes5 expression in the postnatal and adult mouse inner ear and the drug-damaged cochlea. J Assoc Res Otolaryngol. 2009;10:321–340. doi: 10.1007/s10162-009-0162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murata J, Tokunaga A, Okano H, Kubo T. Mapping of notch activation during cochlear development in mice: implications for determination of prosensory domain and cell fate diversification. J Comp Neurol. 2006;497:502–518. doi: 10.1002/cne.20997. [DOI] [PubMed] [Google Scholar]
- 17.White PM, Doetzlhofer A, Lee YS, Groves AK, Segil N. Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature. 2006;441:984–987. doi: 10.1038/nature04849. [DOI] [PubMed] [Google Scholar]
- 18.Oshima K, Grimm CM, Corrales CE, Senn P, Martinez Monedero R, et al. Differential distribution of stem cells in the auditory and vestibular organs of the inner ear. J Assoc Res Otolaryngol. 2007;8:18–31. doi: 10.1007/s10162-006-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rio C, Dikkes P, Liberman MC, Corfas G. Glial fibrillary acidic protein expression and promoter activity in the inner ear of developing and adult mice. J Comp Neurol. 2002;442:156–162. doi: 10.1002/cne.10085. [DOI] [PubMed] [Google Scholar]
- 20.Jin ZH, Kikuchi T, Tanaka K, Kobayashi T. Expression of glutamate transporter GLAST in the developing mouse cochlea. Tohoku J Exp Med. 2003;200:137–144. doi: 10.1620/tjem.200.137. [DOI] [PubMed] [Google Scholar]
- 21.Bok J. Building the mammalian cochlea - an overview. Genes Genom. 2010;32:1–7. [Google Scholar]
- 22.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Amma LL, Goodyear R, Faris JS, Jones I, Ng L, et al. An emilin family extracellular matrix protein identified in the cochlear basilar membrane. Mol Cell Neurosci. 2003;23:460–472. doi: 10.1016/s1044-7431(03)00075-7. [DOI] [PubMed] [Google Scholar]
- 24.Wangemann P. K+ cycling and the endocochlear potential. Hear Res. 2002;165:1–9. doi: 10.1016/s0378-5955(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 25.Sadanaga M, Morimitsu T. Development of endocochlear potential and its negative component in mouse cochlea. Hear Res. 1995;89:155–161. doi: 10.1016/0378-5955(95)00133-x. [DOI] [PubMed] [Google Scholar]
- 26.Rau A, Legan PK, Richardson GP. Tectorin mRNA expression is spatially and temporally restricted during mouse inner ear development. J Comp Neurol. 1999;405:271–280. [PubMed] [Google Scholar]
- 27.Goodyear RJ, Legan PK, Wright MB, Marcotti W, Oganesian A, et al. A receptor-like inositol lipid phosphatase is required for the maturation of developing cochlear hair bundles. J Neurosci. 2003;23:9208–9219. doi: 10.1523/JNEUROSCI.23-27-09208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodyear RJ, Richardson GP. Extracellular matrices associated with the apical surfaces of sensory epithelia in the inner ear: molecular and structural diversity. J Neurobiol. 2002;53:212–227. doi: 10.1002/neu.10097. [DOI] [PubMed] [Google Scholar]
- 29.Richter CP, Emadi G, Getnick G, Quesnel A, Dallos P. Tectorial membrane stiffness gradients. Biophys J. 2007;93:2265–2276. doi: 10.1529/biophysj.106.094474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakai Y, Harvey M, Sokolowski B. Identification and quantification of full-length BK channel variants in the developing mouse cochlea. J Neurosci Res. 2011;89:1747–1760. doi: 10.1002/jnr.22713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang LC, Ryan AF, Cockayne DA, Housley GD. Developmentally regulated expression of the P2X3 receptor in the mouse cochlea. Histochem Cell Biol. 2006;125:681–692. doi: 10.1007/s00418-005-0119-4. [DOI] [PubMed] [Google Scholar]
- 33.Morley BJ, Simmons DD. Developmental mRNA expression of the alpha10 nicotinic acetylcholine receptor subunit in the rat cochlea. Brain Res Dev Brain Res. 2002;139:87–96. doi: 10.1016/s0165-3806(02)00514-x. [DOI] [PubMed] [Google Scholar]
- 34.Hibino H, Horio Y, Inanobe A, Doi K, Ito M, et al. An ATP-dependent inwardly rectifying potassium channel, KAB-2 (Kir4. 1), in cochlear stria vascularis of inner ear: its specific subcellular localization and correlation with the formation of endocochlear potential. J Neurosci. 1997;17:4711–4721. doi: 10.1523/JNEUROSCI.17-12-04711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCallum L, Irvine AE. CCN3–a key regulator of the hematopoietic compartment. Blood Rev. 2009;23:79–85. doi: 10.1016/j.blre.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Whitlon DS, Zhang X, Kusakabe M. Tenascin-C in the cochlea of the developing mouse. J Comp Neurol. 1999;406:361–374. doi: 10.1002/(sici)1096-9861(19990412)406:3<361::aid-cne5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 37.Jagger DJ, Forge A. Assessing PCP in the Cochlea of Mammalian Ciliopathy Models. Methods Mol Biol. 2012;839:239–248. doi: 10.1007/978-1-61779-510-7_19. [DOI] [PubMed] [Google Scholar]
- 38.Morsli H, Choo D, Ryan A, Johnson R, Wu DK. Development of the mouse inner ear and origin of its sensory organs. J Neurosci. 1998;18:3327–3335. doi: 10.1523/JNEUROSCI.18-09-03327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohyama T, Basch ML, Mishina Y, Lyons KM, Segil N, et al. BMP Signaling Is Necessary for Patterning the Sensory and Nonsensory Regions of the Developing Mammalian Cochlea. J Neurosci. 2010;30:15044–15051. doi: 10.1523/JNEUROSCI.3547-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang W, Lin Z, Kulessa H, Hebert J, Hogan BL, et al. Bmp4 is essential for the formation of the vestibular apparatus that detects angular head movements. PLoS Genet. 2008;4:e1000050. doi: 10.1371/journal.pgen.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 42.Helms AW, Johnson JE. Progenitors of dorsal commissural interneurons are defined by MATH1 expression. Development. 1998;125:919–928. doi: 10.1242/dev.125.5.919. [DOI] [PubMed] [Google Scholar]
- 43.Goodyear R, Richardson G. Pattern formation in the basilar papilla: evidence for cell rearrangement. The Journal of neuroscience. 1997;17:6289–6301. doi: 10.1523/JNEUROSCI.17-16-06289.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression patterns of Tectb, Chrna10, and Ptprq in the cochlea during neonatal development. Expression patterns of Tectb (A,B), Chrna1 (C,D) and Ptprq (E, F) were examined by in situ hybridization at P0 (A,C,E) and P8 (B,D,F). (A,B) Tectb was classified in Group E showing increasing gradients towards the apex at both P0 and P8. Consistent with the microarray data, Tectb showed higher expression towards the apex at both P0 (A1–A3) and P8 (B1–B3). In the middle turn of P0 cochlea, Tectb expression was found in the lateral margin of greater epithelial ridge, pillar cells, and the third row of Deiter’s cells, consistent with a previous report [26]. Expression domains of Tectb were either diminished in the base or expanded in the apex (A1–A3). At P8, tectb expression in the pillar cells was disappeared and its increasing gradient towards the apex maintained (B1–B3). (C, D) Chrna10 was classified in Group B showing no gradient at P0 and an increasing gradient towards the apex at P8. Consistently, Chrna10 is expressed in the differentiating hair cells relatively constantly along the tonotopic axis at P0 (C1–C3) and in an increasing gradient towards the apex at P8 (D1–D3). (E, F) Ptprq was also classified in Group B showing no gradient at P0 and an increasing gradient towards the apex at P8. However, Ptprq transcripts are observed in the differentiating hair cells with slightly higher expression in the base at P0 (E1–E3, arrowheads), which represents the qRT-PCR results more rather than the microarray data (Table 3). At P8, Ptprq transcripts are observed in an increasing gradient towards the apex at P8, consistent with the microarray and qRT-PCR results (F1–F3). Both Ptprq and Chrna10 were down-regulated in the base at P8, which may reflect the base-to-apex progression of hair cell maturation.
(TIF)
Expression patterns of A2m and Kcnj10 in the cochlea during embryonic and neonatal development. Expression patterns of A2m (A,B) and Kcnj10 (C,D) were examined by in situ hybridization at P0 (A,C) and P8 (B,D). (A,B) A2m was classified in Group I showing decreasing gradients towards the apex at both P0 and P8. Consistent with the microarray data, A2m transcripts are observed in the Reissner’s membrane in a decreasing gradient toward the apex at P0 (A1–A3). Interestingly, A2m expression is no longer detected in the Reissner’s membrane at P8, but is observed in the spiral ligament area retaining the decreasing gradient toward the apex (B1–B3). Kcnj10 was classified in Group A1 showing no gradient at both P0 and P8 with up-regulation at P8. Consistent with the microarray data, Kcnj10 expression was barely detected in the P0 cochlea, but clearly observed in the stria vascularis and the organ of Corti at P8.
(TIF)
Complete list of the differentially expressed genes between apex and base of the mouse cochlear at P0 or P8.
(XLSX)
Complete list of genes in each group (Groups A to H), classified based on spatial and temporal expression patterns.
(XLSX)
Primer sets used for qRT-PCR.
(DOCX)