Numblike, a negative regulator in glioma cell migration and invasion, was found to mediate nuclear factor kappa B activation by suppressing tumor necrosis factor receptor–associated factor 5.
Abstract
The Notch signaling regulator Numblike (Numbl) is expressed in the brain, but little is known regarding its role in the pathophysiology of glial cells. In this paper, we report that Numbl expression was down-regulated in high-grade human glioma tissue samples and glioblastoma cell lines. To investigate the role of Numbl in glioma migration and invasion, we generated human glioma cell lines in which Numbl was either overexpressed or depleted. Overexpression of Numbl suppressed, while elimination of Numbl promoted, the migration and invasion of glioma cells. Numbl inhibited glioma migration and invasion by dampening NF-κB activity. Furthermore, Numbl interacted directly with tumor necrosis factor receptor–associated factor 5 (TRAF5), which signals upstream and is required for the activation of NF-κB, and committed it to proteasomal degradation by promoting K48-linked polyubiquitination of TRAF5. In conclusion, our data suggest that Numbl negative regulates glioma cell migration and invasion by abrogating TRAF5-induced activation of NF-κB.
INTRODUCTION
Gliomas are the most frequent primary brain tumors, accounting for >50% of all brain tumors (Kleihues et al., 2002). Glioblastoma, the most malignant form, is characterized by increased proliferation and invasion into the surrounding normal brain tissue (Rasheed et al., 1999). In spite of the use of multi-modal therapies, including surgery and radio- and chemotherapy, the mean survival rate in patients with glioma remains extremely low (Maher et al., 2001). The highly lethal nature of this tumor results from the acquisition of an invasive phenotype that allows the tumor cells to infiltrate the surrounding brain tissue (Chintala and Rao, 1996). Therefore innovative approaches that target the invasion of glioma are urgently needed. Although studies have shown that various stimuli promote glioma cell invasion, the underlying mechanisms remain largely unknown.
Nuclear factor kappa B (NF-κB) is a transcription factor that regulates a host of biological events (Karin et al., 2002). During the activation of NF-κB, its inhibitory subunit (IκBα) undergoes phosphorylation, ubiquitylation, and degradation, leading to nuclear translocation of the RelA (p65)–RelB (p50) complex (Ghosh and Karin, 2002; Li and Verma, 2002; Pomerantz and Baltimore, 2002). It has been reported that constitutive activation of NF-κB in glioma plays an important role in the regulation of genes involved in cellular adhesion, migration, and invasion (Wakabayashi et al., 2004; Tran et al., 2006; Sarkar et al., 2008). In contrast, suppressed NF-κB activation impedes glioma migration and invasion. Tumor necrosis factor receptor (TNFR)-associated factors (TRAFs) were identified originally as signal-transducing molecules for TNFR and the interleukin-1 (IL-1) receptor–induced NF-κB activation (Ichikawa et al., 2005; Jackson-Bernitsas et al., 2007). TRAFs, including TRAF1, -2, -3, -5, and -6, have been shown to interact directly or indirectly with members of the TNFR superfamily (Arron et al., 2002; Ha et al., 2009). It has been reported TRAF1 and TRAF2 are involved in glioma proliferation and drug-induced apoptosis (Conti et al., 2005; Angileri et al., 2008). However, whether TRAFs are involved in glioma cell migration and invasion is unknown.
Drosophila Numb (d-Numb) is a key protein involved in neural precursor asymmetric division, segregating preferentially into one of the daughter cells to ensure the two daughters adopt different cell fates (Uemura et al., 1989). In mammals, Numb and Numblike (Numbl) have been identified as homologues of d-Numb. Numbl is primarily localized in the cytoplasm and has redundant functions with Numb in embryonic neurogenesis (Zhong et al., 1996, 1997). Mouse Numb and Numbl can directly bind and inhibit the Notch1 intracellular domain, leading to suppressed Notch signaling (Petersen et al., 2002; Li et al., 2003). Recently Numbl has been found to suppress NF-κB activation via interaction with TRAF6 and TGF-β activated kinase 1 binding protein 2 (TAB2; Phillips et al., 2006; Purow et al., 2008; Brennan et al., 2009).
In the present study, we examined the role of Numbl in glioma cell migration and invasion. We found that a malignant glioma phenotype is associated with lower Numbl expression, as opposed to Numbl expression in normal brain tissue or a more benign glioma phenotype. Numbl directly binds to TRAF5 and promotes its polyubiquitination, followed by proteasomal degradation, thus terminating NF-κB activation. Consequently, invasion and migration of glioma cells is suppressed. Therefore our results suggest that Numbl may play an important role in the invasion and migration of glioma cells.
RESULTS
Numbl suppresses glioma migration and invasion
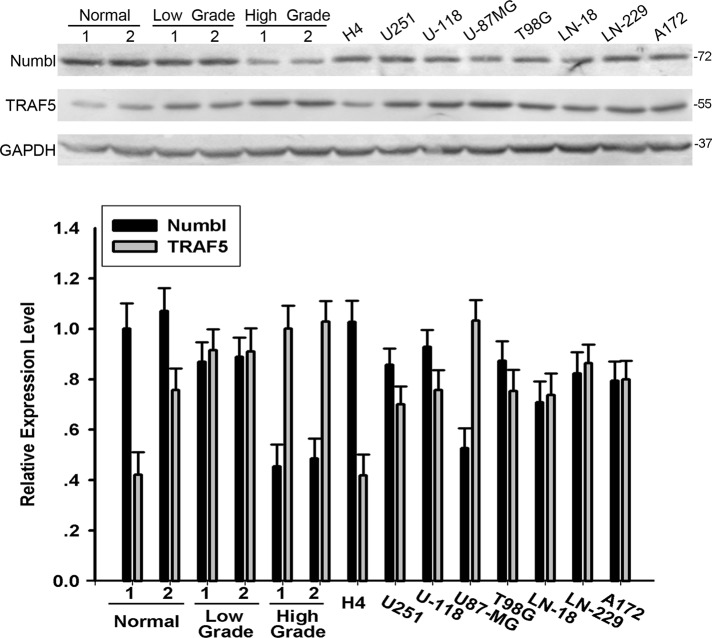
To probe the role of Numbl in the migration and invasion of glioma, we first analyzed the expression levels of Numbl in eight normal brain tissue samples, eight low-grade glioma samples (grades II and III), and eight high-grade glioma samples (grade IV, glioblastoma). As expected, Numbl expression was detectable in all the samples examined. In the eight low-grade glioma tissues, the expression level of Numbl was lower than that observed in the eight normal brain tissue samples (0.29008 ± 0.182531 vs. 1.14861 ± 0.19508) and the high-grade glioma tissue samples (0.29008 ± 0.182531 vs. 0.97915 ± 0.18055) (Figure 1 and Supplemental Figure S1). Migration to the nearby tissue and invasion of basement membrane by tumor cells are thought to be critical for glioma metastasis (Giese et al., 2003). Because glioblastoma is considered more invasive and migratory than other types of glioma, this piece of evidence indicates that Numbl may be involved in the regulation of glioma metastasis.
FIGURE 1:
Western blot analysis of Numbl expression levels in human glioma tissues and cell lines. A representative Western blot image of Numbl and TRAF5 expression in two normal brain tissues, two low-grade glioma tissues, two high-grade glioma tissues, and eight human glioma cell lines. Protein levels of Numbl and TRAF5 were quantified by correcting the average pixel intensity for each band with that of GAPDH as an internal control for equal loading of protein samples. The data are means ± SEM.
At the same time, we also measured Numbl expression in eight glioma cell lines (U251, U-87MG, U-118, T98G, LN-18, LN-229, A172, and H4). Numbl was expressed in all eight cell lines. Among these cell lines, H4 cells expressed the highest amount of intracellular Numbl, whereas U-87MG had the lowest (Figure 1). Previous studies have shown U-87MG cells are more aggressive in cell migration and invasion compared with H4 cells (Jacobs et al., 1997; Mertsch et al., 2009). Therefore, in the following studies, the U-87MG and H4 cell lines were used unless specified otherwise.
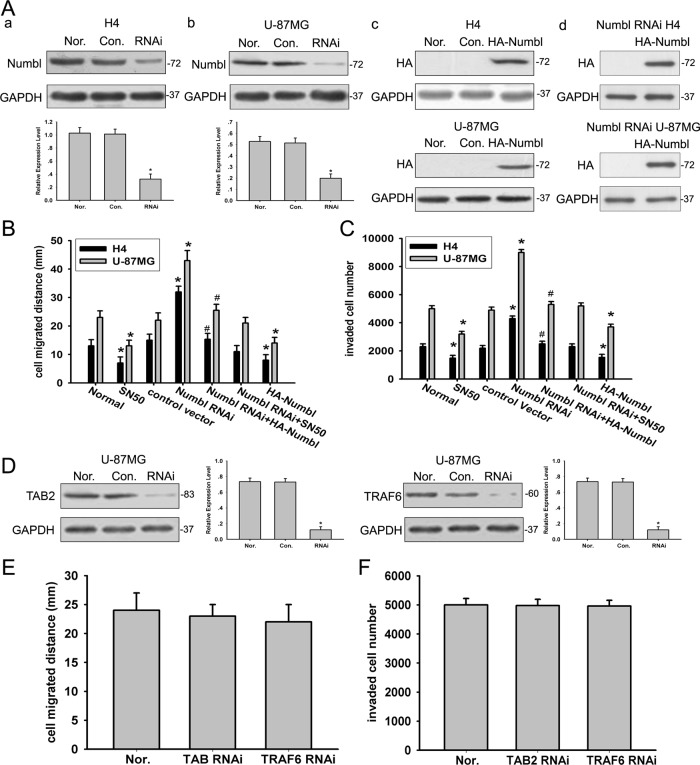
To determine whether the expression of Numbl was associated with glioma cell migration and invasion, we transfected stable cells with Numbl expression plasmid or knockdown plasmid were established. A 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay showed neither knockdown nor overexpression of Numbl impacted cell viabilities (Figure S2), essentially ruling out a possible role for Numbl in regulating apoptosis. On the other hand, a wound-healing assay indicated that the migratory capacity of cells transfected with small interfering RNA (siRNA) targeting Numbl was significantly elevated compared with cells transfected with nonspecific siRNA, whereas it was decreased in both types of glioma cells overexpressing Numbl (Figure 2, A and B). Next we probed the effect of Numbl on the invasion of glioma cell lines by Transwell assay. Depletion of Numbl by siRNA doubled the number of cells that penetrated the Matrigel, whereas ectopic expression of Numbl hampered the invasion of glioma cells (Figure 2C). We then transfected the Numbl-silenced cells with a construct encoding mouse Numbl; glioma migration and invasion were reversed compared with the Numbl knockdown cells (Figure 2, A–C). Taken together, our data indicate that Numbl may play an inhibitory role in glioma migration and invasion.
FIGURE 2:
Effect of Numbl, TRAF6, and TAB2 on glioma cell migration and invasion. (A) H4 and U-87MG glioma cells were stably transfected with the control vector (Con.), Numbl, siRNA (RNAi), and HA-Numbl expression vector as indicated. Protein expression of Numbl or HA-Numbl was analyzed by Western blotting. The data are means ± SEM. *, p < 0.05, compared with the control group. (B) Equal numbers of stably H4 and U-87MG cells were transfected with a plasmid encoding Numbl for 24 h and then seeded into six-well tissue culture plates. A wound-healing assay was performed and analyzed as described in Materials and Methods. The data are means ± SEM. *, p < 0.05, statistically significant compared with the control group; **, p < 0.05, statistically significant compared with Numbl RNAi group. (C) Aliquots of 1 × 106 stably transfected H4 and U-87MG cells were transfected with a plasmid encoding Numbl for 24 h and then cultured in Matrigel chambers. A Transwell assay was performed and analyzed as described in Materials and Methods. The data are means ± SEM. *, p < 0.05, compared with the control group; **, p < 0.05, statistically significant compared with Numbl RNAi group. (D) U-87MG glioma cells were transfected with the control RNAi vector (Con.), TAB2, or TRAF6 siRNA vector, as indicated. Protein expression of TAB2 and TRAF6 was analyzed by Western blotting. The data are means ± SEM. *, p < 0.05, statistically significant compared with the control group. (E and F) U-87MG glioma cells were transfected with knockdown vectors targeting TRAF6 or TAB2. Wound-healing (E) and Transwell (F) assays were performed and analyzed as described in Materials and Methods. The data are means ± SEM.
Numbl suppresses glioma cell invasion and migration by inhibiting NF-κB activation
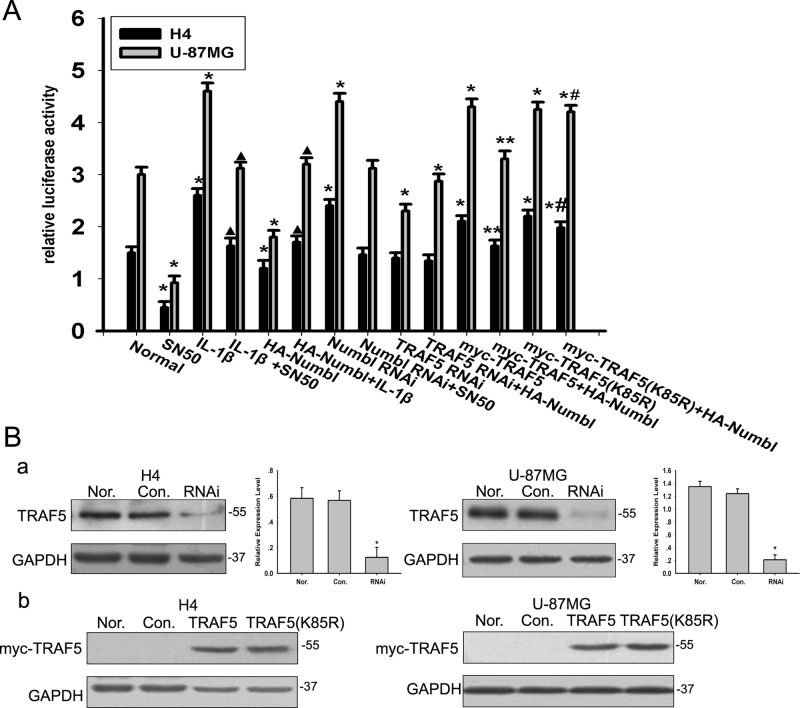
It has been reported that the NF-κB–dependent signaling pathway is involved in glioma migration and invasion (Wisniewski et al., 2010). We confirmed that inhibition of NF-κB activity by SN50, an NF-κB–specific inhibitor could indeed prevent the migration and invasion of glioma cells in vitro (Figure 2, B and C). Huo and colleagues recently reported that Numbl regulates NF-κB activation in HEK 293T cells (Ma et al., 2008). In glioma cells, overexpression of Numbl suppressed NF-κB transcriptional activity in both IL-1β–treated and untreated glioma cells (Figure 3A). Therefore we assessed the possibility that Numbl modulates glioma cell migration and invasion though the NF-κB signaling pathway. Enhanced migration and invasion of glioma cells resulting from Numbl knockdown was blunted in the presence of SN50 (Figure 2, B and C). Collectively, our data suggest that Numbl prevents migration and invasion of glioma cells by suppressing NF-κB activity.
FIGURE 3:
The effect of Numbl and TRAF5 on NF-κB activation. (A) Stable H4 and U-87MG cells were transfected with HA-Numbl, and then treated with SN50 or left untreated. Luciferase activities were detected in these transfected cells. The data are means ± SEM. *, p < 0.05, statistically significant compared with normal cell group; **, p < 0.05, statistically significant compared with TRAF5 overexpression group; #, p < 0.05, statistically significant compared with the Numbl reexpressed TRAF5 overexpressed cells; ▴, p < 0.05, statistically significant compared with the IL-1β–treated cells. (B) H4 and U-87MG glioma cells were transfected with the control RNAi vector (Con.), Numbl, siRNA (RNAi), myc-TRAF5, or myc-TRAF5(K85R) expression vector, as indicated. Protein expression of TRAF5 or myc-TRAF5 was analyzed by Western blotting. The data are means ± SEM. *, p < 0.05, compared with the control group.
Numbl binds to TRAF5 directly and suppresses TRAF5-dependent NF-κB activation
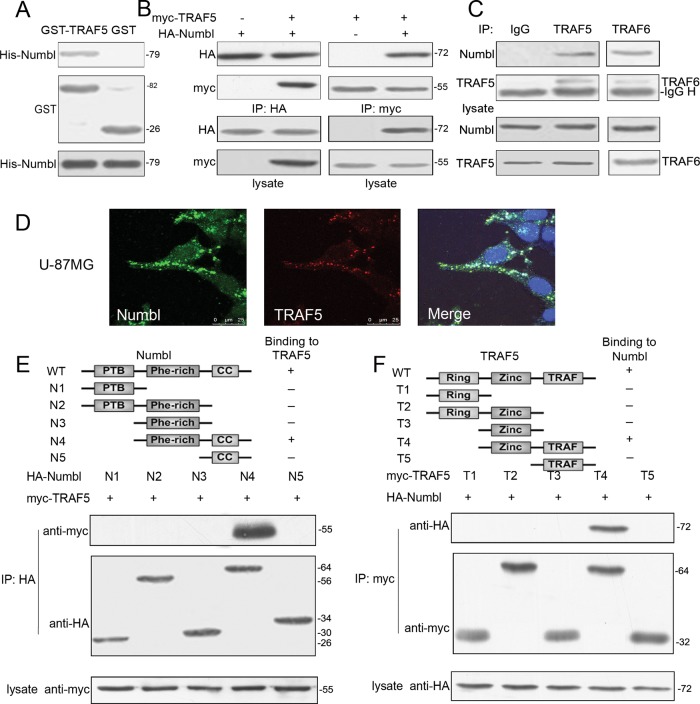
Because we observed that Numbl inhibits glioma migration and invasion by suppressing NF-κB activity, we sought to tackle the underlying mechanism. TRAF6 and TAB2, two molecules involved in NF-κB signaling, have been reported as downstream targets of Numbl. Elimination of endogenous TRAF6 and TAB2 by siRNA, however, had no effect on glioma migration and invasion (Figure 2, D–F), essentially ruling out any role for TRAF6 or TAB2 in Numbl-mediated suppression of glioma cell migration and invasion. A yeast two-hybrid screening revealed that Numbl could also bind to TRAF5, another signaling molecule, during NF-κB activation (unpublished observations). Indeed, there was elevated expression of TRAF5 in high-grade glioma tissue (0.71344 ± 0.16032) when compared with normal brain tissue (0.33327 ± 0.09165) and low-grade glioma tissue (0.48088 ± 0.10391) (Figures 1 and S1). H4 cells also exhibited lower expression of TRAF5 than U-87MG cells, in direct contrast with the expression pattern of Numbl. To further investigate whether Numbl and TRAF5 could directly bind to each other, we performed glutathione S-transferase (GST) pulldown assays. The GST-tagged TRAF5 could bind to His-tagged Numbl directly in vitro (Figure 4A). Furthermore, TRAF5 was found to interact with Numbl in reciprocal coimmunoprecipitations in HEK293T cells transfected with hemagglutinin (HA)-tagged Numbl and myc-tagged TRAF5 (Figure 4B). More importantly, endogenous Numbl and TRAF5 could interact with each other in H4 cells (Figure 4C). Finally, immunofluorescence microscopy illustrated that Numbl and TRAF5 were colocalized in the cytoplasm in U-87MG cells (Figure 4D).
FIGURE 4:
Numbl interacts with TRAF5 in vivo and in vitro. (A) Purified GST-TRAF5 and GST proteins were incubated with His-Numbl in the presence of glutathione beads. The bound proteins were collected and analyzed by Western blotting with anti-His antibody. (B) HA-Numbl and myc-TRAF5 vectors (1.5 μg each) were cotransfected into HEK293T cells. Extracts with equal amount of protein were immunoprecipitated with anti-myc or anti-HA antibody and analyzed by Western blotting. (C) H4 cells were lysed with RIPA buffer and equal amounts of lysates were coimmunoprecipitated with anti-TRAF5 antibody. The normal rabbit immunoglobulin G was used as a negative control, and the TRAF6 antibody was used as a positive control. (D) U-87MG cells were fixed, and immunofluorescence microscopy was performed using the indicated antibodies to show colocalization of endogenous Numbl and TRAF. The nucleus was counterstained with 4′,6-diamidino-2-phenylindole. Scale bar: 25μm. (E) Numbl contains a phosphotyrosine-binding domain (N1), a Phe-rich domain (N3), and a coiled-coil domain (N5). HEK293T cells were transfected with myc-TRAF5 and HA-Numbl mutants (1.5 μg each). Cell lysates were immunoprecipitated with anti-HA antibody and analyzed by Western blotting. (F) TRAF5 contains a finger domain (T1), a zinc finger motif (T3), and a TRAF domain (T5). HEK293T cells were transfected with myc-TRAF5 mutants and HA-Numbl (1.5 μg each). Cell lysates were immunoprecipitated with anti-HA antibody and analyzed by Western blotting.
To further delineate the interaction between Numbl and TRAF5, we performed coimmunoprecipitation assays using deletion mutants of Numbl and TRAF5. The Phe-rich and coiled-coil domains seemed to be required for Numbl to interact with TRAF5, as deletion of either domain abrogated the interaction (Figure 4E). Conversely, the zinc finger domain and TRAF domain of TRAF5 were indispensable for the Numbl-TRAF5 interaction, whereas the Ring domain was neither sufficient nor necessary (Figure 4F). In further support of the notion that Numbl represses NF-κB activity by targeting TRAF5, we found that depletion of endogenous TRAF5 by siRNA normalized NF-κB activity in the presence of exogenous Numbl (Figure 3, A and B). Taken together, these data clearly demonstrate that Numbl suppresses NF-κB activation by interacting with TRAF5.
Numbl promotes K48 polyubiquitination of TRAF5
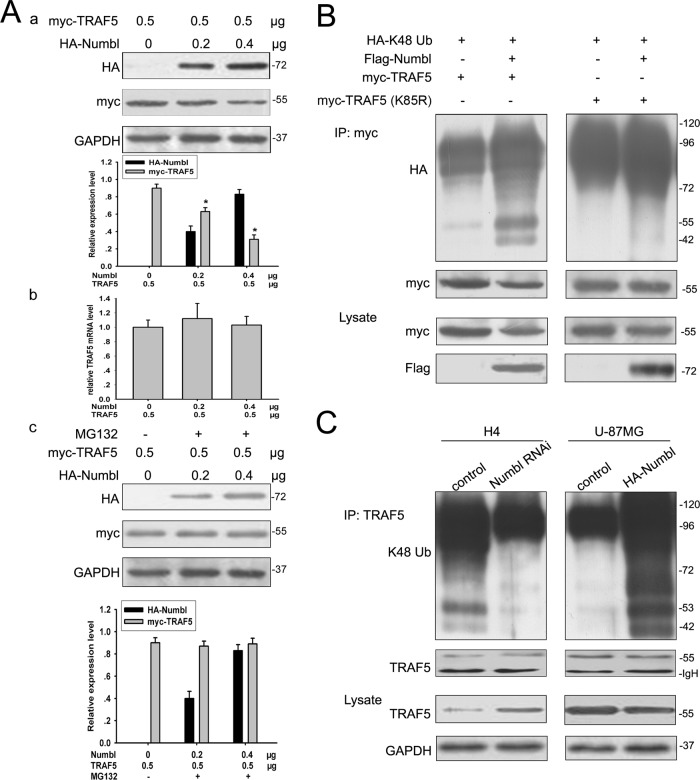
When TRAF5 was cotransfected into HEK293T cells with increasing amounts of Numbl, TRAF5 protein levels were decreased, but TRAF5 mRNA levels remained unchanged (Figure 5Aa and 5Ab). The decrease in TRAF5 protein expression was reversed by the 26S proteasome inhibitor MG132 (Figure 5Ac). These data indicate that Numbl could potentially regulate protein stability of TRAF5. During NF-κB activation, TRAFs undergo polyubiquitination (Nakano et al., 1996). To determine whether Numbl promotes TRAF5 ubiquitination, we examined the ubiquitinated TRAF5 species in HEK293T cells expressing HA-tagged ubiquitin with or without Flag-Numbl. Coexpression of Numbl further increased ubiquitinated TRAF5 and decreased the corresponding unconjugated myc-TRAF5 levels (Figure S3A). Furthermore, TRAF5 polyubiquitination occurred through K48-linked but not K63-linked ubiquitin chains in the presence of Numbl (Figures 5B and S3A). More importantly, Numbl knockdown decreased, while Numbl overexpression increased K48 polyubiquitination of endogenous TRAF5 in glioma cells (Figure 5C). Of note, the basal level of TRAF5 polyubiquitination was higher in H4 cells than in U-87MG cells, consistent with expression patterns of TRAF5 in these cells (Figure 5C).
FIGURE 5:
Numbl promotes K48-linked polyubiquitination of TRAF5. (A) HEK293T cells were transfected with increasing amounts of the HA-Numbl plasmid and myc-TRAF5; this was followed by treatment with or without MG132 (50 μM) for 4 h. After 24 h, cells were collected for Western blot and reverse transcriptase PCR analyses. The data are means ± SEM. *, p < 0.05, statistically significant compared with the first lane. (B) HEK293T cells were transfected with indicated expression vectors (1.5 μg each); this was followed by MG132 treatment for 4 h. Cell lysates were immunoprecipitated with anti-myc antibody and detected by Western blotting. (C) H4 glioma cells were transfected with a Numbl knockdown plasmid (Numbl RNAi), and U-87MG cells were transfected with a Numbl expression plasmid (HA-Numbl). Cell lysates were immunoprecipitated with anti-myc antibody and detected by Western blotting.
To pin down the exact lysine residue conjugated to the K48-linked polyubiquitin chain, we mutated individual lysine residues within TRAF5 (K85, K89, K95, K100, K119) to arginine. Only one mutant (K85R) lost the ability to assemble K48-linked polyubiquitin chains (Figure S3B). Indeed, Numbl promoted K48-linked polyubiquitination of wild type, but not the K85R mutant TRAF5 in HEK293T cells (Figure 5B). The K85R TRAF5 was refractory to Numbl-mediated repression of NF-κB transcriptional activity, acting as a dominant negative mutant (Figure 3, A and B). Taken together, these results strongly suggest that Numbl suppressed NF-κB activation by promoting Lys-48–linked polyubiquitination of TRAF6.
TRAF5 is a downstream target of Numbl for promoting glioma cell migration and invasion
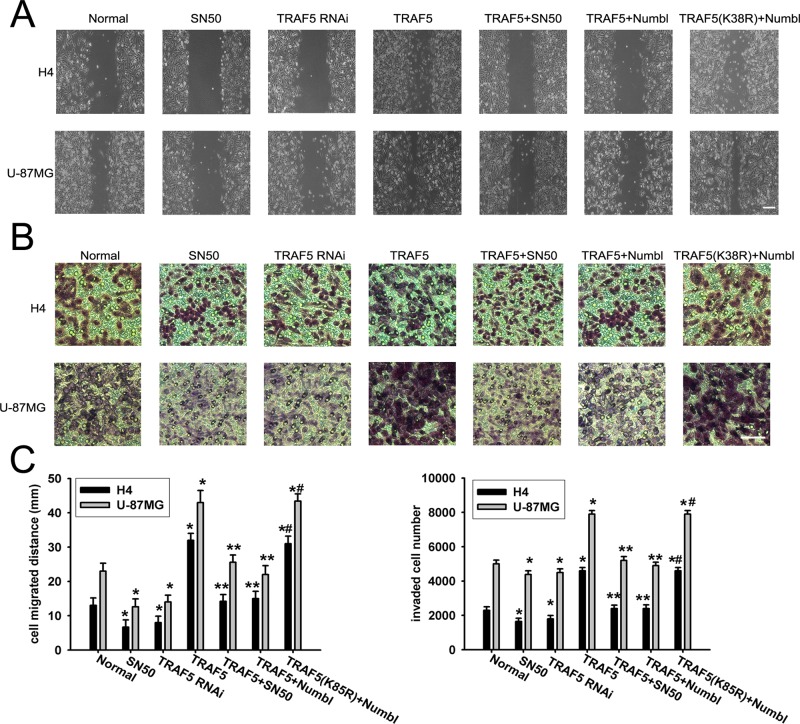
To examine whether the suppression of glioma migration and invasion by Numbl was mediated via TRAF5 degradation, we transfected a TRAF5 siRNA expression vector into H4 and U87-MG cells. As shown in Figure 6, elimination of TRAF5 expression significantly decreased the migration and invasion of the glioma cells and was accompanied by decreased NF-κB activation (Figure 6, A–C). In contrast, overexpression of TRAF5 promoted cell migration and invasion (Figure 6, A–C). Introduction of exogenous Numbl completely negated the increase of cell migration and invasion driven by wild-type TRAF5. Again, the K85R mutant was able to stimulate glioma migration and invasion, even in the presence of Numbl, indicative of its dominant negative nature (Figure 6, A–C). In conclusion, these data suggest that TRAF5 is a downstream target of Numbl in suppressing glioma cell migration and invasion in a ubiquitination-dependent manner.
FIGURE 6:
Numbl suppressed glioma cell migration and invasion through the NF-κB pathway. Stable H4 and U-87MG cells expressing TRAF5 RNAi, myc-TRAF5, or K85R mutant TRAF5 were transfected with or without HA-Numbl and then treated with SN50 or left untreated. Wound-healing (A) and Transwell (B) assays were performed and analyzed as described in Materials and Methods. (C) Statistical analysis of cell migration distances and cell invasion numbers. The data are means ± SEM. *, p < 0.05, statistically significant compared with normal cell group; **, p < 0.05, statistically significant compared with myc-TRAF5 overexpression group; #, p < 0.05, statistically significant compared with the HA-Numbl + TRAF5 overexpressed cells.
DISCUSSION
In the present study, we found that Numbl protein level was attenuated in high-grade glioma tissue samples and glioblastoma cell lines. In vitro studies showed that Numbl could inhibit cell motility and invasiveness. We further showed that TRAF5 degradation and consequent suppression of NF-κB activation were essential for the alleviation of glioma cell invasion induced by Numbl. Taken together, our data provide novel evidence that the Numbl/TRAF5/NF-κB signaling pathway is involved in glioma cell migration and invasion.
Glioma cell invasion involves complex interactions between malignant cells and normal cells (Norden and Wen, 2006). It has been reported that during glioma invasion and migration, growth factors such as vascular endothelial growth factor, human growth factor, and leptin promote glioma migration and invasion by inducing NF-κB activation (Demuth and Berens, 2004; Huang et al., 2009; Norden et al., 2009). Subsequently, activated NF-κB stimulates the transcription of adhesion molecules, photolytic enzymes, and extracellular matrix molecules to facilitate cell migration and invasion (Friedl and Wolf, 2003; Rao, 2003; Tate and Aghi, 2009). However, it remains unclear how NF-κB activity is fine-tuned in this process. Our work reported here suggests that TRAF5-induced NF-κB activation plays an essential role during glioma migration and invasion. TRAF5 was originally identified as an activator in interleukin-induced NF-κB signal transduction via its TRAF domain (Leo et al., 1999; Tada et al., 2001). Previous studies have found that CD40 was overexpressed on the surface of glioma cells and may potentially induce neovascularization by activating the VEGF pathway to promote glioma invasion (Wischhusen et al., 2005; Xie et al., 2010). Recently it has also been reported that IL-1 and IL-6 levels are elevated in glioblastoma, which is linked to increased MMP9 expression via activation of phosphatidylinositol 3-kinase PI3K/Akt and NF-κB (Nozell et al., 2006; Liu et al., 2010). Because TRAF5 can act downstream of CD40 and interleukins, including IL-1β and IL-6 (Nakano et al., 1996; Jackson-Bernitsas et al., 2007), it is possible that TRAF5 plays a key role in CD40- and/or IL1/6-mediated NF-κB activation during glioma migration and invasion.
In the process of activating NF-κB, TRAF proteins undergo ubiquitination (Bradley and Pober, 2001). Although ubiquitination often results in protein degradation, certain types of ubiquitination are important for signaling and/or protein trafficking (Pineda et al., 2007). K63-linked ubiquitination is associated with enhanced signaling transduction and protein trafficking, whereas polyubiquitination through K48 of the ubiquitin chain generally targets proteins for degradation (Pickart, 2001; Mukhopadhyay and Riezman, 2007; Martinez-Forero et al., 2009). In the present study, we found that TRAF5 polyubiquitination was involved in glioma migration and invasion. Specifically, K48-linked polyubiquitination of TRAF5 suppressed glioma migration and invasion by promoting TRAF5 degradation.
It has been shown that Numbl is required for the proper morphogenesis of cerebral cortex, because inactivation of Numbl in radial glial cells leads to progenitor dispersion and disorganized cortical lamination (Shea et al., 2003; Petersen et al., 2004). It is possible that Numbl is involved in brain tumorigenesis. We found that Numbl expression was down-regulated in high-grade glioma tissues and glioblastoma cell lines, and was involved in cell migration and invasion. A recent study has shown that the copy number of Numbl was not altered between normal brain tissues and glioma tissues, indicating that repressed transcription and/or accelerated protein degradation may be responsible for decreased Numbl expression (Blom et al., 2008). Numbl was initially identified as a negative regulator of the Notch signaling pathway. Recently several studies have found that Numbl mediates NF-κB deactivation by preventing TAB2 from interacting with polyubiquitinated TRAF6 and RIPK1 and stimulating the K48-linked polyubiquitination and degradation of TRAF6 (Ma et al., 2008; Zhou et al., 2010). Our data suggest that during glioma migration and invasion, the Notch signaling protein Numbl may act as an inhibitor, targeting TRAF5 to proteasomal degradation and therefore suppressing NF-κB activation. Similar to TRAF6, TRAF5 binds to the C-terminus of Numbl that includes a Phe-rich segment and a coiled-coil domain (Zhou et al., 2010). On the other hand, Numbl interacts with the zinc finger and the TRAF domains of TRAF5. It is possible the K85 site within TRAF5 is exposed during binding to Numbl and may be targeted by a yet unidentified E3 ligase for degradation. We found that Numbl suppresses glioma invasion by promoting TRAF5 ubiquitination at Lys-85. These data indicate that Numbl may serve as an inhibitor in glioma migration and invasion by promoting TRAF5 degradation and suppressing NF-κB signal transduction. Further investigation is warranted to dissect the complex nature of the functional interplay between Numbl and NF-κB.
In conclusion, this study reveals the biological significance of Numbl down-regulation in glioma and illustrates that Numbl is a negative regulator in glioma invasion and migration. Our data presented here provide a viable strategy for the development of novel therapies to prevent glioma invasion.
MATERIALS AND METHODS
Primary human glioma tissue and tissue homogenization
Eight low-grade human glioma samples (WHO grades II and III) and eight high-grade glioma samples (WHO grade IV, glioblastomas) were obtained from the Department of Pathology, Affiliated Hospital of Nantong University, from 2004 to 2008. All tumors were from patients with newly diagnosed glioma who had received no therapy before sample collection. Normal brain specimens were acquired from eight patients undergoing surgery for epilepsy. All samples were reviewed by a panel of three neuropathologists. All the tissues for immunoblot analysis were frozen immediately after surgery. Approximately 100–300 mg of each tissue sample was manually homogenized with a homogenizer in RIPA lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 2 mM sodium pyrophosphate, 25 mM β-glycerophosphate, 1 mM EDTA, 1 mM Na3VO4, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride). After centrifugation, the supernatant was used immediately for Western analysis or stored at −80 ºC.
Plasmids, reagents, and antibodies
The full-length human Numbl (GenBank no. NM_004756.3), mouse Numbl (GenBank no. NM_010950.2), and human TRAF5 (GenBank no. NM_004619.3) cDNA was isolated from the brain cDNA library. The pET28-Numbl, pGEX-6P-1-TRAF5, pCMV-TRAF5-myc, and pCMV-HA-Numbl were generated by standard procedures (primers used are shown in Supplemental Table S1). For RNA interference (RNAi), shRNA sequences targeting the Numbl (AAGGCAAAGCCACTGTAGAGA), TRAF5 (AACGGATAAACGGAGGAACCT), TRAF6 (GenBank no. NM_009424.2, CTTTCCAGAAGTGCCAGGTTA), and TAB2 (GenBank no. NM_015093, 4TGACATTGACTGCTTAACCAA) were cloned into the pCDNA6.2-GW/enhanced green fluorescent protein–micro RNA vector per the instruction manual (Invitrogen). SN50, a cell-permeable inhibitor peptide, was used to suppress NF-κB activation in glioma cell lines. The antibodies used in this study include: mouse monoclonal anti-TRAF5 antibody and rabbit polyclonal anti-GAPDH antibodies (Santa Cruz Biotechnology); mouse monoclonal anti-myc antibody, rabbit polyclonal anti-HA antibody, and rabbit polyclonal anti-Flag antibodies (Sigma-Aldrich); mouse monoclonal anti-HA and rabbit polyclonal anti-myc antibodies (Roche); rabbit polyclonal anti-Numbl antibody (Abcam); mouse monoclonal anti-ubiquitin antibody, mouse monoclonal anti-Lys-63–specific ubiquitin antibody, and mouse monoclonal anti-Lys-48–specific ubiquitin antibody (Millipore). All antibodies for Western blotting were used at a dilution ratio of 1:1000. To detect the localization of Numbl and TRAF5 by immunofluorescence staining, we used mouse monoclonal anti-TRAF5 antibody, rabbit polyclonal anti-Numbl antibody, mouse monoclonal anti-myc antibody, and rabbit polyclonal anti-HA antibody at 1:100 dilution.
Cell culture and transfection
U251, U-118, U-87MG, T98G, LN-18, LN-229, A172 and H4 are all human glioma cell lines derived from patients with glioblastoma. HEK293T is a human embryonic kidney cell line with stably incorporated SV40 large-T antigen. All cell lines were cultured in DMEM (Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies). Transient transfections were performed using Lipofectamine 2000, following the manufacturer's recommendation (Life Technologies). For the creation of stable cells, transfected cells were exposed to 0.5 mg/ml neomycin (for overexpression vector) or 0.01 mg/ml blasticidin (for RNAi vector) for 2 wk.
GST pulldown, immunoprecipitation, and immunoblotting
The interaction between Numbl and TRAF5 was assessed by GST pulldown and coimmunoprecipitation assays as previously reported (Zhou et al., 2010). Briefly, GST-TRAF5 (pGEX-6P-1-TRAF5) and 6His-Numbl (pET28-Numbl) were transformed in BL21 and induced to express by the addition of isopropyl β-d-thiogalactoside (0.1 mM) at 20°C. The fusion proteins GST and GST-TRAF5 were purified with glutathione-Sepharose 4B beads. The 6His-Numbl was purified with Ni–NTA agarose. The GST–bound and GST-TRAF6–bound glutathione-Sepharose 4B beads were incubated with His-Numbl proteins in NETN buffer (100 mM NaCl, 20 mM Tris-Cl, pH 8.0, 0.5 mM EDTA, 0.5% NP-40) for 2 h at 4ºC. Binary complexes were eluted by boiling the beads in 2 × SDS loading buffer (20 mM Tris-HCl, pH 8.0, 100 mM dithiothreitol, 2% SDS, 20% glycerol, 0.016% bromophenol blue) and were separated by SDS–PAGE. For immunoprecipitation assays, cells were collected in phosphate-buffered saline and lysed by RIPA buffer. After being precleared with protein G Sepharose beads, cell lysates were incubated with the specific antibody bound to either protein A or G Sepharose beads (Roche) for 12 h at 4 ºC. Precipitated immune complexes were washed with RIPA buffer four times, eluted by boiling in 2 × SDS sample buffer, resolved by SDS–PAGE gel, and analyzed by immunoblotting, as previously reported (Tao et al., 2009).
Luciferase assay
The pGL3-Basic vector and the NF-κB-dependent luciferase reporter vector (pGL3- NF-κB) were purchased from Promega. Cells were plated 16–24 h prior to transfection and cotransfected with the expression/siRNA vectors, NF-κB luciferase reporter, and pRL-TK (for normalization) using Lipofectamine 2000. After 24 h, aliquots of cell lysate were used to assay NF-κB and Renilla luciferase activities with the dual-luciferase reporter assay system (Promega) on an EG&G Berthold Lumat LB9507 luminometer. Data are reported as the ratio of firefly/Renilla luciferase activity.
Wound-healing assay
Equal numbers of stably transfected cells were seeded into six-well tissue culture plates. When the cells reached 90% confluence, a scratch wound was created in the center of the cell monolayer by gently removing the attached cells with a sterile plastic pipette tip. The debris was removed by washing the cells with serum-free culture medium. Cells boarding the wound were visualized and photographed under an inverted microscope 24 h after the wound was created. The distances cells migrated into the wounded area were calculated by subtracting the distance 24 h after wound healing from the initial distance. A total of nine areas were selected randomly from each well under a 40× objective, and the cells in three wells of each group were quantified in each experiment.
In vitro invasion assays
Cells from stable clones used for invasion assays were performed as previously reported (Tran et al., 2006). Generally, Transwell filters were coated on the upper side with 30 mg of Matrigel (Millipore) for 2 h at 37ºC, and then 1 × 106 cells were subcultured on the Transwell filter. After incubation for 24 h, cells on the upper chamber were fixed with 4% paraformaldehyde and stained with toluidine blue before being counted under an inverted microscope. In all experiments, data were collected from triplicate chambers.
Statistical analysis
All experiments were repeated at least three times. All numerical data are described as mean ± SEM. Data were analyzed using the two-tailed t test. A probability value of 0.05 or less was considered significant.
Supplementary Material
Acknowledgments
The authors thank Yong Xu (Nanjing Medial University) for critical reading of the manuscript and Tianyi Zhang (Nantong University) for his helpful technology support. This work was supported by the National Basic Research Program of China (973 program: no. 2011CB910604 and no. 2012CB822104), the National Natural Science Foundation of China (no. 31070723 and no. 81172879), Key Project Natural Science Foundation of Jiangsu province (no. 11KJA310002), and A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Abbreviations used:
- d-Numb
Drosophila Numb
- GST
glutathione S-transferase
- HA
hemagglutinin
- IκBα
NF-κB inhibitory subunit
- IL-1
interleukin-1
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
- NF-κB
nuclear factor kappa B
- Numbl
Numblike
- RNAi
RNA interference
- siRNA
small interfering RNA
- TAB2
TGF-β activated kinase 1 binding protein 2
- TNFR
tumor necrosis factor receptor
- TRAF
TNFR-associated factor
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-09-0805) on May 16, 2012.
REFERENCES
- Angileri FF, Aguennouz M, Conti A, La Torre D, Cardali S, Crupi R, Tomasello C, Germano A, Vita G, Tomasello F. Nuclear factor-kappaB activation and differential expression of survivin and Bcl-2 in human grade 2-4 astrocytomas. Cancer. 2008;112:2258–2266. doi: 10.1002/cncr.23407. [DOI] [PubMed] [Google Scholar]
- Arron JR, Walsh MC, Choi Y. TRAF-mediated TNFR-family signaling. Curr Protoc Immunol. 2002;87 doi: 10.1002/0471142735.im1109ds51. 11.9D.19. [DOI] [PubMed] [Google Scholar]
- Blom T, Roselli A, Tanner M, Nupponen NN. Mutation and copy number analysis of LNX1 and Numbl in nervous system tumors. Cancer Genet Cytogenet. 2008;186:103–109. doi: 10.1016/j.cancergencyto.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Bradley JR, Pober JS. Tumor necrosis factor receptor-associated factors (TRAFs) Oncogene. 2001;20:6482–6491. doi: 10.1038/sj.onc.1204788. [DOI] [PubMed] [Google Scholar]
- Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, Holland E. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PloS One. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintala SK, Rao JK. Invasion of human glioma: role of extracellular matrix proteins. Front Biosci. 1996;1:d324–d339. doi: 10.2741/a135. [DOI] [PubMed] [Google Scholar]
- Conti A, et al. Expression of the tumor necrosis factor receptor-associated factors 1 and 2 and regulation of the nuclear factor—κB antiapoptotic activity in human gliomas. J Neurosurg. 2005;103:873–881. doi: 10.3171/jns.2005.103.5.0873. [DOI] [PubMed] [Google Scholar]
- Demuth T, Berens ME. Molecular mechanisms of glioma cell migration and invasion. J Neurooncol. 2004;70:217–228. doi: 10.1007/s11060-004-2751-6. [DOI] [PubMed] [Google Scholar]
- Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Karin M. Missing pieces in the NF-κB puzzle. Cell. 2002;109(suppl):S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Giese A, Bjerkvig R, Berens ME, Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol. 2003;21:1624–1636. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- Ha H, Han D, Choi Y. TRAF-mediated TNFR-family signaling. Curr Protoc Immunol. 2009;87 doi: 10.1002/0471142735.im1109ds87. 11.9D.19. [DOI] [PubMed] [Google Scholar]
- Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal. 2009;2 doi: 10.1126/scisignal.287re6. re6. [DOI] [PubMed] [Google Scholar]
- Ichikawa H, Takada Y, Murakami A, Aggarwal BB. Identification of a novel blocker of IκBα kinase that enhances cellular apoptosis and inhibits cellular invasion through suppression of NF-κB-regulated gene products. J Immunol. 2005;174:7383–7392. doi: 10.4049/jimmunol.174.11.7383. [DOI] [PubMed] [Google Scholar]
- Jackson-Bernitsas DG, Ichikawa H, Takada Y, Myers JN, Lin XL, Darnay BG, Chaturvedi MM, Aggarwal BB. Evidence that TNF-TNFR1-TRADD-TRAF2-RIP-TAK1-IKK pathway mediates constitutive NF-κB activation and proliferation in human head and neck squamous cell carcinoma. Oncogene. 2007;26:1385–1397. doi: 10.1038/sj.onc.1209945. [DOI] [PubMed] [Google Scholar]
- Jacobs W, Mikkelsen T, Smith R, Nelson K, Rosenblum ML, Kohn EC. Inhibitory effects of CAI in glioblastoma growth and invasion. J Neurooncol. 1997;32:93–101. doi: 10.1023/a:1005777711567. [DOI] [PubMed] [Google Scholar]
- Karin M, Cao Y, Greten FR, Li ZW. NF-κB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- Kleihues P, Louis DN, Scheithauer BW, Rorke LB, Reifenberger G, Burger PC, Cavenee WK. The WHO classification of tumors of the nervous system. J Neuropathol Exp Neurol. 2002;61:215–225. doi: 10.1093/jnen/61.3.215. discussion, 226–229. [DOI] [PubMed] [Google Scholar]
- Leo E, Zapata JM, Reed JC. CD40-mediated activation of Ig-Cγ1- and Ig-Cε germ-line promoters involves multiple TRAF family proteins. Eur J Immunol. 1999;29:3908–3913. doi: 10.1002/(SICI)1521-4141(199912)29:12<3908::AID-IMMU3908>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Li HS, Wang D, Shen Q, Schonemann MD, Gorski JA, Jones KR, Temple S, Jan LY, Jan YN. Inactivation of Numb and Numblike in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis. Neuron. 2003;40:1105–1118. doi: 10.1016/s0896-6273(03)00755-4. [DOI] [PubMed] [Google Scholar]
- Li Q, Verma IM. NF-κB regulation on the immune system. Nat Rev. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- Liu Q, Li G, Li R, Shen J, He Q, Deng L, Zhang C, Zhang J. IL-6 promotion of glioblastoma cell invasion and angiogenesis in U251 and T98G cell lines. J Neurooncol. 2010;100:165–176. doi: 10.1007/s11060-010-0158-0. [DOI] [PubMed] [Google Scholar]
- Ma Q, Zhou L, Shi H, Huo K. NUMBL interacts with TAB2 and inhibits TNFα and IL-1β-induced NF-κB activation. Cell Signal. 2008;20:1044–1051. doi: 10.1016/j.cellsig.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, DePinho RA. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- Martinez-Forero I, Rouzaut A, Palazon A, Dubrot J, Melero I. Lysine 63 polyubiquitination in immunotherapy and in cancer-promoting inflammation. Clin Cancer Res. 2009;15:6751–6757. doi: 10.1158/1078-0432.CCR-09-1225. [DOI] [PubMed] [Google Scholar]
- Mertsch S, Schurgers LJ, Weber K, Paulus W, Senner V. Matrix gla protein (MGP): an overexpressed and migration-promoting mesenchymal component in glioblastoma. BMC Cancer. 2009;9:302. doi: 10.1186/1471-2407-9-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Nakano H, Oshima H, Chung W, Williams-Abbott L, Ware CF, Yagita H, Okumura K. TRAF5, an activator of NF-κB and putative signal transducer for the lymphotoxin-β receptor. J Biol Chem. 1996;271:14661–14664. doi: 10.1074/jbc.271.25.14661. [DOI] [PubMed] [Google Scholar]
- Norden AD, Drappatz J, Wen PY. Antiangiogenic therapies for high-grade glioma. Nat Rev Neurol. 2009;5:610–620. doi: 10.1038/nrneurol.2009.159. [DOI] [PubMed] [Google Scholar]
- Norden AD, Wen PY. Glioma therapy in adults. Neurologist. 2006;12:279–292. doi: 10.1097/01.nrl.0000250928.26044.47. [DOI] [PubMed] [Google Scholar]
- Nozell S, Laver T, Patel K, Benveniste EN. Mechanism of IFN-β-mediated inhibition of IL-8 gene expression in astroglioma cells. J Immunol. 2006;177:822–830. doi: 10.4049/jimmunol.177.2.822. [DOI] [PubMed] [Google Scholar]
- Petersen PH, Zou K, Hwang JK, Jan YN, Zhong W. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature. 2002;419:929–934. doi: 10.1038/nature01124. [DOI] [PubMed] [Google Scholar]
- Petersen PH, Zou K, Krauss S, Zhong W. Continuing role for mouse Numb and Numbl in maintaining progenitor cells during cortical neurogenesis. Nat Neurosci. 2004;7:803–811. doi: 10.1038/nn1289. [DOI] [PubMed] [Google Scholar]
- Phillips HS, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Pineda G, Ea CK, Chen ZJ. Ubiquitination and TRAF signaling. Adv Exp Med Biol. 2007;597:80–92. doi: 10.1007/978-0-387-70630-6_7. [DOI] [PubMed] [Google Scholar]
- Pomerantz JL, Baltimore D. Two pathways to NF-κB. Mol Cell. 2002;10:693–695. doi: 10.1016/s1097-2765(02)00697-4. [DOI] [PubMed] [Google Scholar]
- Purow BW, et al. Notch-1 regulates transcription of the epidermal growth factor receptor through p53. Carcinogenesis. 2008;29:918–925. doi: 10.1093/carcin/bgn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao JS. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev. 2003;3:489–501. doi: 10.1038/nrc1121. [DOI] [PubMed] [Google Scholar]
- Rasheed BK, Wiltshire RN, Bigner SH, Bigner DD. Molecular pathogenesis of malignant gliomas. Curr Opin Oncol. 1999;11:162–167. doi: 10.1097/00001622-199905000-00004. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Park ES, Emdad L, Lee SG, Su ZZ, Fisher PB. Molecular basis of nuclear factor-κB activation by astrocyte elevated gene-1. Cancer Res. 2008;68:1478–1484. doi: 10.1158/0008-5472.CAN-07-6164. [DOI] [PubMed] [Google Scholar]
- Shea TB, Jung C, Pant HC. Does neurofilament phosphorylation regulate axonal transport? Trends Neurosci. 2003;26:397–400. doi: 10.1016/S0166-2236(03)00199-1. [DOI] [PubMed] [Google Scholar]
- Tada K, et al. Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-κB activation and protection from cell death. J Biol Chem. 2001;276:36530–36534. doi: 10.1074/jbc.M104837200. [DOI] [PubMed] [Google Scholar]
- Tao T, Ji Y, Cheng C, Yang H, Liu H, Sun L, Qin Y, Yang J, Wang H, Shen A. Tumor necrosis factor-alpha inhibits Schwann cell proliferation by up-regulating Src-suppressed protein kinase C substrate expression. J Neurochem. 2009;111:647–655. doi: 10.1111/j.1471-4159.2009.06346.x. [DOI] [PubMed] [Google Scholar]
- Tate MC, Aghi MK. Biology of angiogenesis and invasion in glioma. Neurotherapeutics. 2009;6:447–457. doi: 10.1016/j.nurt.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NL, et al. Increased fibroblast growth factor-inducible 14 expression levels promote glioma cell invasion via Rac1 and nuclear factor-κB and correlate with poor patient outcome. Cancer Res. 2006;66:9535–9542. doi: 10.1158/0008-5472.CAN-06-0418. [DOI] [PubMed] [Google Scholar]
- Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- Wakabayashi K, Kambe F, Cao X, Murakami R, Mitsuyama H, Nagaya T, Saito K, Yoshida J, Seo H. Inhibitory effects of cyclosporin A on calcium mobilization-dependent interleukin-8 expression and invasive potential of human glioblastoma U251MG cells. Oncogene. 2004;23:6924–6932. doi: 10.1038/sj.onc.1207778. [DOI] [PubMed] [Google Scholar]
- Wischhusen J, Schneider D, Mittelbronn M, Meyermann R, Engelmann H, Jung G, Wiendl H, Weller M. Death receptor-mediated apoptosis in human malignant glioma cells: modulation by the CD40/CD40L system. J Neuroimmunol. 2005;162:28–42. doi: 10.1016/j.jneuroim.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Wisniewski P, Ellert-Miklaszewska A, Kwiatkowska A, Kaminska B. Non-apoptotic Fas signaling regulates invasiveness of glioma cells and modulates MMP-2 activity via NFκB-TIMP-2 pathway. Cell Signal. 2010;22:212–220. doi: 10.1016/j.cellsig.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Xie F, et al. CD40 is a regulator for vascular endothelial growth factor in the tumor microenvironment of glioma. J Neuroimmunol. 2010;222:62–69. doi: 10.1016/j.jneuroim.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Zhong W, Feder JN, Jiang MM, Jan LY, Jan YN. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- Zhong W, Jiang MM, Weinmaster G, Jan LY, Jan YN. Differential expression of mammalian Numb, Numblike and Notch1 suggests distinct roles during mouse cortical neurogenesis. Development. 1997;124:1887–1897. doi: 10.1242/dev.124.10.1887. [DOI] [PubMed] [Google Scholar]
- Zhou L, Ma Q, Shi H, Huo K. NUMBL interacts with TRAF6 and promotes the degradation of TRAF6. Biochem Biophys Res Commun. 2010;392:409–414. doi: 10.1016/j.bbrc.2010.01.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.