A newly identified CaMKI substrate, CCT, is a crucial enzyme required for cell membranes. CaMK1 phosphorylates CCT to initiate calcium-induced nuclear entry and binds within a NES. CaMKI vies with exportin 1 for access to the NES, and assembly of a CaMKI–14-3-3–CCT complex is a key mechanism that drives nuclear import of CCT.
Abstract
We identified a new calmodulin kinase I (CaMKI) substrate, cytidyltransferase (CCTα), a crucial enzyme required for maintenance of cell membranes. CCTα becomes activated with translocation from the cytoplasm to the nuclear membrane, resulting in increased membrane phospholipids. Calcium-activated CCTα nuclear import is mediated by binding of its C-terminus to 14-3-3 ζ, a regulator of nuclear trafficking. Here CaMK1 phosphorylates residues within this C-terminus that signals association of CCTα with 14-3-3 ζ to initiate calcium-induced nuclear entry. CaMKI docks within the CCTα membrane-binding domain (residues 290–299), a sequence that displays similarities to a canonical nuclear export signal (NES) that also binds CRM1/exportin 1. Expression of a CFP-CCTα mutant lacking residues 290–299 in cells results in cytosolically retained enzyme. CRM1/exportin 1 was required for CCTα nuclear export, and its overexpression in cells was partially sufficient to trigger CCTα nuclear export despite calcium stimulation. An isolated CFP-290-299 peptide remained in the nucleus in the presence of leptomycin B but was able to target to the cytoplasm with farnesol. Thus CaMKI vies with CRM1/exportin 1 for access to a NES, and assembly of a CaMKI–14-3-3 ζ–CCTα complex is a key effector mechanism that drives nuclear CCTα translocation.
INTRODUCTION
Calcium (Ca2+) is a critical second messenger that is involved in the regulation of multiple cellular processes (Uboha et al., 2007). Many of its effects in cellular signaling are mediated through ubiquitous Ca2+-sensing proteins, such as Ca2+- and calmodulin (CaM)-dependent protein kinase I (CaMKI), a serine/threonine kinase that serves as a key constituent for CaM-dependent signaling (Uboha et al., 2007). This protein belongs to the superfamily of calcium-dependent kinases including CaMKII, CaMKIV, and CaMK kinase (CaMKK; Uboha et al., 2007). CaMKI is a multifunctional protein involved in hormone production, neurite outgrowth, gene transcription, cell survival, and cytoskeletal reorganization (Stedman et al., 2004; Sackmann et al., 2011). CaMKI is a soluble protein in mammalian cells, with predominantly cytosolic substrates that exert diverse functions (Means, 2000). Cytoplasmic localization of CaMKI depends on a nuclear export signal within its regulatory domain (Stedman et al., 2004). The CaMKI gene encodes a 374–amino acid protein that responds directly to Ca2+-CaM with increased activity through relief of intrasteric autoinhibition (Aletta et al., 1996), and its enzymatic behavior also is regulated by reversible phosphorylation (Sackmann et al., 2011). CaMKI phosphorylates a variety of proteins in vitro, but very few physiological targets of CaMKI have been identified. These targets include the synaptic vesicle–associated proteins synapsin I and II (Nairn and Greengard, 1987; Clapperton et al., 2002) and the cAMP response element–binding protein CREB (Sheng et al., 1991). Substrate specificity is regulated by dephosphorylated or phosphorylated forms of the enzyme (Kahl and Means, 2003). The substrate recognition sequence for CaMKI is Hyd-X-Arg-X-X-Ser/Thr-X-X-X-Hyd, where Hyd is a hydrophobic residue at position P − 5 and P + 4 relative to a Ser/Thr residue at the phosphorylation site (Lee et al., 1994). Thus identification of novel molecular targets for CaMKI linked to fundamental processes remains a relatively untapped area of investigation.
Mammalian cells critically rely on the regulatory and rate-limiting enzyme CTP:phosphocholine cytidylyltransferase (CCT) for generation of cell membranes and other vital components, including pulmonary surfactant, lipoproteins, and bile (Jackowski and Fagone, 2005). Deficiency of CCT is linked to apoptosis and embryonic lethality (Wang et al., 2005). CCTα, the major isoform identified in lung cells catalyzes a reaction generating phosphatidylcholine (PtdCho), the major phospholipid of animal membranes. The enzyme has three functional domains—a catalytic core, a membrane-binding domain, and a C-terminal phosphorylation domain—and a classic nuclear localization signal (NLS). CCTα phosphorylation reduces the activity of the enzyme (Jackowski and Fagone, 2005). In lung epithelia CCTα is distributed within the cytoplasm and nucleus. The inactive, soluble, cytoplasmic form of CCTα becomes active after translocation to the endoplasmic reticulum or nuclear membrane (Agassandian et al., 2005). This translocation is reversible, and it is regulated by a variety of intracellular signals. In our recent studies, we demonstrated that 14-3-3 ζ escorts CCTα to the nucleus in a Ca2+-dependent manner (Agassandian et al., 2010). 14-3-3 ζ, a ubiquitously expressed molecular chaperone, binds phosphorylated serine residues within its cargo proteins. Because 14-3-3 ζ binding to cargoes is predominantly phosphorylation dependent, the identification and characterization of a kinase(s) that regulates 14-3-3 ζ engagement with CCTα would be of interest in the context of nuclear trafficking and lipogenesis. In this regard, CaMK-dependent binding of 14-3-3 to phosphoserines 259 and 498 within histone deacetylases activates a nuclear export signal (NES), resulting in export of the deacetylase to the cytoplasm (McKinsey et al., 2000).
The canonical NES within proteins comprises highly conserved hydrophobic residues that interact with CRM1 (exportin 1), a nucleocytoplasmic shuttling protein (Askjaer et al., 1999). This association is stabilized by cooperative binding of nuclear RanGTP (Fornerod et al., 1997; Askjaer et al., 1998; Bogerd et al., 1998). Because a high-affinity NES–CRM1 interaction depends on RanGTP, NES binding to CRM1 is more stable in the nucleus and less stable in the cytoplasm (Fornerod et al., 1997; Kutay and Guttinger, 2005). In addition to RanGTP- and NES-containing cargoes, CRM1 interacts with nucleoporins for passage through the nuclear pore complex (Pante et al., 1994, 1997; Ohno et al., 1998). Another export receptor, calreticulin, also mediates protein nuclear export via binding to the NES or specific DNA-binding domains (Ossareh-Nazari et al., 2001; Holaska et al., 2002); however, CRM1-mediated export of NES-containing proteins remains the most extensively studied export pathway.
In the present study, we demonstrate a role for CaMKI in CCTα cytoplasmic–nuclear trafficking. Our results provide the first evidence that this kinase is required and sufficient for CCTα nuclear translocation in response to calcium stimulation. In mechanistic studies, we show that CaMKI phosphorylates CCTα within its carboxyl-terminus to promote its interaction with 14-3-3 ζ, resulting in nuclear import. Unexpectedly, we identified a docking site for CaMKI within CCTα that resembles a NES (residues 290–299). Using combinatorial approaches, we also demonstrate interactions between CCTα and CRM1, a classic exporter of cargo proteins from the nucleus to the cytoplasm. This interaction is also regulated by calcium. Last, CaMKI and CRM1 in a Ca2+-dependent manner both vie for interaction with CCTα that regulates nucleocytoplasmic trafficking of the lipogenic enzyme.
RESULTS
Ca2+/CaM-dependent protein kinase I phosphorylates CCTα to regulate its binding to 14-3-3 ζ
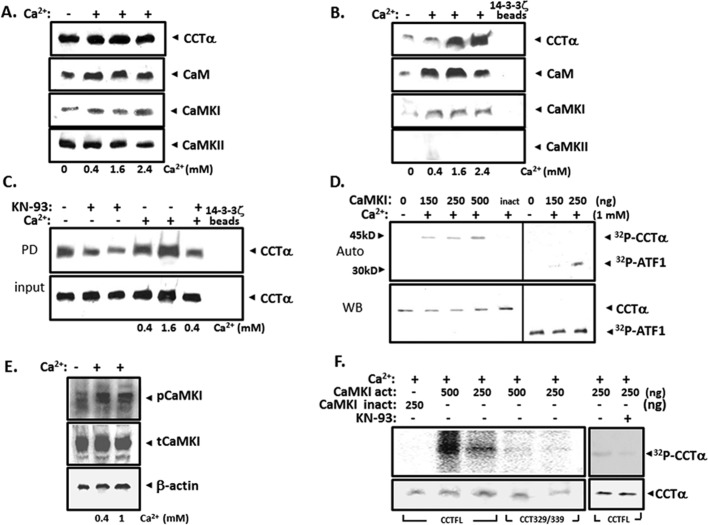
14-3-3 ζ, a binding partner for CCTα, often engages phosphorylated targets. The presence of highly conserved Arg residues within the 14-3-3 ζ binding motif in CCTα suggests that one signal for 14-3-3 ζ interaction with CCTα might be CaMK (Wang et al., 1999). Thus we investigated whether CaMK might induce CCTα phosphorylation and recruitment of 14-3-3 ζ to the enzyme. Total cellular content of CCTα, CaM, CaMKI, and CaMKII was not altered by exogenous Ca2+ (Figure 1A). When these lysates were applied to 14-3-3ζ pull-down assays, we detected CaMKI, CaM, and CCTα as a complex in association with 14-3-3 ζ, whereas CaMKII and CaMKIV (unpublished data) were not detected (Figure 1B). The L-type Ca2+ channel and CaMK inhibitor KN-93 significantly reduced CCTα binding to 14-3-3 ζ in pull-down assays both under native and Ca2+-stimulated conditions (Figure 1C). We observed that KN-93 did not block Ca2+ entry in cells, suggesting that its action might occur by CaMKI inhibition (Agassandian et al., 2010). Recombinant CaMKI increased phosphorylation of the known substrate ATFI but also of purified full-length CCTα in vitro (Figure 1D). Exogenous Ca2+ also increased levels of the phosphorylated or active form of CaMKI (Figure 1E). Last, in vitro phosphorylation assays using as a substrate an internal deletion CCTα variant that lacks the phosphorylation signature needed for 14-3-3 ζ interaction (CCTα Δ329-S339) lacked the ability to be phosphorylated (Figure 1F). KN-93 also inhibited CCTα phosphorylation by CaMKI (Figure 1F).
FIGURE 1:
CaMKI phosphorylates CCTα and regulates its binding to 14-3-3 ζ. MLE cells were incubated without or with Ca2+ at various concentrations for 1 h. Total cell lysates were used for (A) CCTα, CaM, CaMKI, and CaMKII immunoblotting or processed for (B) 14-3-3 ζ pull-down assays, followed by immunoblotting for these individual proteins. (C) Cells were exposed to the CaMKI inhibitor KN-93 (10 μM, lanes 2 and 6; 20 μM, lane 3), for 1 h before addition of Ca2+ (0.4–1.6 mM) for 1 h. Total cell lysates were processed for 14-3-3 ζ pull-down assays, followed by CCTα immunoblotting (top), with control for protein expression (input; bottom). (D) CCTα phosphorylation by CaMKI. Dephosphorylated mouse liver CCTα (2 μg) was incubated with [γ-32P] in the presence or absence of CaMKI (150, 250, and 500 ng) and Ca2+ (1 mM). Heat-inactivated CaMKI (inact) served as a negative control. Reaction products were separated on SDSx10% PAGE, transferred to nitrocellulose, and evaluated by autoradiography (top) and immunoblotting (bottom). (E) Phosphorylation status of CaMKI in cells treated with or without Ca2+ was detected by immunoblot analysis using phospho and total CaMKI antibodies. (F) Site-specific phosphorylation of CCTα by CaMKI. Dephosphorylated full-length CCTα (CCTFL) and CCT329/339 mutant synthesized by in vitro translation were incubated with [γ-32P] in the presence or absence of active or inactive CaMKI (250 and 500 ng), KN-93, and Ca2+ and CaM. Reaction products were separated on SDS–10% PAGE, transferred to nitrocellulose, and evaluated by a phosphoimager (top) and by immunoblotting (bottom). The data in each panel represent n = 3 separate experiments.
CaMKI is required and sufficient for CCTα nuclear transport
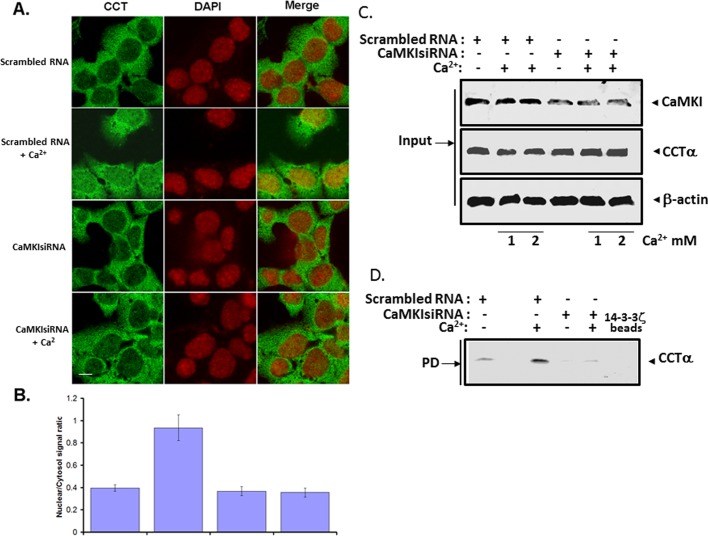
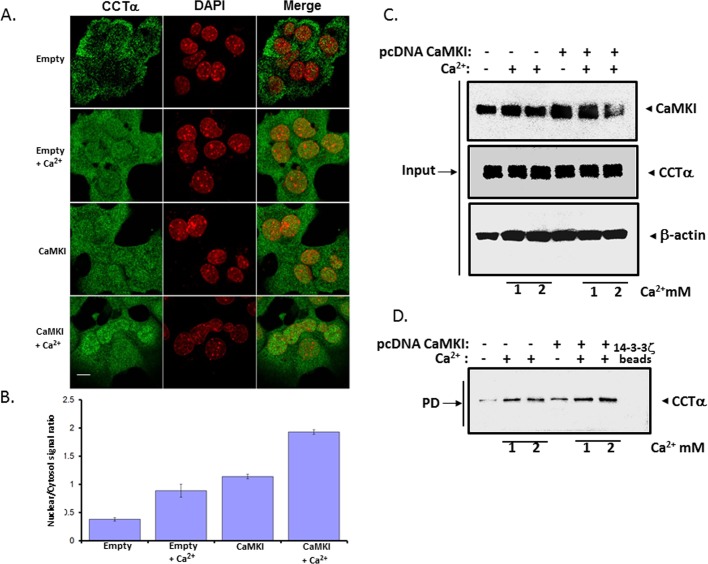
We performed loss-of-function studies in murine lung epithelial (MLE) cells to assess a requirement for CaMKI for CCTα nuclear entry. Cells were transfected with CaMKI small interfering RNA (siRNA) or control RNA (scrambled), and cells were visualized in the presence of exogenous Ca2+. The cation induced CCTα nuclear translocation in the cells, an effect that was remarkably impaired in cells transfected with CaMKI siRNA but not scrambled RNA (Figure 2, A and B). RNA interference of CaMKI was effective in reducing availability of CaMKI protein in cells (Figure 2C). In 14-3-3 ζ pull-down assays, binding of CCTα to 14-3-3 ζ was also inhibited by CaMKI siRNA (Figure 2D). Thus our loss-of-function studies demonstrate that CaMKI is needed for CCTα nuclear translocation induced by Ca2+. As a second approach, we overexpressed CaMKI in cells (Figure 3). Similar to exogenous Ca2+, ectopically expressed CaMKI triggered CCTα nuclear translocation compared with an empty vector (Figure 3, A and B). However, endogenous and exogenously expressed CaMKI predominantly localized in the cytoplasm even upon Ca2+ stimulation (Supplemental Figure S1, A and B). To assess whether CaMKI regulates CCTα binding to 14-3-3 ζ, we performed pull-down assays in which cell lysates transfected with CaMKI or control (empty) vector were purified by 14-3-3 ζ bound to metal affinity resin (Figure 3, C and D). Immunoblot analysis shows that CCTα was effectively associated with 14-3-3 ζ on the beads (Figure 3D). Moreover, binding of CCTα with 14-3-3 ζ was induced by either exogenous Ca2+ or CaMKI overexpression alone (Figure 3D, lanes 2 and 4). Translocation of CCTα to the nucleus was also blocked by KN-93 (Supplemental Figure S2). Thus endogenous and overexpressed CaMKI regulate CCTα and 14-3-3 ζ association in lung epithelia.
FIGURE 2:
Knockdown of CaMKI abrogates CCTα nuclear import. (A, B) MLE cells were plated and transfected with CaMKI siRNA (100 nM) or scrambled RNA (100 nM). At 24 h after transfection, cells were treated with or without Ca2+ (1.0 mM) in serum-free medium for 1 h. One set of these cells was then fixed and immunostained with an antibody recognizing CCTα; cells were also counterstained with DAPI (A). Fluorescence signals within the cytosol and the nucleus were calculated, graphed, and expressed as ratios of nuclear/cytoplasmic intensities (B). (C) Cells were transfected with CaMKI siRNA (100 nM) or scrambled RNA (100 nM) and treated with (+) or without (–) Ca2+ as in A at 1 mM (1) and 2 mM (2) concentrations. Total cell lysates were processed for immunoblotting with CaMKI, CCTα, or β-actin antibodies (Input). (D) Another portion of total lysates was processed for pull-down assays using 14-3-3 beads, followed by CCTα immunoblotting to assess protein interaction. The data in each panel represent n = 3 separate experiments. Scale bars, 10 μM.
FIGURE 3:
CaMKI regulates CCTα nuclear import. (A, B) MLE cells were transfected with CaMKI or empty vector as a control. After transfection (24 h), cells were treated with or without Ca2+ (1 mM) for 1 h. Cells were then fixed and immunostained with CCTα and DAPI antibodies (A). Fluorescent signals within the cytosol and the nucleus were calculated, graphed, and expressed as a ratio of nuclear/cytoplasmic intensities (B). (C) Cells transfected with CaMKI and empty vector were then treated with (+) 1 mM (1) or 2 mM (2) Ca2+ or without (–) Ca2+. Cell lysates were then processed for immunoblotting with CaMKI, CCTα, or β-actin antibodies (Input). (D) Another portion of total lysate was processed for pull-down assays using 14-3-3 ζ beads, followed by CCTα immunoblotting to assess protein interaction. The data in each panel represent n = 3 separate experiments. Scale bars, 10 μM
CaMK1 interacts with CCTα
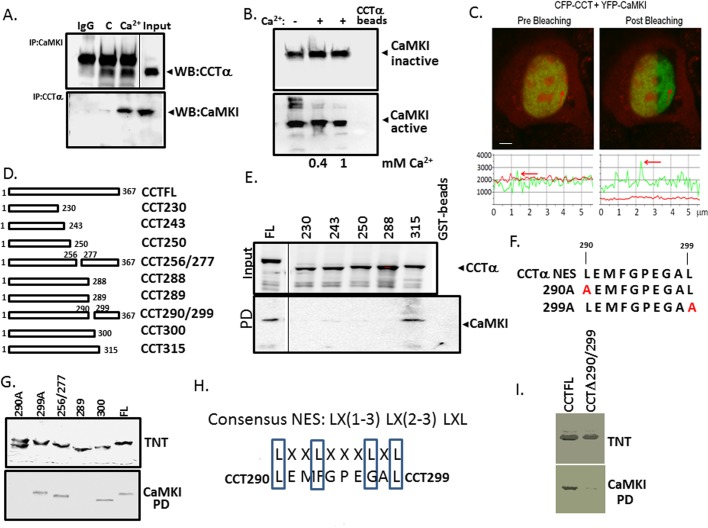
To examine whether CaMKI binds CCTα, we first immunoprecipitated CaMKI and CCTα from lung epithelial cells treated with or without Ca2+, followed by CCTα and CaMKI immunoblotting, respectively (Figure 4A). Binding specificity was monitored using normal rabbit immunoglobulin G (IgG) as a negative control. Our results indicate that endogenous CaMKI associates with CCTα and that this binding is enhanced by Ca2+ (Figure 4A). To support a direct interaction between these partners, we performed pull-down assays applying recombinant active or inactive forms of CaMKI to CCTα affinity resin with or without Ca2+ (Figure 4B). Both forms of CaMKI bound CCTα affinity resin (Figure 4B). Interaction between CaMKI and CCTα was also evaluated by fluorescence resonance energy transfer (FRET) analysis using the acceptor photobleaching technique (Chen and Mallampalli, 2007; Figure 4C). Cells were cotransfected with yellow fluorescent protein (YFP)–CaMKI as the acceptor fluorophore and cyan fluorescent protein (CFP)–CCTα as a donor. Interaction between CaMKI and CCTα was demonstrated at the single-cell level because the donor emission (CFP) signal increased after the nearby acceptor fluorophore (YFP) was inactivated by irreversible photobleaching (Figure 4C).
FIGURE 4:
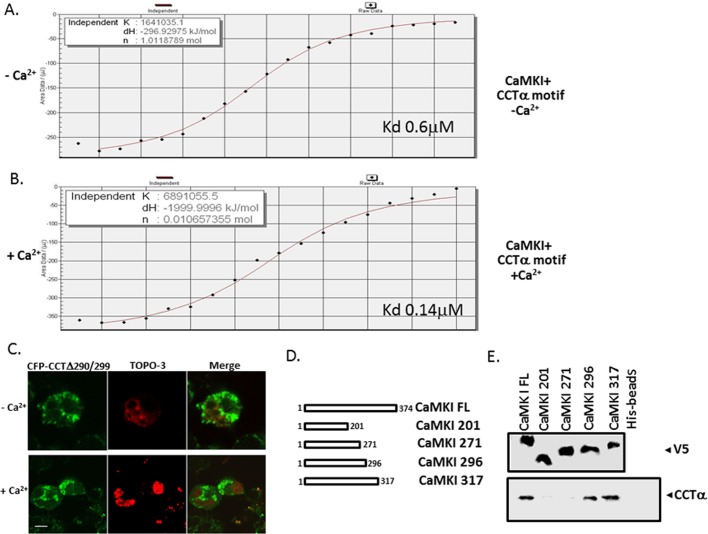
CaMKI interacts with CCTα directly. (A) Coimmunoprecipitation. Lysates from the cells treated with or without Ca2+ were incubated with primary antibody to CaMKI or CCTα for 2 h; normal rabbit IgG was used as a negative control. Cell lysates were then rotated overnight with anti-rabbit IgG beads at 4°C. Proteins were released from the beads by boiling in Laemmli buffer and resolved by SDS–PAGE, followed by immunoblot analysis with CCTα (top) or CaMKI (bottom) antibodies. (B) Purified active or inactive forms of CaMKI were incubated with 0.4 and 1 mM (+) Ca2+ or without Ca2+ (–) for 1 h, followed by CCTα pull-down assays before CaMKI immunoblotting. (C) FRET analysis. Cells were transfected with CFP-CCT and YFP-CaMKI, and CaMKI-CCT interaction at the single-cell level was imaged using laser-scanning microscopy before and after photobleaching. Top, after acceptor photobleaching, the fluorescence intensity of YFP decreased and CFP increased, confirming interaction between CaMKI and CCT. Bottom, graphs showing the quantitative analysis of the fluorescence intensities of FRET images. (D, F) Maps illustrating sequences of individual CCTα constructs used for identification of docking sites for CaMKI. Constructs tested include wild-type CCTFL, a series of mutants progressively truncated within CCTα, internal deletion mutants, and constructs harboring the point mutations within hydrophobic sites. (E) Cells were transfected with GST-tagged CCTα mutants. Top, relative expression. Bottom, levels of individual CCTα mutants bound to CaMKI on GST pull-down assays. (G, I) Top, expression of individual CCTα mutants synthesized using a rabbit reticulocyte lysate system (TnT). Bottom, relative CaMKI binding identified using pull-down assays and CaMKI-conjugated beads followed by autoradiography. (H) A sequence alignment of a canonical NES with the putative CaMKI-docking sequence within CCTα enriched with leucine and hydrophobic residues.
Mapping of interactions between CaMKI and CCTα
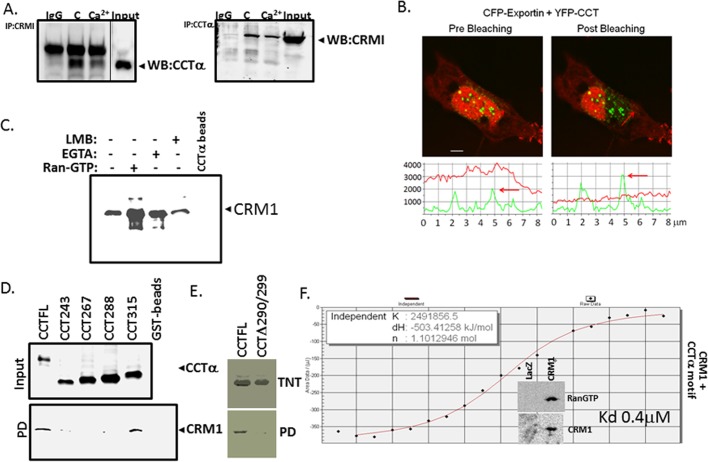
To identify the docking site of CaMKI within CCTα, we expressed progressively truncated glutathione S-transferase (GST)–tagged CCTα constructs in cells, followed by GST-pull down analysis using glutathione–agarose beads (Figure 4, D and E). Lysates from lung epithelial cells transfected with GST-CCTα mutants were processed for CCTα immunoblotting to confirm adequate expression of mutants (Figure 4E, top). Cell lysates applied and proteins eluted from beads were resolved by SDS–PAGE before CaMKI immunoblotting (Figure 4E, bottom). The results show that a potential CaMKI docking site likely resides between residues 288 and 315 within the membrane-binding domain of CCTα. Surprisingly, this region, enriched with leucine and hydrophobic residues, displays some similarities to a classic NES (Figure 4, F and H). To refine our mapping, we synthesized CCTα mutants using in vitro translation before pull-down analysis (Figure 4G). The synthesized CCTα mutants included CCT300 and CCT289 truncated mutants, a CCT256/277 internal deletion mutant that is devoid of key residues important for CCTα cellular localization (267VEEK270; Ridsdale et al., 2010), and two point mutations in which potentially critical leucines within a canonical NES were substituted with alanine (CCT290A and CCT299A; Figure 4G). After synthesis, these mutants were used for CaMKI pull-down assays and then resolved on SDS–PAGE before autoradiography (Figure 4G). The results confirm the GST pull-down data, localizing a potential CaMKI-binding sequence spanning residues 290–299 within CCTα (Figures 4E). Specifically, a point mutation of Leu-290 to Ala (CCT290) and a CCT289 truncated mutant displayed loss of ability to interact with CaMKI (Figure 4G, bottom). This was supported by the observation that an internal deletion mutant, CCTΔ290/299, and CCT290A lacked ability to bind CaMKI and failed to robustly translocate to the nucleus even with exogenous Ca2+ (Figures 4, G and I, and 5C and Supplemental Figure S3). The CCT Δ290/299 mutant also displayed functionality even with the removal of potentially important hydrophobic core residues (Supplemental Figure S4). Isothermal titration calorimetry (ITC) indicated high binding affinities between CaMKI and a peptide fragment from the CCTα membrane-binding sequence (Kd = 0.6 μM); this interaction was enhanced when determining Kd in the presence of Ca2+ (Figure 5, A and B). Thus endogenous CaMKI associates with CCTα both in vivo and in vitro, and this interaction appears to be direct and regulated by Ca2+. Finally, using similar approaches, we identified a CCTα-binding site within the substrate-binding pocket of CaMK1 (residues 271–296; Figure 5, D and E). Thus the results show that the CCTα sequence spanning residues 290–299 is required for CaMKI docking and displays similarity to a universal NES. The data also suggest that CCTα harbors a NES that might engage CRM1/exportin 1 as a mechanistically relevant interaction. Hence the results led us to hypothesize that CRM I is a molecular competitor that vies with CaMKI for access to this motif to regulate CCTα nuclear trafficking.
FIGURE 5:
A motif within CCTα and CaMKI regulates their interaction and CCTα subcellular localization. (A, B) ITC binding analysis of CaMKI and a peptide (LEMFGPEGAL) encoding a CaMKI-binding motif within CCTα without (A) or with (B) Ca2+ (1 mM). (C) The motif between residues 290 and 299 is required for CCTα nuclear import. MLE cells were transfected with a CFP-fused CCTα mutant lacking the region between residues 290 and 299 (CFP-CCT290/299). Cells were then treated with or without Ca2+, and after 1 h of incubation cells were visualized using a confocal microscope. Scale bars, 10 μM. (D, E) Mapping of a CCTα-docking site within CaMKI. (D) A map illustrating V5-His-tagged CaMKI full length (FL) and individual progressively truncated mutants within CaMKI. (E) Top, expression of individual constructs synthesized in vitro using TnT. Bottom, CaMKI FL and mutants synthesized in vitro using TnT that were then processed for His pull-down binding assays. The beads conjugated with individual CaMKI mutants were then rotated with equal aliquots of total cell lysates overnight, followed by immunoblot analysis using CCTα antibody.
CaMKI facilitates Ca2+-regulated CCTα nuclear translocation
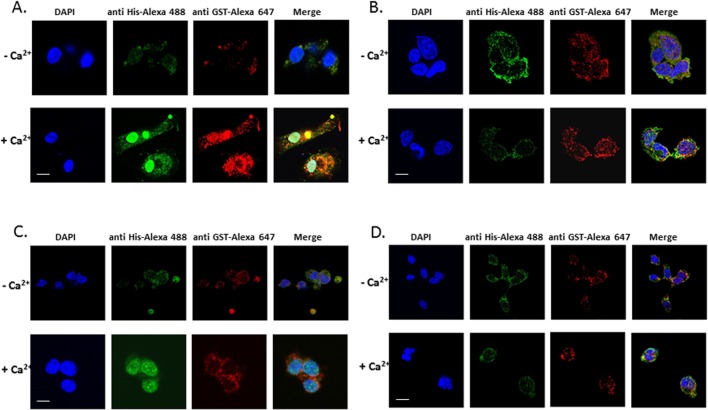
Our prior study (Agassandian et al., 2010) demonstrated that 14-3-3 ζ associates with CCTα to facilitate this enzyme for nuclear translocation in response to Ca2+. CCTα–14-3-3 ζ association occurs in a phosphospecific manner, where CaMKI might provide a phospho-docking site for 14-3-3 ζ. To demonstrate translocation of this protein complex to the nucleus, we performed in vitro nuclear transport assays using purified recombinant proteins (CCTα, 14-3-3 ζ, and CaMKI) in HeLa cells (Figure 6, A and B). In control studies in which we used only two components, CCTα with 14-3-3 ζ or CCTα with CaMKI, CCTα was retained largely in the cytoplasm (Figure 6, C and D). However, CCTα and 14-3-3 ζ both translocate to the nucleus in response to Ca2+ with inclusion of CaMKI, consistent with our prior data. CaMKI was predominantly localized in the cytoplasm (Figure 6, A and B).
FIGURE 6:
Nuclear import assay for CCTα. (A) Purified His-tagged conjugated CCTα, GST-tagged conjugated 14-3-3 ζ , and HeLa cell cytosolic fractions with overexpressd CaMKI were used in a nuclear transport assay in the presence of an ATP-regeneration system with or without 1 mM Ca2+ using digitonin-permeabilized HeLa cells as a model (Materials and Methods). The subcellular distribution of these proteins was visualized by indirect immunofluorescence using anti-His and anti-GST Alexa Fluor–conjugated antibody. (B) Purified His-tagged conjugated CCTα and GST-tagged conjugated 14-3-3 ζ without CaMKI were used as a control. (C) The import reaction containing His-tagged conjugated CCTα, GST-tagged conjugated CaMKI, and the HeLa cell cytosolic fraction with overexpressd 14-3-3 ζ with or without Ca2+ was also applied to the digitonin-permeabilized HeLa cells to assay the nuclear transport as described. The intracellular distribution of these proteins was visualized by indirect immunofluorescence using anti-His and anti-GST Alexa Fluor–conjugated antibody. (D) His-tagged conjugated CCTα and GST-tagged conjugated CaMKI without 14-3-3 ζ were used as a control.
Ca2+ regulates binding of CRM1/exportin 1 and CaMKI to CCTα
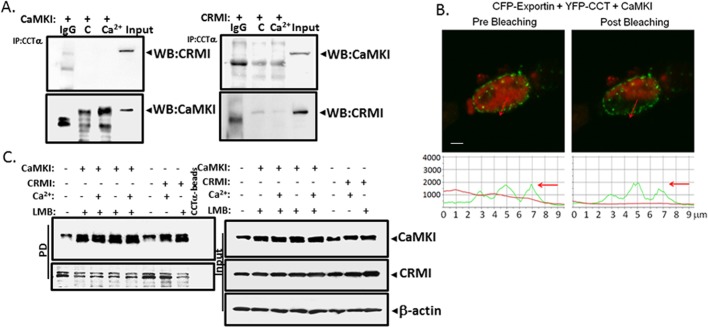
The foregoing data suggest that a molecular signature resembling a NES might serve as a recognition site for both CaMKI and CRM1/exportin 1. To determine whether CRM1 interacts with CCTα in lung epithelia, we first treated cells with or without Ca2+ and then performed coimmunoprecipitations using CCTα and CRM1 antibodies followed by immunoblotting (Figure 7A). Immunoprecipitation with CRM1 and probing with CCTα antibodies revealed that these two proteins interact, an association partially abrogated by Ca2+ (Figure 7A, left). These results were also observed when reversing the antibodies (Figure 7A, right). In contrast, CCTα binding to CaMKI is induced by Ca2+ (Figure 4A). In cells, interaction between CCTα and CRM1 was also seen using FRET analysis (Figure 7B). Furthermore, we observed direct binding between CCTα and CRM1 in pull-down assays using purified recombinant CRM1 and Talon resin–conjugated CCTα that was induced by RanGTP and blocked by leptomycin B (LMB; Figure 7C). The mapping studies using GST pull-downs (Figure 7D) or pull-down studies using CRM1-conjugated beads (Figure 7E) with various CCTα mutants revealed an identical binding motif (CCT290 to CCT299) used by CRM1 to access CCT similar to CaMKI. Hence these data demonstrate that CaMKI and CRM1 might share a unique molecular recognition site within CCTα. These observations were supported with ITC, indicating that binding affinities between CRM1 and a peptide fragment from the CCTα membrane-binding sequence (Kd = 0.4 μM) were comparable with CCTα interaction with CaMKI (Figures 5and 7F). However, in these studies exportin was not purified to homogeneity, as it contained RanGTP and possibly other coregulators that might enhance CRM1 binding affinity to CCTα peptide (Figure 7F, inset). Nevertheless, the tight binding between the interactors observed here is in line with CRM1 interaction with deaminase (Ellyard et al., 2011). Thus endogenous CRM1 directly associates with CCTα in a shared site with CaMKI, and this interaction is also regulated by Ca2+. To evaluate intermolecular competition between CaMKI and CRM1 for access to CCTα, we separately overexpressed these two proteins and then treated these cells with or without Ca2+ (Figure 8A). Immunoprecipitation with CCTα antibody followed by CRM1 immunoblotting did not detect CCTα in association with CRM1 in the cells when CaMKI was overexpressed (Figure 8A, left). Conversely, we did not detect interaction between CCTα and CaMKI in the cells transfected with CRM1 plasmid (Figure 8A, right). Hence there appears to be competition between CaMKI and CRM1 for CCTα binding. These data were supported by FRET analysis; here cells were transfected with CFP-CRM1, YFP-CCTα (Figure 8B), and CaMKI. The data show that CCTα loses its interaction to CRM1 with overexpression of CaMKI (Figure 8B). LMB, which disrupts association of CRM1 with the NES, also regulated CCTα binding to CaMKI. Cells were transfected with CaMKI or CRM1, followed by treatment with Ca2+, LMB, or Ca2+ in combination with LMB. Cell lysates were then used in CCTα-conjugated beads pull-down assays, followed by immunoblot analysis. Proteins were effectively expressed (Figure 8C, right, Input), and combinations of LMB and Ca2+ with CaMKI each induced CaMKI–CCTα association (Figure 8C, left).
FIGURE 7:
CRM1/exportin 1 binds CCTα. (A) Coimmunoprecipitation. Immunoprecipitation with CRM1 antibody and immunoblotting for CCTα (left) and immunoprecipitation with CCTα antibody and immunoblotting for CRM1 (right), using IgG as a negative control. Cells were treated with or without 1 mM of Ca2+ for 1 h. Cell lysates were then processed for immunoprecipitation, followed by immunoblotting using a rabbit TrueBlot kit. (B) FRET was used to assess protein interaction in the cells cotransfected with CFP-exportin 1 and YFP-CCTα. Top, the intense FRET signal confirmed interaction between CCTα and CRM1/exportin 1. Bottom, quantitative analysis of these images. Data represent n = 3 separate experiments. (C) Direct binding of recombinant CRM1 to purified CCTα. Binding reactions were performed using recombinant CRM1 with purified CCTα-conjugated to Talon resin in the presence or absence of RanGTP, LMB, or EGTA, followed by pull-down assays. The results were visualized by immunoblot analysis. (D, E) Identification of a CRM1-docking site within CCTα. GST-fused CCTα mutants were expressed in MLE cells (D, top). Cell lysates were then processed for GST-pull-down assays, followed by immunoblotting (bottom). (E) CCTα full length and an internal deletion mutant were synthesized using TnT (top), followed by pull-down assays using CRM1-conjugated beads (bottom). The beads were prepared using His-tagged CRM1 protein synthesized by TnT. CCTα full length and mutants bound to CRM1 beads were detected using immunoblotting. The results were visualized by immunoblot analysis (D) or autoradiography (E). (F) ITC binding analysis of CRM1 and a peptide (LEMFGPEGAL) encoding a CaMKI-binding motif within CCTα. The insert demonstrates the enrichment of CRM1 fraction with RanGTP. Data in each panel represent n = 2 or 3 separate experiments. *p < 0.05 vs. control.
FIGURE 8:
CaMKI competes with CRM1 for binding to docking site within CCTα. (A) Coimmunoprecipitations with CCTα antibody. Cells transfected with CaMKI (left) or CRM1 (right) and then treated with or without Ca2+ before coimmunoprecipitation with CCTα antibody or IgG as a negative control. Immunoprecipitated samples were then processed for either CRM1 or CaMKI immunoblotting. (B) FRET analysis of the cells cotransfected with CFP-exportin 1, YFP-CCTα, and ectopically expressed CaMKI demonstrates that transfection with CaMKI inhibits CRM1-CCTα interaction at the single-cell level. Bottom, quantitative analysis of the images. (C) Immunoblot analysis. Cells transfected with either His-conjugated CaMKI or His-CRM1 were then treated with or without Ca2+, LMB, or combinations of these factors and processed for CCTα pull-down assays. Right (Input), relative expression of these proteins in cells. β-Actin was used as a loading control. Left, results of pull-down assays and relative binding of individual effectors to CCTα.
Export of CCTα from the nucleus is regulated by CRM1
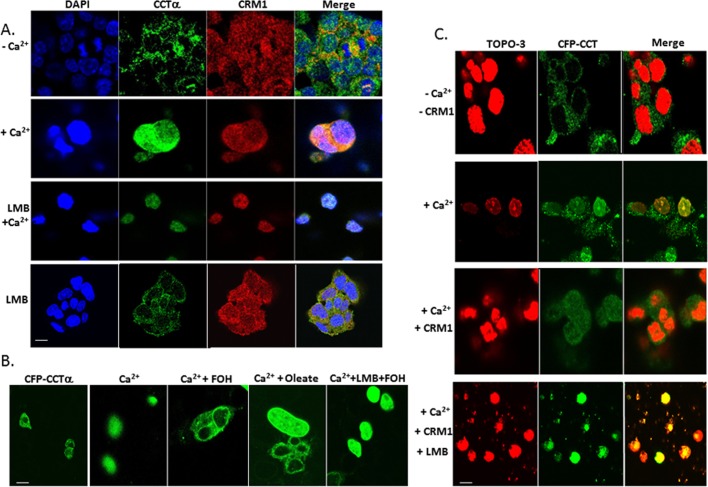
To assess whether CCTα nuclear export is CRM1 dependent, we used a CFP-CCTα reporter construct for cellular expression. First, endogenous CCTα in MLE cells was cytosolically localized, but in the presence of exogenous Ca2+ with or without LMB it relocated to the nucleus. colocalizing with CRM1 (Figure 9A). Effects of farnesol or oleate exposure, known to promote nuclear export of CFP-CCTα, were abrogated by LMB (Figure 9B). Last, Ca2+ was sufficient to trigger nuclear relocation of CFP-CCTα whether or not cells were exposed to LMB (Figure 9, A–C). However, Ca2+-induced nuclear translocation of CFP-CCTα was partially attenuated after expressing CRM1 in cells, evidenced by a reduction in the merged signal (Figure 9C, row 3, right lane). Thus CCTα nuclear export is, in part, CRM1 dependent.
FIGURE 9:
CCTα nuclear export is CRM1 dependent. (A) Intracellular localization of endogenous CCTα and CRM1. Cells were exposed to 1 mM Ca2+ or without Ca2+ for 1 h, or cells were treated with Ca2+ followed by exposure to LMB (200 nM) or exposed to LMB alone for 1 h. (B) CFP-conjugated CCT FL construct was expressed in MLE cells. After 24 h cells were exposed to Ca2+ (1 mM) with or without LMB before exposure to farnesol (FOH; 60 μM for 30 min) or oleate (300 μM for 30 min). (C) Cells were transfected with a CFP-conjugated CCT FL construct as in B before exposure to Ca2+ to induce nuclear entry. Some cells were also transfected with CRM1 or exposed to LMB to modulate CCTα localization. Scale bars, 10 μM.
The CCT 290–299 fragment is sufficient to confer nuclear export
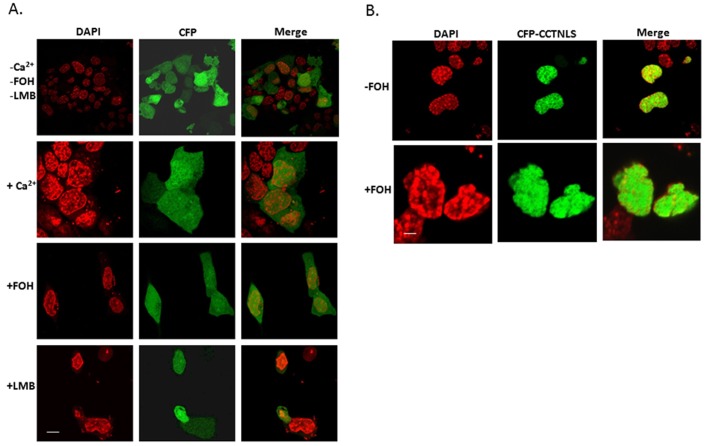
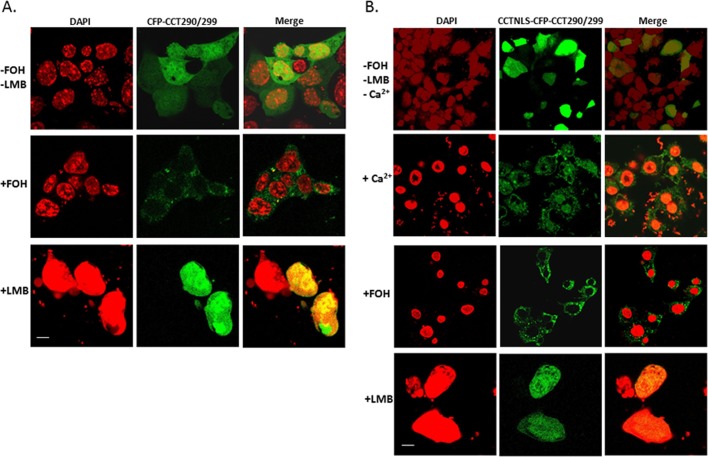
To assess the trafficking behavior of isolated signals within CCTα, we expressed in cells constructs encoding either an isolated NLS (residues 1–40), a isolated NES peptide (290–299), or a product containing both the NLS and NES fused to CFP. As shown in Figure 10A, expression of CFP alone in cells showed diffuse staining because the size of the construct allows for some passive diffusion through the nuclear pore complex. In contrast, expression of a CFP-NLS in cells resulted in an exclusively nuclear distributed fragment regardless of inclusion of farnesol (Figure 10B). In separate studies, farnesol was able to result in expression of a CFP-NES construct that was largely cytosolic (Figure 11A). LMB alone was able to retain the construct in the nucleus (Figure 11A, bottom). Moreover, fusion of the NES with the NLS resulted in a construct that localized to the cytoplasm with farnesol and produced a significant nuclear signal with the addition of Ca2+ or LMB (Figure 11B). Thus the isolated 290–299 fragment binds CRM1 and exhibits LMB sensitivity, and farnesol relocates this fragment to the cytoplasm, which all support that this sequence is a NES within CCTα.
FIGURE 10:
Subcellular distribution of the CFP vector and CFP-CCTNLS. MLE cells were transfected with CFP vector alone (A) or with a CFP-CCTNLS fluorescence construct (B), followed by exposure to FOH (60 μM for 30 min), LMB (200 nM for 1 h), or Ca2+ (1 mM for 1 h). Scale bars, 10 μM.
FIGURE 11:
Cellular expression of a CCT290/299 construct. MLE cells transfected with CFP-CCT290/299 (A) or CCTNLS-CFP-CCT290/299 (B) were then exposed to FOH (60 μM for 30 min), LMB (200 nM for 1 h), or Ca2+ (1 mM for 1 h) and cellular localization examined. Scale bars, 10 μM.
DISCUSSION
In this study we identified a new physiological role for CaMKI in the nuclear trafficking of an indispensable lipogenic enzyme involved in membrane formation and preservation of cell viability. We observed that CaMKI phosphorylates CCTα, which signals 14-3-3 ζ recruitment to facilitate CCTα nuclear entry. CaMKI was both sufficient and required for this process, and the kinase interacts with CCTα via a docking region upstream of a CCTα phosphoacceptor site. Unexpectedly, the CaMKI docking site within the CCTα membrane-binding domain shared some characteristics with a canonical NES, raising the possibility that other critical adaptor molecules also partake in CCTα nuclear–cytoplasmic trafficking. Indeed, these studies are the first to show that exportin 1/CRM1, a known NES adaptor molecule, also associates with CCTα in a shared domain, thereby appearing to exhibit intermolecular competition with CaMKI for association with the lipogenic enzyme under conditions of Ca2+ excess. Of interest, cellular expression of CRM1 was sufficient to override ability of CaMKI to associate with CCTα and triggered CCTα nuclear export despite Ca2+ stimulation (Figure 8C); furthermore, the isolated 290–299 fragment bound CRM1, was LMB sensitive, and was cytosolically expressed in response to farnesol, all indicative of the role of this sequence as a functional NES. However, expression of a CFP-CCTα mutant lacking this putative NES signature in cells resulted in cytosolically retained enzyme, suggesting that the CaMKI–14-3-3 ζ–CCTα partnership is a physiologically predominant interaction mechanism needed for enzyme nuclear trafficking (Figure 5C). The biological role of this pathway requires further investigation, but it is possible that CaMKI-mediated CCTα nuclear import may serve as a cellular survival mechanism by sequestering the enzyme within the nuclear envelope during calcium-activated proteolytic stress (Zhou et al., 2003).
Even though CaMKI is present in all mammalian cells and tissues, knowledge about its biologically relevant substrates is limited (Means, 2000). Furthermore, the role of CaMKI in regulating Ca2+-dependent cellular events is also largely unknown. As shown here, the CaM family of Ca2+/CaM-dependent protein kinases appears to control protein translocation within cells to regulate enzymatic behavior in time and space. In this regard, CaMKI was both required and sufficient to induce CCTα nuclear shift; the kinase, by increasing CCTα phosphorylation, reduces activity of the lipogenic enzyme, with a resultant decrease in phospholipid synthesis (Figures 2 and 3 and Supplemental Figure S5). These results suggest that CaMKI has an important role in inactivating CCTα during its escort to the nuclear compartment, perhaps as a means to reactivate the enzyme later for nuclear membrane formation (Gehrig et al., 2008). The in vitro phosphorylation of CCTα by CaMKI was selective and demonstrates that CCTα might be an endogenous substrate for CaMKI. We detected that CaMKI interacts with CCTα both using recombinant proteins and in cells supportive of a direct molecular association in the context of Ca2+ stimulation. Similar to the behavior of other protein kinases, there exists a specific docking motif that may optimize efficiency of enzyme phosphorylation (Hirschi et al., 2010). Many protein kinases bind with relatively high affinity to these interaction motifs on substrates that are distal to the target phosphorylation sites. These interactions can be crucial for efficient signal transmission (Whisenant et al., 2010). Mutation or deletion of the docking sites within CCTα significantly reduced the ability of CaMKI kinase to associate with the enzyme. These results resemble our findings with the ERK1/2 docking site within the CCTα membrane-binding domain (residues 287–300; Agassandian et al., 2005). This ERK1/2 docking motif represents a “D-site”—a region identified in substrates for mitogen-activated protein kinases (Bardwell, 2006; Whisenant et al., 2010). The D-site consensus motif consists of a basic cluster of one to four residues, a short spacer, and a hydrophobic-X-hydrophobic submotif ((R/K)3-5(X)1-5(Φ-X-Φ)). The D-site within CCTα likely corresponds to residues 281–292, a stretch of amino acids that partially overlaps with the CaMKI docking site (residues 290–299; Figure 4). Thus CCT290 might represent a critical and mutual recognition signal for CaMKI and ERK1/2 signaling pathways used for efficient binding and phosphorylation.
Of note, the CaMKI-docking motif within CCTα spanning residues 290x299 is enriched with hydrophobic residues, with similarities to a NES that could be involved in binding to exportin 1/CRM1. CRM1 is a LMB-sensitive export receptor mediating rapid nuclear exit of proteins via direct binding to a leucine-rich nuclear export signal within cargoes. Prior work suggests that CCTα nuclear export is LMB insensitive but dependent on its membrane-binding domain (Gehrig et al., 2009). This study did not identify and characterize a NES, perhaps because computational results (NetNES) do not predict a NES in the CCTα primary sequence; furthermore, only ∼36% of CRM1-binding sequences fit the wildly accepted canonical NES consensus sequence, L-X(2-3)-[LIVFM]-X(2,3)-L-X-[LI] (la Cour et al., 2003). The disparities between our results and those of others may be related to culture conditions (e.g., Ca2+ concentrations, differing cell lines), as recently shown (Mills and Truong, 2010). Of note, other nuclear exporters, such as calreticulin, did not engage CCTα, supporting a potential role for CRM1 as the primary protein involved in nuclear exit for the enzyme (unpublished data). Although CaMKI is predominantly a cytoplasmic protein (Supplemental Figure S1, A and B), CRM1 exhibits cytoplasmic and nuclear localization, suggesting that its interaction with CCTα may be more widespread within epithelia (Figure 9A). Our results detected CRM1 in association with CCTα, where it also binds within the CaMKI-docking site. CRM1 is able to abrogate CaMKI interaction with CCTα (Figure 8A), and CRM1 expression partially produced retention of CCTα within the cytosol after Ca2+ stimulation (Figure 9C). Thus molecular interplay between these two proteins for access to the NES might regulate CCTα cellular localization. Nevertheless, our findings show that both CaMKI and CRM1 bind to a similar motif within CCTα (Figures 4, and 7, D and E) and that individual expression of CaMKI or CRM1 negates association of the other binding partner with CCTα (Figure 8A), suggesting that distinct and opposing mechanisms govern cytoplasmic–nuclear shuttling in vivo. Last, that LMB was sufficient to impede CCTα nuclear export induced by oleate or farnesol or by CRM1 overexpression supports our hypothesis that CCTα nuclear export is CRM1 dependent, perhaps also mediated by the 290–299 signature. Of importance, however, our data suggest that the 290–299 signature within the entire CCTα cargo primarily serves in nuclear import in concert with CaMKI-mediated phosphorylation of the enzyme C-terminus in the appropriate context of the membrane domain. This is evidenced by our findings that this 290–299 signature is required for nuclear entry (Figure 5C) and that expression of a CCTα fragment encompassing residues 243–367 was sufficient to confer nuclear import to a heterologous protein in cells (Supplemental Figure S6).
The complexity of CCTα nuclear shuttling is evidenced by interplay between NLS and NES signatures, molecular chaperones, and posttranslational modifications. We showed that monoubiquitination masks a canonical NLS within CCTα that results in lysosomal degradation of the enzyme (Chen and Mallampalli, 2009). In the present scenario, CaMKI phosphorylation of specific CCTα residues impairs CRM1 binding and yet recruits 14-3-3 ζ to the enzyme to facilitate nuclear import. An important aspect of this mode of regulatory control is that intracellular Ca2+ concentrations are critical in mediating enzyme trafficking. Elevations of cell Ca2+ concentrations occur during mitosis with disruption of the nuclear envelope, suggesting that CCTα nuclear entry might be synchronized to specific checkpoints during cell cycle progression (Brown and Shoback, 1984; Pszczolkowski et al., 1999). Alternatively, CCTα trafficking into the nucleus could occur pathobiologically during bacterial infections that increase intracellular Ca2+ concentrations (Chen et al., 2011). The complexity of interactions within functional signatures, cell Ca2+ concentrations, and posttranslational modifications in substrates is exemplified by other proteins, such as NF-AT2. This protein contains two NLS, whose interaction with importin depends on its phosphorylation status (Beals et al., 1997). At low Ca2+ concentrations in cells, both NLSs are phosphorylated, inhibiting NF-AT2 nuclear import. With an increase in Ca2+ concentration, calcineurin dephosphorylates NF-AT2, enabling nuclear entry for this cargo (Beals et al., 1997). Similar mechanisms for protein export were identified in which, under osmotic stress, Hog 1p is phosphorylated, which renders NES inaccessible for binding to exportin 1 (Ferrigno et al., 1998). Here Ca2+/CaMKI regulate CCTα nuclear translocation, presumably via a masking mechanism in which CaMKI occupies a putative NES within the membrane-binding domain (Supplemental Figure S7). Because most NESs bind CRM1 with relatively low affinity (Kutay and Guttinger, 2005), the Ca2+/CaMKI interaction with CCTα may be more dominant physiologically, depending on the experimental conditions. This is supported by data showing that a mutant CFP-CCT construct devoid of the putative NES was cytosolically retained (Figure 5C). However, even multipartite recognition of separate weak NES epitopes may occur within CRM1 substrates, enhancing CRM1 binding to cargoes. The identification of such atypical NES signatures within CCTα requires additional investigation.
MATERIALS AND METHODS
Materials
The MLE and HeLa cell lines were obtained from the American Type Culture Collection (Manassas, VA). Recombinant 14-3-3 ζ (YWHAZ) was from GenWay Biotech (San Diego, CA). Rabbit CCTα antiserum was raised against synthetic peptides by Covance Research Products (Richmond, CA). The TnT reticulocyte assay system was obtained from Promega (Madison, WI). The Talon metal affinity resin was from Clontech Laboratories (Valencia, CA). The QuikChange Site-Directed Mutagenesis Kit was from Stratagene (La Jolla, CA). Synthetic peptides used for binding were made by CHI Scientific (Maynard, MA). The pCR4TOPO plasmids from Escherichia coli TOPO-competent cells were from Invitrogen (Carlsbad, CA). FuGENE 6 transfection reagent was purchased from Roche Diagnostics (Indianapolis, IN). CaMKI antibodies were obtained from Millipore (Billerica, MA); rabbit polyclonal was purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and rabbit calreticulin antibody was from Cell Signaling (Danvers, MA). CaMKII antibody was from Santa Cruz Biotechnology. Anti-V5 mouse antibody was from Invitrogen. The Active Ran antibody was purchased from NewEast Biosciences (Malvern, PA). Purified exportin 1 was from BD-Bioscience (Bedford, MA). Nucleic acid and protein purification kits were from Macherey-Nagel (Bethlehem, PA). DNA sequencing was performed by the University of Pittsburgh DNA Core.
Cell culture
MLE cells were maintained in Hite's medium supplemented with 2% fetal bovine serum at 37°C in 5% CO2. MLE cell treatments and transfections were performed in serum-free medium (Agassandian et al., 2010).
In vitro phosphorylation
Purified CCTα from mouse liver or synthesized by TnT was used for in vitro phosphorylation. The ATF1 transcription factor was used as a positive control. First, purified CCTα (2 μg) was dephosphorylated using 5 U of calf alkaline phosphatase in 50 mM Tris-HCl (pH 8.5) at 30°C for 1 h. The reaction included NaF (5 mM) and sodium vanadate (1 mM), followed by freezing at −80°C and thawing at 25°C. Phosphorylation of CCTα and ATF1 was performed using 150–500 ng of CaMKI in the reaction mixture containing kinase buffer (50 mM Tris-HCl, pH 7.5, 5 mM β-glycerol phosphate, and 0.1% β-mercaptoethanol) with 50 μM [γ-32P]ATP in the presence of 1 mM CaCl2. After 20 min of incubation at 30°C, the reaction was terminated with 4× Laemmli protein loading buffer, followed by boiling for 5 min at 95°C. Samples were resolved by 10% SDS–PAGE, transferred to nitrocellulose membranes, and visualized by autoradiography or by using a phosphoimager.
Immunoblot analysis
Immunoblotting was performed using the samples that were separated by SDS–PAGE (10% acrylamide) as described previously (Chen and Mallampalli, 2007; Agassandian et al., 2010). The dilution factor for CaMKI and pCaMKI was 1:500 and for exportin (CRM1) and CCTα was 1:1000. Immunoreactive proteins were detected using a SuperSignal West Femto Substrate reagent (Pierce, Thermo Scientific, Rockford, IL).
Immunoprecipitation
Immunoprecipitation was performed with the rabbit IgG TrueBlot system per the manufacturer's instructions. Cell lysates precleared with anti-rabbit IgG beads were then incubated with primary antibodies to CaMKI, CCTα, CRM1, or normal rabbit IgG for 2 h, followed by overnight rotation with anti-rabbit IgG beads at 4°C. The proteins were released from the beads by boiling in Laemmli buffer for 5 min and then separated by SDS–PAGE before immunoblotting with appropriate antibodies.
Construction of CaMKI and CCTα carboxyl-terminal and deletion mutants
Full-length CCTα and CCTα carboxyl-terminal truncation mutants were constructed as described previously (Agassandian et al., 2010). Additional CCTα and CaMKI mutants were generated by site-directed mutagenesis using similar PCR-based strategies with appropriate forward and reverse primers, using pCR4-CCT as a template (Agassandian et al., 2005; Chen and Mallampalli, 2007). These mutant plasmid constructs were transformed into E. coli TOP10-competent cells for large-scale plasmid preparation. In brief, pCMV5-CCTα was used as a template for PCR, using appropriate forward and reverse primers. All PCR products were gel purified and cloned into pENTR-TOPO. For cloning into GST fusion constructs, pENTR-TOPO plasmids and pDEST27 GST destination vector were incubated with LR Clonase enzyme mix (Invitrogen) at 25°C for 1 h per the manufacturer's instructions. The reaction was terminated by adding proteinase K and heated at 37°C for 10 min. The plasmids were transformed into E. coli Top10-competent cells as described earlier (Agassandian et al., 2010).
In vitro TnT and pull-down assays
For in vitro synthesis, CCTα constructs cloned into pCR4-TOPO4 and added to the rabbit reticulocyte lysate were incubated with T7 RNA polymerase in a 50- μl reaction mix containing [35S]methionine following the manufacturer's instructions. Aliquots (10 μl) of in vitro translation products were then boiled for 5 min in Laemmli buffer and stored at −80°C. The remaining (40 μl) portion of TnT products was processed using pull-down assays with CaMKI-conjugated agarose beads. Proteins eluted from the beads and products of in vitro translation were resolved by SDS–PAGE, followed by autoradiography.
Fluorescence resonance energy transfer analysis
PCR-based strategies were used to construct chimeric cDNAs for FRET as described (Chen and Mallampalli, 2007; Agassandian et al., 2010). The PCR products were gel purified and digested before cloning into a pAmCyan-Cl vector, generating CFP-CCTα and CFP-exportin, or into a pZsYellow1-Cl vector, generating YFP-CaMKI and YFP-CCTα. For analysis of interactions by FRET, cells were plated at 0.12 × 106 cells/well. Cells were then cotransfected with plasmids using FuGENE 6 transfection reagent. Interactions between proteins were detected at the single-cell level using a combination laser-scanning microscope system and a photobleaching method (Nikon A1; Nikon, Tokyo, Japan). FRET quantification of fluorescence images was generated using Zeiss Rel3.2 image software (Carl Zeiss, Jena, Germany). The average fluorescence intensity per pixel was calculated after background subtraction.
Immunofluorescence microscopy
MLE cells were cultured on 35-mm glass-bottom dishes to achieve 50% confluence. Cells were transfected with scrambled RNA or CaMKI siRNA, an empty vector, or psDNACaMKI for 48 h, followed by Ca2+ exposure. Cells were then fixed, processed for immunocytochemical staining, and viewed using a combination laser-scanning microscope system. For quantification of fluorescence, 10 individual cells for each condition were analyzed. After subtraction of background, regions of interest were selected in the nucleus and in the cytosol of each cell, and the average intensity was measured and calculated by using ImageJ analysis software (National Institutes of Health, Bethesda, MD). CFP vector, CFP-CCTFL, CFP-ΔCCT290/299, CFP-CCTNLS, CFP-CCT290/299, and CCTNLS-CFP-CCT290/299 plasmids were transfected on 35-mm glass-bottom dishes with or without CRM1 for 24–48 h, followed by exposure to Ca2+, oleate, farnesol, or leptomycin B for an appropriate amount of time. The nucleus was visualized using 4′,6-diamidino-2-phenylindole (DAPI) or TOPO-3 (Invitrogen). The images were then acquired, and the results were analyzed using Nikon Elements software.
Isothermal titration calorimetry
ITC directly measures the energy associated with chemical reactions triggered by mixing of two components. This method determines the thermodynamic parameters of interaction of the two compounds, which reflect their binding affinity. Here ITC was used to measure the heat of reaction upon titration of a solution containing CaMKI or CRM1 and a peptide (LLEMFGPEGAL) encoding a CaMKI-binding motif within CCTα. For these experiments, 20 5-μl injections of commercially prepared peptide were injected in the cell with CaMKI or CRM1 using NANO ITC (TA Instruments, Lindon, UT). Experiments were conducted at 25°C in phosphate buffer (pH 7.4). The concentration of CaMKI or CRM1 was 0.01 mM, and the concentration of peptide in the syringe was 0.2 mM. The titration curve was fit to determine the binding stoichiometry (n), enthalpy (ΔH), and binding constant (Kd) using the NANO series software provided by the manufacturer.
Enzyme activity
CCTα activity assays were performed as described previously (Agassandian et al., 2010).
PtdCho synthesis
Cells were pulsed with 5 μCi of [methyl-3H]choline for 2 h. Cellular lipids were extracted by the method of Bligh and Dyer (Bligh and Dyer, 1959; Gilfillan et al., 1983) and resolved by thin-layer chromatography (TLC). PtdCho was quantitated by scintillation counting as previously described (Gilfillan et al., 1983; Agassandian et al., 2010).
Nuclear transport assays
MLE cells were cultured on 35-mm glass-bottom dishes to achieve 50% confluency. The cells were then rinsed twice with ice-cold transport buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.3, 110 mM CH3COOK, 2 mM (CH3COO)2Mg, 5 mM CH3COONa, and 0.5 mM ethylene glycol tetraacetic acid [EGTA]) and permeabilized for 5 min in ice-cold transport buffer containing 40 μg/ml digitonin, 2 mM dithiothreitol (DTT), and protease inhibitor, complete EDTA-free. The cells were then washed twice for 10 min with the ice-cold transport buffer containing 2 mM DTT and protease inhibitor. After the last washing, the reaction mixture was applied to each dish. The reaction mixture contained histidine (His)-tagged CCTα purified protein, GST-tagged CaMKI, or 14-3-3 ζ recombinant purified protein and HeLa cytosolic fraction as a source of nuclear transport factors in an ATP regeneration system (1 mM ATP, 5 mM creatine phosphate, and 20 U/ml creatine phosphokinase) for 30 min at 30°C. Cells were then washed twice in the transport buffer, fixed with 4% formaldehyde in phosphate-buffered saline, processed for immunocytochemical staining, and viewed using a combination laser-scanning microscope system. As the secondary antibody to detect indirect immunofluorescence we used specific anti-His and anti-GST tagged antibodies conjugated to Alexa Fluor 488 and Alexa Fluor 647 (Invitrogen), respectively.
Binding assays
Pull-down assays were performed essentially as described (Agassandian et al., 2005, 2010). For in vitro binding assays of purified recombinant CRM1 and purified CCTα in the presence of RanGTP we first loaded purified, untagged Ran (3 μg) with 1 mM GTP by incubation for 1 h at 21°C in the presence of 5 mM (CH3COO)2Mg. The RanGTP was then incubated with 0.25 μM CRM1 and CCTα-conjugated Talon resin in the presence or absence of LMB or EGTA for 1 h at 21°C. Proteins were then released from resin by boiling for 5 min in Laemmli buffer and analyzed by SDS–PAGE.
Statistical analysis
Statistical analysis was performed by one-way analysis of variance or Student's t test. Data are presented as mean ± SE.
Supplementary Material
Acknowledgments
We thank Jeffrey Brodsky for critical review of the manuscript. This material is based on work supported, in part, by the U.S. Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development. This work was supported by a Merit Review Award from the U.S. Department of Veterans Affairs and National Institutes of Health RO1 Grants HL096376, HL097376, and HL098174 (to R.K.M.). The contents presented do not represent the views of the Department of Veterans Affairs of the United States Government.
Abbreviations used:
- ATF-1
cyclic AMP-dependent transcription factor
- Ca2+
calcium
- CaMKI
calmodulin kinase I
- CCTα
CTP:phosphocholine cytidylyl transferase alpha
- CFP
cyan fluorescent protein
- CRM1
chromosome region maintenance 1 (exportin 1)
- DAPI
4′,6,diamidino-2-phenylindole
- EGTA
ethylene glycol tetraacetic acid
- GST
glutathione S-transferase
- ITC
isothermal titration calorimetry
- MLE
murine lung epithelial
- NES
nuclear export signal
- NLS
nuclear localization signal
- LMB
leptomycin B
- PtdCho
phosphatidylcholine
- RanGTP
Ras-related nuclear protein guanosine-5′-triphosphate
- TLC
thin-layer chromatography
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-10-0863) on May 23, 2012.
REFERENCES
- Agassandian M, Chen BB, Schuster CC, Houtman JC, Mallampalli RK. 14-3-3zeta escorts CCTalpha for calcium-activated nuclear import in lung epithelia. FASEB J. 2010;24:1271–1283. doi: 10.1096/fj.09-136044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agassandian M, Zhou J, Tephly LA, Ryan AJ, Carter AB, Mallampalli RK. Oxysterols Inhibit phosphatidylcholine synthesis via ERK docking and phosphorylation of CTP:phosphocholine cytidylyltransferase. J Biol Chem. 2005;280:21577–21587. doi: 10.1074/jbc.M412409200. [DOI] [PubMed] [Google Scholar]
- Aletta JM, Selbert MA, Nairn AC, Edelman AM. Activation of a calcium-calmodulin-dependent protein kinase I cascade in PC12 cells. J Biol Chem. 1996;271:20930–20934. doi: 10.1074/jbc.271.34.20930. [DOI] [PubMed] [Google Scholar]
- Askjaer P, et al. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol Cell Biol. 1999;19:6276–6285. doi: 10.1128/mcb.19.9.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askjaer P, Jensen TH, Nilsson J, Englmeier L, Kjems J. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J Biol Chem. 1998;273:33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- Bardwell L. Mechanisms of MAPK signalling specificity. Biochem Soc Trans. 2006;34:837–841. doi: 10.1042/BST0340837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beals CR, Sheridan CM, Turck CW, Gardner P, Crabtree GR. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science. 1997;275:1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Echarri A, Ross TM, Cullen BR. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EM, Shoback DM. The relationship between PTH secretion and cytosolic calcium concentration in bovine parathyroid cells. Prog Clin Biol Res. 1984;168:139–144. [PubMed] [Google Scholar]
- Chen BB, Coon TA, Glasser JR, Mallampalli RK. Calmodulin antagonizes a calcium-activated SCF ubiquitin E3 ligase subunit, FBXL2, to regulate surfactant homeostasis. Mol Cell Biol. 2011;22:22. doi: 10.1128/MCB.00723-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BB, Mallampalli RK. Calmodulin binds and stabilizes the regulatory enzyme, CTP: phosphocholine cytidylyltransferase. J Biol Chem. 2007;282:33494–33506. doi: 10.1074/jbc.M706472200. [DOI] [PubMed] [Google Scholar]
- Chen BB, Mallampalli RK. Masking of a nuclear signal motif by monoubiquitination leads to mislocalization and degradation of the regulatory enzyme cytidylyltransferase. Mol Cell Biol. 2009;29:3062–3075. doi: 10.1128/MCB.01824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapperton JA, Martin SR, Smerdon SJ, Gamblin SJ, Bayley PM. Structure of the complex of calmodulin with the target sequence of calmodulin-dependent protein kinase I: studies of the kinase activation mechanism. Biochemistry. 2002;41:14669–14679. doi: 10.1021/bi026660t. [DOI] [PubMed] [Google Scholar]
- Ellyard JI, Benk AS, Taylor B, Rada C, Neuberger MS. The dependence of Ig class-switching on the nuclear export sequence of AID likely reflects interaction with factors additional to Crm1 exportin. Eur J Immunol. 2011;41:485–490. doi: 10.1002/eji.201041011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrigno P, Posas F, Koepp D, Saito H, Silver PA. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 1998;17:5606–5614. doi: 10.1093/emboj/17.19.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Gehrig K, Cornell RB, Ridgway ND. Expansion of the nucleoplasmic reticulum requires the coordinated activity of lamins and CTP:phosphocholine cytidylyltransferase alpha. Mol Biol Cell. 2008;19:237–247. doi: 10.1091/mbc.E07-02-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrig K, Morton CC, Ridgway ND. Nuclear export of the rate-limiting enzyme in phosphatidylcholine synthesis is mediated by its membrane binding domain. J Lipid Res. 2009;50:966–976. doi: 10.1194/jlr.M800632-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan AM, Chu AJ, Smart DA, Rooney SA. Single plate separation of lung phospholipids including disaturated phosphatidylcholine. J Lipid Res. 1983;24:1651–1656. [PubMed] [Google Scholar]
- Hirschi A, Cecchini M, Steinhardt RC, Schamber MR, Dick FA, Rubin SM. An overlapping kinase and phosphatase docking site regulates activity of the retinoblastoma protein. Nat Struct Mol Biol. 2010;17:1051–1057. doi: 10.1038/nsmb.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaska JM, Black BE, Rastinejad F, Paschal BM. Ca2+-dependent nuclear export mediated by calreticulin. Mol Cell Biol. 2002;22:6286–6297. doi: 10.1128/MCB.22.17.6286-6297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowski S, Fagone P. CTP: phosphocholine cytidylyltransferase: paving the way from gene to membrane. J Biol Chem. 2005;280:853–856. doi: 10.1074/jbc.R400031200. [DOI] [PubMed] [Google Scholar]
- Kahl CR, Means AR. Regulation of cell cycle progression by calcium/calmodulin-dependent pathways. Endocr Rev. 2003;24:719–736. doi: 10.1210/er.2003-0008. [DOI] [PubMed] [Google Scholar]
- Kutay U, Guttinger S. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 2005;15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- la Cour T, Gupta R, Rapacki K, Skriver K, Poulsen FM, Brunak S. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 2003;31:393–396. doi: 10.1093/nar/gkg101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Kwon YG, Lawrence DS, Edelman AM. A requirement of hydrophobic and basic amino acid residues for substrate recognition by Ca2+/calmodulin-dependent protein kinase Ia. Proc Natl Acad Sci USA. 1994;91:6413–6417. doi: 10.1073/pnas.91.14.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci USA. 2000;97:14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means AR. Regulatory cascades involving calmodulin-dependent protein kinases. Mol Endocrinol. 2000;14:4–13. doi: 10.1210/mend.14.1.0414. [DOI] [PubMed] [Google Scholar]
- Mills E, Truong K. Engineering Ca2+/calmodulin-mediated modulation of protein translocation by overlapping binding and signaling peptide sequences. Cell Calcium. 2010;47:369–377. doi: 10.1016/j.ceca.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Nairn AC, Greengard P. Purification and characterization of Ca2+/calmodulin-dependent protein kinase I from bovine brain. J Biol Chem. 1987;262:7273–7281. [PubMed] [Google Scholar]
- Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Gwizdek C, Dargemont C. Protein export from the nucleus. Traffic. 2001;2:684–689. doi: 10.1034/j.1600-0854.2001.21002.x. [DOI] [PubMed] [Google Scholar]
- Pante N, Bastos R, McMorrow I, Burke B, Aebi U. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J Cell Biol. 1994;126:603–617. doi: 10.1083/jcb.126.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante N, Jarmolowski A, Izaurralde E, Sauder U, Baschong W, Mattaj IW. Visualizing nuclear export of different classes of RNA by electron microscopy. RNA. 1997;3:498–513. [PMC free article] [PubMed] [Google Scholar]
- Pszczolkowski MA, Lee WS, Liu HP, Chiang AS. Glutamate-induced rise in cytosolic calcium concentration stimulates in vitro rates of juvenile hormone biosynthesis in corpus allatum of Diploptera punctata. Mol Cell Endocrinol. 1999;158:163–171. doi: 10.1016/s0303-7207(99)00167-7. [DOI] [PubMed] [Google Scholar]
- Ridsdale R, Tseu I, Wang J, Post M. Functions of membrane binding domain of CTP:phosphocholine cytidylyltransferase in alveolar type II cells. Am J Respir Cell Mol Biol. 2010;43:74–87. doi: 10.1165/rcmb.2009-0231OC. [DOI] [PubMed] [Google Scholar]
- Sackmann S, Lichtenauer U, Shapiro I, Reincke M, Beuschlein F. Aldosterone producing adrenal adenomas are characterized by activation of calcium/calmodulin-dependent protein kinase (CaMK) dependent pathways. Horm Metab Res. 2011;43:106–111. doi: 10.1055/s-0030-1269899. [DOI] [PubMed] [Google Scholar]
- Sheng M, Thompson MA, Greenberg ME. CREB: a Ca(2+)-regulated transcription factor phosphorylated by calmodulin-dependent kinases. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- Stedman DR, Uboha NV, Stedman TT, Nairn AC, Picciotto MR. Cytoplasmic localization of calcium/calmodulin-dependent protein kinase I-alpha depends on a nuclear export signal in its regulatory domain. FEBS Lett. 2004;566:275–280. doi: 10.1016/j.febslet.2004.04.042. [DOI] [PubMed] [Google Scholar]
- Uboha NV, Flajolet M, Nairn AC, Picciotto MR. A calcium- and calmodulin-dependent kinase Ialpha/microtubule affinity regulating kinase 2 signaling cascade mediates calcium-dependent neurite outgrowth. J Neurosci. 2007;27:4413–4423. doi: 10.1523/JNEUROSCI.0725-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yang H, Liu YC, Jelinek T, Zhang L, Ruoslahti E, Fu H. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry. 1999;38:12499–12504. doi: 10.1021/bi991353h. [DOI] [PubMed] [Google Scholar]
- Wang L, Magdaleno S, Tabas I, Jackowski S. Early embryonic lethality in mice with targeted deletion of the CTP:phosphocholine cytidylyltransferase alpha gene (Pcyt1a) Mol Cell Biol. 2005;25:3357–3363. doi: 10.1128/MCB.25.8.3357-3363.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisenant TC, Ho DT, Benz RW, Rogers JS, Kaake RM, Gordon EA, Huang L, Baldi P, Bardwell L. Computational prediction and experimental verification of new MAP kinase docking sites and substrates including Gli transcription factors. PLoS Comput Biol. 2010:6. doi: 10.1371/journal.pcbi.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Ryan AJ, Medh J, Mallampalli RK. Oxidized lipoproteins inhibit surfactant phosphatidylcholine synthesis via calpain-mediated cleavage of CTP:phosphocholine cytidylyltransferase. J Biol Chem. 2003;278:37032–37040. doi: 10.1074/jbc.M304316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.