Telomere maintenance and DNA repair are important processes that protect the genome. The essential helicase mRtel1 functions in homologous recombination repair and replication. In addition, telomeres in mRtel-deficient ES cells appear relatively stable in length, suggesting that mRtel1 is required to allow extension by telomerase.
Abstract
Telomere maintenance and DNA repair are important processes that protect the genome against instability. mRtel1, an essential helicase, is a dominant factor setting telomere length in mice. In addition, mRtel1 is involved in DNA double-strand break repair. The role of mRtel1 in telomere maintenance and genome stability is poorly understood. Therefore we used mRtel1-deficient mouse embryonic stem cells to examine the function of mRtel1 in replication, DNA repair, recombination, and telomere maintenance. mRtel1-deficient mouse embryonic stem cells showed sensitivity to a range of DNA-damaging agents, highlighting its role in replication and genome maintenance. Deletion of mRtel1 increased the frequency of sister chromatid exchange events and suppressed gene replacement, demonstrating the involvement of the protein in homologous recombination. mRtel1 localized transiently at telomeres and is needed for efficient telomere replication. Of interest, in the absence of mRtel1, telomeres in embryonic stem cells appeared relatively stable in length, suggesting that mRtel1 is required to allow extension by telomerase. We propose that mRtel1 is a key protein for DNA replication, recombination, and repair and efficient elongation of telomeres by telomerase.
INTRODUCTION
Telomere maintenance and DNA repair are two essential processes that prevent genome instability and cancer. Telomeres are protective DNA–protein complexes at the end of chromosomes, which in all vertebrates consist of long arrays of TTAGGG repeats and associated proteins. Telomeric nucleoproteins are known as the shelterin complex (de Lange, 2005). Interference with any of the shelterin complex members or with the telomeric sequence itself leads to chromosomal instability and loss of cell viability. In addition to the shelterin complex, many other proteins function at the telomere in processes such as telomere extension and telomere replication. Together these proteins prevent chromosomal instability by promoting telomere maintenance. Nevertheless, telomeric sequences are occasionally lost, resulting in telomere length heterogeneity, which can occur at any telomere in single cells, as observed using quantitative fluorescence in situ hybridization (Q-FISH; Lansdorp et al., 1996; Zijlmans et al., 1997).
Telomeres resemble fragile sites (Sfeir et al., 2009), which are defined as genomic regions that challenge replication, especially when replication is stressed (Durkin and Glover, 2007). There are at least two reasons that telomeres pose problems during replication. First, telomeres are known to form a protective lariat-like structure, referred to as the telomeric (T)-loop (de Lange, 2004), created by invasion of the 3′ single-stranded telomeric DNA end into the duplex region of the telomere repeats. Resolution of the T-loop structure is needed for telomere replication. Second, telomeres are guanine (G) rich and thus are capable of forming stable secondary structures such as guanine quadruplex (G4) DNA structures in vitro (Sen and Gilbert, 1992). In vivo, G4-DNA could form at the single-strand telomeric overhang, the base of the T-loop, or, more generally, during replication, repair, and transcription of telomeric DNA. At telomeres and other genomic sites of G-rich DNA, replication forks could occasionally stall and, if not properly resolved, result in DNA breaks and sporadic loss of telomeric DNA.
Besides occasional loss of telomeric repeats, telomeric DNA is lost in most proliferating cells because of the inability of the replication machinery to copy the very end of the 3′ template DNA strands. Certain cell types, such as embryonic stem cells (ESCs), cells in the germ line, and cells of highly proliferative tissues, can counteract telomere loss by expressing the reverse transcriptase telomerase that adds telomeric repeats onto the 3′ ends of chromosomes (Greider and Blackburn, 1985). To prevent telomere shortening and thereby achieve unlimited replicative lifespan, most cancer cells reactivate telomerase, whereas a minority uses alternative lengthening of telomeres (ALT; Cesare and Reddel, 2010). ALT cells exhibit an increased abundance of extrachromosomal circles of double-stranded telomeric DNA (t-circles), derived from deleterious homologous recombination (HR) events at the T-loop (Wang et al., 2004). Recombination-mediated telomere trimming not only takes place in ALT cells, but it is also likely to be involved in telomere maintenance in normal eukaryotic cells (Bucholc et al., 2001; Pickett et al., 2011).
The average telomere length is determined by the equilibrium between the events that lengthen and shorten telomere arrays. Regulator of telomere length 1 (mRtel1) was found to be a dominant factor in setting telomere length in mice (Ding et al., 2004). mRtel1 encodes an essential DNA helicase, which plays a crucial role in telomere maintenance and DNA repair (Uringa et al., 2011). Chromosomes with low or undetectable telomere repeats are abundant in mouse ESCs lacking mRtel1 (Ding et al., 2004). Furthermore, suppression of mRtel1 in mouse embryonic fibroblasts (MEFs) increased telomere fragility (Sfeir et al., 2009). In addition to its function in telomere maintenance, human RTEL1 was also found to be involved in the repair of DNA double-strand breaks (DSBs; Barber et al., 2008). In vitro, the preferred substrate of RTEL1 is a 3′ single-strand DNA (ssDNA) invaded displacement (D)-loop with a 5′ overhang (Youds et al., 2010). In vivo, Caenorhabditis elegans rtel-1 mutants convert all meiotic DSBs into crossovers, demonstrating that RTEL-1 is required to regulate meiotic recombination in this species (Youds et al., 2010). Despite these important insights, the role of mRtel1 in replication, recombination, DNA repair, and telomere maintenance remains incompletely understood.
In this study, we examined the consequences of mRtel1 deficiency on DNA replication, repair, and recombination, with an emphasis on telomere maintenance. Our results indicate an important role for mRtel1 in replication and the repair of several types of DNA damage. In addition, we show that, depending on the recombination substrate and context, mRtel1 can be either a positive or a negative regulator of HR. Whereas mRtel1 is required for efficient telomere replication in ESCs, we found no evidence for the involvement of mRtel1 in preventing recombination at telomeres. Surprisingly, our data suggest that mRtel1 is required to allow elongation of telomeres by telomerase.
RESULTS
Generation of epitope-tagged and fluorescently tagged mRtel1 knock-in mice
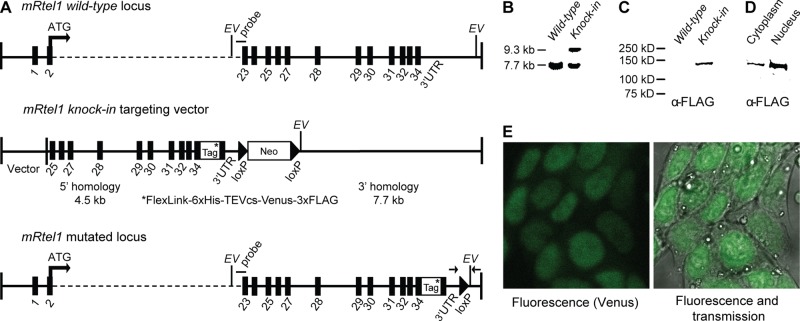
We generated mouse ESCs in which endogenous mRtel1 was replaced with an epitope- and fluorescently tagged mRtel1 fusion protein. Using bacterial artificial chromosome (BAC) recombination-mediated genetic engineering (recombineering; Copeland et al., 2001), we constructed a targeting vector in which the stop codon in exon 34 was deleted and the mRtel1 gene was extended with tag sequences (Figure 1A). Insertion of this knock-in targeting vector at the mRtel1 locus results in the expression of a C-terminally tagged mRtel1 protein at endogenous levels. Correctly targeted clones were identified (Figure 1B), and the fusion protein was expressed (Figure 1C). Subcellular fractioning (Figure 1D) showed that mRtel1 is mainly present in the nucleus.
FIGURE 1:
Generation and characterization of epitope-tagged and fluorescently tagged mRtel1 knock-in ESCs. (A) Schematics of the mRtel1 locus, gene-targeting construct, and targeted locus. The top of the scheme depicts ∼36.6 kb of the mouse mRtel1 locus. Exons are indicated as black boxes. The second line represents the mRtel1-tag targeting construct. Using recombineering, we inserted a multipurpose tag directly 3′ of the gene in front of the termination codon in exon 34. The neomycin resistance marker surrounded by LoxP sites is located 3′ of the mRtel1 3′ UTR. The mutated genomic locus containing the mRtel1-tag fusion is shown at the bottom. The 3′ external probe and diagnostic EcoRV (EV) sites are indicated. Genotyping primers are indicated as arrows at the bottom. (B) Southern blot of EcoRV-digested genomic DNA of mRtel1+/+ and mRtel1+/tag+Neo knock-in ESCs. Wild-type and targeted mouse ESCs show the predicted restriction fragments using the probe indicated in A. (C) Immunoblots of mRtel1+/+ and mRtel1+/tag knock-in ESCs total protein extracts and (D) cytoplasmic and nuclear fractions. mRtel1-tag fusion protein was detected using an anti-FLAG antibody. (E) Fluorescence confocal and transmission images of C-terminally tagged mRtel1 in living mRtel1tag/tag knock-in ESCs.
We then analyzed the localization of fluorescently tagged mRtel1 in mRtel1tag/tag knock-in ESCs. Using confocal microscopy, we detected a diffuse fluorescence signal in the nucleus (Figure 1E). Fluorescence-activated cell sorting (FACS) analysis of wild-type and mRtel1tag/tag knock-in ESCs showed a distinct homogeneous population of cells expressing very low levels of tagged-mRtel1 (unpublished data).
Two positively identified clones were injected into C57bl/6J blastocysts to produce chimeric mice that transmitted the targeted mRtel1 knock-in allele through the germ line. Homozygous mRtel1tag/tag mice were phenotypically wild type and fertile, and no aberrant phenotype has been observed for up to 1 yr of age. In contrast, previously generated mRtel1 knockout mice die around day 10 of embryogenesis (Ding et al., 2004), demonstrating that mRtel1 is essential for embryonic development.
Therefore we conclude that endogenous mRtel1 protein levels are very low, as demonstrated by microscopy and FACS in mRtel1tag/tag knock-in ESCs. In addition, we conclude that C-terminally tagged mRtel1 is a functional protein.
mRtel1 is required during replication
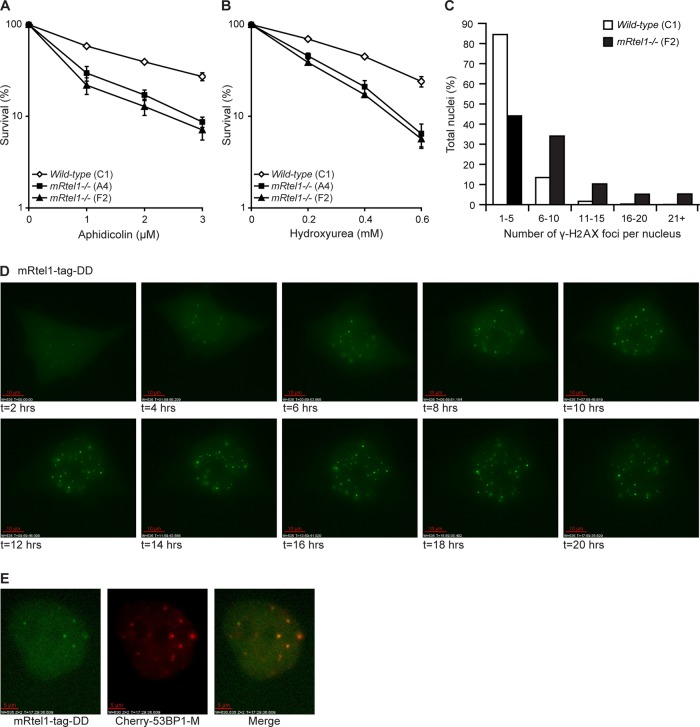
Highest expression of mRtel1 mRNA was detected in actively proliferating cells (Ding et al., 2004). To investigate whether mRtel1 is required during S phase, we determined the DNA-damage sensitivity of mRtel1-deficient ESCs for agents that cause replication fork stalling. Compared to wild-type ESCs, mRtel1-deficient cells were more sensitive to aphidicolin (Figure 2A), an agent that inhibits various DNA polymerases. In addition, sensitivity to hydroxyurea (HU), which inhibits ribonucleotide reductase and thus the synthesis of dNTPs, was increased (Figure 2B). An increase in stalled replication forks and DSBs can be measured by the appearance of phosphorylated H2AX (γ-H2AX; Petermann et al., 2010; Sirbu et al., 2011). We therefore quantified the number γ-H2AX foci in the presence and absence of mRtel1. As depicted in Figure 2C, more γ-H2AX foci per nucleus are present in ESCs lacking mRtel1 compared with wild-type ESCs (p = 0.02). The sensitivity of mRtel1-deficient cells to S phase–specific DNA damage and the increase in γ-H2AX foci suggest that mRtel1 is required during replication.
FIGURE 2:
mRtel1 is required during S phase. (A, B) Sensitivity of wild-type (C1) and two mRtel1-deficient (A4 and F2) ESC lines to the indicated doses of aphidicolin (A) and HU (B). Bars, mean percentage values of four experiments with SDs. (C) Analysis of the number of γ-H2AX foci per nucleus in wild-type (C1) and mRtel1-deficient (F2) ESCs. Foci in 1200 wild-type and 750 mRtel1-deficient nuclei in one focal plane were analyzed. (D) Images from a time-lapse movie of mRtel1-tag-DD expressed in mRtel1-deficient (F2) ESCs and cultured in the presence of aphidicolin (2 μM) and shield-1 (1 μM), both added at t = 0. Three image z-stacks (2.5-μm spacing) were acquired every hour. Projections of three z-stacks are depicted. Scale bar, 10 μm. (E) Colocalization of mRtel1-tag-DD and Cherry-m53BP1-M. Cells were cultured in the presence of aphidicolin (2 μM) and shield-1 (1 μM), and images depicted were taken at 17.5 h. Scale bar, 5 μm.
Next we wanted to know the spatial and temporal localization of mRtel1 upon DNA damage affecting replication fork progression. The very low expression level of mRtel1-Venus in the knock-in mRtel1tag/tag ESCs prevented their use for live-cell imaging studies. Moreover, overexpression of mRtel1 is toxic to mouse ESCs (unpublished data). To circumvent low expression levels and toxicity by constitutive overexpression, we used the inducible, tunable, and reversible Shld1 system (Banaszynski et al., 2006). We generated destabilized mRtel1-tag protein by fusing it to a destabilizing domain (DD) and expressed this in mRtel1-deficient ESCs. The unfolding and subsequent degradation of mRtel1-tag-DD can be blocked by adding the small, cell-permeable stabilizing molecule Shield-1 to the cell culture media. Using this experimental system, we created a short window in time to overexpress and follow mRtel1-tag-DD. mRtel1-tag-DD is present in the nucleus but can also be detected in the cytoplasm (Supplemental Figure S1 and Supplemental Movie S1). In most cells no mRtel1-tag-DD foci could be detected; however, some cells contained a few foci. On addition of aphidicolin (2 μM), an increase in the number of mRtel1 foci over time was observed (Figure 2D and Supplemental Movie S2). To address whether these mRtel1 foci represent sites of DNA damage, we coexpressed mRtel1-tag-DD and the fluorescently tagged m53BP1 minimum domain (m53BP1-M). m53BP1-M is the essential domain of m53BP1 for DNA damage foci formation (Pryde et al., 2005). As visualized in Figure 2E, mRtel1 and m53BP1-M did colocalize upon aphidicolin treatment.
Together these results suggest that mRtel1 is required during S phase and, upon persistent replication stress–induced DNA-damage, forms discrete foci that colocalize with the DNA damage marker m53BP1-M.
mRtel1 is involved in DNA repair
To investigate whether mRtel1 is primarily required for replication or also has a more general role in DNA repair, we determined survival of mRtel1-deficient ESCs upon treatment with various DNA-damaging agents that require the involvement of several DNA repair pathways, including HR, nucleotide excision repair (NER), base excision repair (BER), mismatch repair (MR), and translesion synthesis (TLS).
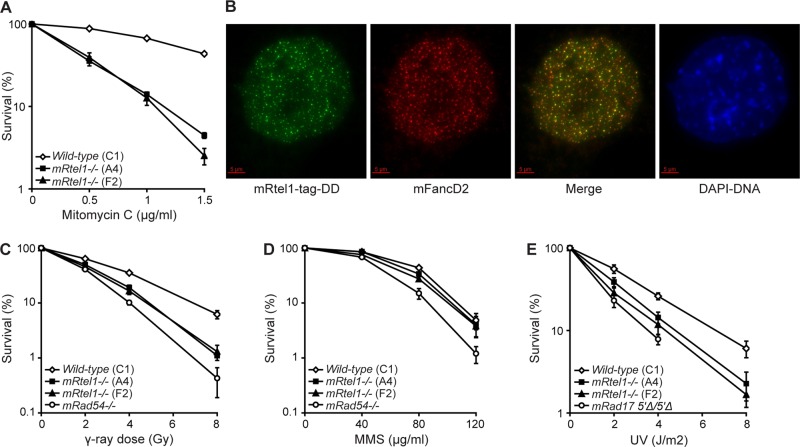
As was shown for C. elegans rtel-1 mutants and RTEL1-depleted human cells (Barber et al., 2008), mRtel1-deficient ESCs are sensitive to DNA interstrand cross-links (ICLs; Figure 3A). ICLs are complex lesions that interfere with transcription and replication. ICL repair requires many proteins from several DNA repair pathways, including HR, NER, MR, and TLS. On treatment with 1 μg/ml mitomycin C (MMC) for 1 h, mRtel1-tag-DD forms foci (Supplemental Figure S2 and Supplemental Movie S3). The number of foci increases in time and reaches a maximum around 13 h after treatment.
FIGURE 3:
mRtel1 promotes DNA repair. (A) Sensitivity of wild-type (C1) and two mRtel1-deficient (A4 and F2) ESC lines to the indicated doses of MMC. (B) mRtel1-tag-DD colocalizes with FancD2. Cells were cultured in the presence of shield-1 (1 μM) and 1 μg/ml MMC for 24 h and paraformaldehyde fixed. FandD2 was detected by immunofluorescence (ab2187; Abcam). Scale bar, 5 μm. (C, D) Sensitivity of wild-type (C1), two mRtel1-deficient (A4 and F2), and mRad54-deficient ESC lines to the indicated doses of γ-rays (C) and MMS (D). (E) Sensitivity of wild-type (C1), mRtel1-deficient (A4 and F2), and mRad175′∆/5′∆ ESC lines to the indicated doses of UV. Bars, mean percentage values of four experiments with SDs.
Sensitivity to MMC is characteristic of cells deficient in any of the genes associated with the human disease Fanconi anemia (FA; Thompson and Hinz, 2009). Fanconi anemia protein D2 (FANCD2) forms DNA repair foci in S phase, which increase in number in response to DNA lesions during replication (Garcia-Higuera et al., 2001; Taniguchi et al., 2002; Hussain et al., 2004). On treatment with MMC, we observed many mRtel1 foci, most of which colocalized with mFancD2 (Figure 3B), suggesting that mRtel1 is involved in ICL repair.
C. elegans rtel-1 mutants and human cells depleted for RTEL1 are not affected by ionizing radiation (Barber et al., 2008). Surprisingly, mRtel1-deficient ESCs are more sensitive to γ-rays than are wild-type cells (Figure 3C). As a positive control for γ-radiation, we used radiation-sensitive ESCs deficient for the DNA repair protein mRad54 (Figure 3C).
In addition, we investigated whether mRtel1 deficiency causes sensitivity to methyl methanesulfonate (MMS) and UV (254 nm) light. MMS lesions are predominantly repaired by BER and alkyltransferases and UV lesions by NER. Unlike MMS-sensitive, mRad54-deficient ESCs, the sensitivity of ESCs lacking mRtel1 to MMS-induced lesions was similar to that of wild-type cells (Figure 3D). In contrast, mRtel1-deficient ESCs are more sensitive to UV light than are wild-type cells (Figure 3E) but are not as sensitive as mRad175′∆/5′∆ ESCs.
From these experiments we conclude that mRtel1 is involved in the repair of DNA lesions induced by MMC, γ-radiation, and UV light.
mRtel1 is a key protein in homologous recombination
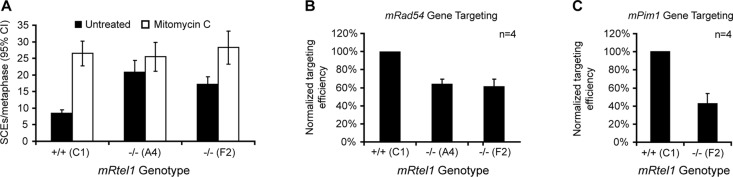
Sister chromatid exchanges (SCEs) depend on HR events between sister chromatids during replication (Sonoda et al., 1999; Wang et al., 2000). Increased spontaneous SCE is a hallmark of defects in the RecQ family helicase BLM (Chaganti et al., 1974). To investigate whether mRtel1 deficiency has an effect on SCE frequency, we measured the spontaneous and DNA damage–induced levels of SCEs in wild-type and mRtel1-deficient ESCs. Under normal cell culture conditions, SCE frequency in ESCs lacking mRtel1 was more than twice that in wild-type cells (Figure 4A). After mild treatment with MMC (0.04 μg/ml), SCE frequencies increased in both mRtel1-deficient and wild-type ESCs but were not significantly different. This finding shows that in the absence of mRtel1, a greater number of DNA strand breaks occurring during DNA replication are repaired using SCE.
FIGURE 4:
mRtel1 is important for HR. (A) Percentage of spontaneous (black bars) and MMC (0.2 μg/ml, 1 h) induced (white bars) SCEs per metaphase measured in wild-type (C1) and two mRtel1-deficient (A4 and F2) ESC lines. Error bars, the 95% confidence interval (CI). (B, C) Gene-targeting efficiency in wild-type (C1) and two mRtel1-deficient (A4 and F2) ESC lines at the mRad54 (B) and mPim1 (C) loci. Error bars, the SD from four independent experiments.
The increase in SCEs suggests that HR between sister chromatids is affected. SCE events, however, only denote crossover recombination when the replication fork is stalled and do not necessarily reflect HR capacity in general. Therefore we also tested the possibility that mRtel1 is involved in the HR subpathway that mediates gene replacement. To assess gene-targeting efficiency in wild-type and mRtel1-deficient ESCs, we measured targeting frequencies at the mRad54 and mPim1 loci. Targeting frequencies at the mRad54 locus were measured using a fluorescence-based targeting assay (Figure 4B; Abraham et al., 2003). Integration of a promoterless Rad54-GFP knock-in targeting construct at the mRad54 locus results in expression of Rad54-GFP from the endogenous promoter, which can be analyzed using flow cytometry. Using this assay, we measured a 40% decrease in targeting efficiency in two independent mRtel1-deficient ESC lines tested. Compromised DNA repair as observed in mRtel1-deficient ESCs could increase the frequency of breaks and thus stimulate random integration at genomic DNA breaks. However, random integration events were not elevated in wild-type and mRtel1-deficient ESCs (unpublished data). In addition to the mRad54 locus, we tested gene replacement at the mPim1 locus (Figure 4C; Bennardo et al., 2008). Compared to wild-type cells, targeting frequencies in mRtel1-deficient ESCs were 60% reduced.
These gene-targeting experiments demonstrate that mRtel1 plays an important role in HR.
mRtel1 is required for telomere replication and localizes transiently at the telomere
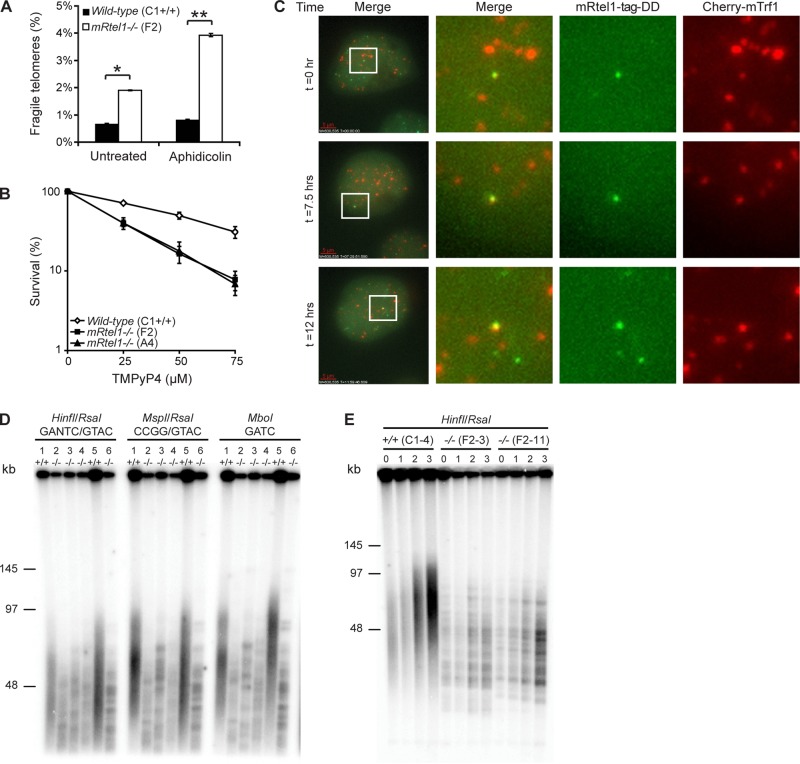
Next we wanted to address the function of mRtel1 more specifically at the telomere. Mammalian telomeres resemble fragile sites (Sfeir et al., 2009). Just like common fragile sites (Glover et al., 1984), these are identified as site-specific breaks or gaps in metaphase chromosomes when treated with low levels of the polymerase inhibitor aphidicolin (Sfeir et al., 2009). Two helicases—BLM and mRtel1—have been implicated in telomere replication in MEFs (Sfeir et al., 2009). To investigate whether mRtel1 is also important for telomere replication in ESCs, we quantified the frequency of fragile telomeres in mRtel1-deficient and wild-type ESCs (Figure 5A). In wild-type ESCs the frequency of fragile telomeres was <1%. However, in mRtel1 null ESCs, the frequency was 2.5-fold increased, consistent with mRtel1-depleted MEFs. Treatment with low levels (0.2 μM) of aphidicolin increased the fragile telomere frequency to nearly 4%, whereas this did not change notably in wild-type cells. These results confirm that mRtel1 is required for telomere replication.
FIGURE 5:
mRtel1 is required for telomere replication and extension. (A) Analysis of fragile telomeres in wild-type (C1) and mRtel1-deficient (F2) ESCs with or without 0.2 μM aphidicolin treatment. Bars, SEM; *p < 0.01, **p < 0.001. (B) Sensitivity of wild-type (C1) and two mRtel1-deficient (A4 and F2) ESC lines to the indicated doses of TMPyP4. Bars, mean percentage values of three experiments with SDs. (C) Images from a time-lapse movie of mRtel1-tag-DD and Cherry-mTrf1 coexpressed in mRtel1-deficient (F2) ESCs and cultured in the presence of shield-1 (1 μM). Imaging started 72 h post Cherry-mTrf1 transfection. Ten image z-stacks (0.5-μm spacing) were acquired every 15 min. Far left, three still images (projection of 10 z-stacks) extracted from a time-lapse movie from a single cell acquired at indicated time points. Merge, zooms on the boxed nuclear region of the left images, showing colocalization of mRtel1-tag-DD (third from left) and Cherry-mTrf1 (far right). Scale bar, 5 μm. (D) TRF analysis of wild-type and mRtel1-deficient ESCs. 1, Wild type (C1); 2, mRtel1 deficient (A4); 3, mRtel1 deficient (E1); 4, mRtel1 deficient (F2); 5, wild type (C1 subclone 4); 6, mRtel1 deficient (F2 subclone 11). (E) TRF analysis of wild-type (C1 subclone 4) and mRtel1-deficient (F2 subclones 3 and 11) ESCs. DNA was extracted from wild-type and mRtel1-deficient clones at t = 0 and after 1, 2, and 3 wk in culture.
To obtain further evidence for a role of mRtel1 in telomere replication, we performed a colony-forming assay using wild-type and mRtel1-deficient cells treated with increasing concentrations of TMPyP4. TMPyP4 binds strongly to G4-DNA, which is likely to be present at many sites in the genome and is believed to be especially abundant at the telomere. G4-DNA could be normally present at the telomere or arise during lagging-strand replication of G-rich telomeric DNA (Ding et al., 2004). Using the colony-forming assay, we found that mRtel1-deficient ESCs were more sensitive to TMPyP4 than were wild-type cells (Figure 5B). Sensitivity for TMPyP4 suggests that mRtel1 is required for replication of G4-DNA. However, it is not known whether mRtel1 unwinds G4-DNA in vivo or can resolve G4-DNA structures in vitro.
In the absence of mRtel1, telomeric replication fork stalling and collapse and subsequent breaks in telomeric DNA could occur more frequently than in normal cells. Chromosome ends with low or undetectable telomere repeats are abundant in mRtel1-deficient ESCs, suggesting significant telomere loss (Ding et al., 2004). To investigate whether telomeres synthesized by leading- and lagging-strand DNA replication were equally affected by mRtel1 deficiency, we performed chromosome orientation FISH on leading- and lagging-strand telomeres (unpublished data; Bailey et al., 1996). We did not find an imbalance in leading/lagging-strand signals, and thus we conclude that mRtel1 is required for both leading- and lagging-strand telomere replication.
Because mRtel1 is involved in telomere replication, we investigated telomeric localization of mRtel1. We performed live-cell imaging on mRtel1-deficient ESCs coexpressing mRtel1-tag-DD and Cherry-mTrf1 (Figure 5C and Supplemental Movie S4). Using Cherry-mTrf1 as a telomere marker, we looked for colocalization of mRtel1 and mTrf1 foci. In Figure 5C, three time points of a movie are depicted in which overlapping mRtel1 and mTrf1 foci are observed. However, at any given time the majority of mRtel1 and mTrf1 foci do not colocalize, suggesting that mRtel1 localizes transiently at the telomere.
No increase in telomeric recombination in the absence of mRtel1
Next we asked whether unscheduled recombination events could cause the abundance of short telomeres in the absence of mRtel1. Because replication-associated recombination events as measured by genomic-SCEs are elevated in mRtel1-deficient ESCs, we investigated whether we could also measure an increase in recombination events at the telomere. Therefore we determined the level of telomeric-SCEs (T-SCEs) using CO-FISH. However, no differences between wild-type and mRtel1-deficient cells were found (unpublished data).
In addition, we investigated the possibility of deleterious HR events at the T-loop in the absence of mRtel1. An enzymatic activity of mRtel1 to unwind T-loops would be in line with the in vitro activity of RTEL1: displacing a 3′ ssDNA invaded D-loop with a 5′ overhang (Youds et al., 2010), which resembles the telomeric D-loop. Deleterious HR events at the T-loop happen frequently in ALT cells, which can be measured as an increased abundance of t-circles (Wang et al., 2004). To investigate whether t-circles were more abundant in mRtel1-deficient ESCs compared to wild-type cells, we analyzed telomeric restriction fragments first by size and then by structure using two-dimensional (2D) gel electrophoresis (Brewer and Fangman, 1987; Cohen and Lavi, 1996). The subsequent 2D telomere blots showed no differences in the amount of t-circles between the ESC lines (unpublished data). In addition, partially single-stranded telomeric (CCCTAA)n DNA circles have been found to be ALT specific (Henson et al., 2009). These circles can be detected using a quantitative c-circle assay. Using this assay, we found no differences in the amount of c-circles between wild-type and mRtel1-deficient cells (unpublished data).
Taken together, these results indicate that increased telomeric recombination cannot explain the variable telomere length in ESCs lacking mRtel1. Furthermore, these results are in agreement with the fact that average telomere length is maintained for many population doublings in mRtel1-deficient ESCs (Ding et al., 2004).
Telomeres are functional but not extended in the absence of mRtel1
Very short telomeres, not detectable by Q-FISH, are relatively abundant in mRtel1-deficient ESCs (Ding et al., 2004). When telomeres are critically short or when telomere maintenance or telomere-protective factors are impaired, this can lead to telomere uncapping. This telomere uncapping results in telomere dysfunction, which is associated with the formation of telomeric DNA damage response factor foci, such as γ-H2AX (Takai et al., 2003). To investigate whether telomeres in ESCs lacking mRtel1 were dysfunctional, we quantified the number of telomere dysfunction–induced foci (TIFs), as visualized by the localization of γ-H2AX at the telomere using immuno-FISH in wild-type and mRtel1-deficient ESCs. The number of γ-H2AX foci at the telomeres was not different from that in wild-type cells in cells lacking mRtel1 (unpublished data), indicating that the undetectable telomeres by Q-FISH are still capped and appear to be functional.
To gain additional insight into the role of mRtel1 at the telomere, we performed high-resolution terminal restriction fragment (TRF) analysis (Kipling and Cooke, 1990). Genomic DNA was digested with HinfI/RsaI, MspI/RsaI, and MboI, resolved using pulsed-field gel electrophoresis, blotted, and probed with a telomeric probe to detect the telomere fragments (Figure 5D). Whereas in wild-type cells the expected telomeric smear representing the heterogeneous telomere length distribution in the ESC population was detected, in cells lacking mRtel1 a striking banding pattern was observed. The observed TRF pattern was independent of the restriction enzymes used, indicating that these are long arrays of telomeric repeats likely not interspersed by other sequences. Therefore the observed banding pattern represents homogeneous telomere length of individual chromosomes within the cell population. From this we conclude that individual telomere length in mRtel1-deficient ESCs is fixed even in the presence of telomerase.
To investigate whether the observed TRF banding pattern could be explained by the inability of telomerase to extent the telomeres, we measured telomere length with increasing population doublings. If telomeres cannot be extended by telomerase, a gradual telomere length decline due to the inability of the replication machinery to replicate the end of the chromosome would be expected, just as in telomerase negative cells. As hypothesized, we observed gradual telomere shortening in mRtel1-deficient ESCs (Figure 5E) over a 3-wk period. These results suggest that mRtel1 is required for telomere extension in mouse ESCs.
DISCUSSION
In this study, we used mouse ESCs lacking mRtel1 to reveal the importance of mRtel1 in replication, DNA repair, HR, and telomere maintenance.
mRtel1-deficient ESCs show a dramatic telomere phenotype, and many chromosomal abnormalities are observed upon differentiation (Ding et al., 2004). Furthermore, constitutive overexpression of mRtel1 in ESCs is not tolerated. Because both mRtel1 deficiency and overexpression give such strong phenotypes, mRtel1 protein levels must be tightly regulated. mRtel1 protein levels are very low, making it difficult to use mRtel1tag/tag knock-in ESCs for fluorescence imaging. However, since tagged-mRtel1 knock-in mouse ESCs and mice do not show any of the phenotypes of mRtel1-deficient ESCs and mice, we expect C-terminally tagged-mRtel1 to be functional. On the basis of this knowledge, we developed an experimental system in which we expressed inducible, tunable, and reversible mRtel1-Venus in mRtel1-deficient ESCs. Using this system, we found that mRtel1 forms foci that increase in number over time upon induction of DNA damage. In addition, these foci colocalize with DNA repair markers, suggesting that mRtel1 is present at sites of active DNA repair.
In accordance with a function of mRtel1 in replication and DNA repair, mRtel1-deficient mouse ESCs contain elevated levels of γ-H2AX foci and are sensitive to aphidicolin, hydroxyurea, γ-radiation, MMC, and UV light, which cause different types of DNA lesions. The sensitivity to a variety of DNA-damaging agents suggests that mRtel1 has separate functions in different DNA repair pathways. Alternatively, mRtel1 could be required in a repair step that is shared among a number of DNA repair pathways. In C. elegans, RTEL-1 is required for the response to DNA damage affecting replication fork progression (Barber et al., 2008). Many DNA-damaging agents form obstacles or breaks in the DNA template that interfere with replication. Because mouse ESCs spend 75% of the cell cycle time in S phase, DNA-damaging agents have a great impact on replication, leading to fork stalling or, when the replisome dissociates, fork collapse. To restart a stalled replication fork or reactivate a collapsed fork, a Holliday junction recombination intermediate might form (Petermann and Helleday, 2010). mRtel1 might be required for strand displacement after Holliday junction–mediated fork restart. In addition, mRtel1 could prevent broken DNA molecules from invading homologous sequences during replication. Both activities would be in agreement with the reported enzymatic activity of purified human RTEL1 in vitro, which prevents the formation of a D-loop structure and displaces a 3′ ssDNA invaded D-loop with a 5′ overhang (Barber et al., 2008; Youds et al., 2010).
In line with an antirecombinogenic function of mRtel1 during replication, SCEs were more abundant in the absence of mRtel1. In addition, gene conversion events in RTEL1-knockdown HeLa cells were increased fourfold as measured using an I-SceI–inducible DSB assay (Barber et al., 2008). Furthermore, all meiotic DSBs created in C. elegans are resolved into crossovers in the absence of rtel-1 (Youds et al., 2010). In contrast, ESCs lacking mRtel1 show a twofold decrease in gene-targeting efficiency at the mRad54 and mPim1 loci. Opposite outcomes in SCEs and gene-targeting assays have been reported previously. For example, FANCC- and FANCD2-deficient DT40 cells show an increase in spontaneous SCEs and a decrease in gene-targeting efficiency (Niedzwiedz et al., 2004; Hirano et al., 2005; Yamamoto et al., 2005). In addition, mErcc1, mRad54, and mRad17 ESC mutants show reduced gene targeting, whereas SCEs are unaffected (Essers et al., 1997; Dronkert et al., 2000; Niedernhofer et al., 2001; Budzowska et al., 2004). One of the differences of HR between sister chromatids and homologous gene replacement is that SCEs are recombination events associated with replication problems, whereas gene replacement is not. Furthermore, the recombination substrates are different; sister chromatids are identical, whereas gene replacement constructs also contain nonidentical sequences. The mechanism of gene replacement is poorly understood. However, the recombination intermediates in replication fork restart resulting in SCEs and gene replacement are likely to be different. For example, the targeting construct for gene replacement reveals two ssDNA ends after exonucleolytic processing, and invasion by both ends into homologous sequences creates two D-loops. For gene replacement, mRtel1 might be required to resolve a recombination intermediate, resulting in a decreased recombination efficiency in the absence of mRtel1. However, mRtel1 deficiency in the context of replication results in recombination intermediates processed into crossing overs.
In this study we show that mRtel1 is not only required for replication in general as discussed, but also for leading- and lagging-strand telomere replication. Using live-cell imaging, we found that mRtel1 is transiently present at the telomere. It is possible that mRtel1 is recruited to the telomere when the replication machinery stalls. Alternatively, very few mRtel1 molecules, not detectable as foci, could be present at every telomere. On the basis of its preferred substrate in vitro, a 3′ ssDNA invaded D-loop with a 5′ overhang (Youds et al., 2010), and the resemblance of this structure with the T-loop, we anticipated that mRtel1 would be required to resolve the T-loop during replication. However, no increase in telomeric HR products, including t-circles and c-circles, were observed in mRtel1-deficient cells. Therefore rapid deletion and extension of telomeres due to recombination cannot explain the telomere length phenotype in ESCs lacking mRtel1.
The increased frequency of chromosomes with undetectable telomeric sequences in mRtel1-deficient ESCs can be explained, however, by the inability of telomeres to be extended by telomerase. In normal cells, telomeric sequences are occasionally lost, presumably due to stalled replication forks and DNA breakage and possibly due to erroneous deletion of the T-loop by HR. In wild-type ESCs, these telomeres are extended by telomerase. However, in ESCs lacking mRtel1, telomeres cannot be extended, so sporadic telomere deletions accumulate over time, resulting in an increased abundance of very short telomeres in mRtel1-deficient cells. The high-resolution TRF confirms this hypothesis. Whereas in wild-type cells a telomeric smear is observed, telomeres of ESCs lacking mRtel1 show a banding pattern. This banding pattern shows that telomere length at individual chromosomes between cells is strikingly uniform. This can be explained by the fact that telomere length was set at the time of fertilization and telomerase was unable to elongate telomeres subsequently. Therefore telomere length gradually decreases in mRtel1-deficient ESCs. From this, we hypothesize that in ESCs lacking mRtel1, telomerase has no access or is not recruited to the telomere. In agreement with this hypothesis, mTerc-deficient ESCs (Niida et al., 1998; Figure 3) and MEFs (Blasco et al., 1997; Figure 4, A and B) show a TRF banding pattern like ESCs lacking mRtel1.
In summary, the present study provides evidence that mRtel1 is required for a variety of HR-mediated DNA repair processes. mRtel1-deficient cells are sensitive to replication inhibitors and are compromised in ICL and DSB repair. In addition, in the absence of mRtel1, gene replacement is less efficient, and SCEs are increased. We hypothesize that mRtel1 is required to resolve a common recombination intermediate in these processes. Our data are in agreement with a role of mRtel1 at stalled or collapsed replication forks by promoting noncrossover repair through the synthesis-dependent strand-annealing subpathway of HR. Furthermore, mRtel1 is transiently present at the telomere and is required for telomere replication and extension in ESCs. It seems possible that recruitment of mRtel1 and telomerase are both transient, for example, via secondary structures at the 3′ end of telomeres that form only rarely. Further experiments will determine whether mRtel1 directly recruits telomerase or promotes telomere accessibility for telomerase.
MATERIALS AND METHODS
Construction of the mRtel1-targeting vector
The mRtel1-targeting construct was made using recombineering (Liu et al., 2003). Briefly, the end-sequenced 129SvEV/AB2.2 BAC (Adams et al., 2005) bMQ-413M4 containing the entire mRtel1 gene was obtained from the Wellcome Trust Sanger Institute (Cambridge, United Kingdom). The BAC was purified from DH10B cells and electroporated in SW102—the bacterial strain designed for BAC recombineering using galK (Warming et al., 2005). All primers used are listed in the Supplemental Materials and Methods. The galK gene was amplified from plasmid pgalK using primers 1140/1141 and inserted in the BAC 3′ of the mRtel1 3′ untranslated region (UTR) using recombineering. The neomycin gene, expressed both from a prokaryotic (em7) and a eukaryotic promoter (PGK) flanked by two loxP sites, was PCR amplified using plasmid PL452 and primers 1142/1143. Subsequently, galK was replaced by the neomycin gene using recombineering. Similarly, using different primer pairs, we introduced the tag sequence (Supplemental Materials and Methods) in exon 34 of mRtel1 just in front of the termination codon. Next we constructed a retrieval vector to retrieve the targeting sequences from the BAC. Using the mRtel1-containing BAC as template and primer pairs 1136/1137 and 1138/1139, we amplified 733 base pairs of the 5′ end and 559 base pairs of the 3′ end of the targeting construct. PCR product 1136/1137 was SpeI/BamHI digested, and 1138/1139 was BamHI/XbaI digested. These products were subsequently cloned into SpeI/XbaI–digested plasmid PL253. Next the retrieval vector was digested with BamHI and electroporated into the SW102 containing the modified mRtel1 BAC. Finally, the BAC sequences between the 5′ and 3′ homology regions were recombined into PL253, resulting in the targeting construct.
Generation and characterization of mRtel1+/tag ES cells and mice
IB10 ESCs were electroporated with 20 μg of the NotI-linearized mRtel1tag knock-in targeting vector (0.4-cm cuvette, 1200 μF, 118 V). Cells were plated, and 24 h after electroporation cells were selected for 7–10 d using 200 μg/ml G418. After selection, clones were picked and expanded. Homologous targeted clones were identified by Southern blotting using a 3′ probe located outside the targeting vector. The mRtel1 3′ probe DNA was amplified by PCR using 129Sv mouse genomic DNA as a template with the following primers: forward, GGCTCTGGGTATTGGATGTGC; reverse, GAACTGCATCTTGAGGACAACACG. This PCR product was 32P labeled. Hybridization and washes were performed using standard procedures. Multiple cell lines containing one mRtel1tag knock-in allele were obtained at a frequency of ∼15%. Subsequently, the neomycin selection marker gene was removed by transient Cre expression. Clones in which the neomycin selection marker was excised were identified by G418 sensitivity and PCR amplification of the remaining loxP site. Two positively identified clones were injected into C57bl/6J blastocysts to produce chimeric mice that transmitted the targeted mRtel1 knock-in allele through the germ line. Male chimeras were bred with C57bl/6J females to produce mRtel1+/tag offspring. Heterozygous mice were intercrossed to produce homozygous mRtel1tag/tag mice. Heterozygous mRtel1+/tag were backcrossed for five generations (N5) to a 129/SvJ background. Subsequently, heterozygous 129/SvJ(N5);mRtel1+/tag mice were intercrossed, and blastocysts from pregnant females were harvested (Bryja et al., 2006). Blastocysts were used for the derivation of mRtel1+/+ (wild type), mRtel1+/tag, and mRtel1tag/tag ESCs.
Mouse embryonic stem cell lines and culture
Wild-type (C1) and mRtel1-deficient (A4, E1, F2; Ding et al., 2004) mouse ESCs were obtained from A. Nagy (Samuel Lunenfeld Research Institute, Toronto, Canada) and cultured in the presence of leukemia inhibitory factor (LIF) and 20% fetal calf serum (FCS) on gelatin-coated plastic culture dishes in DMEM containing 20% FCS in the presence of 100 ng/ml LIF as described previously (Gertsenstein et al., 2002) or in knockout DMEM (10829-018; Life Technologies, Carlsbad, CA) containing 15% Knockout Serum Replacement (10828-028; Life Technologies), 2 mM GlutaMAX-I (35050-038;Life Technologies), 1 mM MEM Non-Essential Amino Acids (11140-035; Life Technologies), 1× penicillin–streptomycin (15140-122; Life Technologies), 0.1 mM 1-Thioglycerol (M6145; Sigma-Aldrich, St. Louis, MO), 3 μM CHIR99021 (Axon 1386; Axon Medchem BV, Groningen, Netherlands), 0.5 μM PD184352 (Axon 1368; Axon Medchem BV), and 1:1000 self-prepared LIF. Aphidicolin (Sigma-Aldrich) treatment (0.2 μM) for fragile telomere analysis was for 16 h.
Plasmids and transfections
The Cherry fluorescent protein was fused to the N-terminus of mTrf1 and m53BP1-M, driven by a CAG promoter. PGK promoter–driven mRtel1-tag-DD was created by PCR-based cloning as described later. All oligo sequences can be found in the Supplemental Material and Methods. Oligo pair 1810/1811 was used to amplify the tag sequence, and the product was subsequently MluI/FseI digested. Similarly, oligo pair 1812/1813 was used to amplify the FKBP DD coding sequences, and the product was FseI/BsshII digested. Subsequently, these two PCR fragments were ligated into the dephosphorylated MluI site of PGK-puro plasmid. Next mRtel1 cDNA (Open Biosystems, Thermo Biosystems, Huntsville, AL) was amplified using oligo pair 1814/1815, and the product was NotI/MluI digested. This digested PCR fragment was ligated into PGK-tag-DD-puro, giving PGK-mRtel1-tag-DD-puro. The mRtel1-tag-DD cell line was generated by transfecting mRtel1-tag-DD-puro plasmid DNA into mRtel1-deficient (F2) ESCs, and upon 24 h transfection, cells were selected with puromycin (Invitrogen). Transfections were performed using Effectene transfection reagent (Qiagen, Mississauga, Canada) to obtain transient or stable expressing cell lines.
Immunoblot analysis
Whole-cell extracts or cyctoplasmic and nuclear protein fractions were separated on 8% SDS-polyacrylamide gels. Subsequently, the separated proteins were transferred onto nitrocellulose membranes (Bio-Rad, Hercules, CA). The mRtel1 fusion protein was detected using a mouse monoclonal anti–FLAG-M2 primary antibody (F1804; 1:4000; Sigma-Aldrich), followed by a secondary antibody coupled to horseradish peroxidase (1:50,000). Immobilized immunoglobulins were visualized using SuperSignal West Pico Chemiluminescent Substrate (Pierce Biotechnology, Rockford, IL).
Cell survival assays
Sensitivity of ESCs to various kind of DNA damage was measured as colony-forming ability after exposure to γ-irradiation, MMC (M4287; Sigma-Aldrich), MMS (129925; Sigma-Aldrich), or UV (254 nm) as described (Essers et al., 1997). In additional cell survival assays, cells were incubated for 24 h with aphidicolin (A0781; Sigma-Aldrich) or HU (H8627; Sigma-Aldrich) and for 48 h with TMPyP4 (613560; Calbiochem, Merck, Darmstadt, Germany). As positive controls previously published mRad54-deficient ESCs (Essers et al., 1997) and mRad175′∆/5′∆ ESCs (Budzowska et al., 2004) were used. All measurements were performed in triplicate. Every experiment was repeated at least three times.
Immunofluorescence and γ-H2AX foci analysis
ESCs were grown on sterilized 0.1% gelatin–coated coverslips. For efficient attachment of ESCs to the glass, coverslips were prepared in the following manner. Dried gelatin-coated coverslips were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 4 h. Subsequently, coverslips were washed six times in PBS to make sure no traces of paraformaldehyde were left. Cells grown using this method had normal morphology compared with standard gelatin-coated plastic dishes. Cells were fixed in 1% paraformaldehyde in PBS for 15 min and blocked with BSA/glycine. Next fixed cells were incubated with an anti–phospho-histone H2AX (Ser-139) primary antibody (γ-H2AX, clone JBW301; 05-636; used 1:1000; Upstate, Millipore, Billerica, MA) for 2 h and subsequently incubated with a secondary goat anti-mouse Alexa 488 antibody (A-11001; used 1:2000; Invitrogen, Carlsbad, CA) for 1 h. Nuclei were stained with 0.2 μg/ml 4′,6-diamidino-2-phenylindole. Slides were mounted using Prolong Gold Antifade Reagent (P36934; Invitrogen). For γ-H2AX, images were made using a TissueFAX and analyzed using TissueQuest cell analysis software (2010) from TissueGnostics (Tarzana, CA). Experiments were done in triplicate. γ-H2AX foci in at least 750 nuclei per experiment were counted and analyzed using TissueQuest software. For mFancd2 immunofluorescence, fixed cells were incubated with a primary rabbit polyclonal anti-FANCD2 antibody (ab2187; Abcam, Cambridge, MA), which was subsequently detected with a secondary goat anti–rabbit Cy5 antibody.
Confocal and time-lapse microscopy
Mouse ES cells were plated in #1.5 Labtek II chambered coverglasses (Nalge Nunc, Rochester, NY). For ESCs to adhere to the glass, the glass was coated with gelatin, air dried, fixed with 4% paraformaldehyde in PBS for 4 h, and rinsed thoroughly with PBS. Time-lapse images were acquired with the DeltaVision RT (Applied Precision, Issaquah, WA) microscope using a 60× Plan Apochromatic/1.4 numerical aperture oil objective (Olympus, Markham, Canada). Confocal microscopy was performed using a LSM 780 and an iLCI Plan-Neofluar 63×/1.3 oil objective (Carl Zeiss, Jena, Germany). ESCs were imaged at 37°C with 5% CO2 perfusion.
Gene-targeting assays at the mRad54 and mPim1 loci
Gene targeting to the mRad54 locus (Essers et al., 1997) and measurement and analysis of Rad54-GFP–targeting events were performed as described (Abraham et al., 2003). Briefly, a promoterless Rad54-GFP knock-in targeting construct was electroporated into ESCs. Targeted integration of this construct at the mRad54 locus results in expression of Rad54-GFP from the endogenous promoter (Abraham et al., 2003). Use of flow cytometry analysis makes it possible to measure homologous integration in individual cells by green fluorescence, and random integration of the construct results in nonfluorescent cells. To measure gene targeting at the mPim1 locus, we used a pim-EJ5-GFP–targeting construct (Bennardo et al., 2008), which was derived from p59xDR-GFP6 (Moynahan et al., 2001). This targeting construct contains a puromycin-selectable marker and a hygromycin-selectable marker, which is in frame with exon 4 of the mPim1 gene. For stable integration of pim-EJ5-GFP, 75 μg of XhoI-linearized DNA was electroporated (0.4-cm cuvette, 250 V, 950 μF) into 1 × 107 ESCs. Subsequently, 48 h after electroporation 10% of the cells were selected in 1.0 μg/ml puromycin and 90% in 110 μg/ml hygromycin. Started clones were counted 6–8 d after selection. Puromycin-resistant clones represent random plus targeted insertion events, whereas hygromycin-positive clones represent correctly targeted clones at the mPim1 locus.
Sister chromatid exchange assay
Wild-type and mRtel1-deficient ESCs were grown in fresh medium containing 10 μg/ml bromodeoxyuridine (BrdU) for two cell cycles (24–30 h) and were then blocked using 0.1 μg/ml Colcemid (Life Technologies) for 2 h. For MMC treatment, ESCs were incubated with 0.2 μg/ml MMC for 1 h before BrdU incorporation. Subsequently, cells were harvested and swollen in hypotonic buffer (55 mM KCl, 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4) at 37°C for 5 min and fixed three times in methanol:acetic acid (3:1) buffer, and cells were dropped onto glass slides to obtain metaphase spreads. Differential staining was carried out as described previously (Perry and Wolff, 1974). Briefly, metaphase spreads were stained with 150 μg/ml Hoechst 33258 solution, exposed to UV light for 1.5 h, rinsed in 2× saline–sodium citrate (SSC; pH 7.0) at 65°C, dried prior to staining in 2% Giemsa solution (Sigma-Aldrich), and mounted. Brightfield imaging was performed on a Zeiss Axioplan 2 Imaging microscope (Carl Zeiss) equipped with an AxioCam MRm digital camera controlled by Isis software (MetaSystems, Altlussheim, Germany). SCE events were scored on metaphase spreads that showed complete sister chromatid differentiation. The number of metaphases analyzed for spontaneous SCEs were as follows: 47, wild type (C1); 39, mRtel1 deficient (A4); and 48, mRtel1 deficient (F2). After MMC treatment the number was as follows: 23, wild type (C1); 25, mRtel1 deficient (A4); and 39, mRtel1 deficient (F2).
TRF analysis
TRFs were prepared by digestion with HinfI/RsaI, MspI/RsaI, or MboI as described previously (Pickett et al., 2011). Mouse TRFs were separated by one-dimensional pulsed-field gel electrophoresis in a 1% agarose gel for 23 h at 6 V/cm, with an initial switch time of 1 and a final switch time of 6. Gels were dried and denatured and subject to in-gel hybridization overnight using a [γ-32P]ATP-labeled (CCCTAA)4 telomeric probe. Gels were washed in 4× SSC and exposed overnight to a PhosphorImage screen.
Telomere-FISH on metaphase spreads
Telomere-FISH on methanol/acetic acid–fixed mouse metaphases spreads with a telomeric C-strand peptide nucleic acid probe was performed as previously described (Lansdorp et al., 1996; Zijlmans et al., 1997).
Supplementary Material
Acknowledgments
We are most grateful to Elizabeth A. Chavez for technical assistance and Shahrokh M. Ghobadloo for his lab contribution. In addition, thank Neal Copeland for providing the plasmids and bacterial strains for recombineering, Jeremy Stark for the pim-EJ5-GFP plasmid, Thomas Wandless for the YFP-L106P plasmid, and Mike Schertzer for the Cherry-m53BP1-M plasmid. We also thank Klaas Sjollema of the University Medical Center Groningen Imaging Center for assisting with γ-H2AX foci microscopy and analysis and Geraldine Aubert and Niek van Wietmarschen for helpful comments and critical reading of the manuscript. In addition, we thank Jackie Schein at the Genome Science Center in Vancouver, as well as the Wellcome Trust Sanger Institute for the BAC clones. Microscopic imaging on the TissueGnostics TissueFAXS system was performed at the University Medical Center Groningen Imaging Center, which is supported by the Netherlands Organisation for Health Research and Development (ZonMW Grant 40-00506-98-9021). This work was supported by a Terry Fox Foundation Program Project Award (Grant 018006) and the Canadian Cancer Society (Grant 105265).
Abbreviations used:
- ALT
alternative lengthening of telomeres
- D-loop
displacement loop
- ESC
embryonic stem cell
- HR
homologous recombination
- MMC
mitomycin C
- MMS
methyl methanesulfonate
- RTEL1
regulator of telomere length 1
- SCE
sister chromatid exchange
- T-loop
telomeric loop
- TRF
terminal restriction fragment
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-03-0179) on May 16, 2012.
REFERENCES
- Abraham J, et al. Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J. 2003;22:6137–6147. doi: 10.1093/emboj/cdg580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DJ, et al. A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics. 2005;82:753–758. doi: 10.1016/j.ygeno.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Bailey SM, Goodwin EH, Meyne J, Cornforth MN. CO-FISH reveals inversions associated with isochromosome formation. Mutagenesis. 1996;11:139–144. doi: 10.1093/mutage/11.2.139. [DOI] [PubMed] [Google Scholar]
- Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LJ, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Bryja V, Bonilla S, Cajanek L, Parish CL, Schwartz CM, Luo Y, Rao MS, Arenas E. An efficient method for the derivation of mouse embryonic stem cells. Stem Cells. 2006;24:844–849. doi: 10.1634/stemcells.2005-0444. [DOI] [PubMed] [Google Scholar]
- Bucholc M, Park Y, Lustig AJ. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:6559–6573. doi: 10.1128/MCB.21.19.6559-6573.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzowska M, et al. Mutation of the mouse Rad17 gene leads to embryonic lethality and reveals a role in DNA damage-dependent recombination. EMBO J. 2004;23:3548–3558. doi: 10.1038/sj.emboj.7600353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc Natl Acad Sci USA. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Lavi S. Induction of circles of heterogeneous sizes in carcinogen-treated cells: two-dimensional gel analysis of circular DNA molecules. Mol Cell Biol. 1996;16:2002–2014. doi: 10.1128/mcb.16.5.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- de Lange T. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol. 2004;19:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;5:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Ding H, et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Dronkert ML, Beverloo HB, Johnson RD, Hoeijmakers JH, Jasin M, Kanaar R. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol Cell Biol. 2000;20:3147–3156. doi: 10.1128/mcb.20.9.3147-3156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- Essers J, Hendriks RW, Swagemakers SM, Troelstra C, de Wit J, Bootsma D, Hoeijmakers JH, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Gertsenstein M, Lobe C, Nagy A. ES cell-mediated conditional transgenesis. Methods Mol Biol. 2002;185:285–307. doi: 10.1385/1-59259-241-4:285. [DOI] [PubMed] [Google Scholar]
- Glover TW, Berger C, Coyle J, Echo B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum Genet. 1984;67:136–142. doi: 10.1007/BF00272988. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, Reddel RR. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol. 2009;27:1181–1185. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- Hirano S, et al. Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. EMBO J. 2005;24:418–427. doi: 10.1038/sj.emboj.7600534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, et al. Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum Mol Genet. 2004;13:1241–1248. doi: 10.1093/hmg/ddh135. [DOI] [PubMed] [Google Scholar]
- Kipling D, Cooke HJ. Hypervariable ultra-long telomeres in mice. Nature. 1990;347:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little MT, Dirks RW, Raap AK, Tanke HJ. Heterogeneity in telomere length of human chromosomes. Hum Mol Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ, Essers J, Weeda G, Beverloo B, de Wit J, Muijtjens M, Odijk H, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 2001;20:6540–6549. doi: 10.1093/emboj/20.22.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Niida H, Matsumoto T, Satoh H, Shiwa M, Tokutake Y, Furuichi Y, Shinkai Y. Severe growth defect in mouse cells lacking the telomerase RNA component. Nat Genet. 1998;19:203–206. doi: 10.1038/580. [DOI] [PubMed] [Google Scholar]
- Perry P, Wolff S. New Giemsa method for the differential staining of sister chromatids. Nature. 1974;251:156–158. doi: 10.1038/251156a0. [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett HA, Henson JD, Au AY, Neumann AA, Reddel RR. Normal mammalian cells negatively regulate telomere length by telomere trimming. Hum Mol Genet. 2011;20:4684–4692. doi: 10.1093/hmg/ddr402. [DOI] [PubMed] [Google Scholar]
- Pryde F, Khalili S, Robertson K, Selfridge J, Ritchie AM, Melton DW, Jullien D, Adachi Y. 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J Cell Sci. 2005;118:2043–2055. doi: 10.1242/jcs.02336. [DOI] [PubMed] [Google Scholar]
- Sen D, Gilbert W. Guanine quartet structures. Methods Enzymol. 1992;211:191–199. doi: 10.1016/0076-6879(92)11012-8. [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011;25:1320–1327. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol Cell Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- Thompson LH, Hinz JM. Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: mechanistic insights. Mutat Res. 2009;668:54–72. doi: 10.1016/j.mrfmmm.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uringa EJ, Youds JL, Lisaingo K, Lansdorp PM, Boulton SJ. RTEL1: an essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic Acids Res. 2011;39:1647–1655. doi: 10.1093/nar/gkq1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Wang W, Seki M, Narita Y, Sonoda E, Takeda S, Yamada K, Masuko T, Katada T, Enomoto T. Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J. 2000;19:3428–3435. doi: 10.1093/emboj/19.13.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, et al. Fanconi anemia protein FANCD2 promotes immunoglobulin gene conversion and DNA repair through a mechanism related to homologous recombination. Mol Cell Biol. 2005;25:34–43. doi: 10.1128/MCB.25.1.34-43.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, Mets DG, McIlwraith MJ, Martin JS, Ward JD, NJ ON, Rose AM, West SC, Meyer BJ, Boulton SJ. RTEL-1 enforces meiotic crossover interference and homeostasis. Science. 2010;327:1254–1258. doi: 10.1126/science.1183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlmans JM, Martens UM, Poon SS, Raap AK, Tanke HJ, Ward RK, Lansdorp PM. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc Natl Acad Sci USA. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.