A signaling pathway downstream of R-Ras is identified that controls axon branching in cortical neurons. Besides the well-known role of R-Ras as an activator of the PI3K signaling pathway, a novel system of cytoskeletal regulation is found by which R-Ras induces axon branching through regulation of the actin-binding protein afadin.
Abstract
Regulation of axon growth, guidance, and branching is essential for constructing a correct neuronal network. R-Ras, a Ras-family small GTPase, has essential roles in axon formation and guidance. During axon formation, R-Ras activates a series of phosphatidylinositol 3-kinase signaling, inducing activation of a microtubule-assembly promoter—collapsin response mediator protein-2. However, signaling molecules linking R-Ras to actin cytoskeleton–regulating axonal morphology remain obscure. Here we identify afadin, an actin-binding protein harboring Ras association (RA) domains, as an effector of R-Ras inducing axon branching through F-actin reorganization. We observe endogenous interaction of afadin with R-Ras in cortical neurons during the stage of axonal development. Ectopic expression of afadin increases axon branch number, and the RA domains and the carboxyl-terminal F-actin binding domain are required for this action. RNA interference knockdown experiments reveal that knockdown of endogenous afadin suppressed both basal and R-Ras–mediated axon branching in cultured cortical neurons. Subcellular localization analysis shows that active R-Ras–induced translocation of afadin and its RA domains is responsible for afadin localizing to the membrane and inducing neurite development in Neuro2a cells. Overall, our findings demonstrate a novel signaling pathway downstream of R-Ras that controls axon branching.
INTRODUCTION
Neurons are highly polarized cells with two distinct processes—a single long axon and multiple dendrites. In the development of the nervous system, neurons project axons to distant target cells. To construct a correct neuronal network, axonal growth, guidance, and branching are strictly regulated by extrinsic cues and intrinsic signals (O'Donnell et al., 2009; Bilimoria and Bonni, 2011). Extensive studies have revealed that Ras- and Rho-family small GTPases are involved in control of axon morphology (Hall and Lalli, 2010). Small GTPases function as molecular switches by cycling between a GDP-bound inactive state and a GTP-bound active state. When activated, they interact with their specific effectors to induce various biological events in cells.
The Ras family comprises a huge number of small GTPases. Among the Ras-family proteins, H-Ras, K-Ras, and N-Ras are well known as regulators of cell growth and differentiation through the kinase-mediated pathways of extracellular signal-regulated kinase (ERK) and phosphatidylinositol 3-kinase (PI3K; Kinbara et al., 2003; Arimura and Kaibuchi, 2007). The R-Ras–subfamily proteins R-Ras, TC21 (R-Ras2), and M-Ras (R-Ras3) show relatively similar homology and form a distinct branch of classic Ras proteins (Matsumoto et al. 1997). In spite of their structural similarity, R-Ras–subfamily proteins have different functions owing to distinct effector coupling. R-Ras activates PI3K but not ERK, regulating integrin activation, cell migration, and angiogenesis (Keely et al., 1999; Zhang et al., 1996; Komatsu and Ruoslahti, 2005), whereas M-Ras stimulates the ERK pathway by activating B-Raf, thereby promoting neurite outgrowth in PC12 cells (Kimmelman et al., 2002).
A well-established model system for studying the mechanism of neuronal morphogenesis is the hippocampal or cortical pyramidal neuron (Bradke and Dotti, 2000). Cultured neurons first extend several undifferentiated neurites (at stages 1 and 2). At stages 2 and 3, one of the neurites begins to elongate rapidly to become the axon, and subsequent axonal branching occurs. The remaining minor neurites develop into dendrites at stage 4. R-Ras selectively accumulates in a single neurite of stage 2 neurons and controls axon formation through PI3K-mediated signaling (Oinuma et al., 2007). By contrast, the expression of endogenous M-Ras in cortical neurons is low during the stage of axonal development and up-regulated during the stage of dendritic development, and M-Ras regulates dendrite morphology through activation of the ERK pathway (Saito et al., 2009). The activity of R-Ras increases after plating and peaks between stages 2 and 3, when neuronal polarization and axon formation occur (Oinuma et al., 2007). Active R-Ras stimulates a series of PI3K signaling, inducing activation of a microtubule-assembly promoter, collapsin response mediator protein-2 (CRMP-2; Ito et al., 2006). Dynamic morphological changes of neurons are accompanied by actin cytoskeletal reorganization. However, signaling molecules linking R-Ras to axonal actin cytoskeleton remain obscure.
Afadin, also known as AF-6, is a multidomain adaptor protein having two Ras association (RA) domains and a PSD-95/Dlg/ZO-1 domain (Takai et al., 2008). Afadin was originally identified as a filamentous-actin (F-actin)–binding protein localized at cell–cell contact sites (Mandai et al., 1997). In epithelial cells, afadin serves as a linker of nectins and actin cytoskeleton and controls adherence junction formation (Takahashi et al., 1999; Kurita et al., 2011). In addition to nectins, afadin interacts with various proteins involved in maintenance of adherence junctions and stabilizes cell–cell connections (Tachibana et al., 2000; Pokutta et al., 2002, Asada et al., 2003). Recent studies have shown that afadin regulates cell migration in fibroblasts and breast cancer cells (Miyata et al., 2009; Fournier et al., 2011). In neurons, several reports have shown that afadin controls dendritic spine morphology in vitro and in vivo (Xie et al., 2005, Beaudoin et al., 2012). Although afadin localizes not only in dendrites but also in axons (Lim et al., 2008), roles of afadin in axonal morphogenesis have been obscure. Afadin has two RA domains in the amino-terminal region. RA domains were found in various Ras effectors such as Raf-1 and PI3K, and Ras-family GTPases have been implicated as an upstream regulator of afadin (Rodriguez-Viciana et al., 2004). It has been reported that the Ras-family GTPase Rap1 binds to afadin to control leading-edge formation and dendritic spine plasticity in vitro and apical–basal polarity in neuroepithelial cells in vivo (Xie et al., 2005; Miyata et al., 2009; Carmena et al., 2011). However, little is known about signaling systems of Ras-family GTPases and afadin during axonal morphogenesis.
In this study, we identify afadin as an effector of R-Ras in cortical neurons and demonstrate a novel signaling pathway involved in regulation of axon branching.
RESULTS
Active R-Ras interacts with the RA domain of afadin
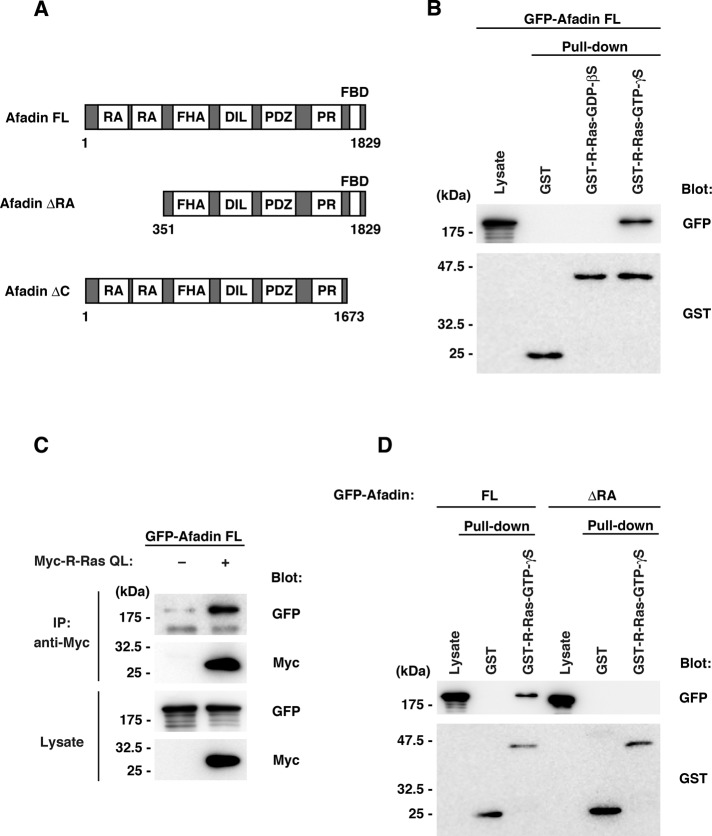
We carried out yeast two-hybrid screening of a rat brain cDNA library to isolate potential effectors for R-Ras in neurons. Through the screening, we found that one of the positive clones encoded the amino-terminal region of afadin, a multidomain adapter protein having two Ras-association domains (Figure 1A). To determine whether R-Ras interacts with afadin in a GTP-dependent manner, we expressed green fluorescent protein (GFP)–tagged afadin full length (FL) in HEK293T cells and performed a pull-down assay with purified glutathione S-transferase (GST)–fused R-Ras preloaded with GDP-βS or GTP-γS. As shown in Figure 1B, the GTP-γS–preloaded active form but not the GDP-βS–preloaded inactive form of R-Ras interacted with afadin. To further examine whether active R-Ras interacts with afadin in cells, we performed immunoprecipitation analysis. Myc-tagged R-Ras QL (constitutively active mutant) and GFP-tagged afadin FL were expressed in HEK293T cells, and the cell lysates were immunoprecipitated with anti-Myc antibody. Afadin was coimmunoprecipitated with R-Ras QL (Figure 1C), indicating that active R-Ras binds to afadin in mammalian cells. To define the R-Ras–interacting region within afadin, we expressed GFP-tagged afadin FL or afadin ΔRA, a mutant of afadin lacking amino-terminal RA domains (Figure 1A), in HEK293T cells and used it in a pull-down assay. Afadin FL bound to the active form of R-Ras, but afadin ΔRA did not (Figure 1D). Collectively these results suggest that active R-Ras interacts with afadin through the RA domains.
FIGURE 1:
Active R-Ras interacts with the RA domains of afadin. (A) Schematic structures of afadin constructs used in this study. DIL, dilute domain; FBD, F-actin–binding domain; FHA, forkhead-associated domain; PR, proline-rich region; RA, Ras association domain. Numbers indicate amino acid positions within the sequence. (B) Lysates from HEK293T cells transfected with GFP-tagged afadin FL were used in pull-down assays with GST or GST-fused R-Ras preloaded with GDP-βS or GTP-γS. Total and bound proteins were analyzed by immunoblotting. (C) Lysates from HEK293T cells transfected with GFP-tagged afadin FL and HA-tagged R-Ras QL (constitutively active mutant) were immunoprecipitated with anti-HA antibody. Lysate inputs and immunoprecipitates were analyzed by immunoblotting. (D) Lysates from HEK293T cells transfected with GFP-tagged afadin FL or ΔRA were used in pull-down assays with GST or GST-fused R-Ras preloaded with GTP-γS. Lysate inputs and bound proteins were analyzed by immunoblotting.
R-Ras interacts with afadin in cortical neurons during axonal development
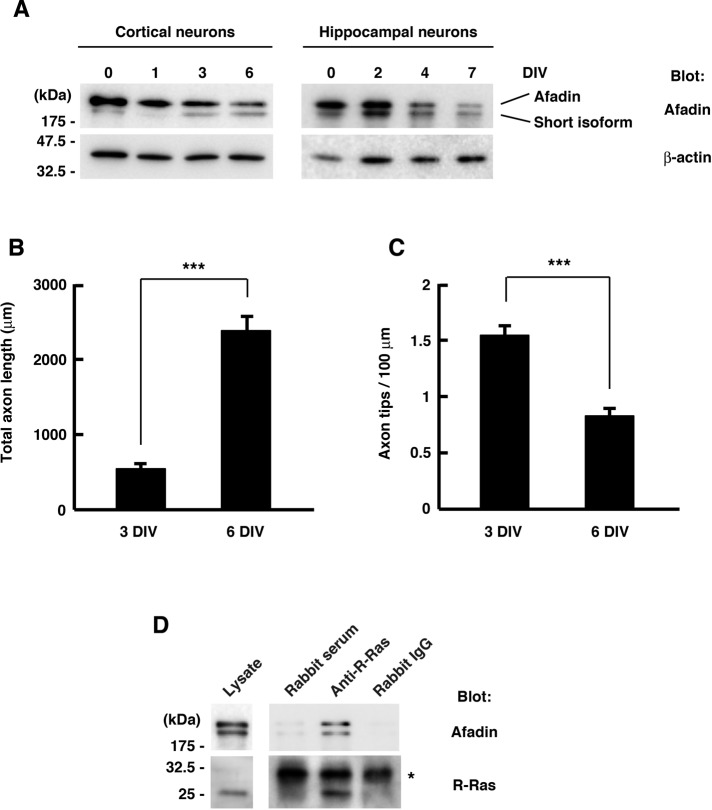
We previously reported a role of R-Ras in axon formation. R-Ras is selectively accumulated to axons and activated during axon formation in cultured cortical or hippocampal neurons (Oinuma et al., 2007). Therefore we next examined whether afadin was expressed in neurons during early developmental stages. Western blot analysis with primary cultured rat cortical or hippocampal neurons showed that the expression of afadin was relatively high around 0–3 d in vitro (DIV), when axonal development occurred, and thereafter declined when dendrites began to grow (Figure 2A), implicating a role of afadin in axonal development. We also observed the time course of the axonal development of cultured cortical neurons more precisely. Cortical axons branched more actively during 0–3 DIV, and axon elongation rather than branching was observed during the later period of 3–6 DIV (Figure 2, B and C). These results suggest that the level of endogenous afadin matches with the observed time course of active axon branching. To determine whether endogenous R-Ras interacts with afadin in neurons during axonal development, we lysed cultured cortical neurons at 2 DIV and immunoprecipitated them with rabbit antiserum against R-Ras. As shown in Figure 2D, endogenous afadin was coimmunoprecipitated with R-Ras by anti–R-Ras antiserum but not by nonimmunized control rabbit serum or purified rabbit immunoglobulin G (IgG) used as a control, indicating that R-Ras interacts with afadin in cortical neurons during axonal development.
FIGURE 2:
R-Ras interacts with afadin in cortical neurons during axonal development. (A) Primary cultured rat cortical (left) and hippocampal (right) neurons were lysed at the indicated time points, and the cell lysates were analyzed by immunoblotting. Both the long and short afadin isoforms were detected. (B, C) The development of axonal morphology of cultured cortical neurons was examined. Total axon length (B) and axon tip number per 100 μm (C) were measured at 3 and 6 DIV. The results are means ± SEM of three independent experiments (n = 45; ***p < 0.001, one-way ANOVA with Dunnett's post hoc test). (D) Cortical neurons were lysed at 2 DIV and immunoprecipitated with rabbit anti–R-Ras antiserum. Nonimmunized rabbit serum and purified rabbit IgG were used as controls. Lysate inputs and immunoprecipitates were analyzed by immunoblotting. Asterisk indicates the position of the immunoglobulin light chains.
Knockdown of afadin suppresses axon branching, but not elongation, in cortical neurons
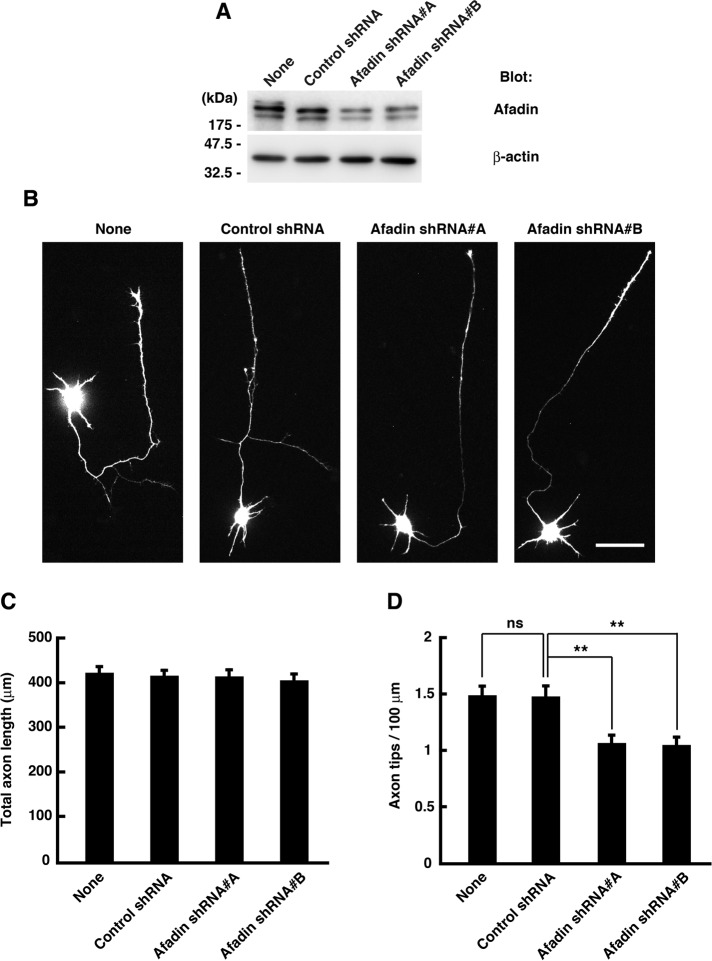
Next, to assess the role of afadin in axonal development, we generated two short hairpin RNA (shRNA) expression vectors designed to target two different regions of the afadin transcript. These shRNAs effectively reduced the amount of endogenous afadin in cortical neurons (Figure 3A). Cortical neurons were transfected with yellow fluorescent protein (YFP) and shRNAs before plating, and their axonal morphology was analyzed at 3 DIV (Figure 3B). Expression of the control shRNA did not affect axonal morphology, but two afadin-specific shRNAs reduced axon branching without affecting total axon length (Figure 3, C and D). These results suggest that afadin regulates axon branching, but not elongation, in cortical neurons.
FIGURE 3:
Knockdown of afadin suppresses axon branching in cultured neurons. (A) Cortical neurons were transfected with YFP and control shRNA or afadin shRNA (#A or #B) before plating using nucleofection technology and lysed at 3 DIV, and the whole-cell lysates were analyzed by immunoblotting. (B) Cortical neurons were transfected with YFP and shRNAs before plating using nucleofection technology and fixed at 3 DIV. The fluorescence images of YFP are shown. Scale bar, 50 μm. (C, D) Total axon length (C) and axon tip number per 100 μm (D) were measured. The results are means ± SEM of three independent experiments (n = 60; ns, not significant; **p < 0.01, one-way ANOVA with Dunnett's post hoc test).
The RA domains are required for inducing axon branching in cortical neurons
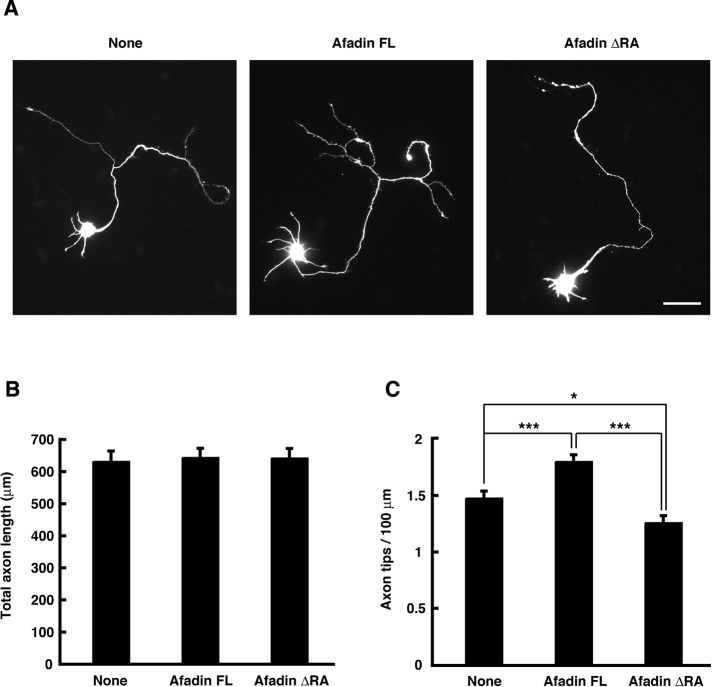
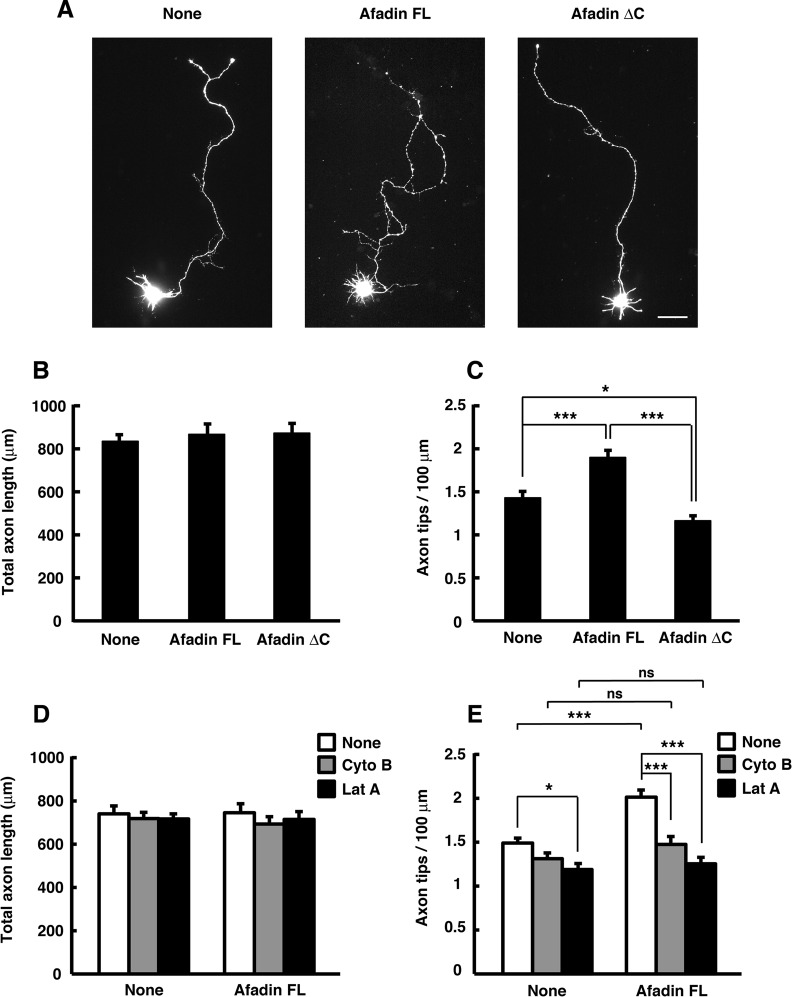
To determine whether the RA domains are required for afadin to induce axon branching, we examined the effect of ectopic expression of afadin FL or ΔRA mutant on axonal morphology. Cortical neurons were transfected with YFP and either Myc-tagged afadin FL or ΔRA mutant at 1 DIV, and their axonal morphology was observed at 3 DIV. As shown in Figure 4, ectopic expression of afadin FL promoted axon branching, but not elongation, compared with expression of YFP alone. On the other hand, the overexpression of afadin ΔRA suppressed basal axon branching without affecting axon length. These results suggest that the amino-terminal RA domains are necessary for afadin to induce axon branching.
FIGURE 4:
Ectopic expression of afadin FL, but not ΔRA mutant, induces axon branching. (A) Cortical neurons were transfected with YFP and Myc-tagged afadin FL or ΔRA mutant at 1 DIV and fixed at 3 DIV. The fluorescence images of YFP are shown. Scale bar, 50 μm. (B, C) Total axon length (B) and axon tip number per 100 μm (C) were measured. The results are means ± SEM of three independent experiments (n = 60; *p < 0.05, ***p < 0.001; one-way ANOVA with Dunnett's post hoc test).
Active R-Ras promotes axon branching through afadin
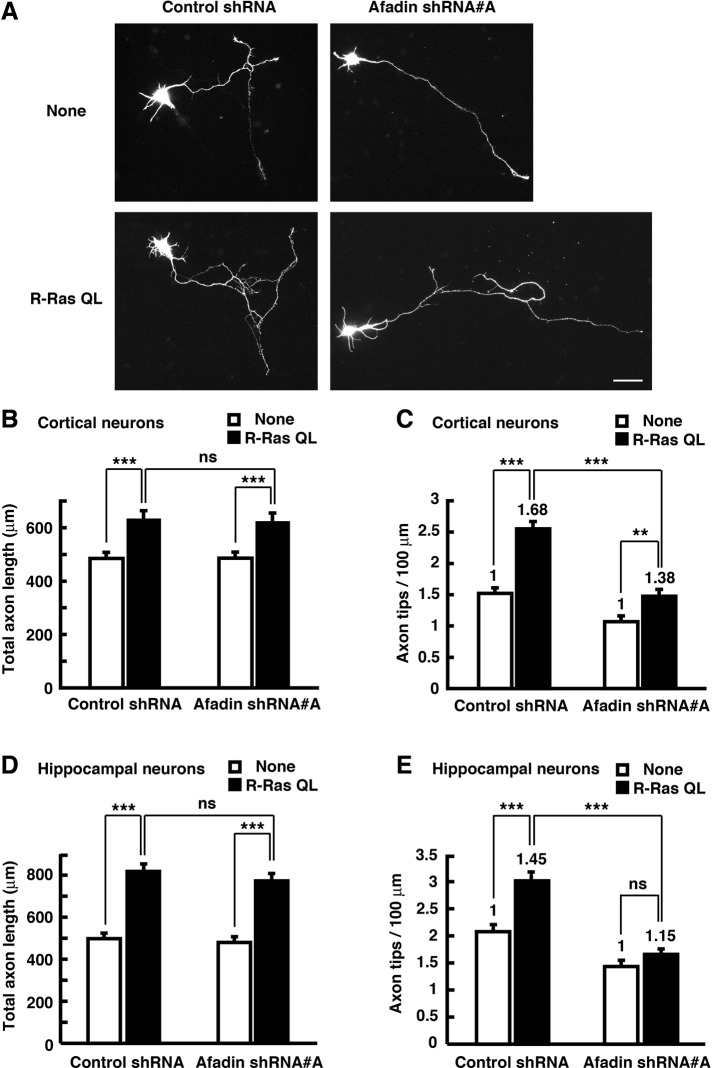
We next investigated whether afadin regulates axon branching downstream of R-Ras. Cortical neurons were transfected with YFP, Myc-tagged R-Ras QL, and afadin-specific shRNA before seeding by nucleofection, which enables proteins to be expressed in a widespread and modest condition, and their axon length and branch number were measured at 3 DIV. Ectopic expression of R-Ras QL increased both total axon length and branch number. Knockdown of afadin suppressed both basal and R-Ras QL–mediated axon branching but not total axon length (Figure 5, A–C). The same results were observed in cultured hippocampal neurons (Figure 5, D and E). These results suggest that afadin contributes to regulation of axon branching, but not axon outgrowth, downstream of R-Ras in both cortical and hippocampal neurons.
FIGURE 5:
Knockdown of afadin suppresses axon branching induced by active R-Ras. (A) Cortical neurons were transfected with YFP and shRNAs together with Myc-tagged R-Ras QL before plating using nucleofection technology and fixed at 3 DIV. The fluorescence images of YFP are shown. Scale bar, 50 μm. (B, C) Total axon length (B) and axon tip number per 100 μm (C) were measured. (D, E) Hippocampal neurons were transfected as described in A, and total axon length (D) and axon tip number per 100 μm (E) were measured. The results are means ± SEM of three independent experiments (n = 60; ns, not significant; **p < 0.01, ***p < 0.001; one-way ANOVA with Dunnett's post hoc test). The fold increase of axon branching by ectopic expression of R-Ras is indicated in the columns.
R-Ras induces membrane translocation of afadin
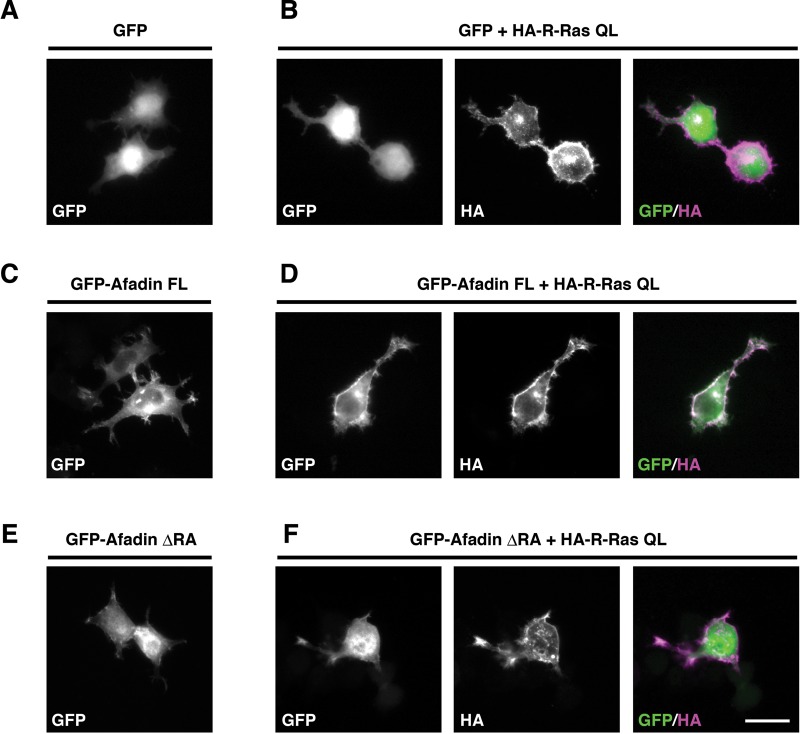
Previous studies report that regulation of subcellular localization of afadin is important for its various cellular functions, such as directional cell migration in fibroblasts and dendritic spine morphogenesis in neurons (Xie et al., 2005; Miyata et al., 2009). Therefore we next examined whether R-Ras controls the subcellular distribution of afadin. Mouse neuroblastoma Neuro2a cells were transfected with GFP-tagged afadin constructs and hemagglutinin (HA)-tagged R-Ras QL, and subcellular localization of afadin was observed with immunofluorescent microscopy. We found that R-Ras QL was localized in the cell periphery (Figure 6B), but afadin FL or afadin ΔRA by itself was distributed mainly in the cytosol (Figure 6, C and E). When coexpressed with R-Ras QL, afadin FL was translocated to the cell periphery, where it was colocalized with R-Ras QL (Figure 6D). By contrast, this translocation was not observed with GFP or GFP-afadin ΔRA (Figure 6, B and F).
FIGURE 6:
Active R-Ras translocates afadin FL, but not afadin ΔRA, to the cell periphery. Neuro2a cells were transiently transfected with GFP (A), GFP + HA-R-Ras QL (B), GFP-afadin FL (C), GFP-afadin ΔRA + HA-R-Ras QL (D), GFP-afadin ΔRA (E), and GFP-afadin ΔRA + HA-R-Ras QL (F). The cells were fixed at 1 d after transfection. The fluorescence images of GFP (A–F, green in merge) and the immunofluorescence images with anti-HA antibody (B, D, F, magenta in merge) are shown. Scale bar, 25 μm.
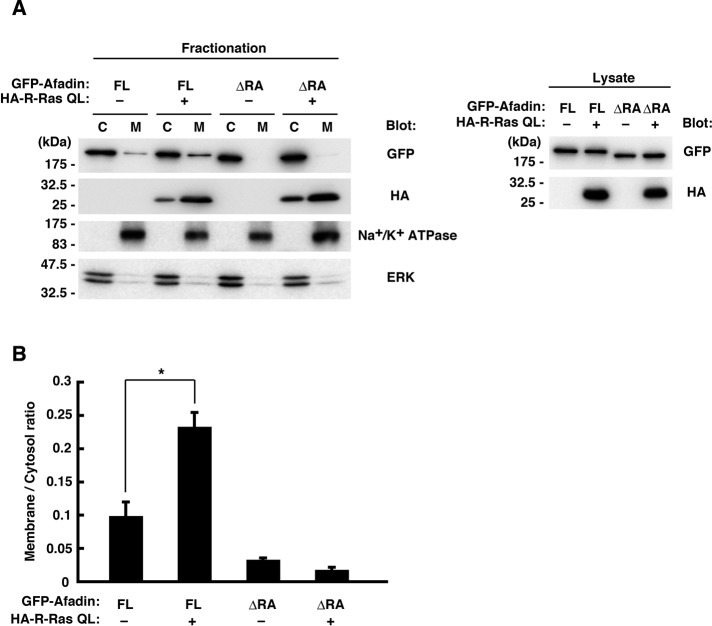
We further performed biochemical fractionation analysis to confirm the R-Ras–dependent membrane translocation of afadin. We prepared membrane and cytosol fractions from Neuro2a cells expressing GFP-tagged afadin constructs and HA-tagged R-Ras QL and analyzed the distribution of afadin by immunoblotting. Consistent with the immunofluorescence analysis in Figure 6, afadin FL or afadin ΔRA by itself was scarcely detected in the membrane. When coexpressed with R-Ras QL, afadin FL was partially translocated to the membrane fraction, whereas such translocation was not observed with afadin ΔRA (Figure 7). These results suggest that R-Ras binding to the RA domains of afadin induces its membrane translocation.
FIGURE 7:
Afadin is localized to the membrane by active R-Ras in a RA domain–dependent manner. (A) Neuro2a cells were transfected with GFP-tagged afadin FL or ΔRA mutant together with HA-tagged R-Ras QL, and the cytosol (C) and the membrane (M) fractions were analyzed by immunoblotting. Na+/K+-ATPase and ERK were used as markers of membrane and cytosol, respectively. Expression levels of the constructs were verified by immunoblotting of the cell lysates (right). (B) The membrane/cytosol ratio of GFP-tagged afadin was analyzed. Data are presented as the means ± SEM of three independent experiments (*p < 0.05, one-way ANOVA with Dunnett's post hoc test).
R-Ras and afadin coordinately promote neurite development
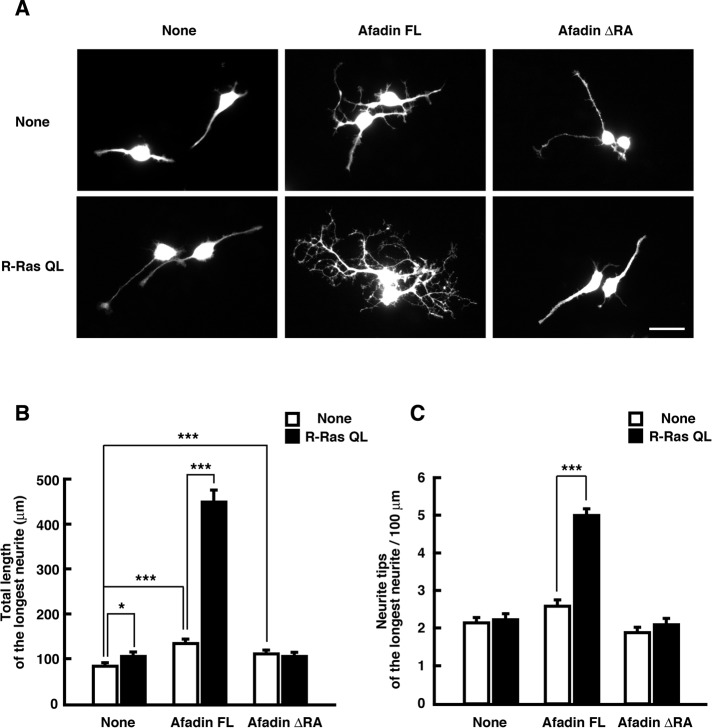
Neuro2a cells differentiate and extend neurites in response to several stimuli such as dibutyryl cAMP (dbcAMP) and retinoic acid (Prasad and Hsie, 1971; Shea et al., 1985), and differentiated Neuro2a cells are used as a model system of neurite development. We examined whether afadin induces neurite development in a R-Ras–dependent manner. Neuro2a cells were transfected with YFP, Myc-tagged afadin constructs, and HA-tagged R-Ras QL and then differentiated with dbcAMP. After 2 d, the longest neurite of the differentiated cells was analyzed. Ectopic expression of afadin FL induced neurite elongation and branching weakly, and these effects were enhanced by ectopic expression of R-Ras QL. On the other hand, although ectopic expression of afadin ΔRA alone slightly induced neurite elongation, the R-RasQL–induced enhancement was not observed (Figure 8). These results suggest that R-Ras and afadin coordinately regulate neurite morphology.
FIGURE 8:
R-Ras and afadin coordinately promotes neurite development. (A) Neuro2a cells were transfected with YFP and Myc-tagged afadin constructs (top) or YFP and Myc-tagged afadin constructs together with HA-tagged R-Ras QL (bottom). Then the cells were differentiated with 1 mM dbcAMP for 48 h and fixed. The fluorescence images of YFP are shown. Scale bar, 50 μm. (B, C) Total length of the longest neurite (B) and neurite tip number per 100 μm of the longest neurite (C) in differentiated cells were measured. The results are the means ± SEM of three independent experiments (n = 60; *p < 0.05, **p < 0.01, ***p < 0.001; one-way ANOVA with Dunnett's post hoc test).
Afadin induces axon branching in an actin-dependent manner
Afadin is an F-actin–binding multidomain adaptor protein and interacts with F-actin in the carboxyl-terminal region (Mandai et al., 1997). To explore the underlying mechanism of afadin-mediated axon branching, we observed the intracellular distribution of endogenous afadin in cultured cortical neurons at 2 DIV. As shown in Figure 9, endogenous afadin was accumulated at the sites of budding growth cones located at the axonal tips and shafts, where it was well colocalized with F-actin. This suggests the involvement F-actin in regulating afadin-mediated axon branching. To determine whether the carboxyl-terminal F-actin–binding domain is required for afadin-mediated axon branching, we assessed the ability of the truncated form of afadin, afadin ΔC (Figure 1A), which lacks the ability to bind F-actin (Mandel et al., 1997), to affect axon branching in cortical neurons. We transfected cortical neurons with afadin ΔC at 1 DIV and observed them at 3 DIV and found that afadin FL–induced promotion of axon branching was impaired by deletion of the carboxyl-terminal F-actin–binding domain (Figure 10, A–C). We further examined whether axon branching is affected by pharmacological treatments that attenuate actin dynamics. Treatment of cortical neurons with reagents that inhibit polymerization of actin filaments has been reported to selectively inhibit initiation of axon branching without significantly reducing axon length (Dent and Kalil, 2001). As shown in Figure 10, D and E, treatment with cytochalasin B (1 μM) or latrunculin A (0.25 μM), an inhibitor for actin polymerization, completely blocked afadin FL–induced promotion of axon branching without affecting the total axon length. These results suggest that afadin induces axon branching through the carboxyl-terminal F-actin–binding domain in an actin-dependent manner.
FIGURE 9:
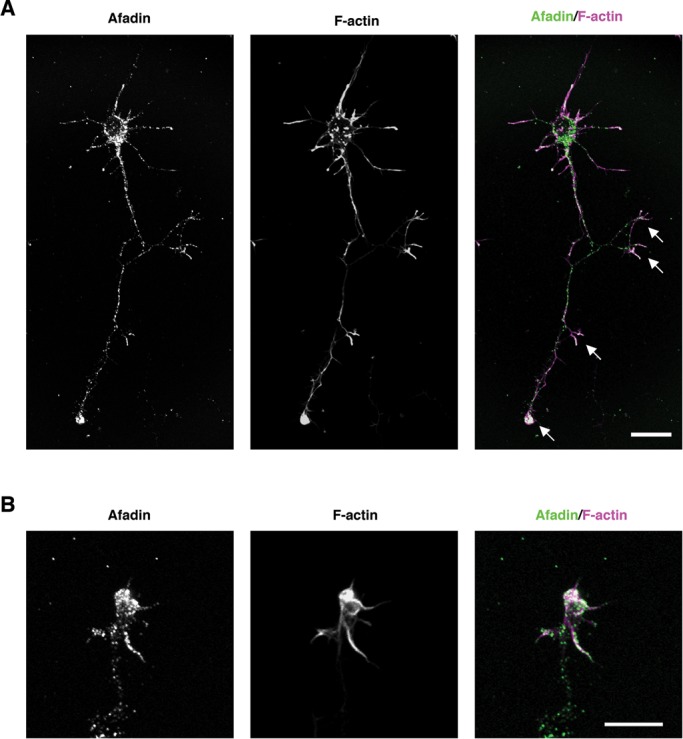
Afadin accumulates at the sites of budding growth cones located at the axonal tips and shafts, where it is well colocalized with F-actin. (A, B) Intracellular distribution of endogenous afadin in cultured cortical neurons at 2 DIV observed with confocal microscopy. Neurons were costained with anti-afadin antibody (green in merge) and phalloidin (magenta in merge) to visualize F-actin. Arrows represent budding growth cones. (B) A high-power image of a representative budding growth cone. Scale bar, 20 µm (A), 10 μm (B).
FIGURE 10:
Afadin induces axon branching in an actin-dependent manner. (A) Cortical neurons were transfected with YFP and Myc-tagged afadin FL or ΔC mutant at 1 DIV and fixed at 3 DIV. The fluorescence images of YFP are shown. Scale bar, 50 μm. (B, C) Total axon length (B) and axon tip number per 100 μm (C) were measured. The results are means ± SEM of three independent experiments (n = 45; *p < 0.05, ***p < 0.001; one-way ANOVA with Dunnett's post hoc test). (D, E) Cortical neurons transfected with YFP and Myc-tagged afadin FL at 1 DIV were treated with cytochalasin B (Cyto B; 1 μM), latrunculin A (Lat A; 0.25 μM), or DMSO (vehicle control) for 3 h before fixation at 3 DIV. Total axon length (D) and axon tip number per 100 μm (E) were measured. The results are means ± SEM of three independent experiments (n = 33; ns, not significant; *p < 0.05, ***p < 0.001; one-way ANOVA with Dunnett's post hoc test).
DISCUSSION
Axonal growth, guidance, and branching are strictly regulated by extrinsic cues and intrinsic signals (O'Donnell et al., 2009; Bilimoria and Bonni, 2011), and the morphogenetic events of axons require cytoskeletal reorganization, including microtubules and actin filaments. It was recently found that R-Ras, a Ras-family GTPase, has essential roles in axon formation and elongation (Oinuma et al., 2007). Active R-Ras reorganizes microtubules through activation of a series of PI3K signaling, inducing activation of CRMP-2, a microtubule-assembly promoter (Ito et al., 2006). However, signaling molecules linking R-Ras to axonal actin cytoskeleton remain obscure. In this study, we identified afadin, an actin-binding multidomain adaptor protein, as an effector of R-Ras inducing axon branching.
Ectopic expression of active R-Ras induced both axon branching and elongation, and RNA interference knockdown of afadin suppressed R-Ras–mediated axon branching but not R-Ras–mediated axonal elongation. In addition, ectopic expression of afadin increased axon branch number but not elongation. These results suggest a specific role of afadin in axon branching rather than axonal outgrowth in cultured cortical neurons, and another mechanism contributes to R-Ras-mediated axonal elongation. Axon elongation and branching are cooperative but distinct processes, and a variety of molecular systems are related to these two processes. These processes use either common systems or distinct mechanisms, depending on their respective features of morphological changes. R-Ras is a critical key player in all steps of axon maturation, including axon elongation and branching, as well as specification (Ivins et al., 2000; Oinuma et al., 2007). We showed that R-Ras binds to afadin in an activity-dependent manner, indicating that afadin is a downstream effector of R-Ras. Among various effectors of R-Ras, afadin may be included in the system of R-Ras–regulated axon branching. Furthermore, in the Neuro2a cell model system, coexpression of active R-Ras and afadin increased not only neurite branch number, but also neurite length, suggesting that R-Ras/afadin pathway can potentially regulate neurite elongation. Because neurites of Neuro2a cells are immature protrusions, molecular systems participated in neurite formation may be in part undifferentiated in usage.
The suppression of R-Ras–induced axon branching by afadin knockdown is partial. This might be due to the knockdown efficiency of the shRNAs against afadin, but there may be another mechanism regulating axon branching downstream of R-Ras in addition to the R-Ras/afadin pathway. In fact, it was reported that R-Ras activation induces phosphorylation and inactivation of glycogen synthase kinase-3b (GSK-3β) at Ser-9 through the PI3K/Akt pathway (Oinuma et al., 2007), and activation of the PI3K/Akt pathway and inactivation of GSK-3β have also been implicated in axon branch formation (Yoshimura et al., 2005; Drinjakovic et al., 2010). R-Ras organizes axon branching through coordinate operation of multiple effectors, including afadin and the PI3K pathway.
Many extracellular cues and neuronal activity are implicated in axon formation (Dent and Kalil, 2001; Yamada et al., 2010; Bilimoria and Bonni, 2011). It has been reported that netrin-1 and fibroblast growth factor-2 promote axon branching and that semaphorin-3A, a repulsive axonal guidance factor, suppresses it without affecting axon outgrowth in cortical neurons (Dent et al., 2004). Ephrin, another repulsive guidance molecule, has been reported to represent another factor with a clear influence on axon branching (Bilimoria and Bonni, 2011). Plexins—receptors for semaphorins—directly inactivate R-Ras in cultured neurons (Oinuma et al., 2004a, 2004b; Toyofuku et al., 2005; Uesugi et al., 2009). In addition, ephrin has also been reported to inactivate R-Ras activity to induce axonal repulsion (Dail et al., 2006). Given that these guidance factors regulate R-Ras activity, R-Ras/afadin pathway may participate in intracellular signaling of these extracellular cues for axon branching.
Two types of cell adhesion—cell–substrate and cell–cell adhesion—contribute to cell morphogenesis. Cell–substrate adhesion is mediated by the binding of integrins at the cell surface to the extracellular matrix (ECM) proteins, and integrins must be activated and tethered to the actin cytoskeleton inside the cell in order to be able to bind the ECM proteins with high affinity (Liddington and Ginsberg, 2002). The ECM proteins have been reported to regulate axon branching (Féréol et al., 2011), and R-Ras contributes to activation of β1 integrins and strengthens cell adhesion in nonneuronal cells (Zhang et al., 1996). R-Ras also promotes integrin-mediated neurite outgrowth and cell attachment of retinal neurons (Ivins et al., 2000). In addition, afadin, together with the intracellular molecule nectin, contributes to cell–cell adhesion (Takai et al., 2008), and the role of afadin in the control of cell–substrate adhesion remains obscure. Of interest, recent work has demonstrated the mechanochemical mechanisms regulating the sensitivity of growing axons to ECM ligands. Modulation of sensitivity toward ECM ligands occurs through force-regulated adhesion, and higher intracellular traction forces induced by the actin cytoskeleton result in reinforcement of adhesion sites on the ECM (Féréol et al., 2011). We speculate that the R-Ras/afadin system might regulate cell–substrate adhesion by linking integrins to the actin cytoskeleton in growing axons.
We also examined how R-Ras regulates afadin activity. Afadin is a scaffold protein, and its membrane translocation is a critical step for its functioning. Previous studies showed that translocation of afadin by the small GTPase Rap1 is required in directional cell migration and dendritic spine plasticity (Xie et al., 2005; Miyata et al., 2009). We found that afadin is translocated to the membrane by active R-Ras through the amino-terminal RA domains, and this membrane translocation is correlated with the activity of afadin to induce neurite outgrowth and branching in Neuro2a cells. Afadin interacts with F-actin in the carboxyl-terminal region (Mandai et al., 1997), and it was reported that F-actin accumulation occurs prior to invasion of microtubules at the site of axon branch formation (Kalil et al., 2000). Cortical axon branching occurs in vitro through changes in growth cone morphologies (Szebenyi et al., 1998). Szebenyi et al. (1998) performed high-resolution time-lapse imaging on cultured cortical neurons and found that growth cones tend to pause in regions where axon branches later develop. They showed that remnants of reorganized growth cones are left behind on the axon shaft as filopodial or lamellar protrusions, and axon branches subsequently emerge from these regions of the axon shaft. In the present study, using cultured cortical neurons, we observed that endogenous afadin accumulates at the sites of budding growth cones located at the axonal tips and shafts, where it is well colocalized with F-actin. Furthermore, the carboxyl-terminal F-actin–binding domain of afadin is required for axon branching, and pharmacological inhibitors for actin polymerization block afadin FL–induced promotion of axon branching, suggesting that afadin-mediated axon branching requires F-actin reorganization. In addition to F-actin, many binding partners of afadin have been identified, including regulators of cytoskeletal reorganization such as Src kinase and α-catenin (Tachibana et al., 2000; Pokutta et al., 2002; Radziwill et al., 2007). It is inferred that R-Ras induces membrane translocation of afadin-tethering F-actin and a variety of signaling molecules required for axon branch formation, and changes in localization or activity of afadin-binding proteins might contribute to axon branching induced by R-Ras/afadin. Further work will be required to clarify the molecular mechanisms upstream and downstream of R-Ras/afadin signaling involved in regulating axon branching.
To construct a correct and complex neuronal network, axons form elaborate branches allowing neurons to make synaptic connections with multiple targets and establish unique patterns of connectivity. Understanding molecular systems of axon branching is an important issue toward which the present work should provide useful information.
MATERIALS AND METHODS
DNA constructs and mutagenesis
GST-fused human R-Ras and HA-tagged R-Ras QL (Q87L), a constitutively active mutant, have been described previously (Oinuma et al., 2004a). Myc-tagged R-Ras QL was subcloned into pcDNA3 (Invitrogen, Carlsbad, CA). Expression plasmids of GFP-tagged rat afadin FL (1–1829) and ΔRA mutant (351–1829) were kindly provided by Y. Takai (Kobe University, Kobe, Japan), and afadin ΔC (1–1673), a deletion mutant of afadin lacking the F-actin–binding domain, was generated by PCR-mediated mutagenesis. Myc-tagged afadin constructs were subcloned into pCXN2. pCAG vector encoding YFP was a generous gift from J. Miyazaki (Osaka University, Osaka, Japan) and T. Saito (Chiba University, Chiba, Japan). For two-hybrid screening, a constitutively active mutant of mouse TC21, TC21 (G23V), was generated by PCR-mediated mutagenesis. Then the CAAX motifs of R-Ras QL and TC21 GV were deleted to produce R-Ras QL ΔC and TC21 GV ΔC by PCR and subcloned into pGBKT7 (Clontech, Mountain View, CA). The shRNAs for afadin were designed to target 19 nucleotides of rat transcript and were expressed by using pSinencer-2.1 (Invitrogen). The target sequences are as follows: afadin shRNA#A (nucleotides 308–326, 5´-GATTGGACATTGATGAGAA-3´), and afadin shRNA#B (nucleotides 2862–2880, 5´-CATTCCAAATGGTTTACAA-3´). The shRNA for rat R-Ras (nucleotides 426–444, 5´-CAAGGCAGATCTGGAGACA-3´), which has no effect on R-Ras expression and axonal morphology (Oinuma et al., 2007), was used as a control shRNA.
Antibodies and reagents
We used the following antibodies: mouse monoclonal antibodies against Myc (9E10), GFP (B-2), and afadin (clone-35); rabbit polyclonal antibodies against HA (Y-11) and GST (Santa Cruz Biotechnology, Santa Cruz, CA); a rabbit polyclonal anti-ERK antibody (Cell Signaling Technology, Beverly, MA); a rabbit polyclonal anti-Myc antibody (MBL); a rat monoclonal anti-HA antibody (3F10; Roche Applied Science, Indianapolis, IN); a mouse monoclonal anti–Na+/K+ ATPase α subunit antibody (Millipore, Billerica, MA); a mouse monoclonal anti–β-actin antibody (Sigma-Aldrich); a rabbit anti–R-Ras antiserum (BD Biosciences, San Diego, CA); horseradish peroxidase (HRP)–conjugated secondary antibodies (DakoCytomation, Hamburg, Germany); HRP-conjugated Clean-Blot IP Detection Reagent (ThermoFisher Scientific, Waltham, MA); and Alexa Fluor 594 or 647–conjugated secondary antibodies (Invitrogen). Poly-l-lysine, dbcAMP, and cytochalasin B were from Sigma-Aldrich, and latrunculin A was from Invitrogen.
Yeast two-hybrid screening
A rat brain cDNA library fused to the GAL4 activation domain of the pACT2 vector (Clontech) was screened using pGBKT7/R-Ras QL ΔC and pGBKT7/TC21 GV ΔC as baits in the yeast strain AH109 according to the manufacturer's protocols. Interaction between the baits and library proteins activates transcription of the reporter genes HIS3, Ade, and lacZ. From 1.4 × 107 transformants, 728 colonies grew on selection medium lacking histidine and adenine and were also positive for β-galactosidase activity. One of them was found to encode the amino-terminal region of afadin. β-Galactosidase activity assay was performed as described previously (Katoh et al., 2002).
Cell culture and transfection
HEK293T cells and Neuro2a cells were cultured in DMEM containing 10% fetal bovine serum (FBS), 4 mM glutamine, 100 U/ml penicillin, and 0.2 mg/ml streptomycin under humidified air containing 5% CO2 at 37°C. Transient transfections were carried out with Lipofectamine Plus (Invitrogen) for HEK293T cells or Lipofectamine 2000 (Invitrogen) for Neuro2a cells, according to the manufacturer's instructions. To observe neurite morphology, we suspended Neuro2a cells transfected with the indicated plasmids and mixed them with parental cells at a 1:4 ratio. After they were reseeded on poly-l-lysine–coated glass cover slips (circular, 13 mm in diameter; Matsunami Glass, Osaka, Japan), the cells were differentiated with 1 mM dbcAMP in DMEM containing 5% FBS for 48 h.
Primary cortical and hippocampal neurons were prepared from the embryonic day 19 rat brains as described previously (Ishikawa et al., 2003). The dissociated neurons were seeded on poly-l-lysine–coated glass cover slips (circular, 13 mm in diameter) at a density of 2 × 104 cells or plastic dishes (60 or 100 mm in diameter) in DMEM containing 10% FBS, 4 mM glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin and cultured under humidified air containing 5% CO2 at 37°C. After 4 h, the medium was replaced with Neurobasal medium (Invitrogen) containing 2 or 4% (for immunofluorescence analyses of shRNA-transfected cells) B27 supplement (Invitrogen), 0.5 mM GlutaMAX (Invitrogen), 50 U/ml penicillin, and 0.05 mg/ml streptomycin, and neurons were cultured under humidified air containing 5% CO2 at 37°C. For overexpression experiments, cortical neurons were transfected with the indicated plasmids at 1 DIV with Lipofectamine 2000 according to the manufacturer's instructions. For knockdown experiments, transfection was performed by Rat Neuron Nucleofector Kit (Lonza, Basel, Switzerland) following the manufacturer's instructions. Cytochalasin B and latrunculin A were dissolved in dimethyl sulfoxide (DMSO), and pharmacological treatment (1 μM cytochalasin B, 0.25 μM latrunculin A) was performed for 3 h before fixation. All animal experiments were conducted according to the guidelines of the Kyoto University Research Committee.
Immunoblotting
Proteins were separated by SDS–PAGE and were electrophoretically transferred onto a polyvinylidene difluoride membrane (Millipore). The membrane was blocked with 3% low-fat milk in TBS (Tris-buffered saline) and then incubated with primary antibodies. The primary antibodies were detected with HRP–conjugated secondary antibodies and a chemiluminescence detection kit (ECL; GE Healthcare, Piscataway, NJ). Images were captured using an LAS3000 analyzer (Fujifilm, Tokyo, Japan) equipped with Science Lab software (Fujifilm).
Pull-down assays
Recombinant GST-fusion proteins were purified from Escherichia coli as described (Katoh et al., 2002). We loaded GST-fused R-Ras with guanine nucleotides by incubating the protein with 100 μM GDP-βS or GTP-γS in loading buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5 mg/ml bovine serum albumin, 1 mM dithiothreitol, and 10% glycerol) at 30°C for 10 min. The reaction was stopped by the addition of MgCl2 to a final concentration of 10 mM.
For pull-down assays, HEK293T cells transfected with GFP-tagged afadin FL or ΔRA mutant were washed with TBS and lysed with the ice-cold cell lysis buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 4 mM MgCl2, 1% NP-40, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mM dithiothreitol). The cell lysates were then centrifuged for 10 min at 16,000 × g at 4°C. The supernatants were mixed with either 5 μg GST or GST-fused R-Ras preloaded with guanine nucleotides and incubated for 1.5 h at 4°C with glutathione–Sepharose beads (GE Healthcare). After the beads were washed with the ice-cold wash buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 4 mM MgCl2, 0.5% NP-40, and 10% glycerol), the bound proteins were eluted in Laemmli sample buffer and analyzed by SDS–PAGE and immunoblotting.
Immunoprecipitation
HEK293T cells transfected with indicated plasmids were washed with TBS and lysed with the ice-cold IP buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 4 mM MgCl2, 1% NP-40, 10% glycerol, 1 mM PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin). The cell lysates were then centrifuged for 5 min at 16,000 × g at 4°C. The supernatants were incubated with anti-Myc antibody (9E10) for 2 h and subsequently with protein G–Sepharose (GE Healthcare) for 1 h. The beads were washed with ice-cold IP buffer, and bound proteins were analyzed by SDS–PAGE and immunoblotting. For the detection of endogenous protein binding, primary cultured cortical neurons were lysed at 2 DIV with ice-cold IP buffer. After centrifugation for 10 min at 16,000 × g at 4°C, the supernatants were incubated with anti–R-Ras antiserum for 1 h and then with protein G–Sepharose beads for 1 h. Nonimmunized rabbit serum and rabbit IgG (anti-HA, Y-11) were used as negative controls. The beads were washed with ice-cold IP buffer, and bound proteins were analyzed by SDS–PAGE and immunoblotting.
Separation of membrane and cytosol fractions
Separation of membrane and cytosol fractions was performed as described (Hiramoto-Yamaki et al., 2010). Briefly, Neuro2a cells transfected with indicated plasmids were washed with TBS and suspended in ice-cold buffer A (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 50 mM NaF, 1 mM Na3VO4, and 1 mM PMSF). After rapid freezing in liquid nitrogen and thawing in a water bath, cells were centrifuged for 10 min at 16,000 × g at 4°C. The supernatants were removed and used as cytosol fractions. The pellets were washed with buffer A and lysed with buffer A containing 1% Triton X-100. After centrifugation for 10 min at 10,000 × g at 4°C, the supernatants were used as membrane fractions. Both cytosol and membrane fractions were analyzed by SDS–PAGE and immunoblotting.
Immunofluorescence microscopy
Neuro2a cells and primary cultured neurons on cover slips were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min. After residual paraformaldehyde had been quenched with 50 mM NH4Cl in PBS for 10 min, cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min and incubated with 10% FBS in PBS for 30 min. Cells were incubated with the primary antibodies for 1 h, followed by incubation with Alexa 594 or 647–conjugated secondary antibodies for 1 h. The cells on coverslips were mounted in 90% glycerol containing 0.1% p-phenylenediamine dihydrochloride in PBS and photographed with a Leica DC350F digital camera system (Leica, Wetzlar, Germany) equipped with a Nikon Eclipse E800 microscope (Nikon, Tokyo, Japan).
For immunostaining of endogenous afadin in cultured cortical neurons, fixed and permeabilized neurons were incubated with 1% Blocking Reagent (Roche Applied Science) for 30 min. Then cells were incubated with anti-afadin antibody (clone-35, 1:200 dilution) diluted with Can Get Signal Immunostain Immunoreaction Enhancer Solution A (Toyobo, Osaka, Japan), followed by incubation with Alexa 488–conjugated anti-mouse IgG (1:2000 dilution) for 1 h. For visualizing F-actin, neurons were incubated for 1 h with Alexa 594–conjugated phalloidin (Invitrogen) with Can Get Signal Immunostain Immunoreaction Enhancer Solution A. The cells on coverslips were mounted in Prolong Gold antifade reagent (Invitrogen). To obtain a z-plane image, we captured optical sections of images through the cell in 0.20-μm steps using a laser-scanning confocal imaging system (FluoView FV1000-D; Olympus, Tokyo, Japan) and a microscope equipped with spectral system (IX81-S; Olympus) with a 60×/numerical aperture (NA) 1.35 or 100×/NA 1.40 oil objective (Olympus). The images were arranged and labeled using Photoshop 7.0 (Adobe, San Jose, CA).
Data analysis
Densitometry analysis was performed with Multi Gauge, version 3.1 (Fujifilm). For quantification of axonal morphology, a process that was at least twice as long as the other processes and was more than twice the cell body diameter was defined as an axon, as described elsewhere (Schwamborn and Püschel 2004; Yin et al., 2008; Sepúlveda et al., 2009), and total length and tip number were measured. For quantification of neurite morphology of Neuro2a cells, the cells that contained a process at least twice as long as the cell diameter were determined as differentiated cells, as described elsewhere (Cowley et al., 1994; Sepúlveda et al., 2009), and total length and tip number of the longest neurite were measured.
The number of processes >5 μm in length was counted and defined as “tip number” (Marler et al., 2008). The length of each neurite was measured from the edge of the cell body (or its branching point) to the tip of the neurite, and total axon length means the sum of the lengths of all of the axons of the neuron. Statistical significance was determined by the analysis of variance (ANOVA) and post hoc test (Dunnett's T3) using SPSS software, version 16.0 (IBM, Armonk, NY). Differences at the level of p < 0.05 were considered statistically significant.
Acknowledgments
We are grateful to Y. Takai for expression plasmids for afadin and J. Miyazaki and T. Saito for the enhanced YFP dual-promoter expression plasmid. This research was supported by the Science and Technology Agency of Japan, Precursory Research for Embryonic Science and Technology Project (Development and Function of Neuronal Networks; to I.O.); Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Scientific Research (B) 23390019; to M.N.); Young Scientists (A) 23689005 (to I.O.); and Challenging Exploratory Research 23657127 (to I.O.); and grants from the Uehara Memorial Foundation and Daiichi-Sankyo Foundation of Life Science (to I.O.).
Abbreviations used:
- ANOVA
analysis of variance
- CRMP-2
collapsin response mediator protein-2
- dbcAMP
dibutyryl cAMP
- DIV
days in vitro
- DMSO
dimethyl sulfoxide
- ECM
extracellular matrix
- ERK
extracellular signal-regulated kinase
- F-actin
filamentous actin
- FBS
fetal bovine serum
- FL
full length
- GFP
green fluorescent protein
- GSK-3b
glycogen synthase kinase-3b
- GST
glutathione S-transferase
- HRP
horseradish peroxidase
- PBS
phosphate-buffered saline
- PI3K
phosphatidylinositol 3-kinase
- PMSF
phenylmethylsulfonyl fluoride
- RA
Ras association
- shRNA
short hairpin RNA
- TBS
Tris-buffered saline
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-02-0103) on May 16, 2012.
The authors declare that they have no conflict of interest.
REFERENCES
- Arimura N, Kaibuchi K. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat Rev Neurosci. 2007;8:194–205. doi: 10.1038/nrn2056. [DOI] [PubMed] [Google Scholar]
- Asada M, Irie K, Morimoto K, Yamada A, Ikeda W, Takeuchi M, Takai Y. ADIP, a novel afadin- and alpha-actinin-binding protein localized at cell-cell adherens junctions. J Biol Chem. 2003;278:4103–4111. doi: 10.1074/jbc.M209832200. [DOI] [PubMed] [Google Scholar]
- Bradke F, Dotti CG. Establishment of neuronal polarity: lessons from cultured hippocampal neurons. Curr Opin Neurobiol. 2000;10:574–581. doi: 10.1016/s0959-4388(00)00124-0. [DOI] [PubMed] [Google Scholar]
- Beaudoin GM 3rd, Schofield CM, Nuwal T, Zang K, Ullian EM, Huang B, Reichardt LF. Afadin, a ras/rap effector that controls cadherin function, promotes spine and excitatory synapse density in the hippocampus. J Neurosci. 2012;32:99–110. doi: 10.1523/JNEUROSCI.4565-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilimoria PM, Bonni A. Molecular regulation of axon branching. Neuroscientist, 2011 doi: 10.1177/1073858411426201. doi: 10.1177/1073858411426201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena A, Makarova A, Speicher S. The Rap1-Rgl-Ral signaling network regulates neuroblast cortical polarity and spindle orientation. J Cell Biol. 2011;195:553–562. doi: 10.1083/jcb.201108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley S, Paterson H, Kemp P, Marshall CJ. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- Dail M, Richter M, Godement P, Pasquale EB. Eph receptors inactivate R-Ras through different mechanisms to achieve cell repulsion. J Cell Sci. 2006;119:1244–1254. doi: 10.1242/jcs.02842. [DOI] [PubMed] [Google Scholar]
- Dent EW, Barnes AM, Tang F, Kalil K. Netrin-1 and semaphorin 3A promote or inhibit cortical axon branching, respectively, by reorganization of the cytoskeleton. J Neurosci. 2004;24:3002–3012. doi: 10.1523/JNEUROSCI.4963-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Kalil K. Axon branching requires interactions between dynamic microtubules and actin filaments. J Neurosci. 2001;21:9757–9769. doi: 10.1523/JNEUROSCI.21-24-09757.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinjakovic J, Jung H, Campbell DS, Strochlic L, Dwivedy A, Holt CE. E3 ligase Nedd4 promotes axon branching by downregulating PTEN. Neuron. 2010;65:341–357. doi: 10.1016/j.neuron.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Féréol S, Fodil R, Barnat M, Georget V, Milbreta U, Nothias F. Micropatterned ECM substrates reveal complementary contribution of low and high affinity ligands to neurite outgrowth. Cytoskeleton. 2011;68:373–388. doi: 10.1002/cm.20518. [DOI] [PubMed] [Google Scholar]
- Fournier G, et al. Loss of AF6/afadin, a marker of poor outcome in breast cancer, induces cell migration, invasiveness and tumor growth. Oncogene. 2011;30:3862–3874. doi: 10.1038/onc.2011.106. [DOI] [PubMed] [Google Scholar]
- Hall A, Lalli G. Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb Perspect Biol. 2010;2:a001818. doi: 10.1101/cshperspect.a001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramoto-Yamaki N, Takeuchi S, Ueda S, Harada K, Fujimoto S, Negishi M, Katoh H. Ephexin4 and EphA2 mediate cell migration through a RhoG-dependent mechanism. J Cell Biol. 2010;190:461–477. doi: 10.1083/jcb.201005141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa Y, Katoh H, Negishi M. A role of Rnd1 GTPase in dendritic spine formation in hippocampal neurons. J Neurosci. 2003;23:11065–11072. doi: 10.1523/JNEUROSCI.23-35-11065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Oinuma I, Katoh H, Kaibuchi K, Negishi M. Sema4D/plexin-B1 activates GSK-3β through R-Ras GAP activity, inducing growth cone collapse. EMBO Rep. 2006;7:704–709. doi: 10.1038/sj.embor.7400737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivins JK, Yurchenco PD, Lander AD. Regulation of neurite outgrowth by integrin activation. J Neurosci. 2000;20:6551–6560. doi: 10.1523/JNEUROSCI.20-17-06551.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalil K, Szebenyi G, Dent EW. Common mechanisms underlying growth cone guidance and axon branching. J Neurobiol. 2000;44:145–158. [PubMed] [Google Scholar]
- Katoh H, Harada A, Mori K, Negishi M. Socius is a novel Rnd GTPase-interacting protein involved in disassembly of actin stress fibers. Mol Cell Biol. 2002;22:2952–2964. doi: 10.1128/MCB.22.9.2952-2964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keely PJ, Rusyn EV, Cox AD, Parise LV. R-Ras signals through specific integrin alpha cytoplasmic domains to promote migration and invasion of breast epithelial cells. J Cell Biol. 1999;145:1077–1088. doi: 10.1083/jcb.145.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmelman AC, Nuñez Rodriguez N, Chan AM. R-Ras3/M-Ras induces neuronal differentiation of PC12 cells through cell-type-specific activation of the mitogen-activated protein kinase cascade. Mol Cell Biol. 2002;22:5946–5961. doi: 10.1128/MCB.22.16.5946-5961.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinbara K, Goldfinger LE, Hansen M, Chou FL, Ginsberg MH. Ras GTPases: integrin's friends or foes? Nat Rev Mol Cell Biol. 2003;4:767–776. doi: 10.1038/nrm1229. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Ruoslahti E. R-Ras is a global regulator of vascular regeneration that suppresses intimal hyperplasia and tumor angiogenesis. Nat Med. 2005;11:1346–1350. doi: 10.1038/nm1324. [DOI] [PubMed] [Google Scholar]
- Kurita S, Ogita H, Takai Y. Cooperative role of nectin-nectin and nectin-afadin interactions in formation of nectin-based cell-cell adhesion. J Biol Chem. 2011;286:36297–36303. doi: 10.1074/jbc.M111.261768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddington RC, Ginsberg MH. Integrin activation takes shape. J Cell Biol. 2002;158:833–839. doi: 10.1083/jcb.200206011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ST, Lim KC, Giuliano RE, Federoff HJ. Temporal and spatial localization of nectin-1 and l-afadin during synaptogenesis in hippocampal neurons. J Comp Neurol. 2008;507:1228–1244. doi: 10.1002/cne.21608. [DOI] [PubMed] [Google Scholar]
- Mandai K, et al. Afadin: a novel actin filament-binding protein with one PDZ domain localized at cadherin-based cell-to-cell adherens junction. J Cell Biol. 1997;139:517–528. doi: 10.1083/jcb.139.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marler KJ, Becker-Barroso E, Martínez A, Llovera M, Wentzel C, Poopalasundaram S, Hindges R, Soriano E, Comella J, Drescher U. A TrkB/EphrinA interaction controls retinal axon branching and synaptogenesis. J Neurosci. 2008;28:12700–12712. doi: 10.1523/JNEUROSCI.1915-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Asano T, Endo T. Novel small GTPase M-Ras participates in reorganization of actin cytoskeleton. Oncogene. 1997;15:2409–2417. doi: 10.1038/sj.onc.1201416. [DOI] [PubMed] [Google Scholar]
- Miyata M, et al. Localization of nectin-free afadin at the leading edge and its involvement in directional cell movement induced by platelet-derived growth factor. J Cell Sci. 2009;122:4319–4329. doi: 10.1242/jcs.048439. [DOI] [PubMed] [Google Scholar]
- O'Donnell M, Chance RK, Bashaw GJ. Axon growth and guidance: receptor regulation and signal transduction. Annu Rev Neurosci. 2009;32:383–412. doi: 10.1146/annurev.neuro.051508.135614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinuma I, Ishikawa Y, Katoh H, Negishi M. The semaphorin 4D receptor plexin-B1 is a GTPase activating protein for R-Ras. Science. 2004a;305:862–865. doi: 10.1126/science.1097545. [DOI] [PubMed] [Google Scholar]
- Oinuma I, Katoh H, Negishi M. Molecular dissection of the semaphorin 4D receptor plexin-B1-stimulated R-Ras GTPase-activating protein activity and neurite remodeling in hippocampal neurons. J Neurosci. 2004b;24:11473–11480. doi: 10.1523/JNEUROSCI.3257-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinuma I, Katoh H, Negishi M. R-Ras controls axon specification upstream of glycogen synthase kinase-3beta through integrin-linked kinase. J Biol Chem. 2007;282:303–318. doi: 10.1074/jbc.M607979200. [DOI] [PubMed] [Google Scholar]
- Pokutta S, Drees F, Takai Y, Nelson WJ, Weis WI. Biochemical and structural definition of the l-afadin- and actin-binding sites of alpha-catenin. J Biol Chem. 2002;277:18868–18874. doi: 10.1074/jbc.M201463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad KN, Hsie AW. Morphologic differentiation of mouse neuroblastoma cells induced in vitro by dibutyryl adenosine 3′:5′-cyclic monophosphate. Nat New Biol. 1971;233:141–142. doi: 10.1038/newbio233141a0. [DOI] [PubMed] [Google Scholar]
- Radziwill G, Weiss A, Heinrich J, Baumgartner M, Boisguerin P, Owada K, Moelling K. Regulation of c-Src by binding to the PDZ domain of AF-6. EMBO J. 2007;26:2633–2644. doi: 10.1038/sj.emboj.7601706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Sabatier C, McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol Cell Biol. 2004;24:4943–4954. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Oinuma I, Fujimoto S, Negishi M. Plexin-B1 is a GTPase activating protein for M-Ras, remodelling dendrite morphology. EMBO Rep. 2009;10:614–621. doi: 10.1038/embor.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn JC, Püschel AW. The sequential activity of the GTPases Rap1B and Cdc42 determines neuronal polarity. Nat Neurosci. 2004;7:923–929. doi: 10.1038/nn1295. [DOI] [PubMed] [Google Scholar]
- Sepúlveda MR, Vanoevelen J, Raeymaekers L, Mata AM, Wuytack F. Silencing the SPCA1 (secretory pathway Ca2+-ATPase isoform 1) impairs Ca2+ homeostasis in the Golgi and disturbs neural polarity. J Neurosci. 2009;29:12174–12182. doi: 10.1523/JNEUROSCI.2014-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea TB, Fischer I, Sapirstein VS. Effect of retinoic acid on growth and morphological differentiation of mouse NB2a neuroblastoma cells in culture. Brain Res. 1985;353:307–314. doi: 10.1016/0165-3806(85)90220-2. [DOI] [PubMed] [Google Scholar]
- Szebenyi G, Callaway JL, Dent EW, Kalil K. Interstitial branches develop from active regions of the axon demarcated by the primary growth cone during pausing behaviors. J Neurosci. 1998;18:7930–7940. doi: 10.1523/JNEUROSCI.18-19-07930.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana K, Nakanishi H, Mandai K, Ozaki K, Ikeda W, Yamamoto Y, Nagafuchi A, Tsukita S, Takai Y. Two cell adhesion molecules, nectin and cadherin, interact through their cytoplasmic domain-associated proteins. J Cell Biol. 2000;150:1161–1176. doi: 10.1083/jcb.150.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, et al. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with afadin, a PDZ domain-containing protein. J Cell Biol. 1999;145:539–549. doi: 10.1083/jcb.145.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y, Ikeda W, Ogita H, Rikitake Y. The immunoglobulin-like cell adhesion molecule nectin and its associated protein afadin. Annu Rev Cell Dev Biol. 2008;24:309–342. doi: 10.1146/annurev.cellbio.24.110707.175339. [DOI] [PubMed] [Google Scholar]
- Toyofuku T, Yoshida J, Sugimoto T, Zhang H, Kumanogoh A, Hori M, Kikutani H. FARP2 triggers signals for Sema3A-mediated axonal repulsion. Nat Neurosci. 2005;8:1712–1719. doi: 10.1038/nn1596. [DOI] [PubMed] [Google Scholar]
- Uesugi K, Oinuma I, Katoh H, Negishi M. Different requirement for Rnd GTPases of R-Ras GAP activity of plexin-C1 and plexin-D1. J Biol Chem. 2009;284:6743–6751. doi: 10.1074/jbc.M805213200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Huganir RL, Penzes P. Activity-dependent dendritic spine structural plasticity is regulated by small GTPase Rap1 and its target AF-6. Neuron. 2005;48:605–618. doi: 10.1016/j.neuron.2005.09.027. [DOI] [PubMed] [Google Scholar]
- Yamada A, Uesaka N, Hayano Y, Tabata T, Kano M, Yamamoto N. Role of pre- and postsynaptic activity in thalamocortical axon branching. Proc Natl Acad Sci USA. 2010;107:7562–7567. doi: 10.1073/pnas.0900613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin DM, Huang YH, Zhu YB, Wang Y. Both the establishment and maintenance of neuronal polarity require the activity of protein kinase D in the Golgi apparatus. J Neurosci. 2008;28:8832–8843. doi: 10.1523/JNEUROSCI.1291-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T, Kawano Y, Arimura N, Kawabata S, Kikuchi A, Kaibuchi K. GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity. Cell. 2005;120:137–149. doi: 10.1016/j.cell.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Vuori K, Wang H, Reed JC, Ruoslahti E. Integrin activation by R-ras. Cell. 1996;85:61–69. doi: 10.1016/s0092-8674(00)81082-x. [DOI] [PubMed] [Google Scholar]