Abstract
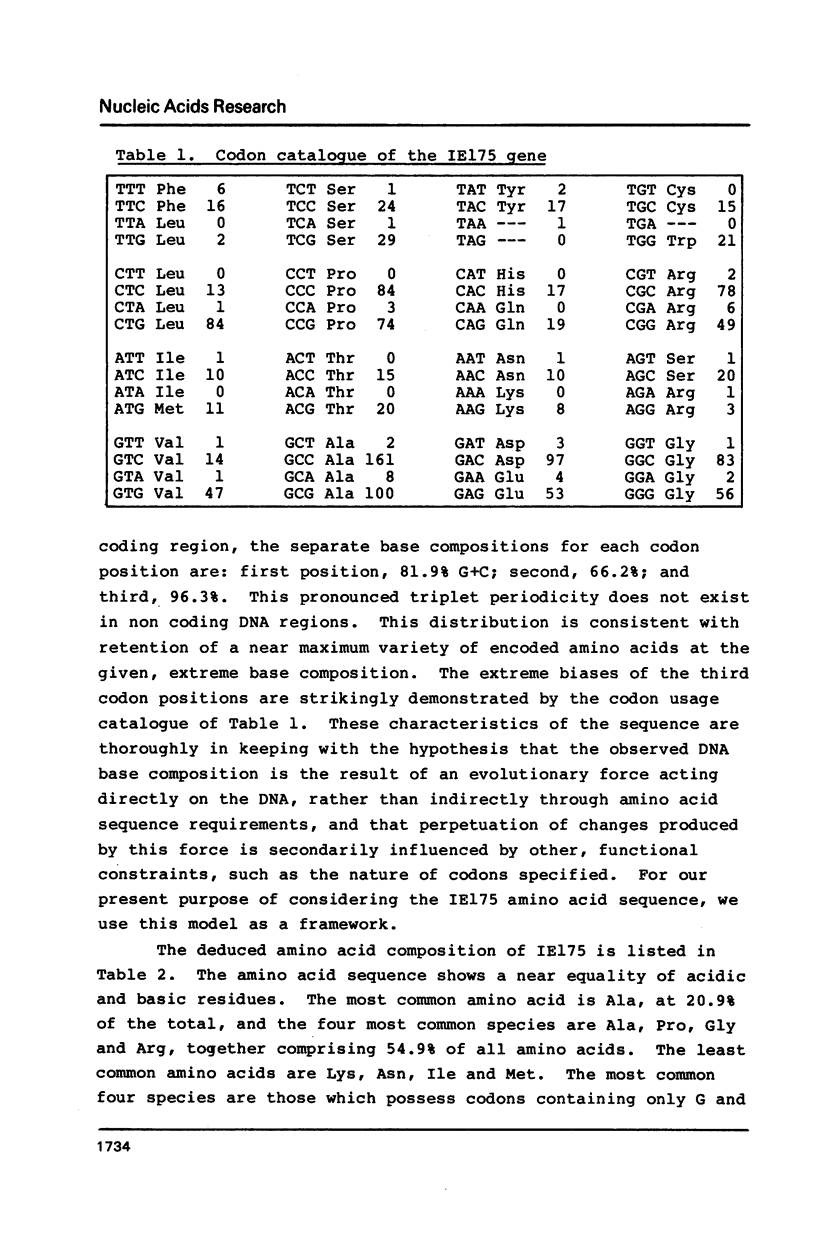
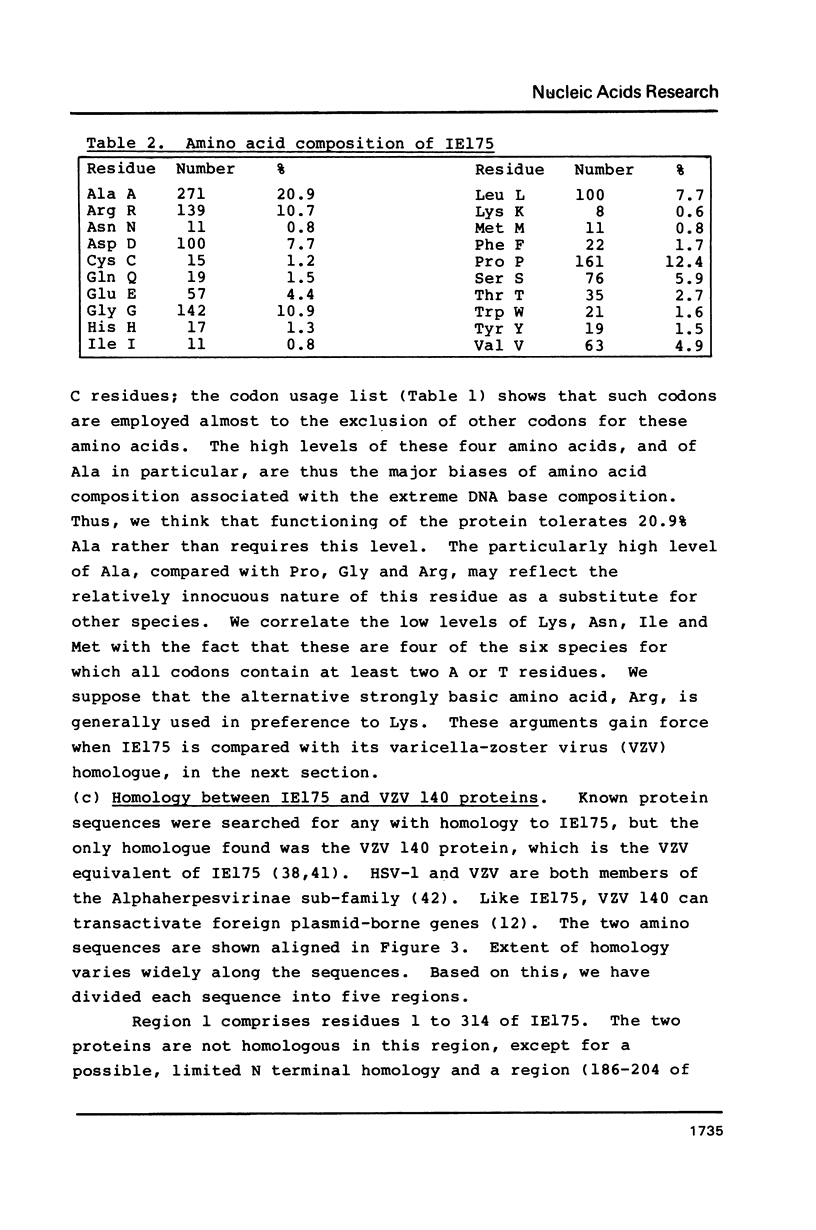
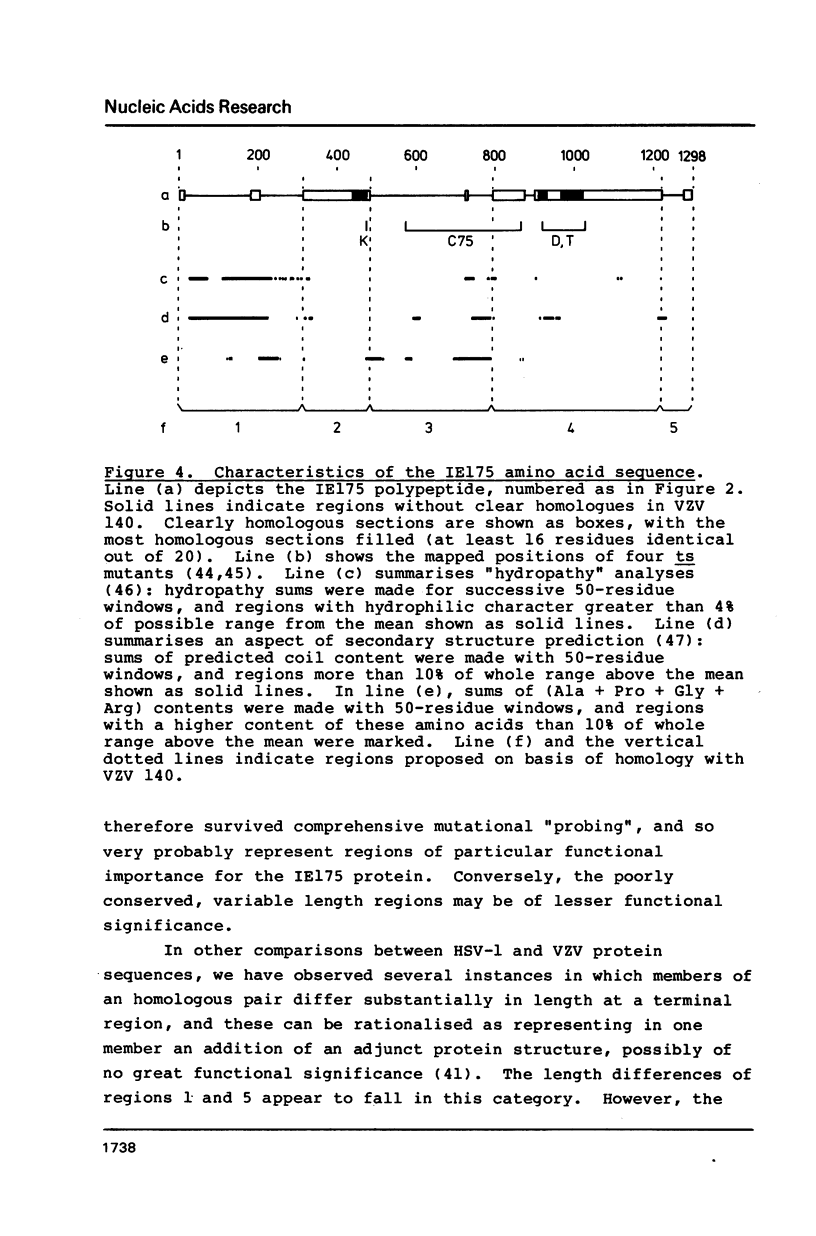
We report the complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1, as 6633 base pairs of composition 79.5% G+C. This contains immediate early gene 3, encoding the IE175 protein, an important transcriptional activator of later virus genes. The IE175 coding region was identified as a 3894 base sequence of 81.5% G+C DNA. The base composition of this gene is thus the most extreme yet determined, and the IE175 predicted amino acid composition is correspondingly biased, most notably with an alanine content of 20.9%. Functionally important regions of the IE175 polypeptide were tentatively identified by comparison with the sequence of the homologous protein from varicella-zoster virus and from locations of ts mutations, and were correlated with properties of the amino acid sequence. Aspects of the evolution of such an extreme composition DNA sequence were discussed.
Full text
PDF















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classification and nomenclature of viruses. Fourth report of the International Committee on Taxonomy of Viruses. Intervirology. 1982;17(1-3):1–199. doi: 10.1159/000149278. [DOI] [PubMed] [Google Scholar]
- Cordingley M. G., Campbell M. E., Preston C. M. Functional analysis of a herpes simplex virus type 1 promoter: identification of far-upstream regulatory sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2347–2365. doi: 10.1093/nar/11.8.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A. J., Scott J. E. DNA sequence of the major inverted repeat in the varicella-zoster virus genome. J Gen Virol. 1985 Feb;66(Pt 2):207–220. doi: 10.1099/0022-1317-66-2-207. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Wilkie N. M. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J Gen Virol. 1981 Aug;55(Pt 2):315–331. doi: 10.1099/0022-1317-55-2-315. [DOI] [PubMed] [Google Scholar]
- Davison M. J., Preston V. G., McGeoch D. J. Determination of the sequence alteration in the DNA of the herpes simplex virus type 1 temperature-sensitive mutant ts K. J Gen Virol. 1984 May;65(Pt 5):859–863. doi: 10.1099/0022-1317-65-5-859. [DOI] [PubMed] [Google Scholar]
- Deininger P. L. Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Anal Biochem. 1983 Feb 15;129(1):216–223. doi: 10.1016/0003-2697(83)90072-6. [DOI] [PubMed] [Google Scholar]
- Delius H., Clements J. B. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976 Oct;33(1):125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- Dixon R. A., Schaffer P. A. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate early protein VP175. J Virol. 1980 Oct;36(1):189–203. doi: 10.1128/jvi.36.1.189-203.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982 Sep 9;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- ElKareh A., Murphy A. J., Fichter T., Efstratiadis A., Silverstein S. "Transactivation" control signals in the promoter of the herpesvirus thymidine kinase gene. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1002–1006. doi: 10.1073/pnas.82.4.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R. D. A detailed analysis of an HSV-1 early promoter: sequences involved in trans-activation by viral immediate-early gene products are not early-gene specific. Nucleic Acids Res. 1984 Apr 11;12(7):3037–3056. doi: 10.1093/nar/12.7.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R. D. DNA sequence elements required for regulated expression of the HSV-1 glycoprotein D gene lie within 83 bp of the RNA capsites. Nucleic Acids Res. 1983 Oct 11;11(19):6647–6666. doi: 10.1093/nar/11.19.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R. D., Dunlop M. Trans activation of plasmid-borne promoters by adenovirus and several herpes group viruses. Nucleic Acids Res. 1984 Aug 10;12(15):5969–5978. doi: 10.1093/nar/12.15.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R. D. Trans activation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 1984 Dec 20;3(13):3135–3141. doi: 10.1002/j.1460-2075.1984.tb02270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. J., Powell K. L. DNA-binding properties of a herpes simplex virus immediate early protein. J Virol. 1982 Dec;44(3):1084–1087. doi: 10.1128/jvi.44.3.1084-1087.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoff H., Ansorge W. Improvements of DNA sequencing gels. Anal Biochem. 1981 Aug;115(2):450–457. doi: 10.1016/0003-2697(81)90031-2. [DOI] [PubMed] [Google Scholar]
- Hall J. D., Coen D. M., Fisher B. L., Weisslitz M., Randall S., Almy R. E., Gelep P. T., Schaffer P. A. Generation of genetic diversity in herpes simplex virus: an antimutator phenotype maps to the DNA polymerase locus. Virology. 1984 Jan 15;132(1):26–37. doi: 10.1016/0042-6822(84)90088-6. [DOI] [PubMed] [Google Scholar]
- Honess R. W. Herpes simplex and 'the herpes complex': diverse observations and a unifying hypothesis. The eighth Fleming lecture. J Gen Virol. 1984 Dec;65(Pt 12):2077–2107. doi: 10.1099/0022-1317-65-12-2077. [DOI] [PubMed] [Google Scholar]
- Honess R. W., Watson D. H. Unity and diversity in the herpesviruses. J Gen Virol. 1977 Oct;37(1):15–37. doi: 10.1099/0022-1317-37-1-15. [DOI] [PubMed] [Google Scholar]
- Kagawa Y., Nojima H., Nukiwa N., Ishizuka M., Nakajima T., Yasuhara T., Tanaka T., Oshima T. High guanine plus cytosine content in the third letter of codons of an extreme thermophile. DNA sequence of the isopropylmalate dehydrogenase of Thermus thermophilus. J Biol Chem. 1984 Mar 10;259(5):2956–2960. [PubMed] [Google Scholar]
- Kieff E. D., Bachenheimer S. L., Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971 Aug;8(2):125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Ludwig H., Haines H. G., Biswal N., Benyesh-Melnick M. The characterization of Varicella-zoster virus DNA. J Gen Virol. 1972 Jan;14(1):111–114. doi: 10.1099/0022-1317-14-1-111. [DOI] [PubMed] [Google Scholar]
- Mackem S., Roizman B. Differentiation between alpha promoter and regulator regions of herpes simplex virus 1: the functional domains and sequence of a movable alpha regulator. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4917–4921. doi: 10.1073/pnas.79.16.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden H. S., Crombie I. K., Subak-Sharpe J. H. Control of protein synthesis in herpesvirus-infected cells: analysis of the polypeptides induced by wild type and sixteen temperature-sensitive mutants of HSV strain 17. J Gen Virol. 1976 Jun;31(3):347–372. doi: 10.1099/0022-1317-31-3-347. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Dolan A., Donald S., Rixon F. J. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J Mol Biol. 1985 Jan 5;181(1):1–13. doi: 10.1016/0022-2836(85)90320-1. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982 Nov;31(1):89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- Murchie M. J., McGeoch D. J. DNA sequence analysis of an immediate-early gene region of the herpes simplex virus type 1 genome (map coordinates 0.950 to 0.978). J Gen Virol. 1982 Sep;62(Pt 1):1–15. doi: 10.1099/0022-1317-62-1-1. [DOI] [PubMed] [Google Scholar]
- O'Hare P., Hayward G. S. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol. 1985 Mar;53(3):751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palfreyman J. W., Maclean J. B., Messeder E., Sheppard R. C. Successful use of oligopeptides as immunogens in the preparation of antisera to immediate-early gene products of herpes simplex virus type 1. J Gen Virol. 1984 May;65(Pt 5):865–874. doi: 10.1099/0022-1317-65-5-865. [DOI] [PubMed] [Google Scholar]
- Pereira L., Wolff M. H., Fenwick M., Roizman B. Regulation of herpesvirus macromolecular synthesis. V. Properties of alpha polypeptides made in HSV-1 and HSV-2 infected cells. Virology. 1977 Apr;77(2):733–749. doi: 10.1016/0042-6822(77)90495-0. [DOI] [PubMed] [Google Scholar]
- Persson R. H., Bacchetti S., Smiley J. R. Cells that constitutively express the herpes simplex virus immediate-early protein ICP4 allow efficient activation of viral delayed-early genes in trans. J Virol. 1985 May;54(2):414–421. doi: 10.1128/jvi.54.2.414-421.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston C. M. Abnormal properties of an immediate early polypeptide in cells infected with the herpes simplex virus type 1 mutant tsK. J Virol. 1979 Nov;32(2):357–369. doi: 10.1128/jvi.32.2.357-369.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston C. M. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J Virol. 1979 Jan;29(1):275–284. doi: 10.1128/jvi.29.1.275-284.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston C. M., Cordingley M. G., Stow N. D. Analysis of DNA sequences which regulate the transcription of a herpes simplex virus immediate early gene. J Virol. 1984 Jun;50(3):708–716. doi: 10.1128/jvi.50.3.708-716.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston V. G. Fine-structure mapping of herpes simplex virus type 1 temperature-sensitive mutations within the short repeat region of the genome. J Virol. 1981 Jul;39(1):150–161. doi: 10.1128/jvi.39.1.150-161.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A high speed, high capacity homology matrix: zooming through SV40 and polyoma. Nucleic Acids Res. 1982 Aug 11;10(15):4765–4782. doi: 10.1093/nar/10.15.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea T. J., Timmins J. G., Long G. W., Post L. E. Mapping and sequence of the gene for the pseudorabies virus glycoprotein which accumulates in the medium of infected cells. J Virol. 1985 Apr;54(1):21–29. doi: 10.1128/jvi.54.1.21-29.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rixon F. J., Campbell M. E., Clements J. B. The immediate-early mRNA that encodes the regulatory polypeptide Vmw 175 of herpes simplex virus type 1 is unspliced. EMBO J. 1982;1(10):1273–1277. doi: 10.1002/j.1460-2075.1982.tb00024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rixon F. J., Clements J. B. Detailed structural analysis of two spliced HSV-1 immediate-early mRNAs. Nucleic Acids Res. 1982 Apr 10;10(7):2241–2256. doi: 10.1093/nar/10.7.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979 Mar;16(3):481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Schröder C. H., DeZazzo J., Knopf K. W., Kaerner H. C., Levine M., Glorioso J. A herpes simplex virus type 1 mutant with a deletion in the polypeptide-coding sequences of the ICP4 gene. J Gen Virol. 1985 Jul;66(Pt 7):1589–1593. doi: 10.1099/0022-1317-66-7-1589. [DOI] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R., McLachlan A. D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982 Jan 11;10(1):141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D., McMonagle E. C. Characterization of the TRS/IRS origin of DNA replication of herpes simplex virus type 1. Virology. 1983 Oct 30;130(2):427–438. doi: 10.1016/0042-6822(83)90097-1. [DOI] [PubMed] [Google Scholar]
- Stow N. D., McMonagle E. C., Davison A. J. Fragments from both termini of the herpes simplex virus type 1 genome contain signals required for the encapsidation of viral DNA. Nucleic Acids Res. 1983 Dec 10;11(23):8205–8220. doi: 10.1093/nar/11.23.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P. A fast homology program for aligning biological sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):447–455. doi: 10.1093/nar/12.1part2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson R. J., Clements J. B. A herpes simplex virus type 1 function continuously required for early and late virus RNA synthesis. Nature. 1980 May 29;285(5763):329–330. doi: 10.1038/285329a0. [DOI] [PubMed] [Google Scholar]
- Watson R. J., Preston C. M., Clements J. B. Separation and characterization of herpes simplex virus type 1 immediate-early mRNA's. J Virol. 1979 Jul;31(1):42–52. doi: 10.1128/jvi.31.1.42-52.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitton J. L., Clements J. B. Replication origins and a sequence involved in coordinate induction of the immediate-early gene family are conserved in an intergenic region of herpes simplex virus. Nucleic Acids Res. 1984 Feb 24;12(4):2061–2079. doi: 10.1093/nar/12.4.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]



