Abstract
Phosphatidylinositol (4,5)-bisphosphate (PIP2) is best known as a plasma membrane-bound regulatory lipid. While PIP2 and phosphoinositide-modifying enzymes coexist in the nucleus, their roles in the nucleus remain unclear. Here we show that the nuclear inositol polyphosphate multikinase (IPMK), which functions both as an inositol- and a PI3-kinase, interacts with the nuclear receptor SF-1 (NR5A1) and phosphorylates its bound ligand, PIP2. IPMK failed to recognize SF-1/PIP2 after blocking or displacing PIP2 from SF-1’s large hydrophobic pocket. In contrast to IPMK, p110 catalytic subunits of type 1 PI3-kinases were inactive on SF-1/PIP2. These and other in vitro analyses demonstrated specificity of IPMK for the SF-1/PIP2 protein/lipid complex. Once generated, SF-1/PIP3 is readily dephosphorylated by the lipid phosphatase PTEN. Importantly, decreasing IPMK or increasing PTEN expression greatly reduced SF-1 transcriptional activity. This ability of lipid kinases and phosphatases to alter the activity and directly remodel a non-membrane protein/lipid complex such SF-1/PIP2, establishes a new pathway for promoting lipid-mediated signaling in the nucleus.
Introduction
The coexistence of phosphoinositide phosphates (PIPn) and their modifying enzymes in the nucleus (1–3) suggests that similar to cytosolic signaling, lipid second messengers are generated to affect nuclear events. In turn, nuclear effector proteins are postulated to interpret or decode nuclear lipid signals by interacting with phosphoinositide headgroups, or in some instances, by binding the lipid tails of phosphoinositides (1). In addition to the presence of phosphoinositides on the inner nuclear envelop (4), others report that phosphoinositides, especially PI(4,5)P or PIP2 are present in detergent-treated nuclei stripped of their membrane (5). The existence of an intranuclear pool of PIP2 not associated with a classic lipid bilayer is further supported by colocalization of PIP2 and lipid kinases to nuclear speckles (6, 7). However, the importance of phosphoinositides in nuclear events remains enigmatic, in part because very few nuclear effector proteins have been identified. Nuclear proteins known to interact with PIP2 include the poly (A) polymerase Star-PAP that controls expression of select mRNAs (8), ALY, which mediates mRNA export (9), and BRG1, a component of the BAF chromatin remodeling complex (10). Also included in this short list are members of the NR5A subfamily of nuclear receptors, steroidogenic factor-1 (SF-1, NR5A1) and liver receptor homolog (LRH-1, NR5A2). Both are capable of binding PIP2 as well as other phospholipids, which are thought to act as regulatory ligands (11–15).
SF-1 is a key factor in regulating endocrine transcriptional networks whose targets include a host of steroidogenic enzymes and peptide hormones during both embryonic and adult life. NR5A1 human mutations are frequent, accounting for several monogenic forms of human disorders in sexual development (DSS), male infertility and premature ovarian failure (16). In our previous study, one such missense mutation (R255L) positioned near the entry of the ligand binding pocket diminished PIP2 binding and reduced SF-1 activity (12, 17). While many proteins recognize phospholipids by either interacting with the headgroup (AKT (18)) or enveloping the entire phospholipid (sfh-1, (19)), SF-1 binds phospholipids with the acyl chains buried deep in the hydrophobic ligand binding domain (LBD) leaving the head group exposed at the mouth of the pocket (Fig. 1A). This protein-lipid arrangement prompted us to ask if phosphoinositide 3-kinases (PI3-kinases) might target the solvent exposed 3 hydroxyl present in an SF-1/PIP2 complex. Among PI3-kinases, the best studied are p110s, which participate in cytosolic lipid signaling. More recently, the evolutionarily conserved inositol polyphosphate multikinase (IPMK/Ipk2/Arg82) was also shown to convert PI(4,5)P to PI(3,4,5)P in vitro and in vivo (20, 21). IPMK belongs to the large inositol kinase superfamily (22), and is essential for generating higher-ordered phosphorylated inositols (23, 24). Prior to the realization that IPMK possessed kinase activity, yeast genetic screens identified IPMK as a nuclear factor required for adaptive transcriptional responses (23, 25–27). More recently, mouse genetic studies revealed that IPMK is essential for normal embryonic development (21, 28). Here, we assessed whether PI3-kinases and phosphatases directly modify the exposed phospholipid headgroup in an SF-1/PIPn complex, and asked how such modifications might affect SF-1-mediated transcription.
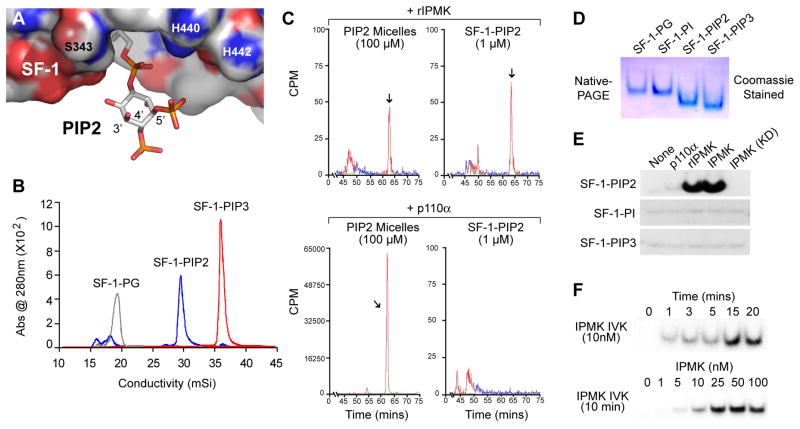
Fig. 1. IPMK Generates SF-1/PIP3 From SF-1/PIP2.
(A) SF-1/PIP2 model based on the crystal structure of SF-1/PC (PDB ID 3F7D) with SF-1 surface charges represented as: blue = basic, red = acidic, and PIP2 shown as stick representation. (B) Anion-exchange chromatography of purified mouse SF-1/PIPn complexes, as indicated. (C) In vitro 32P-labeled kinase (IVK) assays carried out with rIPMK and p110α/p85α (p110α) on SF-1/PIP2 complexes or PIP2-micelle substrates, analyzed by SAX-HPLC. Elution time of the PIP3 species is indicated (arrow). (D) Coomassie-stained SF-1/PIPn complexes from (B) resolved by 4–20% native-PAGE. (E) Autoradiography of IVK reactions analyzed by native-PAGE, using indicated SF-1/PIPn substrates (1 μM) and indicated enzymes (100nM). (F) IVK reactions with SF-1/PIP2 (1 μM) carried out at indicated times and concentrations of IPMK.
Results
IPMK Specifically Phosphorylates PIP2 when bound to SF-1
To determine if IPMK and/or p110 PI3-kinases phosphorylate PIP2 when bound to SF-1, SF-1 LBD-PIP2 (SF-1/PIP2) complexes were formed from purified recombinant protein as described previously (12) (Fig. 1B). Using in vitro kinase (IVK) assays and SF-1/PIP2 as substrate, rat IPMK (rIPMK) was able to generate PI(3,4,5)P (PIP3) as monitored by SAX-HPLC (Fig. 1C, upper panel). PIP3 signals were undetectable with p110 PI3-kinases (α or γ) or with a kinase-dead rIPMK (D127A) (Fig. 1C lower panels and Fig. S1), illustrating that IPMK, but not p110 PI3-kinases phosphorylate PIP2 when bound to SF-1. Using native-PAGE, which preserves the association between SF-1 and PIPn (Fig 1D), we confirmed that SF-1/PIP3 could be generated only from SF-1/PIP2 (Fig. 1E). Moreover, neither p110α nor the kinase-dead IPMK (D144A) exhibited such activity (Fig. 1E). IPMK phosphorylates SF-1/PIP2 in a dose-dependent manner, with saturation observed in typical reaction times (15 min) (Fig. 1F). Not surprisingly, the structurally related inositol kinase IP3K lacking PI3-kinase activity (29), failed to act on SF-1/PIP2 (Fig S1E).
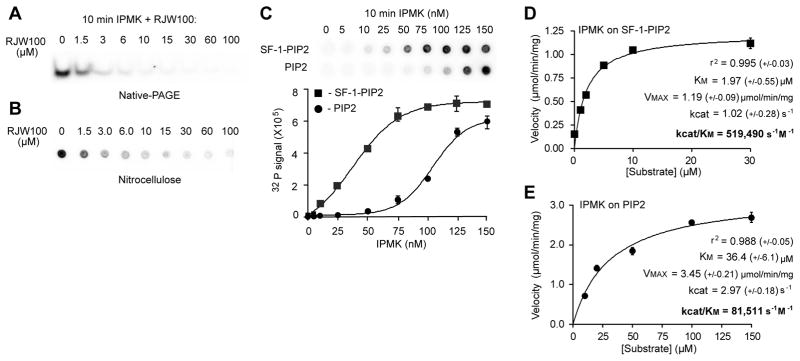
To better define the activity of IPMK on SF-1/PIP2 we used the newly described SF-1 ligand RJW100, which efficiently displaces PIPs from SF-1 (30). An RJW100-dependent drop in labeled SF-1/PIP3 after analyzing IVK reactions by native-PAGE (Fig 2A) and by nitrocellulose capture (Fig 2B); this later method detects PIP3 as well as SF-1/PIP3 (31). These data establish two important points: first, PIP2 and not the SF-1 protein is phosphorylated by IPMK, and second, in these IVK conditions, IPMK recognizes PIP2 only when bound to SF-1. Comparison of IPMK activities on SF-1/PIP2 and PIP2-micelles revealed a preference for the protein/lipid substrate, with activity saturating at lower concentrations than observed for PIP2 micelles (Fig. 2C). For these assays, the PIP2-micelle and SF-1/PIP2 substrates contained equal concentrations of PIP2 (1 μM) and preparation of the PIP2-micelle followed standard protocols as described in Materials and Methods. Kinetic parameters describing IPMK activity on SF-1/PIP2 (Fig 2D) are comparable to values obtained for inositol phosphates (32). When compared to PIP2-micelles, the specificity constant for SF-1/PIP2 was found to be higher (Fig 2E); for these latter studies the total phospholipid content was held constant at 100 μM and the ratio of PS carrier lipid to PIP2 adjusted accordingly.
Fig. 2. SF-1/PIP2 Is A Bona Fide Substrate Of IPMK In Vitro.
(A) Autoradiography of IVK reactions of IPMK on SF-1/PIP2 substrate (1 μM) with increasing concentrations of RJW100 (SF-1 ligand, μM) analyzed by native-PAGE. (B) IPMK IVK assays on SF-1/PIP2 (1 μM) with increasing concentrations of RJW100 ligand added to displace PIP2, analyzed by nitrocellulose capture assay and quantified by phosphorimaging as described in Materials and Methods. (C) Comparison of IVK reactions using increasing amounts of IPMK and either 1 μM SF-1/PIP2 or PIP2-micelles, which were generated with phosphatidylserine carrier lipid as described in Methods. (D) IPMK reaction velocities plotted against increasing concentrations of SF-1/PIP2 or (E) PIP2-micelles. Data were fit to both non-linear (Michaelis-Menton) and linear double-reciprocal (Lineweaver-Burke) curves by GraphPad Prism software, refer to Fig S2 for double reciprocal plots. Kinetic parameters describing each enzyme/substrate pair, as indicated, (+/− represents standard error except r2, where +/− represents the absolute sum of squares for the curve fit).
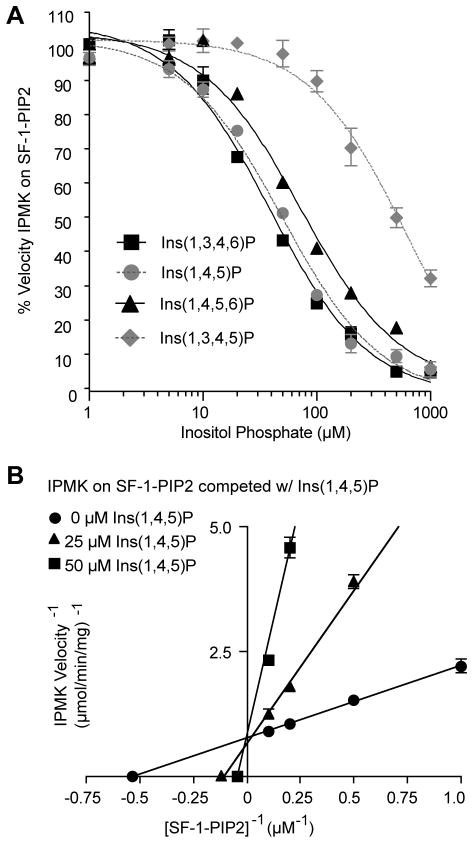
We next compared IPMK activity on SF-1/PIP2 with that of inositol phosphates (InsPs). Inositol phosphates compete with SF-1/PIP2 for IPMK activity, with the best InsPs substrates competing at a 5-fold molar concentration. IC50 values for four different InsPs correlated well with the rank order of the Vmax/Km values published for human IPMK (32), and competed for SF-1/PIP2 in the following order from best to worst competitor: Ins(1,3,4,6)P ≈ Ins(1,4,5)P > Ins(1,4,5,6)P ≫ Ins(1,3,4,5)P (Fig 3A). Further kinetic analyses with Ins(1,4,5)P, one of the higher affinity InsP substrates, established that SF-1/PIP2 and Ins(1,4,5)P bind the identical active site in IPMK (Fig. 3B). Collectively, these in vitro data show that SF-1/PIP2 is a valid substrate for IPMK.
Fig. 3. SF-1/PIP2 binds to the identical active site in IPMK as phosphorylated inostiols.
(A) IPMK reaction velocities on SF-1/PIP2 (10 μM) in the presence of competitor phosphorylated inositols (InsP), as indicated. (B) Double-reciprocal competition plots of IPMK reaction velocities determined with indicated concentrations of Ins(1,4,5))P and SF-1/PIP2, refer to Fig S2 for full data set.
Reducing IPMK Activity Impairs SF-1 Transcriptional Activity
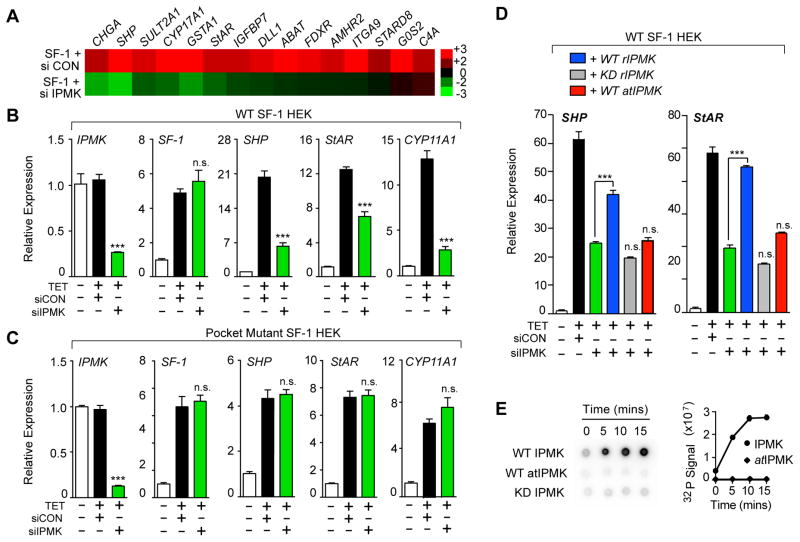
To assess whether recognition of SF-1/PIP2 by IPMK is biologically relevant in a cellular setting, we asked if expression of SF-1 target genes is altered after knocking-down or chemically inhibiting endogenous IPMK in human embryonic kidney HEK 293 cells. siIPMK resulted in a significant drop in established SF-1 targets including SHP, StAR and other transcripts, as judged by microarray profiling (Fig. 4A) and by RT-qPCR (Fig 4B). Importantly, the residual activity of an SF-1 pocket mutant (A270W, L345F), which prevents PIP2 from binding the LBD (12), is unaffected by siIPMK treatment (Fig 4C). As expected, overexpressing rIPMK restored SF-1 target gene expression after knocking-down endogenous human IPMK (siIPMK) in HEK 293 cells; SF-1 transcripts were unaffected (Fig S3). However, neither the rIPMK KD mutant nor the Arabidopsis thaliana IPMK (atIPMK) were able to rescue (Fig. 4D). Unlike human or rat IPMK, atIPMK lacks all PI3-kinase activity, as shown here for SF-1/PIP2 (Fig 4E), and previously for PIP2 vesicles (21).
Fig. 4. Reducing IPMK Levels Diminishes SF-1 Transcription.
(A) Heat map of selective SF-1 targets after profiling HEK 293 SF-1 cells transfected with control siRNA (siCON) or siIPMK followed by induction of SF-1 by tetracycline (TET). (B) Changes in transcript levels of indicated genes after siRNA treatment of HEK 293 cells expressing SF-1 wild type (WT) or (C) an SF-1 pocket mutant (A270W, L345F). (D) Relative expression of SHP and StAR transcripts as in (B), following transfection wild type or kinase dead (KD) rat IPMK (rIPMK), or Arabidopsis IPMK (atIPMK). (E) Phosphorimage and quantification of IPMK IVK reactions on SF-1/PIP2 analyzed by nitrocellulose capture, using different IPMK enzymes as indicated. Data represent mean +/− SEM, *** = p < 0.001 here and in all figures.
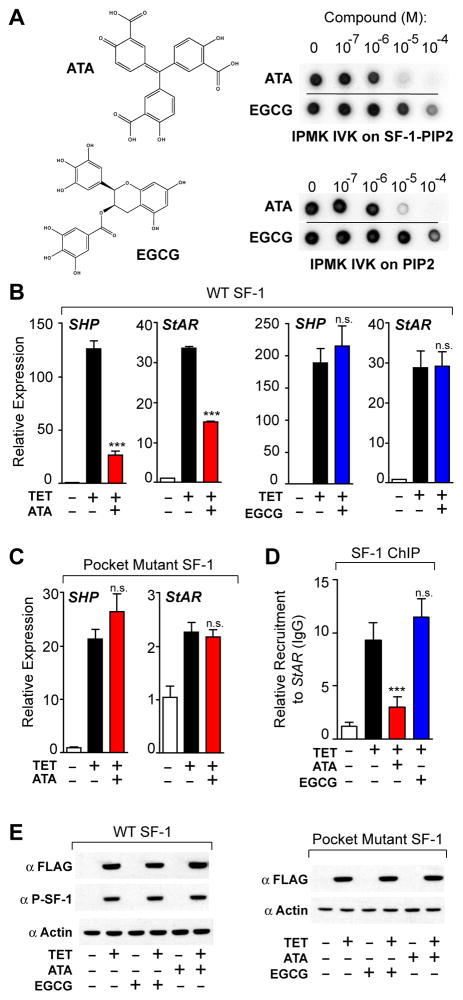
Activation of SF-1 targets was also monitored after adding a known inhibitor of IPMK, aurintricarboxcylic acid (ATA) and the related polyphenolic compound, epigallocatechin gallate (EGCG) (33), which acts as a broad-spectrum inhibitor of IP3 inositol kinases. Consistent with their pharmacological profiles, ATA but not EGCG inhibited IPMK in IVK assays (Fig. 5A, right panels), and selectively decreased SF-1 target genes (Fig 5B). Similar to results with siPMK, activity of the SF-1 pocket mutant was unaffected by ATA (Fig 5C). The PI3-kinase inhibitor wortmannin failed to inhibit IPMK in vitro (Fig S4C) and generated a significantly different pattern of SF-1 target gene expression compared to ATA or siIPMK (Fig S4D). ATA exposure also decreased recruitment of SF-1 to the StAR promoter by chromatin immunoprecipitation (ChIP, Fig. 5D). Levels of total SF-1 protein, phospho-S203 SF-1, and SF-1 transcripts, as well as SF-1 subcellular localization were unchanged following ATA exposure (Figs 5E and S4A, B). Taken together, these results suggest strongly that generation of SF-1/PIP3 from SF-1/PIP2 by IPMK enhances SF-1-mediated transcription.
Fig. 5. Chemical Inhibition Of IPMK Diminishes SF-1 Transcription.
(A) Structures of aurintricarboxcylic acid (ATA) and epigallocatechin gallate (EGCG) and their effect on IPMK IVK activity. (B) qPCR analysis of SHP and StAR transcripts following 50 μM ATA (red) or EGCG (Blue) treatment of HEK 293 cells expressing wild type or (C) pocket mutant SF-1. (D) Recruitment of SF-1 to the endogenous promoter of StAR as measured by ChIP-qPCR in identical cellular conditions as (B). (E) Western analysis of total SF-1 (anti-FLAG) and phosphorylated SF-1 (anti-P-SF-1) after ATA or EGCG treatment as in (B).
IPMK Phosphorylates and Interacts with Cellular SF-1/PIP2
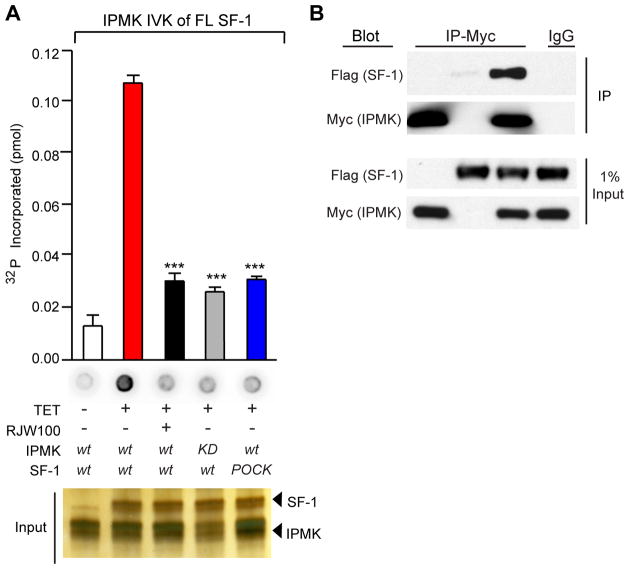
We next determined whether full-length (FL) SF-1 purified from cultured cells is associated with PIP2, and like the SF-1 LBD/PIP2 substrate, is recognized by IPMK. Indeed, wild type SF-1 isolated from HEK 293 cells (Fig. S5A) is readily phosphorylated by IPMK (Fig 6A). Significantly less phosphorylation was detected when the IPMK KD mutant, the SF-1 pocket mutant, or excess RJW100 ligand are used in IVK reactions (Fig 6A). These results, coupled with the fact that IPMK fails to phosphorylate SF-1 protein itself (Fig S5B), imply that SF-1/PIP2 is present in vivo, and is available for phosphorylation by IPMK. Finally, co-immunoprecipitation experiments showed that IPMK and SF-1 are capable of interacting in cells (Fig. 6B), further facilitating IPMK recognition of SF-1/PIP2.
Fig. 6. IPMK Interacts With And Phosphorylates Cellular SF-1.
(A) Nitrocellulose capture assays and quantification of IVK reactions carried out with SF-1 WT or pocket mutant purified from HEK 293 cells, and using wild type or kinase-dead IPMK (100nM). RJW100 SF-1 ligand (10 μM) was added to indicated reactions 10 min prior to addition of IPMK. (B) Anti-Myc immunoprecipitation (IP) of MYC-tagged-IPMK transiently transfected into HEK 293 cells expressing Flag-tagged SF-1, with control IgG shown in last lane. Corresponding input of total SF-1 or IPMK protein is shown in lower panel.
PTEN Hydrolyzes SF-1/PIP3 and Attenuates SF-1 Activity
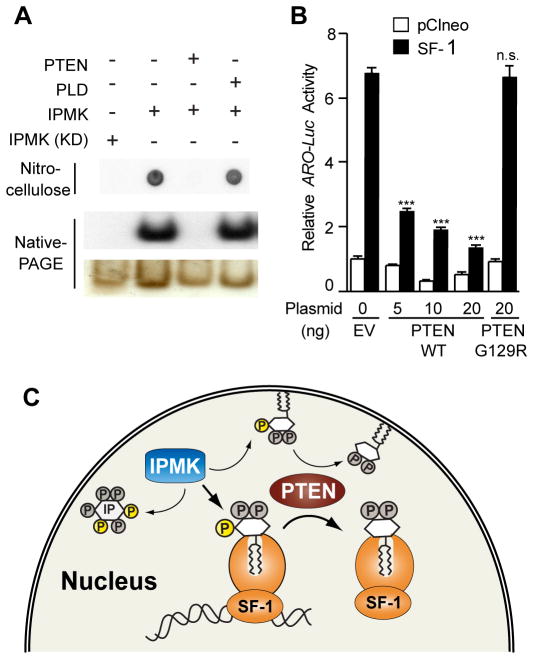
The emerging roles of PTEN in the nucleus (34) raised the possibility that this lipid phosphatase might regenerate SF-1/PIP2 by hydrolyzing PIP3 bound to SF-1. Indeed, PTEN efficiently removed the radiolabeled 3′-phosphate from IPMK IVK reactions on SF-1/PIP2 resulting in an upward shift on native-PAGE, whereas incubation with phospholipase D had no effect (Fig. 7A). Kinetic studies of PTEN activity on SF-1/PIP3 revealed apparent Km and Vmax values of 1.0 μM and 0.7 μmol/min/mg (Fig S2), compared with the published values of 5 μM and 1.8 μmol/min/mg for PIP3 in phosphatidylcholine vesicles (35). Moreover, coexpression of wild type PTEN and SF-1 in human PTEN-negative Ishikawa cells severely compromised SF-1 activity (Fig. 7B), suggesting that IPMK and PTEN oppose each other to enhance or reduce the functional output of SF-1, respectively. Thus, similar to membrane bound phospholipids, the headgroups of PIP2/PIP3 bound to SF-1 appear perfectly posed for modification by both lipid kinases and lipid phosphatases.
Fig. 7. PTEN Opposes IPMK Activity on SF-1/PIP2.
(A) Radiolabeled SF-1/PIP3, analyzed by nitrocellulose capture or native-PAGE after incubating with phosphatase PTEN or phospholipase D (PLD). (B) Aromatase-luciferase (ARO-Luc) reporter activity after transfecting Ishikawa cells with wild type SF-1 or control vector, plus PTEN or an inactive PTEN mutant (G129R). (C) Schematic depicting the multiple functions of IPMK as a soluble InsPs kinase, and as a PI3-kinase acting on both membrane bound PIP2 and SF-1/PIP2. SF-1 transcriptional output is affected after IPMK and PTEN modify the solvent-exposed phospholipid head group present in the bound PIP2 or PIP3 ligands.
Discussion
Our findings suggest that in addition to regulating inositol phosphate and phosphoinositide metabolism, IPMK also modulates gene expression by selective phosphorylation of PIP2 when bound to a nuclear protein, such as SF-1 (Fig. 7C). IPMK was found to efficiently and selectively phosphorylate PI(4,5)P when complexed with recombinant SF-1 LBD, or when bound to purified full length SF-1. The ability of IPMK to act on cell-derived SF-1 illustrates for the first time that a substantial fraction of SF-1 is normally bound to PIP2 in cells. By contrast, IPMK no longer recognizes SF-1/PIP2 after displacing the phospholipid ligand with a synthetic ligand for NR5A receptors, or after introducing bulky amino acid substitutions that fill the large hydrophobic ligand binding cavity of SF-1. Thus, in the context of SF-1, IPMK targets SF-1/PIP2 only when PIP2 is incorporated into the LBD of SF-1. In vitro competition studies also showed that the solvent exposed PIP2 headgroup in a SF-1/PIP2 complex competes with inositol polyphosphates for the same binding site on IPMK. When taken together with our cellular data, we posit that protein-phosphoinositide complexes, such as SF1/PIP2 represent a new bona fide class of substrates for the multikinase, IPMK.
Our results add to the remarkable versatility of this kinase, which is proposed to be the oldest member of the inositol kinase superfamily (36). Based on the previously elucidated NR5A/phospholipid structures, we suggest that the 3′ hydroxyl in the phospholipid headgroup is optimally positioned for recognition by IPMK. In contrast to IPMK, we found that other kinases known to phosphorylate the 3′-hydroxy group were inactive on the SF-1/PIP2 substrate; these include human IP3-kinase, Arabidopsis IPMK, and p110 PI3-kinases. Thus, the 3′ hydroxyl group in SF-1/PIP2 cannot be the sole determinant for IPMK recognition of this protein/lipid substrate. Alignment of human IPMKs with other members of inositol phosphate kinase superfamily identified an IPMK-specific loop (S260-E375) that resides outside of the highly conserved core catalytic domain (32, 36). Interestingly, this loop is absent in Arabidopsis IPMK. Structure-function studies on yeast IPMK also found that this loop is required for protein-protein interactions and normal transcriptional responses (26). Clearly, it will be of interest to determine if these IPMK-specific regions mediate interaction between IPMK and SF-1 and facilitate recognition of the SF-1/PIP2 substrate.
The multi-faceted nature of IPMK raises the possibility that the transcriptional effects of IPMK on SF-1 reflect indirect mechanisms of IPMK on phospholipid and/or inositol metabolism. However, as mentioned above, PI3-kinase inhibitor wortmannin elicited a significantly different transcriptional response than ATA or siIPMK, consistent with its failure to inhibit IPMK. The possibility that siIPMK simply alters inositol metabolism to lower SF-1 activity is a valid suggestion, especially in light of the recent discovery that Ins(1,4,5,6)P4 functions as an modulatory bridging factor for HDAC3 and the histone deacetylase activation domain of the NCOR2/SMRT corepressor (37). However, despite the fact that atIPMK retains the inositol phosphate kinase activity of mammalian IPMKs, this plant IPMK fails to restore SF-1 activity after human siIPMK knockdown. Moreover, SF-1 activity is inert to the pan IP3-kinase inhibitor EGCG, again showing that SF-1 is fully functional even after disturbing general inositol metabolism. Finally, given our data with the SF-1 pocket mutant and the fact that IPMK interacts with SF-1, we favor a model whereby IPMK regulates SF-1 activity by directly phosphorylating its bound phospholipid ligand, PIP2.
This new type of IPMK activity might help explain the more intricate roles of IPMK in transcription, especially in aspects of chromatin remodeling (38). That IPMK and PTEN directly modify a protein-bound lipid to affect transcription is a clear departure from their established roles in membrane phosphoinositide and inositol metabolism (Fig 7C). As outlined by Anderson (1), these new activities would be predicted to compartmentalize nuclear PIP2 and PIP3 lipid signals, and analogous to the docking roles of membrane-bound phosphoinositides, may help to bridge SF-1 to other PIPn-interacting proteins, such as the BAF remodeling complex (39), or chromatin associated PH-domain proteins (40). Thus, we speculate that the exposed inositol moiety in SF-1/PIPn plays a similar role as inositol tetraphosphate does in bridging corepressor complexes. Such interactions may account for the observed decrease in chromatin recruitment of SF-1 after chemically inhibiting IPMK with ATA. Interestingly, silencing DAG kinase θ also reduced SF-1 activity and recruitment to target gene promoters (41). Beyond the phosphoinositide cycle, others have shown that nuclear sphingolipids and their kinases also affect histone acetylation and chromatin recruitment (41, 42). It remains to be determined whether these and other lipid modifying enzymes regulate nuclear factors by directly remodeling their bound lipid cargo, as shown here for IPMK and SF-1/PIP2. Although further research is needed to understand how nuclear proteins, such as SF-1 decode nuclear lipid signaling to affect gene expression, our study begins to unravel the longstanding enigma of nuclear phosphoinositides.
Materials and Methods
Purification of LBD SF-1/PIPn or Cell-Derived FL SF-1/PIPn
SF-1 ligand binding domain (LBD) protein was purified to homogeneity as previously described (11). Briefly, mouse SF-1 constructs spanning amino acids 219–462 were expressed in E. coli, purified and complexed with dipalmitoyl (C16, fully saturated) PI, PI(4,5)P (PIP2), or PI(3,4,5)P (PIP3) to generate SF-1/PIPn complexes. Protein/lipid complexes were purified from free PIPs and unbound SF-1 LBD using Mono Q chromatography on an AKTA FPLC system (Pharmacia, GE biosciences), as described (12). All PIPs were purchased from Cayman Chemical (Ann Arbor, MI), and stored under a vacuum as lyophilized powder or under N2 as a 1g/L solution in ddH2O.
To purify full length SF-1 from cells, 3X-FLAG tagged full-length WT SF-1 was immunopurified from two-15 cm plates, each containing 15×106 HEK 293 WT or pocket mutant (A270W, L345F) SF-1 cells using M2-Anti-FLAG agarose resin (Sigma). Cells were harvested at 4°C in native lysis buffer (50 mM HEPES pH7.5, 400 mM NaCl, 0.2 mM EDTA, 5 mM MgCl2, 2 mM CHAPS, 50 mM DTT, and protease inhibitors [Roche]). Cleared lysates were incubated with M2 FLAG resin (Sigma, 30 μL of 50% slurry) for 45 minutes at 4°C, washed and eluted (50 mM HEPES pH7.5, 100 mM NaCl, 2 mM CHAPS, 50 mM DTT and protease inhibitors) with 150 μg/mL 3X FLAG peptide (Sigma). Eluates (~47 ng SF-1) were used in IPMK IVK assays, as described below, except that 1 μM cold ATP was used. For RJW100-treated IVK reactions, 20 μM RJW100 SF-1 ligand was added to IVK reactions for 15 minutes at 37°C, followed by addition of ATP to initiate IVK reactions, which were analyzed by nitrocellulose capture and by SDS-PAGE (BioRad, Hercules, CA).
IPMK purification and IVK reactions
Wild type human (accession Q8NFU5) and rat (accession AY014898) 6XHis-tagged IPMK constructs in pTRCb (gift of A. Resnick, U Penn) were expressed in E. coli, and purified as described (20). Kinase dead (KD) mutants included rat IPMK (rIPMK, D127A), and human IPMK (IPMK, D144A) (20). IPMK (EC = 2.7.1.151) IVK reactions were carried out in 50 mM HEPES pH 7.5, 5 mM MgCl2, 10 μCi of 32P-γATP (Perkin Elmer, Boston, MA) and 10 μM cold ATP (Roche, Mannheim, Germany) at 37°C in either 25 or 50 μL reactions, except in kinetic studies (see below). IVK using human p110α/p85α (Millipore, Billerica, MA) or human p110γ (Sigma, St. Louis, MO) were performed according to manufacturer’s recommendations. PIP2 micelles substrates were prepared using phosphatidylserine (PS) carrier lipid with the total lipid content equal to 100 μM. Lyophilized C16 saturated (dipalmitoyl) PIPs (Cayman) were kept under vacuum in borosilicate glass and directly resuspended in ddH2O to 1 g/L, sonicated and stored under N2 at 4°C.
For HPLC analysis of IVK reactions, lipids were extracted using methanol:chloroform (2:1), de-acylated in methylamine:methanol:n-butanol:H2O (26:45:11:16), then polar glyceroinositol head-groups subjected to strong anion exchange HPLC using a Whatman PartiSphere SAX column over a gradient of NH4H2PO4, monitored in real time using a Packard flow scintillation detector. Reference retention times included the product of p110α/p85α and p110γ activity on PIP2 micelles and free radiolabeled 32P-γATP.
For native-PAGE analysis, total IVK reactions were loaded and resolved on TBE-buffered 4–20% PAGE polyacrylamide gels (Lonza, Basel, Switzerland) followed by autoradiography or phosphorimaging. Nitrocellulose capture analysis of IVK reactions was performed essentially as described (31) with 10% of the entire IVK reaction spotted onto nitrocellulose (0.2 micron pore size), followed by 1M NaCl and 1% phosphoric acid washes. Membranes were phosphorimaged to quantify signal.
For PTEN assays, IPMK IVK reactions were stopped with excess ATP (10 mM) and PTEN (0.3μg) and 10 mM DTT added, and incubated for 15 minutes at 37°C. Reactions were analyzed by nitrocellulose capture assay and by native-PAGE. For kinetic analyses, PTEN reactions were coupled to p110α to quantify PTEN generation of PIP2 from SF-1/PIP3. PTEN reactions on SF-1/PIP3 were stopped by a methanol:chloroform (2:1) extraction, dried down, and the PIP2 content indirectly quantified by standard p110α IVK reactions, followed by nitrocellulose capture analyses.
IPMK kinetic parameters were measured in the linear range with respect to time and enzyme concentrations as described above using the indicated substrate concentrations and 10 mM cold ATP, analyzed by nitrocellulose capture, then subjected to both linear and non-linear curve fits using GraphPad Prism; both fits yielded similar a value. Competition with IPns (Cayman Chemical) freshly suspended in 50 mM HEPES (7.5) and 5 mM MgCl2, were analyzed by nitrocellulose capture; IPn are not retained.
Cell Lines, siRNA and Luciferase Assays
3XFLAG N-terminally tagged mSF-1 cDNA (wild type or pocket mutant (A270W, L345F) constructs were expressed in Flip-in, T-Rex tetracycline (TET)-inducible human embryonic kidney 293 (HEK 293) cells (Invitrogen Corporation, Carlsbad, CA), maintained at 37°C in 5% CO2 in DMEM supplemented with 10% tetracycline-certified-free FBS (Hyclone, Logan, UT), SF-1 protein was induced for 18 hrs using 50 ng/mL TET in 70% EtOH.
For siRNA experiments, HEK 293 WT or pocket mutant SF-1 cells were reverse transfected with 20 nM SmartPool Silencer siRNA directed against human IPMK, or control siRNA #1, (Dharmacon, Lafayette, CO), using RNAiMAX transfection reagent (Invitrogen), according to the manufacturer’s protocol. Fifty-four hours post-siRNA transfection, SF-1 was induced for 18 hours, cells harvested and analyzed as described below. For rescue experiments, cells were transfected with 20 ng of Rattus norvegicus or Arabidopsis thaliana IPMK constructs (gift of A. Resnick) using RNAiMAX transfection reagent. HEEBO microarrays or RT-qPCR was carried out using cDNA as described below.
For cellular PTEN experiments, PTEN-negative Ishikawa cells (human endometrial carcinoma) were cotransfected with an ARO-Luc reporter (250 ng), 3X FLAG-mSF-1 (pCINEO) and WT or phosphatase dead (G129R) human PTEN (in pCDNA3) using FuGENE 6 (Roche) and analyzed 24 hrs after transfection for luciferase activity using a Ventas Microplate Luminometer (Turner Biosystems, Sunnyvale, CA). PTEN vectors were a generous gift from D. Stokoe.
Small Molecules
RJW100 SF-1 ligand was generously provided by Richard J. Whitby (Southampton University, UK) (30), and was used in a 4% final DMSO (Sigma Ultrapure) solution for all IVK reactions; PIP2 binding to SF-1 is unaffected by 4% DMSO (30). SF-1/PIP2 complexes (1 μM) were incubated with serially diluted RJW100 for 30 minutes at 37°C in IPMK assay buffer (see above), followed by addition of IPMK (141 nM) plus 10 μCi 32P-ATP. After 15 minutes IVK reactions quenched by spotting onto nitrocellulose and washing, as described above. Light-sensitive aurintricarboxcylic acid (ATA, Sigma) was kept in the dark. Epigallocatechin gallate (EGCG) and wortmannin were also from Sigma. Four hours after SF-1 induction with TET (50 ng/mL), ATA or EGCG was added to HEK 293 SF-1 cells in 6 well plates for 14 hours at indicated concentrations, then harvested and analyzed.
Other Methods
HEEBO (Human Exonic Evidence Based Open-source) microarray analysis and qPCR were carried out as described previously (43). All primer sets were validated and designed to cross an intron/exon boundary, primer sequences are provided in Table S1. ChIP was carried out as described (43) using HEK 293 SF-1 cells treated with TET (50 ng/ml) followed by addition of DMSO vehicle, 50 μM ATA or EGCG for 14 hours. Primer sets are listed in Table S2.
For Co-IP studies, HEK 293 SF-1 cells grown in a 10 cm dish were transfected with 5 μg of Myc-tagged human IPMK expression vector using Fugene HD (Roche), and 36 hrs later SF-1 was induced with TET (50 ng/ml) for 16 hours. Cells were lysed in 50 mM Tris-Cl (8.0), 150 mM NaCl, 1mM EDTA, 0.5% NP-40 with protease inhibitors, cleared, and protein lysate (1 μg) was immunoprecipitation used for anti-c-Myc agarose (Sigma) and analyzed by Western blotting (anti-Myc, 1:2,500 [Bishop lab, UCSF]; anti-Flag M2, 1:10,000 [Sigma]). Other antibodies for Western blots included anti-phospho S203 SF-1 (custom antibody) at 1:1000 and anti-actin (Sigma) at 1:4000 using the Pierce Super Signal West Pico Enhanced chemiluminescence detection kit (Thermo Scientific, Waltham, MA).
Supplementary Material
Acknowledgments
We especially thank Drs. R. Whitby, A. Resnick, J. York, and G. Miller for supplying critical reagents, and Drs. M. Feldman, E. Sablin, E. Faivre, G. Narlikar, C. Craik and D. Julius for advice and for critically evaluating the manuscript. This work was supported by NIGMS K12 IRACDA Postdoctoral Fellowship to R.D.B. and NIDDK DK072246 to H.A.I.
References
- 1.Barlow CA, Laishram RS, Anderson RA. Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends Cell Biol. 2010;20:25. doi: 10.1016/j.tcb.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Divecha N, Banfic H, Irvine RF. The polyphosphoinositide cycle exists in the nuclei of Swiss 3T3 cells under the control of a receptor (for IGF-I) in the plasma membrane, and stimulation of the cycle increases nuclear diacylglycerol and apparently induces translocation of protein kinase C to the nucleus. The EMBO journal. 1991;10:3207. doi: 10.1002/j.1460-2075.1991.tb04883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irvine RF. Nuclear lipid signalling. Nat Rev Mol Cell Biol. 2003;4:349. doi: 10.1038/nrm1100. [DOI] [PubMed] [Google Scholar]
- 4.Smith CD, Wells WW. Phosphorylation of rat liver nuclear envelopes. II. Characterization of in vitro lipid phosphorylation. J Biol Chem. 1983;258:9368. [PubMed] [Google Scholar]
- 5.Vann LR, Wooding FB, Irvine RF, Divecha N. Metabolism and possible compartmentalization of inositol lipids in isolated rat-liver nuclei. Biochem J. 1997;327(Pt 2):569. doi: 10.1042/bj3270569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boronenkov IV, Loijens JC, Umeda M, Anderson RA. Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Molecular biology of the cell. 1998;9:3547. doi: 10.1091/mbc.9.12.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osborne SL, Thomas CL, Gschmeissner S, Schiavo G. Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. Journal of cell science. 2001;114:2501. doi: 10.1242/jcs.114.13.2501. [DOI] [PubMed] [Google Scholar]
- 8.Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- 9.Okada M, Jang SW, Ye K. Akt phosphorylation and nuclear phosphoinositide association mediate mRNA export and cell proliferation activities by ALY. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8649. doi: 10.1073/pnas.0802533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rando OJ, Zhao K, Janmey P, Crabtree GR. Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:2824. doi: 10.1073/pnas.032662899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, Bynum JM, Madauss K, Montana V, Lebedeva L, Suzawa M, Williams JD, Williams SP, Guy RK, Thornton JW, Fletterick RJ, Willson TM, Ingraham HA. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 12.Sablin EP, Blind RD, Krylova IN, Ingraham JG, Cai F, Williams JD, Fletterick RJ, Ingraham HA. Structure of SF-1 bound by different phospholipids: evidence for regulatory ligands. Mol Endocrinol. 2009;23:25. doi: 10.1210/me.2007-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Choi M, Cavey G, Daugherty J, Suino K, Kovach A, Bingham NC, Kliewer SA, Xu HE. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Molecular cell. 2005;17:491. doi: 10.1016/j.molcel.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Mullaney BC, Blind RD, Lemieux GA, Perez CL, Elle IC, Faergeman NJ, Van Gilst MR, Ingraham HA, Ashrafi K. Regulation of C. elegans fat uptake and storage by acyl-CoA synthase-3 is dependent on NR5A family nuclear hormone receptor nhr-25. Cell metabolism. 2010;12:398. doi: 10.1016/j.cmet.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JM, Lee YK, Mamrosh JL, Busby SA, Griffin PR, Pathak MC, Ortlund EA, Moore DD. A nuclear-receptor-dependent phosphatidylcholine pathway with antidiabetic effects. Nature. 2011;474:506. doi: 10.1038/nature10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bashamboo A, Ferraz-de-Souza B, Lourenco D, Lin L, Sebire NJ, Montjean D, Bignon-Topalovic J, Mandelbaum J, Siffroi JP, Christin-Maitre S, Radhakrishna U, Rouba H, Ravel C, Seeler J, Achermann JC, McElreavey K. Human male infertility associated with mutations in NR5A1 encoding steroidogenic factor 1. Am J Hum Genet. 2010;87:505. doi: 10.1016/j.ajhg.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biason-Lauber A, Schoenle EJ. Apparently normal ovarian differentiation in a prepubertal girl with transcriptionally inactive steroidogenic factor 1 (NR5A1/SF-1) and adrenocortical insufficiency. Am J Hum Genet. 2000;67:1563. doi: 10.1086/316893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas CC, Deak M, Alessi DR, van Aalten DM. High-resolution structure of the pleckstrin homology domain of protein kinase b/akt bound to phosphatidylinositol (3,4,5)-trisphosphate. Current biology: CB. 2002;12:1256. doi: 10.1016/s0960-9822(02)00972-7. [DOI] [PubMed] [Google Scholar]
- 19.Schaaf G, Ortlund EA, Tyeryar KR, Mousley CJ, Ile KE, Garrett TA, Ren J, Woolls MJ, Raetz CR, Redinbo MR, Bankaitis VA. Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Molecular cell. 2008;29:191. doi: 10.1016/j.molcel.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Resnick AC, Snowman AM, Kang BN, Hurt KJ, Snyder SH, Saiardi A. Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12783. doi: 10.1073/pnas.0506184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maag D, Maxwell MJ, Hardesty DA, Boucher KL, Choudhari N, Hanno AG, Ma JF, Snowman AS, Pietropaoli JW, Xu R, Storm PB, Saiardi A, Snyder SH, Resnick AC. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1391. doi: 10.1073/pnas.1017831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes W, Jogl G. Crystal structure of inositol phosphate multikinase 2 and implications for substrate specificity. J Biol Chem. 2006;281:38109. doi: 10.1074/jbc.M606883200. [DOI] [PubMed] [Google Scholar]
- 23.Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 24.Saiardi A, Caffrey JJ, Snyder SH, Shears SB. Inositol polyphosphate multikinase (ArgRIII) determines nuclear mRNA export in Saccharomyces cerevisiae. FEBS Lett. 2000;468:28. doi: 10.1016/s0014-5793(00)01194-7. [DOI] [PubMed] [Google Scholar]
- 25.Dubois E, Messenguy F. Pleiotropic function of ArgRIIIp (Arg82p), one of the regulators of arginine metabolism in Saccharomyces cerevisiae. Role in expression of cell-type-specific genes. Molecular & general genetics: MGG. 1994;243:315. doi: 10.1007/BF00301067. [DOI] [PubMed] [Google Scholar]
- 26.El Alami M, Messenguy F, Scherens B, Dubois E. Arg82p is a bifunctional protein whose inositol polyphosphate kinase activity is essential for nitrogen and PHO gene expression but not for Mcm1p chaperoning in yeast. Mol Microbiol. 2003;49:457. doi: 10.1046/j.1365-2958.2003.03562.x. [DOI] [PubMed] [Google Scholar]
- 27.Steger DJ, Haswell ES, Miller AL, Wente SR, O’Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frederick JP, Mattiske D, Wofford JA, Megosh LC, Drake LY, Chiou ST, Hogan BL, York JD. An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8454. doi: 10.1073/pnas.0503706102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller GJ, Hurley JH. Crystal structure of the catalytic core of inositol 1,4,5-trisphosphate 3-kinase. Molecular cell. 2004;15:703. doi: 10.1016/j.molcel.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Whitby RJ, Stec J, Blind RD, Dixon S, Leesnitzer LM, Orband-Miller LA, Williams SP, Willson TM, Xu R, Zuercher WJ, Cai F, Ingraham HA. Small molecule agonists of the orphan nuclear receptors steroidogenic factor-1 (SF-1, NR5A1) and liver receptor homologue-1 (LRH-1, NR5A2) J Med Chem. 2011;54:2266. doi: 10.1021/jm1014296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knight ZA, Feldman ME, Balla A, Balla T, Shokat KM. A membrane capture assay for lipid kinase activity. Nat Protoc. 2007;2:2459. doi: 10.1038/nprot.2007.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang SC, Miller AL, Feng Y, Wente SR, Majerus PW. The human homolog of the rat inositol phosphate multikinase is an inositol 1,3,4,6-tetrakisphosphate 5-kinase. J Biol Chem. 2002;277:43836. doi: 10.1074/jbc.M206134200. [DOI] [PubMed] [Google Scholar]
- 33.Mayr GW, Windhorst S, Hillemeier K. Antiproliferative plant and synthetic polyphenolics are specific inhibitors of vertebrate inositol-1,4,5-trisphosphate 3-kinases and inositol polyphosphate multikinase. J Biol Chem. 2005;280:13229. doi: 10.1074/jbc.M500545200. [DOI] [PubMed] [Google Scholar]
- 34.Planchon SM, Waite KA, Eng C. The nuclear affairs of PTEN. Journal of cell science. 2008;121:249. doi: 10.1242/jcs.022459. [DOI] [PubMed] [Google Scholar]
- 35.McConnachie G, Pass I, Walker SM, Downes CP. Interfacial kinetic analysis of the tumour suppressor phosphatase, PTEN: evidence for activation by anionic phospholipids. Biochem J. 2003;371:947. doi: 10.1042/BJ20021848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shears SB. How versatile are inositol phosphate kinases? Biochem J. 2004;377:265. doi: 10.1042/BJ20031428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watson PJ, Fairall L, Santos GM, Schwabe JW. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481:335. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen X, Xiao H, Ranallo R, Wu WH, Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- 39.Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 40.Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Molecular cell. 2004;13:677. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 41.Li D, Urs AN, Allegood J, Leon A, Merrill AH, Jr, Sewer MB. Cyclic AMP-stimulated interaction between steroidogenic factor 1 and diacylglycerol kinase theta facilitates induction of CYP17. Mol Cell Biol. 2007;27:6669. doi: 10.1128/MCB.00355-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, Luo C, Marmorstein R, Kordula T, Milstien S, Spiegel S. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 2009;325:1254. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee FY, Faivre EJ, Suzawa M, Lontok E, Ebert D, Cai F, Belsham DD, Ingraham HA. Eliminating SF-1 (NR5A1) sumoylation in vivo results in ectopic hedgehog signaling and disruption of endocrine development. Developmental cell. 2011;21:315. doi: 10.1016/j.devcel.2011.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.