Abstract
Salmonella specifically localize to malignant tumors in vivo, a trait potentially exploitable as a delivery system for cancer therapeutics. To characterize mechanisms and genetic responses of Salmonella during interaction with living neoplastic cells, we custom designed a promoterless transposon reporter containing bacterial luciferase. Analysis of a library containing 7,400 independent Salmonella transposon insertion mutants in co-culture with melanoma or colon carcinoma cells identified five bacterial genes specifically activated by cancer cells: adiY, yohJ, STM1787, STM1791, and STM1793. Experiments linked acidic pH, a common characteristic of the tumor microenvironment, to a strong, specific and reversible stimulus for activation of these Salmonella genes in vitro and in vivo. Indeed, a Salmonella reporter strain encoding a luciferase transgene regulated by the STM1787 promoter, which contains a tusp motif, showed tumor-induced bioluminescence in vivo. Furthermore, Salmonella expressing Shiga toxin from the STM1787 promoter provided potent and selective anti-tumor activity in vitro and in vivo, demonstrating the potential for a conditional bacterial-based tumor-specific therapeutic.
Keywords: Salmonella, cancer, transposon, screen, reporter, bioluminescence
Introduction
The evolving and highly heterogeneous landscape of tumor genetics and the tumor microenvironment pose a significant challenge for treating advanced solid tumors (1). Many characteristics of the tumor microenvironment, such as hypoxia, acidic pH, and a disorganized vascular architecture, limit delivery and efficacy of therapeutics and radiation treatments (2). Additionally, tumors undergoing targeted molecular therapy often relapse due to the utilization of autonomous parallel-redundant signaling pathways (3). Beyond the primary tumor, identifying disseminated disease that has metastasized to various organ sites is challenging, and systemically treating cancer often produces off-target toxicities. The ultimate anti-tumor therapy is one that overcomes these physiologic obstacles while simultaneously targeting tumors and avoiding normal tissue toxicity.
The remarkable ability of commensal and pathogenic bacterial strains to localize and preferentially grow within tumors has been well documented (4). The immune-privileged, hypoxic and nutrient-rich ‘tumor soil’ facilitates colonization by facultative anaerobic bacteria (5). These observations have spurred research into the diagnostic and therapeutic potential of genetically engineered and attenuated therapeutic strains of bacteria such as Salmonella, Listeria and Clostridium (5). Salmonella is one of the most studied of therapeutic bacteria, and upon systemic administration, is able to colonize xenograft tumors at rates 1,000 times greater than that of other organs, thereby abrogating tumor growth (6, 7). A firm understanding of the genetic programs involved in normal pathogenesis, characterization of spatiotemporal kinetics and dynamics during intra-tumoral colonization in vivo, genetic tractability, as well as the oncolytic capacity of Salmonella typhimurium have made Salmonella strains ideal candidates for anti-cancer bacterial development (8).
S. typhimurium by itself can illicit an anti-tumoral response through several potentially separate but synergistic mechanisms. First, as a pathogenic and cytotoxic bacterium, S. typhimurium can induce apoptosis of cancer cells (9). Second, pathogen-associated molecular patterns (PAMPs) of S. typhimurium, such as lipopolysaccharide (LPS) and flagellin, are capable of activating innate immunity by initiating pro-inflammatory TLR-MyD88/TRIF-NF-κB signaling cascades (10). Third, intracellular Salmonella flagellin can also enhance an anti-tumor adaptive immune response caused by the associative recognition with cancer cell antigen. The resulting signaling cascades ultimately augment antigen presentation of dendritic cells (DC), thereby promoting T cell clonal expansion and differentiation which leads to an associative recognition of the cancer cell with the PAMPs of Salmonella (11, 12). Finally, despite the initial tumor regression, these tumors may eventually relapse, which has spurred the development of Salmonella as a delivery vehicle for anti-cancer co-therapies (13). Indeed, Salmonella have been used as tumor-specific vectors for gene transfer of RNAi or suicide genes, as well as targeted expression of apoptosis-inducing biologics, such as TRAIL, FASL and the bacterial toxin, Cytolysin A, all of which display pronounced anti-tumor affects in vivo (5).
However, few studies have investigated the specific genetic responses of Salmonella to tumor cells and bacterial mechanisms regulating these atypical “host” interactions. To address these quires, we engineered a bioluminescent transposon reporter-trap to screen a S. typhimurium library for genes specifically regulated by co-culture with malignant cells in vitro. Five genes were identified by the screen and their promoter sequences were found to be specifically activated by the acidic microenvironment associated with cancer cells in vitro and tumors in vivo. Finally, we utilized the most pH-sensitive promoter sequence to demonstrate the utility of tumor-regulated Salmonella promoters to conditionally regulate the expression of a toxic tumor transgene in vitro and in vivo.
Results
A high-throughput screen to identify tumor cell-induced gene activation events in Salmonella
To conduct a large-scale, unbiased screen for genes up-regulated by contact with malignant cells, we used a Tn5-based transposon as the backbone of a luxAB reporter construct. We chose to use the bacterial luciferase enzyme genes (luxAB) only, in contrast to the full bacterial luciferase operon (luxCDABE), because the size of the transposon containing the full operon prohibited efficient chromosomal integration, while using only the luxAB genes allowed for efficient genomic insertion of the transposon. The transposon was designed to restrict reporter gene expression to only those chromosomal integration sites downstream of an active promoter. A kanamycin resistance cassette with a constitutive promoter was also included to select for integration into the chromosome (Figure 1a). After construction, the purified transposon was electroporated into S. typhimurium strain SB300A1(14) for random chromosomal integration, producing a 7,400 clone bacterial library.
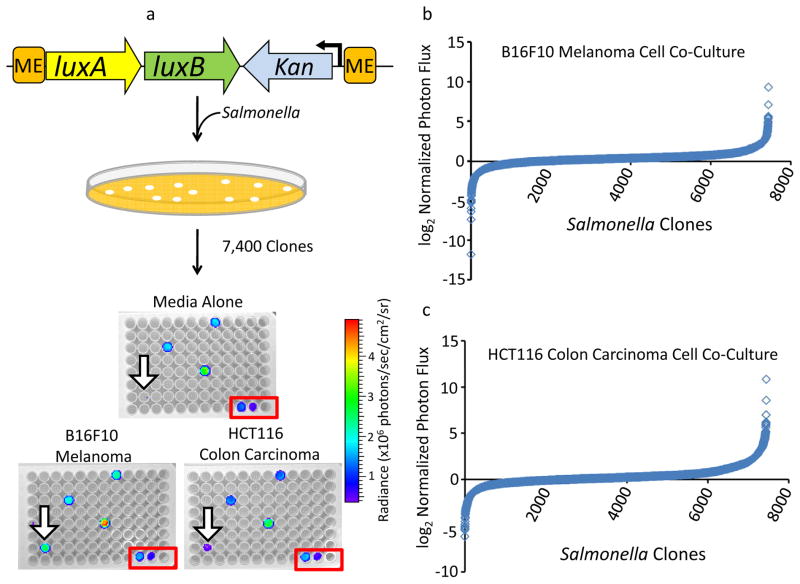
Figure 1. Design and utilization of a high throughput screen to identify tumor cell-induced gene activation events in Salmonella.
(a) A schematic of the promoter trap system using Tn5-based luxAB chromosomal integration. Expression of the promoterless luxAB reporter vector, and resulting Salmonella bioluminescence, is dependent on “trapping” an active promoter upstream of the chromosomal integration site. The transposon was randomly integrated into strain SB300A1, and kanamycin-resistant colonies were selected and arrayed into 96-well plates for library screening. Representative primary screening plates in triplicate show responses of Salmonella library strains to three separate co-culture conditions: media alone (top), B16F10 melanoma cells (bottom left), HCT116 colon carcinoma cells (bottom right). Hit 47.74, showing selective activation in co-culture with cancer cells, is indicated by the black open arrowhead, while the signals in the upper and central wells represent non-selective activation of clones. In each plate, wells H10, H11, and H12 (red box) contain media and bacteria constitutively expressing luxCDABE, bacteria constitutively expressing luxAB, and no bacteria, respectively, as controls. Primary library screening data from Salmonella promoter trap clones co-cultured with B16F10 melanoma cells (b) or HCT116 colon carcinoma cells (c). Data are reported as the log2 of the normalized signal for each library clone, where normalized signal was the ratio of the signal in the condition of interest to the signal in media alone.
Initially, the entire Salmonella library was subjected to a primary screen in the context of three conditions: tissue culture media alone, B16F10 melanoma cells and HCT116 colon carcinoma cells, both of the latter in monolayer co-culture with the Salmonella reporter library. The tumor cells were grown in 96-well plate format overnight and then bacterial clones added to wells corresponding to each of the two co-culture conditions and media alone. After a two-hour incubation, bioluminescence imaging of plates enabled identification of clones specifically up-regulating genes in the context of exposure to melanoma and/or colon carcinoma cells (Figure 1a). Results of the screen from co-culture with melanoma and colon carcinoma cells are shown in Figures 1b and 1c, respectively. In each case, data are shown as a rank-ordered S-plot of the log2 of the normalized signal for each clone of the library, where normalized signal was the ratio of the signal in the condition of interest to the signal in media alone. The majority of data points clustered around zero, indicating that most mutants interrogated in the assay did not show tumor-specific gene regulation. However, quartile analysis with a boundary for hit selection corresponding to a high stringency targeted error rate (α = 0.0027) identified five candidate mutants wherein the transposon reporter was specifically up-regulated during co-culture with malignant cells.
Verification and characterization of Salmonella gene activation events in the context of tumor cell co-culture
Following the primary screen, we utilized inverse touchdown PCR to map the specific location of each transposon in the Salmonella genome (15). Table 1 documents the site of chromosomal integration for the transposon and candidate gene up-regulated in each isolate. All genes were novel in that they have not been previously reported to be involved in Salmonella-host interactions, nor involved in Salmonella colonization of neoplasia. Interestingly, the genomic insertion sites of the transposon in three of the clones inserted in a cluster in the chromosomal sequence. Mapped to three different, but closely linked genes (STM1787, STM1791 and STM1793, respectively), two are known hydrogenases, and all three genes are likely co-regulated and involved in the same Salmonella function. Sequencing showed that in one high stringency hit, the transposon had inserted into adiY, a Salmonella gene known to be involved in an acid tolerance response (16). The transposon in the fifth clone was identified to have landed in yohJ, a putative membrane protein (17).
Table 1.
Transposon chromosomal insertion locations in Salmonella reporter mutants.
| Strain Name | Transposon Insertion Location | Base pairs Downstream of Start Codon | Function (Putative) (17) |
|---|---|---|---|
| Tn:1787 | STM1787 | 1,189 | Hydrogenase |
| Tn:1791 | STM1791 | 505 | Hydrogenase |
| Tn:1793 | STM1793 | 661 | Cytochrome oxidase |
| Tn:adiY | adiY | 439 | araC-like transcriptional activator; arginine- dependent acid tolerance |
| Tn:yohJ | yohJ | 205 | Hypothetical membrane protein |
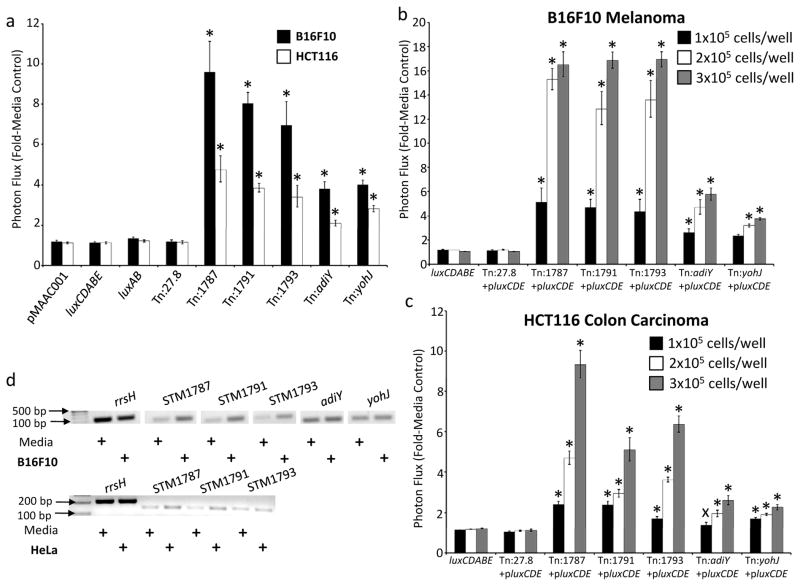
To validate cancer cell co-culture-specific gene activation events identified in the primary screen, we first repeated the co-culture assay in quadruplicate in at least three independent experiments for each clone. Figure 2a shows the data from one representative experiment for clones verified by this assay. Again, all five clones showed statistically significant enhancement of bioluminescence in the presence of tumor cells, with a trend toward greater gene up-regulation when co-cultured with B16F10 melanoma cells. Then, to further characterize tumor cell-induced response of Salmonella, we utilized the tumor cells in a dose-response assay (Figures 2b, c). Additionally, to verify that reporter activation seen in the Salmonella reporter-trap clones was not an effect of differing substrate permeability due to mutations in bacterial genes, bacteria were generated that contained the original chromosomal luxAB insertion as well as a plasmid constitutively expressing luxCDE, the biosynthetic genes for the long-chain aldehydes that act as the optical substrates of the bacterial luciferase operon. Therefore, for this assay, it was not necessary to add decanal to the media. Identical inoculations of bacteria showed greater up-regulation of the reporter when exposed to greater numbers of tumor cells in co-culture conditions, indicating that the stimuli from tumor cells instigated a graded response from the bacteria. Because expression of the lux operon genes fully complemented the use of exogenous decanal in the system, the data confirmed that the effect was not an artifact of exogenous decanal permeability in the primary screen.
Figure 2. Verification of Salmonella gene activation events in the context of tumor cell co-culture.
(a) Salmonella reporter clones displaying gene activation signals during co-culture with tumor cell lines (black bars, B16F10 cells; open bars, HCT116 cells). Salmonella strains luxAB and Tn:27.8 contain chromosomal luxAB genes under constitutive promoter control; luxCDABE Salmonella contain the full luciferase operon inserted into the chromosome; pMAAC001 constitutively expresses plasmid-encoded luxCDABE. (b, c) Salmonella reporter clones display dose-responsive gene activation in co-culture with B16F10 and HCT116 cells. Bacteria were co-cultured with 1×105, 2×105, or 3×105 B16F10 or HCT116 cells/well. Data were normalized as the ratio of the signal in the condition of interest to signal in media alone. Error bars correspond to SEM. All p value calculations are between luxCDABE and the group indicated by the symbol: * p ≤ 1×10−7; xp ≤ 0.06. (d) Semi-quantitative reverse transcriptase PCR with wild-type SB300A1 bacteria verifies that genes identified by the reporter transposon screen in Salmonella are activated during co-culture with B16F10 melanoma and HeLa tumor cells. rrsH = ribosomal RNA.
Finally, to verify that the reporters in fact reflected mRNA transcriptional regulation in wild-type Salmonella during co-culture with tumor cells, we utilized semi-quantitative PCR. Following a three-hour co-culture of wild-type (SB3001A1) bacteria with B16F10 cells or in tissue culture media alone, isolated RNA was reverse transcribed to cDNA. Semi-quantitative PCR of cDNA showed that co-culture with B16F10 melanoma cells enhanced the intensity of target gene transcripts, but not control ribosomal RNA transcripts (rrsH) (Figure 2d). The effect was generalizable, as co-culture with HeLa tumor cells produced similar results (Figure 2d).
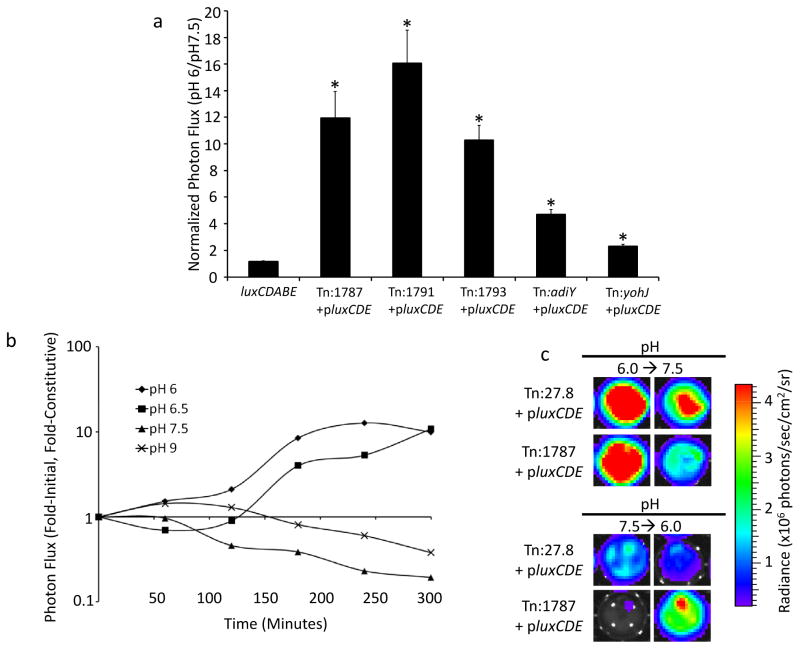
Notably, of the genes identified in this screen, at least one, adiY, has previously been reported to be up-regulated in acidic pH conditions (16). One characteristic of tumor microenvironments in vivo is an abnormally acidic pH (18). In fact, due to the Warburg effect, cancer cells are constitutively glycolytic, even in high oxygen conditions, releasing lactic acid and thereby creating a particularly acidic tumor microenvironment (19). For these reasons, the Salmonella transposon insertion mutants were further investigated for reporter signal activation in acidic conditions. Figure 3a shows that reporter signals increased in acidic pH media compared to neutral media. Each of the clones up-regulated the reporter gene at pH 6.0 compared to the physiological pH of normal body tissue (pH 7.5), suggesting that the stimulus Salmonella responded to in the context of neoplastic cells was microenvironment acidification.
Figure 3. Acidic pH specifically and reversibly stimulates the Tn:1787 trapped promoter.
(a) Bacteria were cultured in media of different pH values and reporter activation by Salmonella library clones in low pH media (pH 6) were compared to reporter activation in normal pH (7.5). Genes identified in the tumor cell co-culture screen were activated in the context of acidic pH compared to pH 7.5. pMAAC001 and luxCDABE constitutively express plasmid-encoded and chromosomally-encoded luxCDABE, respectively. Data were normalized as the ratio of the signal in media pH 6.0 to signal in media pH 7.5. Error bars correspond to standard error. The data show one representative experiment with 4 replicates per condition tested. All p-value calculations are between luxCDABE and the group indicated by the asterisk, *p ≤ 2×10−14. (b) Mice bearing B16F10 flank tumor xenografts were injected intratumorally with tumor-activated (Tn:1787+pluxCDE) or constitutively bioluminescent (Tn:27.8+pluxCDE) Salmonella. The excised tumors were imaged hourly and data are presented as the normalized signal at each time point. The normalized signal represents the ratio of the mean of the fold-initial signal of two Tn:1787+pluxCDE-colonized tumors to the mean of the fold-initial signal of two constitutive Tn:27.8+pluxCDE-colonized tumors. The data presented are from a representative experiment; the experiment was performed independently two times, each with two mice per bacterial treatment group. (c) Representative ex vivo tumor imaging shows reversibility of the bioluminescent signal in the tumor-activated Salmonella. Images on the left show Salmonella-infected tumor explants after 6 hours of incubation at the indicated pH (pH 6.0, top; pH 7.5, bottom). Two hours later (8 hours total), media was removed and replaced with media of the indicated pH (pH 7.5, top; pH 6.0, bottom). Images on the right show Salmonella-infected tumor explants 4 hours after the pH of the media was changed.
To determine whether the activated genes were required for localization to tumors or required for colonization and growth within tumors in vivo, Salmonella strains null for genes identified in the screen were constructed. Selected genes were deleted using a lambda red recombinase insertional deletion strategy, which inserted a chloramphenicol resistance cassette into the targeted genes. The deletion mutants were created from a parental Salmonella strain (luxCDABE msbB-) containing a chromosomally-integrated and constitutively-expressed bacterial luciferase operon for imaging bacterial localtization in vivo in real time. The strain also contained a msbB gene deletion, which causes a less immunogenic LPS structure and minimizes septic shock effects when the strain is administered intravenously (20). Based on the analysis that the identified STM1787, STM1791 and STM1793 genes were contained in a single operon, we targeted a large region of this operon for deletion in a single mutant strain, 1789−1793−. The gene adiY also appeared to be a part of a larger operon of co-regulated genes and was therefore targeted along with the adjacent genes adi and yjdE. The gene yohJ was targeted individually. In a B16F10 melanoma tumor xenograft model, all bacterial strains were injected via mouse tail vein and deletion mutants compared to the parental strain for localization to and persistence within the tumor using bioluminescence imaging (Supplementary Figure 1). All mutant strains and the parent strain were capable of tumor localization and persistence, indicating that although the identified genes were activated by tumor cell co-culture in vitro, they were not essential for bacterial colonization of the tumor. The experiment was also performed in an HCT116 colon carcinoma xenograft model with similar results. Supplementary Table 1 summarizes the numbers of mice with colonized tumors on or before day 10 in each experiment. Additionally, in pilot competitive infection studies, there was no significant difference between the STM1789-1793 mutant and the parental Salmonella strain (luxCDABE msbB-) in tumor colonization (CFU/ml; data not shown).
Specificity and reversibility of the Salmonella STM1787 promoter in vivo
We next sought to demonstrate the specificity of the STM1787 promoter activation in the tumor microenvironment in vivo. We chose this promoter because it displayed the highest acidic pH induction in vitro (Figure 2a). Here, we used the constitutively bioluminescent Salmonella strain Tn:27.8+pluxCDE or the conditionally bioluminescent strain Tn:1787+pluxCDE, each of which constitutively express plasmid-encoded luxCDE, but the latter strain will only bioluminesce upon activation of the chromosomally-encoded luxAB reporter. In a B16F10 melanoma tumor xenograft model, bacteria were injected via mouse tail vein or intratumorally and allowed two days to localize and adapt to tumors in vivo. Tumors were then excised, incubated in solutions of various pH values and imaged periodically for six hours. Initially, all tumors showed bioluminescent bacteria ex vivo. Over time, constitutive Tn:27.8 Salmonella showed a gradual increase in signal consistent with bacterial growth in the tumor explants. This behavior was also observed in the Tn:1787 Salmonella-infected tumor explants incubated in low pH media. By contrast, when the Tn:1787 Salmonella-infected tumor explants were maintained in basic media conditions throughout, the signal initially increased, but then plateaued around 4 hours and decreased in comparison to the constitutively bioluminescent Tn:27.8 strain (Figure 3b,c). This finding suggested that bacterial gene expression was initially engaged by the low pH conditions of the in vivo tumor microenvironment, but after exposure to a higher pH environment ex vivo, the promoter driving the reporter was repressed and signal declined. Further, this ex vivo effect was reversible. When the medium on the Tn:1787 Salmonella-infected tumor explant was changed from pH 6.0 to pH 7.5, the bioluminescent signal decreased. Conversely, when the media was changed from pH 7.5 to pH 6.0, the signal increased (Figure 3b, c). These effects were not seen with the constitutive Tn:27.8 Salmonella-infected tumor explants, and provided further evidence in support of the specificity of the trapped Salmonella promoter in the Tn:1787 transposon mutant for the tumor microenvironment.
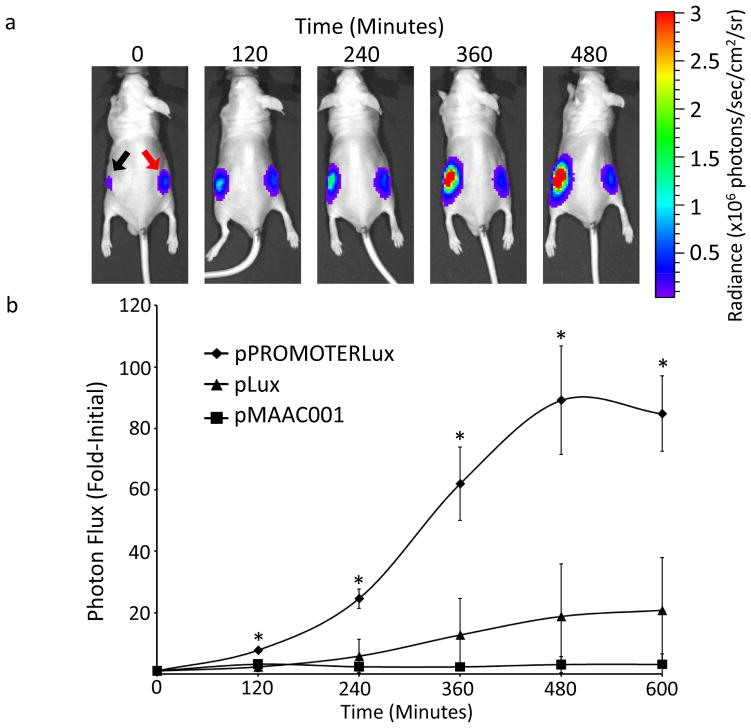
Because the identified promoters were highly activated in the tumor microenvironment ex vivo, utilization of these promoters provided a unique opportunity to design tumor-targeting bacterial vectors subject to multiple levels of controlled specificity in vivo. Thus, we sought to determine if the acidic pH of the tumor microenvironment could be exploited to specifically activate a target transgene during tumor localization. As proof of principle, we constructed Salmonella reporter strains expressing plasmids encoding the bacterial luciferase operon driven by either constitutive promoters or an inducible promoter to demonstrate tumor-mediated transgene activation in vivo. The plasmids pMAAC001 and pLux both encoded constitutively-expressed luciferase operons, while the pPROMOTERLux plasmid was engineered to contain the luciferase operon driven by the Salmonella candidate promoter (STM1787) comprising 500 base pairs upstream of the putative transcription start site of tumor-activated genes STM1787, STM1793 and STM1791 (which we will now refer to as the STM1787 promoter). Bacteria expressing these plasmids were identically injected into mice bearing HCT116 tumor xenografts on each flank (Figure 4). We chose to utilize intratumoral injection to directly compare reporter gene activation from two different bacterial strains, one inducible and the other constitutive, over time in the same mouse. Although reporter signals from pPROMOTERLux-expressing bacteria were low immediately after injection into the tumor, the bacteria quickly induced a 90-fold enhanced expression of the reporter after an 8 hr exposure to the tumor microenvironment (Figure 4a). Concurrently, bacteria constitutively expressing pLux- or pMAAC001-luciferase showed <20-fold or no reporter activation, respectively, after exposure to the tumor microenvironment (Figures 4a and 4b). These data directly demonstrated tumor-specific induction of a transgene from the Salmonella STM1787 promoter in an in vivo system. Therefore, the STM1787 promoter could be used as a platform to design tumor-targeting Salmonella strains capable of specifically delivering a therapeutic gene or toxin to the site of a tumor in vivo.
Figure 4. The STM1787 promoter in Salmonella is rapidly activated in vivo by the tumor microenvironment.
(a) A representative mouse with two HCT116 flank tumor xenografts. The left tumor (black arrow) was injected with STM1787 pPROMOTERLux-expressing Salmonella, while the right tumor (red arrow) was injected with constitutive pMAAC001-expressing Salmonella, and the mouse imaged at the indicated times post-injection. (b) The mean photon flux for each set of Salmonella-injected tumors, normalized to the initial signal in each tumor, plotted as a function of time. Error bars represent SEM; pPROMOTERLux (n=6); pLux (n=3); pMAAC001 (n=3), *p<0.025
Selective anti-tumor therapy in vivo
We utilized the cancer cell-activated STM1787 promoter to regulate the expression of Shiga toxin 2 (Stx2), a toxic transgene of bacterial origin, in a wild type strain of S. typhimurium (SB300A1) to selectively induce tumor cell death in vitro and in vivo. Stx2 is a secreted AB5 holotoxin composed of a single N-glycosidase A subunit that is directed to target eukaryotic cell membranes through interaction of the pentameric B subunits and the host receptor, glycosphingolipid globotriaosylceramide (Gb3) (21). Once inside the host cell, the A subunit cleaves the 28S RNA of the 60S ribosomal subunit, thereby inhibiting peptide elongation and inducing apoptosis. Stx2B has been extensively studied for its tumor targeting potential as many invasive tumors display high levels of Gb3 (22).
Using bioluminescence as a reporter of total tumor cell mass, we performed a co-culture experiment with HeLaCMV-Fluc cells in vitro. First, plated HeLa cells were grown to confluency to acidify the media and then co-cultured with strain SB300A1 transformed with P1787 (empty vector) or P7187-Stx2. Both SB300A1 transformants were also grown in media alone. The supernatant was then filtered from each of the groups and aliquoted onto separately plated HeLaCMV-Fluc cells in increasing volumes: 1) +media+P1787; 2) +media+P1787-Stx2; 3) +HeLa+P1787; and 4) +HeLa+P1787-Stx2. After 24 hours of treatment, major toxicity was only observed in HeLaCMV-Fluc cells treated with the supernatant of +HeLa+P1787-Stx2 (Figure 5a); a general concentration-response trend was observed (Figure 5b). Stx2 expression was verified using mRNA PCR amplification (Figure 5b inset). No overt cytotoxicity was observed in HeLaCMV-Fluc cells treated with supernatant from any of the other conditioned media groups.
Figure 5. P1787-driven Stx2 cytoxicity is selectively activated by the cancer cell environment in vitro.
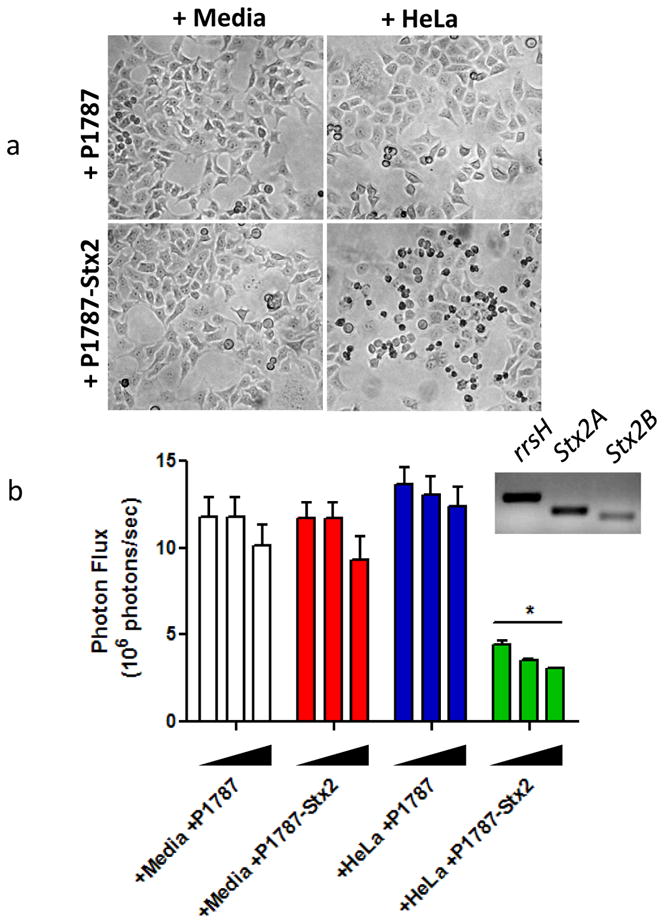
(a) Representative brightfield microscopy of HeLaCMV-FLuc cells treated with 4 different conditioned, filtered media for 24 hours (+media+P1787; +media+P1787-Stx2; +HeLa+P1787; and +HeLa+P1787-Stx2). Note the dramatic membrane blebbing and apoptotic morphology of +HeLa+P1787-Stx2 conditioned media-treated cells. (b) Bioluminescence imaging of HeLaCMV-FLuc cells treated with increasing amounts of 4 different conditioned, filtered media for 24 hours (bar groups, left to right: 17%, 29%, 44% of total volume per well). *p<0.0005 compared to all other treatments. Inset represents PCR amplification of Stx2A/B mRNA from P1787-Stx2 transformed SB300A1 co-cultured with HeLa cells.
Given the selective regulation and associated toxicity of P1787-Stx2 in vitro, we next desired to demonstrate the tumor-targeting potential in vivo using established s.c. flank HeLaCMV-Fluc xenograft tumors and bioluminescence imaging. In two independent proof-of-principle experiments, intratumoral injection of a single high-dose of SB300A1 transformed with P1787-Stx2 resulted in an 80% mean reduction in initial viable tumor mass five days after treatment (Figure 6a). Furthermore, when tumors were treated with a single low-dose of SB300A1 transformed with P1787-Stx2, a robust anti-tumoral effect was observed after two weeks (Figure 6b). We also observed that treatment with P1787 resulted in tumor stasis, consistent with previous reports that S. typhimurium alone can block tumor growth in vivo, while LB broth alone showed no inhibitory effect (23). To verify tumor cell death independent of bioluminescence signal, tumor H&E sections from the high-dose treatment were analyzed (Figure 6c). Sections through LB-treated (control) tumors showed a broad rim of viable tumor cells with focal necrotic regions centrally. In the P1787 tumors, a thin rim of viable tumor cells was present in most areas, with fibroinflammatory reaction at the periphery of the mass. The central necrotic zone was larger and contained more neutrophils than in the LB-treated tumors. In the P1787-Stx2-treated tumors, viable tumor cells were difficult to find, and in most sections, only a central necrotic zone surrounded by fibroinflammatory reaction was present. Note that mice treated with high-dose P1787-Stx2 eventually succumbed to the combined bacterial and Stx2 toxin load. However, mice receiving low-dose P1787-Stx2 were healthy for two weeks, at which point the experiment was concluded, but each still displayed a significant reduction in tumor size compared to P1787 alone (Figure 6b).
Figure 6. Enhanced anti-tumor response with P1787-Stx2 in vivo.
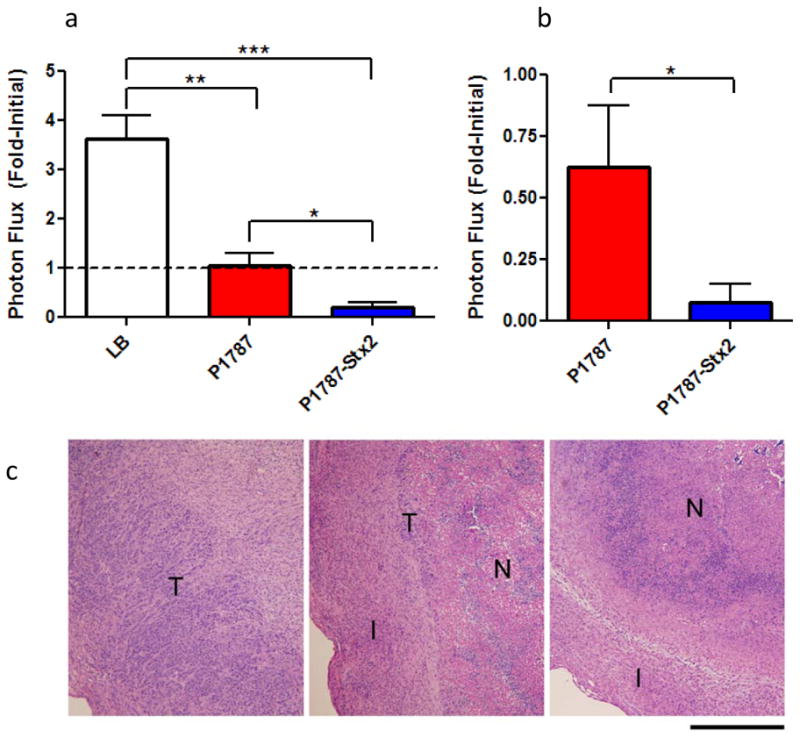
(a) Viable cell mass of HeLaCMV-FLuc tumors from mice treated with LB (n = 14), or high-dose SB300A1 transformed with P1787 (n= 12) or P1787-Stx2 (n = 9) at five days post treatment. Results are combined from two independent experiments and presented as fold-initial photon flux. Dotted line demarks lack of fold-change in tumor bioluminescence. Error bars indicate standard error of the mean ***p<0.0002,**p<0.0003, *p<0.007). (b) Fold-initialized photon flux of HeLaCMV-FLuc tumors from mice treated with low-dose SB300A1 transformed with P1787 (n= 7) or P1787-Stx2 (n = 7) at 14 days post treatment. Error bars indicate standard error of the mean.*p <0.04. (c) H&E staining of HeLaCMV-FLuc tumors from mice treated with LB (left), or high-dose SB300A1 transformed with P1787 (middle) or SB300A1 transformed with P1787-Stx2 (right) after five days. Regions of tumor are denoted as tumor (T), fibroinflammatory reaction (I), and necrotic zone (N). Scale bar, 500 μm.
Discussion
Salmonella typhimurium bacteria are typically classified as human gastrointestinal pathogens and a common cause of modern food-borne illness. However, another noted characteristic of Salmonella is the capacity to colonize tumor tissue. In fact, in the 1800’s, physicians began to intentionally use bacteria as tumor therapeutics, but due to significant toxicity and lack of consistent, reliable results, these practices were abandoned. However, modern studies using attenuated strains and longitudinal imaging have demonstrated colonization of tumors by Salmonella in real time and have sparked a renewed interest in this concept using Salmonella (23, 24) as well as various other tumor-localizing microbes as an option for cancer treatment (25–31). These observations along with the current intense focus on developing PAMP/TLR-based anti-cancer immunotherapies offer unique opportunities for combinatorial strategies in tumor targeting.
Both wild-type and genetically engineered Salmonella are capable of inducing tumor regression in mouse cancer models (4), as was observed in our experiments (Figure 6). A number of studies utilize bacteria as treatment vectors per se or as drug delivery vehicles by exploiting their potentially low toxicity and high genetic tractability to maximize therapeutic efficacy (5). In this regard, various attenuated Salmonella strains have been developed for use in tumor-targeting studies, including specific amino acid auxotrophs and LPS mutants (20, 32). However, the greatly reduced toxicity of Salmonella LPS mutants (msbB-) observed in swine models has not been observed in mouse models (33, 34). In more than one instance, attenuated Salmonella have even been used in a clinical trial to treat cancer in humans (35, 36). However, trials so far show relatively low rates of tumor colonization in human hosts, which may be due to excessive attenuation of the bacterial strains (5, 34). Indeed, one study indicates that induction of TNFα by bacteria is necessary for optimal colonization of tumors (37). Nonetheless, few studies have investigated the phenotypic and gene expression patterns of these tumor-targeting bacteria following exposure to tumor cells.
In this study, we utilized an engineered transposon to interrogate the Salmonella genome for genes activated during exposure to cancer cells. Toward this objective, we generated a library of greater than 7,400 independent transposon insertions, which, assuming random integration, would predict genomic transposon insertion into each of Salmonella’s 4,620 genes at least once. From this library, we identified five Salmonella genes specifically up-regulated during co-culture with cancer cells: STM1787, STM1791, STM1793, adiY and yohJ. Following identification of these tumor cell-activated genes, verification in secondary assays and confirmation in wild-type Salmonella, we determined that the common stimulus for up-regulation of target gene expression was acidic pH. In another study aimed at identifying Salmonella promoters involved in tumor colonization in vivo, Salmonella genomic DNA was digested and ligated randomly upstream of a GFP reporter. In this study, the major stimulus identified in reporter activation was hypoxia, but no pH-regulated promoters were identified (38). Another recent study performed a similar in vivo screen utilizing a promoter-trap GFP based system and indentified a conserved ‘tumor specific’ DNA motif (tusp) in the promoters of Salmonella genes specifically activated in a tumor xenograft model (33). While pH and hypoxia are physiologically linked, the five genes identified herein show no overlap with the promoters identified by Leschner et al. (33) nor Arrach et al. (38). However, the STM1787 promoter located upstream of three of our own target genes (STM1787, STM1791, and STM1793) did contain the conserved tusp motif identified by Leschner et al. (tattttatataaa). The discrepancy in promoter identification may stem from the different bacterial strains or strategies utilized for gene identification in the two studies. Whereas Arrach et. al. utilized a plasmid-based overexpression system, the present study identified genes by chromosomal integration of a transposon. Nonetheless, hydrogenase genes are noted in some cases to be up-regulated in low oxygen conditions, indicating that hypoxia may serve as a further stimulus for the pH-induced promoters identified in the present study (39). However, in pilot studies with an incubation pouch system used for growing anaerobic bacteria, we did not observe any significant changes in transposon reporter activity under hypoxic conditions (KF, unpublished data). While these data do not necessarily rule out oxygen-independence, pH appeared to be the dominant signal inducing responses in the promoters identified by our bioluminescent transposon reporter-trap screen. It will also be of interest in future studies to determine if in addition to hypoxia, pH is another regulator of Salmonella promoters that contain the largely uncharacterized tusp motif.
In view of the usual pathophysiology of Salmonella, it is not surprising that Salmonella strains have gained the ability to precisely regulate genes in response to different pH environments. Salmonella encounter low pH conditions regularly during human infection, for example, during transit through the stomach, and later during intracellular trafficking through the phagosome (40, 41). Interestingly, the acidic pH of the tumor environment in vivo has long been noted as an important microenvironmental condition when designing effective tumor treatments (18, 42). Additionally, the low pH environment of the tumor inhibits host defense. Cytotoxic immune cell activity and cytokine secretion has been shown to be impaired by a low extracellular pH (43). In contrast, with a bacterial-driven tumor therapeutic, low pH may become an exploitable advantage, by adding another level of selectivity to bacterial gene activation. Indeed, the utility of a low pH-activated bacterial therapeutic will avoid toxicity to the liver and spleen which are the other major off-target organ sites of bacterial colonization, but which generally have a neutral pH (33). In this case, a bacterial-based system may succeed, while both conventional therapeutics and host defenses fail.
When using bacteria as a vector for drug delivery studies, tumor-specific colonization and subsequent expression is a major concern. The genes identified herein are highly expressed in an acidic tumor environment, but are not required for bacterial tumor targeting (Supplemental Figure 1). Therefore, the promoters regulating these genes and further dissection of the complex regulation of the tusp motif may generate ideal chromosomal insertion site candidates or synthetic promoter systems for utilization in therapeutic gene, pro-drug or toxin delivery studies. We have identified the STM1787 promoter as an ideal bacterial sequence capable of driving tumor-specific expression of a transgene, and demonstrated this in vivo using bioluminescent imaging. We further applied the STM1787 promoter to conditionally regulate the expression of Stx2 in wildtype S. typhimurium in tumor targeting toxicity models in vitro and in vivo. In proof-of-principle studies, we observed dramatic cancer cell death in a co-culture model in vitro and dramatic tumor response over a relatively short time scale with a robust therapeutic effect in vivo. Future pharmacokinetic studies with P1787-Stx2 will be required to optimize mode of delivery, dose, and efficacy. In addition, it will be of interest to take advantage of the recent discovery that manganese treatment protects the host against lethal levels of Shiga toxin (44). Clearly, other relevant tumor toxins could be explored downstream of STM1787.
In summary, by adapting the STM1787 promoter in Salmonella to drive expression of an appropriate therapeutic transgene, the resulting bacterial vector would provide two independent mechanisms for specifically targeting tumors. First, Salmonella specifically localize to and accumulate in primary tumors and metastases in vivo. Second, the STM1787 promoter is preferentially activated in the acidic tumor microenvironment. The combined effect of these two levels of specificity provides a potential option to design more successful PAMP/TLR-based immunotherapeutic bacterial systems in the future.
Methods
Bacterial strains and culture conditions
The Salmonella typhimurium strains SB300A1 (14), SB300A1FL6 (luxCDABE) (45), luxAB and AM3 (luxCDABE msbB-) were grown in LB broth with appropriate antibiotics. SB300A1FL6 is modified by chromosomal integration of luxCDABE and is constitutively bioluminescent. The luxAB strain consists of SB300A1FL6 with the integrated luxE gene disrupted. This strain does not bioluminesce without addition of exogenous decanal substrate. The AM3 strain has the SB300A1FL6 background, but also has an msbB gene disruption, giving it a less immunogenic LPS structure. The Tn:27.8 strain, specifically identified from the screen as a non-inducible mutant, phenocopies luxAB with constitutive bioluminescence that requires exogenous decanal.
Tissue culture cell lines and culture conditions
B16F10 murine melanoma and HeLa cells were obtained from ATCC and cultured according to ATCC directions. HCT116 human colon carcinoma cells were a gift from Bert Vogelstein and were cultured according to ATCC methods. Cell lines were not further authenticated.
Plasmids
The plasmid pMAAC001 contains the full bacterial luciferase operon luxCDABE driven by a T7 promoter and an ampicillin resistance cassette. The plasmid pLuxCDE consists of the pMAAC001 backbone amplified using the forward primer cccgggattggggaggttggtatgtaa and the reverse primer cccgggtgaatgatttgatgagccaaa (XmaI sites underlined). This product was then XmaI digested and re-ligated to exclude the majority of the luxA and luxB genes. pLux and pPROMOTERLux plasmids were constructed by inserting the full bacterial luciferase operon between the KpnI and BamHI restriction sites in the vector pUC19. The pPROMOTERLux plasmid additionally had a 500 base pair promoter region (STM1787) from the Salmonella genome inserted upstream of the luciferase operon between the SacI and KpnI restriction enzyme sites. The 500 base pair sequence was amplified from the Salmonella genome using the forward primer aaagagctcacgccctctttcaaacagtc and the reverse primer aaaggtaccgcttgataaaaggtctcctcgt (SacI and KpnI sites underlined). To construct the P1787-based vectors, 500bp of the endogenous SB300A1 1787 promoter was cloned into the BglII and NdeI sites of pET3a (Novagen) using the following primers: forward-gagagagaagatct gggacgccctctttcaaacagtctc, reverse-ccttcctgcccatatgaacgcgtattttttctcctttttgcacc. This cloning strategy conserved the endogenous 1787 Shine-Dalgarno sequence and removed the T7 promoter and synthetic RBS of pET3a. The P1787-Stx2A/B vector was constructed by inserting the Stx2A/B operon downstream of the 1787 promoter using NdeI and BamHI with the following primers: forward-gagagagacatatgaagtgtatattatttaaatgggtactgtgcctgttactgggtttttcttcggtatcc, reverse-ccttccttccggatccttatcaatggtgatggtgatggtgg.
Construction of a Salmonella typhimurium reporter-trap library
Salmonella strain SB300A1 was used to construct a bacterial library comprising approximately 7,400 clones of unique chromosomal integrations of our reporter transposon (14). The custom Tn5-based transposon was designed with the EZ-Tn5 system (Epicentre, Madison, WI) using the pMOD4 transposon construction vector. A kanamycin-resistance cassette and promoter from EZ-Tn5<KAN-2> was amplified using the forward primer acgacaaagcttggacgcgatggatatgttct and the reverse primer agcttttctagaggtggaccagttggtgattt (HindIII and XbaI restriction sites underlined) and inserted into the HindIII and XbaI restriction sites of pMOD4. The luciferase enzyme genes luxAB from Photorhabdus luminescens were amplified with the forward primer acagtcgaattccgccgaatgagaattgagat and the reverse primer aagctgggtacctgttggctgctttcactcac (EcoRI and KpnI sites underlined) and inserted between the EcoRI and KpnI sites in pMOD4 (45). The plasmid contained an R6Kγ origin of replication and therefore was amplified in E. coli DH5α λpir, purified, digested with PvuII, and the transposon fragment recovered by gel purification. The purified transposon was combined with transposase (Epicentre). After bench top incubation for 30 minutes, followed by 48 hours at 4°C, the transposon DNA was electroporated into bacteria as per the vendor’s instructions. Bacteria were plated on LB kanamycin plates to select for transformants containing the chromosomally-integrated transposon. Each clone was expanded and stored in 60% glycerol in 96-well plates at −80°C.
Screening the library
To screen for gene activation events occurring in the context of malignant cells, Salmonella library clones were cultured under three different conditions: co-culture with B16F10 mouse melanoma cells, co-culture with HCT116 human colon carcinoma cells and culture in media alone. Each of the two tumor cell lines were seeded into 96-well white plates at approximately 70–80% confluency in DMEM with 10% FBS. In the plate containing media alone, each well contained 100 μl of DMEM with 10% FBS only. Plates were incubated overnight to allow tumor cell adhesion to the 96-well white plates. Independently, bacterial clones were grown overnight in LB broth with kanamycin in 96-well plates and subcultured the following day 1:10 into LB broth. Five to six hours after subculturing, 30 μl of bacterial culture were added to three replicate plates, each corresponding to a separate culture condition. Bacteria were allowed to co-incubate with the malignant cells or media alone for 2 hours. Subsequently, bacteria were imaged by adding 30 μl of decanal solution, waiting 10 minutes, and imaging with an IVIS 100 imaging system (Caliper; acquisition time, 60 sec; binning, 4; filter, < 510; f stop, 1; FOV, 23 cm) (46). Because white plates were used to maximize signal intensity, images were aquired utilizing a <510 filter to reduce phosphorescence from the plates. Three control wells were included on every plate comprising: luxCDABE Salmonella (SB300A1FL6), which contain the full luciferase operon inserted into the chromosome; luxAB strain, which contains the luciferase enzyme genes only and therefore requires addition of exogenous substrate to image reporter activity in the assay; and a blank well, which contained media, but was not inoculated with bacteria, to serve as a control for background luminescence. Imaged plates were analyzed with Living Image (Caliper) and Igor (Wavemetric) analysis software packages as described (47). Data were normalized by dividing the photon flux of experimental wells by media alone wells and presented as the log2 of the normalized photon flux data.
Identification of hits
Library screening data representing photon flux from each well of a library plate were analyzed with Image J software (48). To identify statistically significant hits from the primary screens, we utilized a set of statistical requirements. First, a threshold was set to identify active clones. Clones that did not produce photon signals greater than three standard deviations above the signal in the un-inoculated, media alone wells were not further analyzed. A quartile method of statistical analysis was then applied to the remaining clonal data (49). For quartile analysis, plates of clones were grouped by assay date into sets for data analysis. For each set, we normalized data by calculating the log2 of the fold-change of photon flux signal between the condition of interest (co-culture with B16F10 or HCT116 cells) and media alone. From this data, we calculated the median (Q2), first (Q1), and third (Q3) quartile values. The boundary for hit selection was calculated as Q3 + c(ICQ), where ICQ = Q3-Q1 and c = 1.7239, corresponding to a high stringency targeted error rate of α = 0.0027 (49).
Verification of primary screen hits
To verify hits identified by the primary screen, clones were tested again in a similar manner, in quadruplicate. The assay followed the same steps as those in the primary screen, except each clone was tested in 4 wells under each of three conditions across a 12-well row in a black 96-well plate. Imaging was done with an IVIS 100 imaging system (acquisition time, 60 sec; binning, 4; filter, open; f stop, 1; FOV, 23 cm).
Identification of transposon insertion site
To map sites of transposon integration in the chromosome of clones of interest, an inverse touchdown PCR strategy was used (15). Genomic DNA was isolated from bacteria using DNAzol (Molecular Research Center, Cincinatti, Ohio). PCR was performed using bacterial chromosomal DNA, 20 pmols of a primer specific to the 5′ end of the transposon (atggctcataacaccccttg), and 100 pmols of a degenerate primer (cggaatccggatngayksnggntc). Reactions were initiated with a 95°C preparation step for 5 minutes, followed by 25 cycles comprising denaturation at 95°C for 45 seconds, annealing at various temperatures for 45 seconds and extension at 72°C for 2 minutes. The annealing temperature started at 60°C and decreased 0.5°C per cycle for the subsequent 24 cycles. Then PCR proceeded with 25 cycles of 95°C for 45 seconds, 50°C for 45 seconds and 72°C for 2 minutes. PCR reaction products were fractionated on a 1% agarose gel, and the most prominent bands in each lane were excised and gel purified (Qiagen kit). For some reactions, PCR products were purified (Qiagen) and the resulting purified PCR product was used as a template for a second round of PCR using a different transposon-specific primer (aacatcagagattttgagacacc) before gel purification of products. The cycling conditions and degenerate primer used in the second round of PCR were the same as round one.
Semi-quantitative RTPCR
Salmonella strains SB300A1, P1787 or P1787 transformed SB300A1 were subcultured from a stationary phase culture 1:10 and grown for 6 hours. Bacteria were then diluted 1:20 and 30 μl added to 96-well plates containing tissue culture media alone, B16F10 melanoma cells or HeLa cells, seeded 24 hours previously at 100,000 cells/well and 50,000 cells/well, respectively. After three and a half hours of co-culture, extracellular media containing bacteria was removed from the 96-well plates and triplicates pooled. Media were centrifuged to pellet bacteria and pellets were frozen at −80°C. After thawing, pellets were resuspended in 200 μl water with 5 mg/ml lysozyme and incubated at room temperature for 5 minutes. Then, 700 μl of RLT buffer was added and bacterial RNA was purified using the Qiagen RNeasy kit (Qiagen Inc, Valencia, CA). Samples were then treated with DNase I at room temperature for 15 minutes, after which EDTA was added and samples were incubated for 10 minutes at 65°C to inactivate the DNase. Samples were then ethanol precipitated and resuspended in 30 μl water. For reverse transcriptase PCR, 1 μg of total RNA was used as a template and reverse transcribed using Superscript II Reverse Transcriptase and 300 ng random primers as per the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Following RTPCR, samples were treated with RNase H for 25 minutes at 37°C. To perform semi-quantitative PCR, samples were amplified using primers specific to each gene target or to ribosomal RNA: STM1787 (forward: tcggtagatcgcatgatgtc, reverse: ggttggtcataagcctgtcg), STM1791 (forward: acacgggaacatccagattc, reverse: cggcaaaggacaaatctcat), STM1793(forward: ttcggcaacctgtttttagg, reverse: acgcctccttgcataatcac), adiY (forward: ccttattgaccgccaactgt, reverse: gtggtcaagaaagcgggata), yohJ (forward: caggcatttttcttgcatca, reverse: cgccatataacgaatcagca), rrsH (forward: cagccacactggaactgaga, reverse: gttagccggtgcttcttctg), Stx2A (forward: atgacgccgggagacgtgga, reverse: ggccacagtccccagtatcgct) and Stx2B (forward: gcaatggcggcggattgtgc, reverse: acaatccgccgccattgcat). PCR cycling conditions were: 95°C for 5 minutes, 30 cycles (or 20 cycles for rrsH reactions) of denaturation at 95°C for 45 seconds, annealing at 50°C for 45 seconds and extension at 72°C for 1 minute. PCR products were fractionated on a 1% agarose gel.
Construction of deletion mutants
Mutant strains deficient for the identified target genes were constructed in Salmonella strain luxCDABE msbB- (AM3), which contains a constitutively active, chromosomally-encoded bacterial luciferase operon as well as a mutation in msbB to create a less immunogenic LPS structure. Mutants were constructed using a lambda red recombinase strategy (50). First, primers were designed to amplify the chloramphenicol-resistance cassette in pKD3 with tails flanking the targeted locus of the Salmonella genome to be deleted. Primer sequences specifically targetting the genome for each mutant were used (adi forward targetting primer: atgaaagtattaattgttgaaagtgagtttctgcatcaggacacctgggtgtgtaggctggag-ctgcttc, adi reverse targetting primer: atcctgtttaaccggcgcatccagcggatacgggtttttgtgaatgc-ggtcatatgaatatcctccttag; yohJ forward targetting primer: agtaagtcactgaatattatctg-gcaatatatacgcgcttgtgtaggctggagctgcttc, yohJ reverse targetting primer: ttttttcgttcc-cttctgcccaaccactttacgctcaccgcatatgaatatcctccttag; STM1789-1793 forward targetting primer: atgaatgcgcaacgcgtagtggtgatggggttaggaaaccgtgtaggctggagctgcttc, STM1789-1793 reverse targetting primer: ctaataaagttcatgatcgttgcggcggagggtccccaggcatatgaa-tatcctccttag). PCR fragments were then electroporated into AM3 bacteria expressing plasmid-encoded red recombinase. Following electroporation, growth on chloramphenicol plates at 37°C selected for strains that had lost the temperature-sensitive recombinase plasmid and inserted the chloramphenicol-resistance cassette into the targeted genomic loci. Deletion of the genes was confirmed by PCR.
Dose-response to tumor cells
To test the dose-response of hits from the screen to tumor cell co-culture, the assay was performed as described, except that either B16F10 or HCT116 cells were plated at 1×105; 2 × 105; or 3 × 105 cells per well 24 hours before co-culture with bacteria. Stationary phase bacteria were diluted 1:50 and incubated for 6 hours before identical aliquots were allowed to co-culture with the malignant cells. Growth curves performed for each mutant strain at different pH values showed no significant differences. Imaging was done with an IVIS 100 imaging system (acquisition time, 10 sec; binning, 8; filter, open; f stop, 1; FOV, 20 cm). Imaged plates were analyzed with Living Image (Caliper) and Igor (Wavemetrics) analysis software packages as described (47).
Assaying promoter activation in different pH media
Stationary phase bacteria were subcultured 1:100 into LB broth. Five to six hrs after subculturing, 10 μl of bacterial culture were added to 190 μl pre-warmed HEPES-buffered media in black 96-well plates adjusted to different pH values, and allowed to incubate for three and a half hours. Bacteria were then imaged with an IVIS 100 imaging system (acquisition time, 60 sec; binning, 8; filter, open; f stop, 1; FOV, 20 cm).
Mouse imaging studies
To generate tumor xenografts, 6-week old nu/nu mice (Taconic) were injected subcutaneously in the right flank with 1 × 106 B16F10 cells or 2.5 × 106 HCT116 cells in 100 μl PBS. Tumors were allowed to grow for two (B16F10) or three (HCT116) weeks before bacterial challenge. Saturated cultures of strain AM3 and deletion mutant bacteria were subcultured 1:100 into LB and grown for 3 hours. Bacteria were then diluted to 1 × 106 bacteria/ml (based on OD600 readings) and 100 μl were injected via tail vein. Mice were imaged as indicated using an IVIS 100 imaging system (acquisition time, 60 sec; binning, 8; filter, open; f stop, 1; FOV, 20 cm). Photon flux data were calculated by utilizing user-determined regions of interest (ROIs) around bioluminescent tumors with Living Image software.
For in vivo promoter inducibility experiments, 6-week old nu/nu mice (Taconic) were injected subcutaneously in the right and left flanks with 1 × 107 HCT116 cells in 100 μl PBS. Tumors were allowed to grow for one week. Saturated cultures of Salmonella strain SB300A1 containing plasmids pMAAC001, pPROMOTERLux, or pLux were subcultured 1:100 into LB and grown for 3 hours. Twenty microliters of bacterial culture were injected intratumorally. Mice were imaged as indicated using an IVIS 100 imaging system (acquisition time, 180 or 60 sec; binning, 8; filter, open; f stop, 1; FOV, 25 cm). Photon flux data were calculated by utilizing software-determined regions of interest (ROIs) around bioluminescent tumors with Living Image software.
Tumor ex vivo imaging
6-week old nu/nu mice (Taconic) were injected subcutaneously in the right flank with 1 × 105 B16F10 cells and tumors allowed to grow for two and a half weeks. Saturated cultures of bacteria were diluted and 5 × 105 bacteria (based on OD600 readings) were injected intratumorally. At 24 and 48 hours following bacterial injections, mice were sacrificed, and tumors excised and dissected into 4 sections each. The bacterial-colonized tumor sections were incubated in HEPES/Tris-buffered media at the indicated pH values and imaged using an IVIS 100 imaging system at the indicated times (acquisition time, 180 sec; binning, 8; filter, open; f stop, 1; FOV, 12 cm).
In vitro toxicity assays
Confluent HeLa cells or mock media alone (DMEM + 10% FBS) were inoculated at 1:100 with a stationary culture of SB300A1 transformed with P1787 or P1787-Stx2A/B and cultured at 37 C for 18 hours. The cultured media was then separately filtered through a 0.22 μm filter to remove the bacteria and subsequently aliquoted at various volumes onto HeLaCMV-FLuc cells pre-plated in a 96 well plate in quintuplicate. 24 hours later, bioluminescence of the conditioned media-treated HeLaCMV-FLuc cells was imaged using an IVIS 100. Phase contrast microscopy (TMS-F, Nikon) was used in parallel to qualitatively confirm loss of cell viability.
In vivo toxicity assays
6 week old male homozygous CrTac:NCr-Foxn1nu mice (Taconic) were subcutaneously injected in the right flank with of 4.5 × 106 HeLaCMV-FLuc cells in 20 μL DMEM. When tumor volumes reached approximately 100 mm2 (5 days later), mice were injected i.p. with 150mg/kg of D-luciferin and 10 minutes later imaged using an IVIS 100. Immediately following imaging, mice were injected intratumorly with LB broth, or SB300A1 transformed with P1787, or SB300A1 transformed with P1787-STx2A/B at either 2.5 × 105 (low-dose) or 2 × 106 (high-dose) CFU/injection. Mice (n= 9–14 in each group) were weighed and imaged for bioluminescence every five days for 2 weeks. Viable tumor mass is presented as fold-initial photon flux (pre-treatment/post-treatment).
Histology
Tumors were excised and immediately frozen at −80°C. Frozen tumors were fixed in 10% neutral buffered formalin for 24 hours. Prior to paraffin embedding, histology sectioning and H&E staining, fixed tumors were washed with 30%, 50% and then 70% ethanol for 5 minutes each.
Statistics
Error bars represent the standard error of the linearly regressed data or the standard error of the mean where noted.
Supplementary Material
Statement of significance.
Salmonella, which often encounter acidic environments during classical host infection, may co-opt evolutionarily conserved pathways for tumor colonization in response to the acidic tumor microenvironment. We identified specific promoter sequences that provide a platform for targeted Salmonella-based tumor therapy in vivo.
Acknowledgments
The authors thank colleagues of the Molecular Imaging Center for helpful discussions and Reece Goiffon for statistical assistance and David Haslam for the Stx2 plasmid. This study was supported in part by a grant from the National Institutes of Health to the Molecular Imaging Center at Washington University (P50 CA94056), NIH Training Grants T32 GM007067 for stipend support to K.F. and T32 CA113275 for stipend support to B.K., and The Siteman Cancer Center supported in part by a NCI Cancer Center Support Grant (P30 CA91842).
Footnotes
Conflict of Interest: None of the authors have a COI.
References
- 1.Ellis M, Ding L, Shen D, Luo J, Suman V, Wallis J, et al. Whole genome sequencing to characterise breast cancer response to aromatase inhibition. Nature. 2012 doi: 10.1038/nature11143. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cairns R, Papandreou I, Denko N. Overcoming physiologic barriers to cancer treatment by molecularly targeting the tumor microenvironment. Mol Cancer Res. 2006;4:61–70. doi: 10.1158/1541-7786.MCR-06-0002. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Gardlik R, Behuliak M, Palffy R, Celec P, Li CJ. Gene therapy for cancer: bacteria-mediated anti-angiogenesis therapy. Gene Ther. 2011;18:425–431. doi: 10.1038/gt.2010.176. [DOI] [PubMed] [Google Scholar]
- 5.Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer. 2010;10:785–794. doi: 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forbes NS, Munn LL, Fukumura D, Jain RK. Sparse initial entrapment of systemically injected Salmonella typhimurium leads to heterogeneous accumulation within tumors. Cancer Res. 2003;63:5188–5193. [PubMed] [Google Scholar]
- 7.Zhao M, Yang M, Li XM, Jiang P, Baranov E, Li S, et al. Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proc Natl Acad Sci U S A. 2005;102:755–760. doi: 10.1073/pnas.0408422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganai S, Arenas RB, Sauer JP, Bentley B, Forbes NS. In tumors Salmonella migrate away from vasculature toward the transition zone and induce apoptosis. Cancer Gene Ther. 2011;18:457–466. doi: 10.1038/cgt.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasinskas RW, Forbes NS. Salmonella typhimurium lacking ribose chemoreceptors localize in tumor quiescence and induce apoptosis. Cancer Res. 2007;67:3201–3209. doi: 10.1158/0008-5472.CAN-06-2618. [DOI] [PubMed] [Google Scholar]
- 10.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 11.Garaude J, Kent A, van Rooijen N, Blander JM. Simultaneous targeting of toll-and nod-like receptors induces effective tumor-specific immune responses. Sci Transl Med. 2012;4:120ra116. doi: 10.1126/scitranslmed.3002868. [DOI] [PubMed] [Google Scholar]
- 12.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 13.Pawelek JM, Low KB, Bermudes D. Bacteria as tumour-targeting vectors. Lancet Oncol. 2003;4:548–556. doi: 10.1016/s1470-2045(03)01194-x. [DOI] [PubMed] [Google Scholar]
- 14.McKinney J, Guerrier-Takada C, Galan J, Altman S. Tightly regulated gene expression system in Salmonella enterica serovar Typhimurium. J Bacteriol. 2002;184:6056–6059. doi: 10.1128/JB.184.21.6056-6059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levano-Garcia J, Verjovski-Almeida S, da Silva AC. Mapping transposon insertion sites by touchdown PCR and hybrid degenerate primers. Biotechniques. 2005;38:225–229. doi: 10.2144/05382ST03. [DOI] [PubMed] [Google Scholar]
- 16.Kieboom J, Abee T. Arginine-dependent acid resistance in Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:5650–5653. doi: 10.1128/JB.00323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tannock IF, Rotin D. Acid pH in tumors and its potential for therapeutic exploitation. Cancer Res. 1989;49:4373–4384. [PubMed] [Google Scholar]
- 19.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Low KB, Ittensohn M, Le T, Platt J, Sodi S, Amoss, et al. Lipid A mutant Salmonella with suppressed virulence and TNFalpha induction retain tumor-targeting in vivo. Nat Biotechnol. 1999;17:37–41. doi: 10.1038/5205. [DOI] [PubMed] [Google Scholar]
- 21.O’Loughlin EV, Robins-Browne RM. Effect of Shiga toxin and Shiga-like toxins on eukaryotic cells. Microbes Infect. 2001;3:493–507. doi: 10.1016/s1286-4579(01)01405-8. [DOI] [PubMed] [Google Scholar]
- 22.Engedal N, Skotland T, Torgersen ML, Sandvig K. Shiga toxin and its use in targeted cancer therapy and imaging. Microb Biotechnol. 2011;4:32–46. doi: 10.1111/j.1751-7915.2010.00180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pawelek JM, Low KB, Bermudes D. Tumor-targeted Salmonella as a novel anticancer vector. Cancer Res. 1997;57:4537–4544. [PubMed] [Google Scholar]
- 24.Yu YA, Shabahang S, Timiryasova TM, Zhang Q, Beltz R, Gentschev I, et al. Visualization of tumors and metastases in live animals with bacteria and vaccinia virus encoding light-emitting proteins. Nat Biotechnol. 2004;22:313–320. doi: 10.1038/nbt937. [DOI] [PubMed] [Google Scholar]
- 25.Dang LH, Bettegowda C, Huso DL, Kinzler KW, Vogelstein B. Combination bacteriolytic therapy for the treatment of experimental tumors. Proc Natl Acad Sci U S A. 2001;98:15155–15160. doi: 10.1073/pnas.251543698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang LH, Bettegowda C, Agrawal N, Cheong I, Huso D, Frost P, et al. Targeting vascular and avascular compartments of tumors with C. novyi-NT and anti-microtubule agents. Cancer Biol Ther. 2004;3:326–337. doi: 10.4161/cbt.3.3.704. [DOI] [PubMed] [Google Scholar]
- 27.Weibel S, Stritzker J, Eck M, Goebel W, Szalay AA. Colonization of experimental murine breast tumours by Escherichia coli K-12 significantly alters the tumour microenvironment. Cell Microbiol. 2008;10:1235–1248. doi: 10.1111/j.1462-5822.2008.01122.x. [DOI] [PubMed] [Google Scholar]
- 28.Stritzker J, Weibel S, Hill PJ, Oelschlaeger TA, Goebel W, Szalay AA. Tumor-specific colonization, tissue distribution, and gene induction by probiotic Escherichia coli Nissle 1917 in live mice. Int J Med Microbiol. 2007;297:151–162. doi: 10.1016/j.ijmm.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 29.Agrawal N, Bettegowda C, Cheong I, Geschwind JF, Drake CG, Hipkiss EL, et al. Bacteriolytic therapy can generate a potent immune response against experimental tumors. Proc Natl Acad Sci U S A. 2004;101:15172–15177. doi: 10.1073/pnas.0406242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bettegowda C, Foss CA, Cheong I, Wang Y, Diaz L, Agrawal N, et al. Imaging bacterial infections with radiolabeled 1-(2′-deoxy-2′-fluoro-beta-D-arabinofuranosyl)-5-iodouracil. Proc Natl Acad Sci U S A. 2005;102:1145–1150. doi: 10.1073/pnas.0408861102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hajitou A, Trepel M, Lilley CE, Soghomonyan S, Alauddin MM, Marini FC, 3rd, et al. A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell. 2006;125:385–398. doi: 10.1016/j.cell.2006.02.042. [DOI] [PubMed] [Google Scholar]
- 32.Zhao M, Yang M, Ma H, Li X, Tan X, Li S, et al. Targeted therapy with a Salmonella typhimurium leucine-arginine auxotroph cures orthotopic human breast tumors in nude mice. Cancer Res. 2006;66:7647–7652. doi: 10.1158/0008-5472.CAN-06-0716. [DOI] [PubMed] [Google Scholar]
- 33.Leschner S, Deyneko IV, Lienenklaus S, Wolf K, Bloecker H, Bumann D, et al. Identification of tumor-specific Salmonella Typhimurium promoters and their regulatory logic. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leschner S, Weiss S. Salmonella-allies in the fight against cancer. J Mol Med (Berl) 2010;88:763–773. doi: 10.1007/s00109-010-0636-z. [DOI] [PubMed] [Google Scholar]
- 35.Toso JF, Gill VJ, Hwu P, Marincola FM, Restifo NP, Schwartzentruber DJ, et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J Clin Oncol. 2002;20:142–152. doi: 10.1200/JCO.2002.20.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heimann DM, Rosenberg SA. Continuous intravenous administration of live genetically modified salmonella typhimurium in patients with metastatic melanoma. J Immunother. 2003;26:179–180. doi: 10.1097/00002371-200303000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leschner S, Westphal K, Dietrich N, Viegas N, Jablonska J, Lyszkiewicz M, et al. Tumor invasion of Salmonella enterica serovar Typhimurium is accompanied by strong hemorrhage promoted by TNF-alpha. PLoS One. 2009;4:e6692. doi: 10.1371/journal.pone.0006692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arrach N, Zhao M, Porwollik S, Hoffman RM, McClelland M. Salmonella promoters preferentially activated inside tumors. Cancer Res. 2008;68:4827–4832. doi: 10.1158/0008-5472.CAN-08-0552. [DOI] [PubMed] [Google Scholar]
- 39.Hayes ET, Wilks JC, Sanfilippo P, Yohannes E, Tate DP, Jones BD, et al. Oxygen limitation modulates pH regulation of catabolism and hydrogenases, multidrug transporters, and envelope composition in Escherichia coli K-12. BMC Microbiol. 2006;6:89. doi: 10.1186/1471-2180-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ibarra JA, Steele-Mortimer O. Salmonella--the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell Microbiol. 2009;11:1579–1586. doi: 10.1111/j.1462-5822.2009.01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Foster JW, Spector MP. How Salmonella survive against the odds. Annu Rev Microbiol. 1995;49:145–174. doi: 10.1146/annurev.mi.49.100195.001045. [DOI] [PubMed] [Google Scholar]
- 42.Gerweck LE, Seetharaman K. Cellular pH gradient in tumor versus normal tissue: potential exploitation for the treatment of cancer. Cancer Res. 1996;56:1194–1198. [PubMed] [Google Scholar]
- 43.Muller B, Fischer B, Kreutz W. An acidic microenvironment impairs the generation of non-major histocompatibility complex-restricted killer cells. Immunology. 2000;99:375–384. doi: 10.1046/j.1365-2567.2000.00975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mukhopadhyay S, Linstedt AD. Manganese blocks intracellular trafficking of Shiga toxin and protects against Shiga toxicosis. Science. 2012;335:332–335. doi: 10.1126/science.1215930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flentie KN, Qi M, Gammon ST, Razia Y, Lui F, Marpegan L, et al. Stably integrated luxCDABE for assessment of Salmonella invasion kinetics. Mol Imaging. 2008;7:222–233. [PMC free article] [PubMed] [Google Scholar]
- 46.Pfeifer CG, Marcus SL, Steele-Mortimer O, Knodler LA, Finlay BB. Salmonella typhimurium virulence genes are induced upon bacterial invasion into phagocytic and nonphagocytic cells. Infect Immun. 1999;67:5690–5698. doi: 10.1128/iai.67.11.5690-5698.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gross S, Piwnica-Worms D. Real-time imaging of ligand-induced IKK activation in intact cells and in living mice. Nat Methods. 2005;2:607–614. doi: 10.1038/nmeth779. [DOI] [PubMed] [Google Scholar]
- 48.Rasband W. ImageJ. 1.3.1_03. Bethesda, Maryland: National Institutes of Health; 2005. [Google Scholar]
- 49.Zhang XD, Yang XC, Chung N, Gates A, Stec E, Kunapuli P, et al. Robust statistical methods for hit selection in RNA interference high-throughput screening experiments. Pharmacogenomics. 2006;7:299–309. doi: 10.2217/14622416.7.3.299. [DOI] [PubMed] [Google Scholar]
- 50.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.