Apoptosis is an ordered cascade of enzymatic events that culminates in cell death and the cleavage of DNA into characteristic nucleosomal fragments. It is operative during embryonic development and, during adult life, plays a key role in various physiological processes, such as tissue remodeling and execution and regulation of the immune response. The consequences of inappropriate apoptotic responses are profound; for example, the failure of cells to initiate apoptosis in response to DNA damage has been implicated in the development and progression of cancer, whereas inappropriate activation of apoptosis is thought to be a contributing factor in Alzheimer's disease and Parkinson's disease. Rigorous signaling requirements and a complex network of inhibitory molecules maintain tight control of the apoptotic process, while permitting rapid and effective responses to diverse extracellular and intracellular signals. Growth-factor withdrawal, specific peptide hormones (e.g., tumor necrosis factor α, Fas ligand, and TRAIL), changes in intracellular or extracellular calcium, drugs (e.g., staurosporine and phorbol esters), DNA damage, and mitochondrial poisons can all induce apoptosis, with each of these triggers activating a different portion of the pathway. As our knowledge of the apoptotic cell death cascade increases, it is becoming apparent that the system is more complex than first thought. In a recent issue of PNAS, Khaled and colleagues (1) have addressed how cells might accomplish this fine-tuning of the apoptosis cascade by analyzing the regulation of the proapoptotic molecule Bax in response to the withdrawal of growth factors.
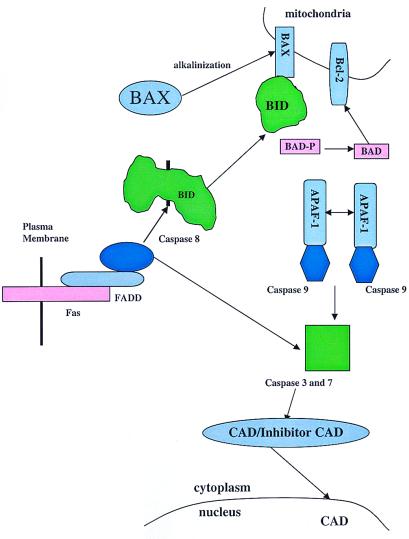
The marked similarities in apoptosis among all metazoans have contributed greatly to our understanding of the mammalian pathway. In the nematode Caenorhabditis elegans, three gene products are central to the regulation of the death cascade, CED-3, CED-4, and CED-9. CED-4 binds to CED-3, thereby regulating the activation of CED-3 and the death cascade, whereas CED-9 blocks the activity of CED-4 (2). In the mammalian system, caspase 8, a cysteine protease, shares homology with the C. elegans protein CED-3. Caspase 8 is an initiator caspase that activates effector caspases, including caspase 6, caspase 3, and caspase 7, through selective cleavage (Fig. 1). Caspase 8 contains a death effector domain, which is capable of interacting with the death effector domains contained in the adaptor molecules that link it to specific extracellular and intracellular signals. For example, the peptide hormone Fas ligand binds to a specific transmembrane receptor, Fas. On trimerization, Fas interacts with a specific adaptor protein FADD, which contains a death effector domain, and activates caspase 8. Similar initiation pathways have been established for tumor necrosis factor α and TRAIL.
Figure 1.
Cascade of the apoptotic response. CAD, caspase-activated deoxyribonuclease.
The effector caspases 3 and 7 also are regulated by the APAF-1 protein, a homolog of CED-4, in a manner analogous to that of the nematode CED-3–CED-4 interaction. APAF-1 contains an N-terminal caspase recruitment domain. When the APAF-1 protein is homodimerized in the presence of dATP and cytochrome C released from mitochondria, APAF-1 brings two caspase 9 molecules together, thereby activating this initiator caspase. The activated caspase 9 then induces the cleavage of the effector proteases, caspase 3 and 7, activating their protease activity (Fig. 1). Interestingly, caspase 3 and 7 also may cleave the initiator caspase 8, which, in its position at the head of this pathway, can activate a positive feedback loop for this cascade.
The effector caspases are responsible for the controlled degradation process that is characteristic of apoptotic cell death. In addition to degrading specific proteins, such as laminin, the effector caspases target the complex containing the nuclease responsible for DNA cleavage (CAD). CAD resides in the cytoplasm in a complex with its inhibitor (ICAD). During apoptosis, the effector caspases cleave ICAD, thereby allowing the active CAD to migrate from the cytoplasm into the nucleus and to initiate DNA degradation.
Because the cellular consequences of errant activation of the apoptotic cascade are considerable, the enzymes that play an important role in regulating this cascade must be under strict inhibitory controls. The caspases cannot be activated in the absence of either trimerization of a receptor or dimerization of APAF-1. In addition, mammalian cells make use of a set of regulatory proteins, the bcl-2 family, which show similarities to the nematode protein CED-9. Consistent with its role in blocking cell death, Bcl-2 was identified initially through analysis of chromosomal translocations in follicular and diffuse lymphomas. The Bcl-2 family of prosurvival proteins includes proteins Bcl-XL, Bcl-w, Mcl-1, etc., which share multiple amino acid domains, notably, the BH1, BH2, BH3, and BH4 domains and a hydrophobic membrane anchor. Bcl-2 family members are located in the outer membrane of the mitochondria and function, at least in part, by blocking the release of cytochrome C from the mitochondria (Fig. 1).
The cloning of these Bcl-2 prosurvival proteins revealed related proteins that, in contrast to the prototype Bcl-2 family members, are proapoptotic when overexpressed. One such proapoptotic protein, Bax, contains BH1, BH2, and BH3 domains and a hydrophobic membrane anchor but lacks the BH4 domain. Bax has similarities to Egl-1, a protein of C. elegans that inhibits CED-9. Other Bax family members, including Bak, Bok, Bik, Bad, Bid, etc., all contain BH3 domains, although the majority lack the BH1 and BH2 domains and the hydrophobic membrane domain. In contrast to the Bcl-2 family members, insertion of Bax family members into the mitochondrial membrane induces the release of cytochrome C and the induction of apoptotic cell death.
The regulation of this diverse proapoptotic Bax family seems to be quite complex. The assorted mechanisms used by this family of proteins to achieve activation suggest that this system may permit the triggering of the apoptotic pathway in response to specific sets of multiple weak signals rather than to sufficiently strong individual stimuli. Members of the Bax family of proteins are activated variously by cleavage by caspases, inhibition of protein kinases and/or activation of phosphatases, and an increase in intracellular pH. For example, Bid, which does not contain a hydrophobic membrane insertion sequence, is activated by cleavage by caspase 8 (3). This cleavage exposes its BH3 domain, allowing dimerization with Bax and Bcl-2, stimulating Bax and inhibiting Bcl-2 activity. The BH3 domain-containing proapoptotic protein Bad uses yet another regulatory mechanism. In cells treated with growth factor, it is phosphorylated by Akt (4), c-AMP-dependent protein kinase (5), and p90RSK (6) kinases creating 14-3-3 binding sites and allowing Bad to dimerize with 14-3-3, which results in sequestration of the protein away from the mitochondria. Removal of growth factors, such as IL-7 and IL-3, would likely free Bad, possibly through the activation of a phosphatase, from 14-3-3 interaction and allow the binding of Bad to Bcl-2 through its BH3 domain, thereby inhibiting Bcl-2 activity. The Bax molecule seems to reside in the cytoplasm and responds to various stimuli by migration to the mitochondria (7) where it is capable of causing cytochrome C release (8), thereby activating APAF-1 dimerization and the apoptotic cascade. Khaled and colleagues (1) have shown that growth-factor starvation induced by removal of IL-7 from the D1 T cell line or IL-3 from the pro B cell line, BaF3, can cause translocation of Bax from the cytoplasm to the mitochondria. They further show that withdrawal of growth factor causes an increase in the intracellular pH that allows the unfolding of Bax and the insertion of its C-terminal hydrophobic domain into the mitochondrial membrane, thereby promoting apoptosis. Given the number of members of this family and their diversity, the mechanisms involved in controlling these proteins may be even more complex. This complexity could allow modulation of the apoptotic response. Thus, cells that are starved of growth factors for short periods of time would be prevented from executing the full apoptotic program, even though individual members of this proapoptotic family are activated.
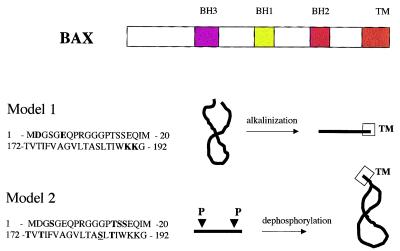
In their paper, Khaled and colleagues (1) have shown that high pH induces an increase in the sensitivity of Bax to proteases, the availability of Bax for immunoprecipitation by an antibody specific for the N terminus, and the partitioning of the protein into Triton X-114 detergent. Taken together, these results suggest that a conformational change in Bax is induced by an increase in cellular pH. In addition, the authors have shown that mutation of the C-terminal lysine residues 189 or 190 to leucines or the N-terminal aspartic acid residue 2 and glutamic acid residue 6 to alanines also renders the protein more susceptible to protease digestion and implies that such changes confer a more open configuration on the protein. However, increased insertion into the mitochondrial membrane of these mutants was not shown. These data suggest that the N and C termini are bound by charged residues that may be neutralized at high pH, thus opening up the protein (Fig. 2). The C termini of the Bcl-2 and Bax families of proteins contain a domain that is necessary for mitochondrial insertion; its deletion blocks apoptosis induced by Bax (9). This domain (amino acids 171–192) contains multiple hydrophobic residues interspersed with threonines and serines. Others have shown that the mutation of serine 184 to an acidic residue (S184E, S184K, or S184D) blocks the ability of Bax to insert into the mitochondrial membrane, whereas mutation of this residue to a hydrophobic amino acid (S184A or S184V) enhances localization of Bax to the mitochondrial membrane (10). Similarly, the N terminus of Bax contains multiple serines and threonines interspersed with hydrophobic residues. Because both serines and threonines are commonly phosphorylated residues and because acidic residues can mimic this effect, it is possible to posit an alternative hypothesis. At the time of growth-factor starvation, dephosphorylation of the N and C termini of a phosphorylated Bax molecule would allow the two hydrophobic ends of the protein to associate and make for easier insertion into mitochondrial membranes (Fig. 2).
Figure 2.
Mechanism of BAX structural regulation necessary for mitochondrial insertion. The domains of Bax are shown, including a C-terminal transmembrane domain (TM). Model 1 shows N-terminal amino acids 1–20 and C-terminal amino acids 172–192 of Bax with specific charged amino acids identified as important for structural unfolding (1) in bold. Model 2 identifies threonines and serines in the N- and C-terminal regions that are possible phosphorylation sites. Serine 184 (20), which has been identified as regulatory amino acid for mitochondrial insertion, is underlined.
Khaled and colleagues (1) have shown that Bax translocates to the mitochondrial membrane after 6 hours of growth-factor starvation, although apoptotic cell death does not occur until 24 hours. The authors suggest that this delay reflects the levels of Bcl-2 protein (but not mRNA, which is short lived), which remain elevated until this time. Addition of IL-7 to the growth-factor-starved cells after translocation of Bax to the mitochondrial membrane but before apoptosis can reinitiate cell growth. This reversal suggests that Bax translocation and insertion into the mitochondrial membrane alone are not sufficient to induce apoptotic cell death.
It has been hypothesized that Bax induces the release of cytochrome C by inhibiting Bcl-2 function through binding of the BH1, BH2, and BH3 domains. There is, however, considerable evidence to support the alternative hypothesis that Bax and Bcl-2 function independently in regulating apoptosis. Both Bcl-2 and Bax are capable of forming ion channels in artificial membranes (11), although Bcl-2 inhibits the activity of Bax at neutral pH. These data would suggest that although Bax function can be inhibited by Bcl-2, it does not require this interaction to function. Moreover, it has been shown that the removal of a portion of the BH1, BH2, and BH3 domains does not prevent Bax from enhancing chemotherapy-induced apoptosis (12), and the enforced dimerization of Bax with the FKBP protein induces translocation of Bax to the mitochondria and cell death even in the presence of Bcl-2 (13). Likewise, Bax mutant proteins that fail to bind to Bcl-2 are capable of inducing apoptosis (14), and the phenotype of Bax knockout mice differs from that of the Bcl-2-deficient strain (15).
If the inhibition of Bax by Bcl-2 does not depend on the binding of Bcl-2 to Bax, how then does Bcl-2 inhibit Bax? In cell lines that overexpress Bcl-2, Bax is unable to insert into the mitochondrial membrane and remains in the cytosol (7), suggesting the possibility that the number of insertion sites are limited. However, when Bax was titrated to determine the receptor number, no limitation could be found. Khaled and colleagues (1) have reported that on IL-7 withdrawal, a good portion of the Bax remains in the cytosol, which is consistent with a limitation on insertion sites. Thus, it is possible that Bcl-2 levels in the mitochondrial membrane must decrease if Bax levels in the mitochondria are to rise. Alternatively, additional Bax family members may play an essential role in the process. It has been reported previously that growth-factor starvation leads to intracellular acidification at later time points (16), although acidification was not evident during T cell starvation in the work of Khaled and colleagues (1). Acidic pH enhances the dimerization of the Bax family of proteins (17) and induces caspase activation (16). As described above, Bid can then be cleaved by caspases and bind to Bax through its BH3 domain. Bid/Bax binding markedly enhances the ability of Bax to induce the release of cytochrome C from mitochondria (18). In addition, as growth-factor starvation proceeds, it is possible that Bad, freed from the 14-3-3 interaction, could bind simultaneously to Bcl-2 and inhibit its channel regulatory function. Early growth-factor readdition may occur before the cleavage of Bid and the dephosphorylation of Bad, making reversal of the harmful effects of Bax insertion into the mitochondrial membrane possible.
Finally, there is a growing body of evidence that supports the concept that the protein regulators of apoptosis play an important role in the regulation of the cell cycle. Survivin, an antiapoptotic protein, increases markedly in the G2/M phase of the cell cycle and may regulate cell-cycle progression through this checkpoint (19). Mature T cells that overexpress Bax have a lower level of p27 and enter the S phase more rapidly than do T cells that overexpress Bcl-2. T cells that overexpress Bcl-2 progress through S more slowly than wild-type cells (20). Therefore, Bax may play an important role regulating the G1/S checkpoint. It can be suggested, albeit hypothetically, that the primary effect of translocation of Bax to the mitochondria in response to withdrawal of growth factors is sequestration of Bax, which, in turn prevents the transition of cells through G1/S. It may be only on prolonged starvation that Bax, through interaction with other family members, becomes the initiator of the apoptotic death pathway. As one knows, skipping lunch alone may be uncomfortable, but it is not terminal.
Footnotes
See companion article on page 14476 in issue 25 of volume 96.
References
- 1.Khaled A R, Kim K, Hofmeister R, Muegge K, Durum S K. Proc Natl Acad Sci USA. 1999;96:14476–14481. doi: 10.1073/pnas.96.25.14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chinnaiyan A M, Chaudhary D, O'Rourke K, Koonin E I, Dixit V M. Nature (London) 1997;388:728–729. doi: 10.1038/41913. [DOI] [PubMed] [Google Scholar]
- 3.Li H, Zhu H, Xu C-J, Yuan J. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 4.Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 5.Harada H, Becknell B, Wilm M, Mann M, Huang L J, Taylor S S, Scott J D, Korsmeyer S J. Mol Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- 6.Bonni A, Brunet A, West A E, Datta S R, Takasu M A, Greenberg M E. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 7.Wolter K G, Hsu Y-T, Smith C L, Nechushtan A, Xi X-G, Youle R J. J Cell Biol. 1997;139:1281–1292. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jurgensmeier J M, Xie Z, Quinn D, Ellerby L, Bredesen D, Reed J C. Proc Natl Acad Sci USA. 1998;95:4997–5002. doi: 10.1073/pnas.95.9.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nechustan A, Smith C L, Hsu Y-T, Youle R J. EMBO J. 1999;18:2330–2341. doi: 10.1093/emboj/18.9.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goping I S, Gross A, Lavoie J N, Nguyen M, Jemmerson R, Roth K, Korsmeyer S J, Shore G C. J Cell Biol. 1998;143:207–215. doi: 10.1083/jcb.143.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlesinger P H, Gross A, Yin X-M, Yamamoto K, Saito M, Waksman G, Korsmeyer S J. Proc Natl Acad Sci USA. 1997;94:11357–11362. doi: 10.1073/pnas.94.21.11357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simonian P L, Grillot D A, Andrews D W, Leber B, Nunez G. J Biol Chem. 1996;271:32073–32077. doi: 10.1074/jbc.271.50.32073. [DOI] [PubMed] [Google Scholar]
- 13.Gross A, Jockel J, Wei M C, Korsmeyer S J. EMBO J. 1998;17:3878–3885. doi: 10.1093/emboj/17.14.3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zha H, Reed J C. J Biol Chem. 1997;272:31482–31488. doi: 10.1074/jbc.272.50.31482. [DOI] [PubMed] [Google Scholar]
- 15.Knudson C M, Korsmeyer S J. Nat Genet. 1997;16:358–363. doi: 10.1038/ng0897-358. [DOI] [PubMed] [Google Scholar]
- 16.Furlong I J, Ascaso R, Rivas A L, Collins M K. J Cell Sci. 1997;110:653–661. doi: 10.1242/jcs.110.5.653. [DOI] [PubMed] [Google Scholar]
- 17.Xie Z, Schendel S, Matsuyama S, Reed J C. Biochemistry. 1998;37:6410–6418. doi: 10.1021/bi973052i. [DOI] [PubMed] [Google Scholar]
- 18.Desagher S, Osen-Sand A, Nichols A, Eskes R, Montessuit S, Lauper S, Maundrell K, Antonssson B, Martinou J-C. J Cell Biol. 1999;144:891–901. doi: 10.1083/jcb.144.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F, Ambrosini G, Chu E Y, Plescia J, Tognin S, Marchisio P C, Altieri D C. Nature (London) 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 20.Brady H J, Gil-Gomez G, Kirberg J, Berns A J. EMBO J. 1996;115:6991–7001. [PMC free article] [PubMed] [Google Scholar]